Martin Graef highlights Yamashita et al.’s finding that mitophagy can occur independently of the canonical mitochondrial fission apparatus.
Abstract
Whether or not mitophagy depends on prior mitochondrial fragmentation by the canonical mitochondrial division machinery is controversial. In this issue, Yamashita et al. (2016. J. Cell Biol. https://doi.org/10.1083/jcb.201605093) report that mitochondrial fragments start to bud and divide from mitochondrial tubules when in tight association with forming autophagosomes, but independently of the mitochondrial division factor Drp1/Dnm1.
Macroautophagy, hereafter autophagy, is a highly conserved catabolic pathway critically involved in intracellular homeostasis, quality control, and stress responses. Double-membraned vesicular structures, so-called autophagosomes, are a hallmark of autophagy and form de novo and transfer portions of the cytoplasm for degradation and recycling to lysosomes or vacuoles (Feng et al., 2014). A multicomponent core autophagy machinery seems to coopt membranes and membrane-remodeling components of diverse sources to drive the nucleation, expansion, and closure of a single-membrane structure, the isolation membrane, to form the double-membrane autophagosome. Autophagy can proceed in a nonselective manner, in which autophagosomes appear to randomly enwrap portions of the cytoplasm. However, selective modes of autophagy also exist, allowing cells to target specific cargoes preferentially or even exclusively for degradation. Selectivity is achieved by receptor-mediated physical interactions linking the autophagy machinery to the cargo, which subsequently drives the formation of an isolation membrane closely surrounding the bound cargo. The selective turnover of mitochondria by autophagy, so-called mitophagy, is critically involved in the quality control of mitochondria.
Mitochondria form highly dynamic networks of interconnected tubules, which undergo constant fusion and division. To maintain structural and functional integrity, fusion and division of mitochondrial outer and inner membrane occur in a spatially and temporally coordinated manner mediated by evolutionarily conserved dynamin-related GTPases: Mfn1/2 (Fzo1) and Opa1 (Mgm1) for fusion and Drp1 (Dnm1) for fission in mammals (yeast). During mitochondrial division, Drp1/Dnm1 is recruited to mitochondria by the effectors/adaptors, Mdv1 and Fis1 in yeast and Fis1, Mff, MiD49, and MiD51 in mammals. Drp1 assembles in its GTP-bound state into helical oligomeric structures wrapping around mitochondrial constriction sites established and marked by the endoplasmic reticulum (Lackner, 2014). GTP hydrolysis-driven conformational changes in Drp1/Dnm1 assemblies result in helical constriction and scission of the two mitochondrial membranes. Mitochondrial architecture is intimately linked to the functional state of the organelle and the cell, and mitochondrial fragmentation resulting from increased Drp1/Dnm1-mediated division is generally associated with mitochondrial stress and dysfunction (Nunnari and Suomalainen, 2012).
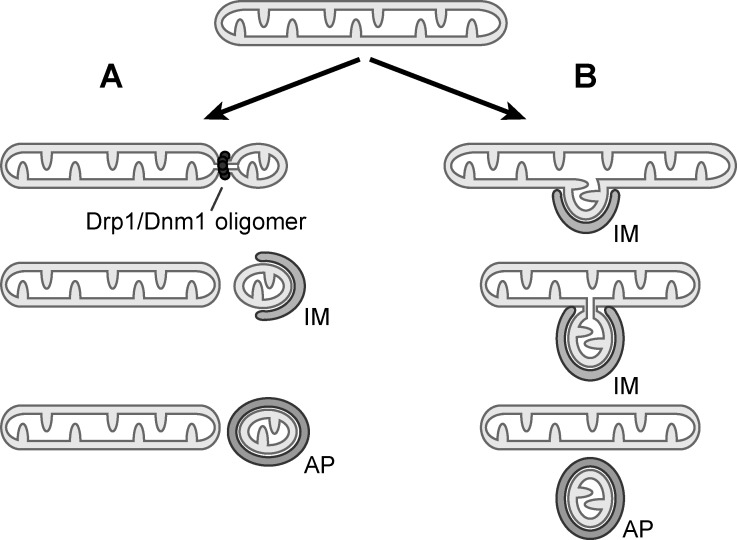
In mammalian cells, dysfunctional mitochondria activate the ubiquitin ligase parkin and the ubiquitin kinase PINK1 (PINK1–parkin pathway), which generate phospho-ubiquitinated mitochondrial outer membrane proteins. Phospho-ubiquitin serves as an autophagy signal and recruits the core autophagy machinery through the action of two mitophagy receptors, NDP52 and optineurin (Narendra et al., 2008; Lazarou et al., 2015). In addition, mitochondrial outer membrane proteins Bcl2-L-13, the mammalian homologue of Atg32 in yeast, also recruits the autophagy machinery upon mitochondrial dysfunction, indicating redundant pathways exist to intiate mitophagy (Kanki et al., 2009; Okamoto et al., 2009; Murakawa et al., 2015). Given that mitochondria are engulfed by autophagosomes of a seemingly limited size, a critical role for Drp1/Dnm1-mediated division to generate mitochondrial fragments of appropriate size for turnover was anticipated and supported by several studies in yeast and mammalian cells (Fig. 1 A; Kanki et al., 2009; Tanaka et al., 2010; Rambold et al., 2011; Abeliovich et al., 2013; Mao et al., 2013; Kageyama et al., 2014; Ikeda et al., 2015). However, a few studies indicated that Drp1/Dnm1-independent mitophagy might occur in yeast and mammals (Mendl et al., 2011; Bernhardt et al., 2015; Murakawa et al., 2015). Whether or not Drp1/Dnm1-mediated mitochondrial division is required for mitophagy remained a controversial topic.
Figure 1.
Drp1/Dnm1-dependent and -independent mitophagy. (A) Drp1/Dnm1-mediated mitochondrial division generates a mitochondrial fragment, which is subsequently targeted by an isolation membrane (IM) and engulfed in an autophagosome (AP). (B) Drp1/Dnm1-independent mitophagy. Autophagosome biogenesis is initiated on mitochondrial tubules and leads to budding and division of a mitochondrial fragment concurrently with the expansion of the isolation membrane. The autophagy machinery might drive the scission event directly or through closure of the isolation membrane before the mitochondrial fragment is encapsulated by an autophagosome.
In this issue, Yamashita et al. examine the role of Drp1/Dnm1-mediated mitochondrial division for mitophagy in an extensive study covering two yeast systems, Saccharomyces cerevisiae and Pichia pastoris, as well as three different cell culture systems, including HeLa and SH-SY5Y cells and mouse embryonic fibroblasts, under several mitophagy-inducing conditions. In both yeasts, Yamashita et al. (2016) detected slightly delayed but significant turnover of mitochondria during starvation in the absence of Dnm1 itself or the Dnm1 anchor protein Fis1. Confirming a minor role for Dnm1-mediated fission during mitophagy, dnm1Δ and fis1Δ yeast cells accumulated small mitochondrial fragments in autophagosomes when analyzed by fluorescence or electron microscopy, indicating that bite-sized mitochondrial fragments are generated and taken up by autophagosomes independently of the canonical mitochondrial division machinery. Next, the authors tested different cell culture systems for the effect of loss of Drp1-mediated mitochondrial division on mitophagy. For this, mitophagy was induced primarily by exposing cells to hypoxic conditions as well as by treating cells with deferiprone, an iron-chelating drug, or by dissipation of the mitochondrial membrane potential using carbonyl cyanide m-chlorophenylhydrazone. Under all tested conditions, Yamashita et al. (2016) observed turnover of small mitochondrial fragments by mitophagy in the absence of the Drp1-based mitochondrial division machinery. Collectively, these data indicate that Drp1/Dnm1-independent mitophagy is evolutionarily conserved and occurs under a variety of mitophagy-inducing conditions.
These observations raised two important questions: How are the mitochondrial fragments found in autophagosomes generated in the absence of Drp1/Dnm1, and does such a Drp1/Dnm1-independent mechanism still occur when the canonical mitochondrial division machinery is present? To address these questions, Yamashita et al. (2016) performed fluorescence-based live-cell imaging and followed the formation of autophagosomes and the generation of mitochondrial fragments over time in wild-type and Drp1-knockout HeLa cells during hypoxia-induced mitophagy. Remarkably, autophagosome biogenesis occurred on mitochondrial tubules and small mitochondrial fragments started to bud and divide from mitochondrial tubules concurrently with the expansion and closure of the isolation membrane of forming autophagosomes, irrespective of whether Drp1 was present in these cells or not (Fig. 1 B). In fact, using triple color imaging, (Yamashita et al., 2016) showed that whereas Drp1 formed foci at canonical division sites on mitochondrial tubules as expected, it was not detected at the sites of mitochondrial budding associated with autophagosome formation. Hence, Drp1/Dnm1 is not only dispensable for mitophagy, it also seems to be absent from sites of mitochondrial constriction and division during mitophagy. These key findings provide a novel perspective on the process of mitophagy. In contrast to previous sequential models proposing that mitochondrial fragments form in a Drp1/Dnm1-dependent manner before they can be subsequently targeted by autophagy, data presented by Yamashita et al. (2016) support a model in which autophagosome formation and mitochondrial budding and division are spatially and temporally coordinated events (Fig. 1). In fact, they showed that the generation of these mitochondrial fragments depended on the integrity of the autophagy machinery, raising the possibility that the autophagy machinery itself drives mitochondrial budding and division through nucleation and expansion of the isolation membrane at these sites. Alternatively, an as-yet-unidentified machinery might promote membrane scission during autophagosome formation for mitophagy.
The model proposed by Yamashita et al. (2016) of Drp1/Dnm1-independent mitochondrial division during mitophagy poses some interesting new questions. Foremost, how are sites of autophagosome formation on mitochondrial tubules for mitophagy determined and what are the potential signals from mitochondria to the autophagy machinery? It was shown previously that the autophagy machinery is recruited in a punctate pattern that coincides with focal ubiquitination sites after parkin recruitment along mitochondrial tubules that are producing elevated levels of reactive oxygen species (ROS; Yang and Yang, 2013). Thus, generation of a short-ranged ROS signal could initiate formation of autophagosomes by selectively targeting ROS-producing regions on mitochondrial tubules. Future work is required to dissect the detailed mechanisms that control autophagosome formation on damaged mitochondria.
How can we reconcile this new study by Yamashita et al. (2016) with previously published data suggesting a role for Drp1/Dnm1 in mitophagy into a unifying model? Although Drp1/Dnm1-mediated mitochondrial division is not essential, it might facilitate mitophagy by generating mitochondrial fragments that are easily targeted and degraded. Perhaps the severity or nature of the stress may determine if mitochondrial fragmentation driven by Drp1/Dnm1 becomes the dominant factor for mitochondrial turnover by mitophagy. Under stress conditions that do not induce Drp1/Dnm1-mediated fragmentation, or only lower rates, Drp1/Dnm1-independent mitochondrial fragmentation driven by the autophagy machinery might be the rate-limiting process for mitochondrial turnover by mitophagy. Careful analysis of mitochondrial dynamics, and the contribution of these two partially redundant pathways for mitophagy, in the presence of different cellular stresses will be required to clarify these issues.
Acknowledgments
I would like to apologize to all my colleagues who have contributed critical work to the topics discussed here, but whose work I could not cite because of space limitations.
This work was supported by the Max Planck Society.
The authors declare no competing financial interests.
References
- Abeliovich H., Zarei M., Rigbolt K.T., Youle R.J., and Dengjel J.. 2013. Involvement of mitochondrial dynamics in the segregation of mitochondrial matrix proteins during stationary phase mitophagy. Nat. Commun. 4:2789 10.1038/ncomms3789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt D., Müller M., Reichert A.S., and Osiewacz H.D.. 2015. Simultaneous impairment of mitochondrial fission and fusion reduces mitophagy and shortens replicative lifespan. Sci. Rep. 5:7885 10.1038/srep07885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., and Klionsky D.J.. 2014. The machinery of macroautophagy. Cell Res. 24:24–41. 10.1038/cr.2013.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y., Shirakabe A., Maejima Y., Zhai P., Sciarretta S., Toli J., Nomura M., Mihara K., Egashira K., Ohishi M., et al. 2015. Endogenous Drp1 mediates mitochondrial autophagy and protects the heart against energy stress. Circ. Res. 116:264–278. 10.1161/CIRCRESAHA.116.303356 [DOI] [PubMed] [Google Scholar]
- Kageyama Y., Hoshijima M., Seo K., Bedja D., Sysa-Shah P., Andrabi S.A., Chen W., Höke A., Dawson V.L., Dawson T.M., et al. 2014. Parkin-independent mitophagy requires Drp1 and maintains the integrity of mammalian heart and brain. EMBO J. 33:2798–2813. 10.15252/embj.201488658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanki T., Wang K., Cao Y., Baba M., and Klionsky D.J.. 2009. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell. 17:98–109. 10.1016/j.devcel.2009.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner L.L. 2014. Shaping the dynamic mitochondrial network. BMC Biol. 12:35 10.1186/1741-7007-12-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., and Youle R.J.. 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 524:309–314. 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao K., Wang K., Liu X., and Klionsky D.J.. 2013. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev. Cell. 26:9–18. 10.1016/j.devcel.2013.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendl N., Occhipinti A., Müller M., Wild P., Dikic I., and Reichert A.S.. 2011. Mitophagy in yeast is independent of mitochondrial fission and requires the stress response gene WHI2. J. Cell Sci. 124:1339–1350. 10.1242/jcs.076406 [DOI] [PubMed] [Google Scholar]
- Murakawa T., Yamaguchi O., Hashimoto A., Hikoso S., Takeda T., Oka T., Yasui H., Ueda H., Akazawa Y., Nakayama H., et al. 2015. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat. Commun. 6:7527 10.1038/ncomms8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., and Youle R.J.. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803. 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J., and Suomalainen A.. 2012. Mitochondria: In sickness and in health. Cell. 148:1145–1159. 10.1016/j.cell.2012.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto K., Kondo-Okamoto N., and Ohsumi Y.. 2009. Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell. 17:87–97. 10.1016/j.devcel.2009.06.013 [DOI] [PubMed] [Google Scholar]
- Rambold A.S., Kostelecky B., Elia N., and Lippincott-Schwartz J.. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 108:10190–10195. 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A., Cleland M.M., Xu S., Narendra D.P., Suen D.F., Karbowski M., and Youle R.J.. 2010. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. J. Cell Biol. 191:1367–1380. 10.1083/jcb.201007013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita S.-i., Jin X., Furukawa K., Hamasaki M., Nezu A., Otera H., Saigusa T., Yoshimori T., Sakai Y., Mihara K., and Kanki T.. 2016. Mitochondrial division occurs concurrently with autophagosome formation but independently of Drp1 during mitophagy. J. Cell Biol. 10.1083/jcb.201605093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J.Y., and Yang W.Y.. 2013. Bit-by-bit autophagic removal of parkin-labelled mitochondria. Nat. Commun. 4:2428 10.1038/ncomms3428 [DOI] [PubMed] [Google Scholar]