Abstract
The paucity of traditional epidemiological data during epidemic emergencies calls for alternative data streams to characterize the key features of an outbreak, including the nature of risky exposures, the reproduction number, and transmission heterogeneities. We illustrate the potential of Internet data streams to improve preparedness and response in outbreak situations by drawing from recent work on the 2014–2015 Ebola epidemic in West Africa and the 2015 Middle East respiratory syndrome (MERS) outbreak in South Korea. We show that Internet reports providing detailed accounts of epidemiological clusters are particularly useful to characterize time trends in the reproduction number. Moreover, exposure patterns based on Internet reports align with those derived from epidemiological surveillance data on MERS and Ebola, underscoring the importance of disease amplification in hospitals and during funeral rituals (associated with Ebola), prior to the implementation of control interventions. Finally, we discuss future developments needed to generalize Internet-based approaches to study transmission dynamics.
Keywords: transmission chain, cluster, Internet data stream, big data, transmission patterns, exposure setting, contact network, reproduction number, Ebola, MERS
Mathematical models of disease transmission have the potential to guide public health control strategies during epidemic emergencies [1]. However, transmission models require careful ground-truthing in epidemiologic data to generate accurate incidence forecasts, estimate key transmission parameters such as the reproduction number, and assess the strength of interventions required for control. Detailed epidemiological data are typically scarce, however, during the early stages of an emerging infection, owing to delays in identification of early transmission events, especially in regions with limited surveillance, and reluctance to rapidly release data in the public domain [2].
In the absence of detailed epidemiological information rapidly available from traditional surveillance systems, alternative data streams are worth exploring to gain a reliable understanding of disease dynamics in the early stages of an outbreak. The world of social media and the Internet offers great opportunities to explore the performances of nontraditional surveillance systems in outbreak situations [3–6]. Of particular interest is the reconstruction of transmission chains between successive cases (termed “transmission trees” or “clusters”), which is critical to understand the nature of exposure events and transmission heterogeneities, and the temporal evolution of the reproduction number over disease generations. Indeed, the effective reproduction number estimated during the early epidemic growth phase quantifies the transmission potential of an infectious pathogen, in turn informing the likelihood of large-scale outbreaks and the intensity of control interventions needed to stamp out the outbreak [7, 8]. Early estimates of >1.0 for the reproduction number indicate the potential for a major outbreak, and estimates of <1.0 indicate that small transmission chains are possible but that the infection will quickly die out before a large-scale epidemic can be generated. The reproduction number is a dynamic and complex quantity, however, that depends on local conditions that may change over the course of the outbreak, including behavioral and environmental factors, and control interventions.
Here we review recent efforts to collate and analyze Internet reports from authoritative media outlets and public health authorities to gain reliable information on exposure patterns and transmission chains for emerging infections, in the near absence of granular epidemiological reports. We illustrate the potential of Internet data streams by drawing from recent work on the 2014–2015 Ebola epidemic in West Africa and the 2015 Middle East Respiratory Syndrome (MERS) outbreak in South Korea and discuss how to expand this work in the context of the big-data revolution.
THE 2015 MERS OUTBREAK IN SOUTH KOREA
Our first example of the use of Internet-based information draws from a recent large-scale outbreak of infection due to MERS coronavirus, a zoonotic virus that has caused sporadic but recurrent outbreaks in humans since March 2012, particularly in the Middle East [9]. The concentration of human infections has been linked to the local population of dromedary camels, which may serve as an intermediate host for MERS [10, 11]. The human-to-human transmission potential of MERS in the community at large appears to remain subcritical; however, outbreaks tend to be amplified via nosocomial transmission [9, 12, 13]. Case importation from the Middle East continues to represent a substantial risk for outbreaks, as recently exemplified by the 2015 MERS outbreak in South Korea. We concentrate on this large outbreak, sparked by a single index case who arrived in South Korea on 4 May 2015. The index case developed symptoms 7 days later and did not receive a diagnosis of MERS until 20 May 2015, after having sought treatment in several healthcare facilities [14].
In the case of the 2015 South Korean MERS outbreak, epidemiological information was available from traditional surveillance systems, but detailed, high-resolution data had to be parsed out from online reports emanating from disparate health authorities, including the Korean Centers for Disease Control, the Ministry of Health and Welfare of South Korea, and the World Health Organization [14–17]. Systematic near real-time analysis of these online reports allowed reconstruction of MERS transmission chains, which can be considered a single giant cluster in this outbreak (Supplementary Figure 1) [18]. The full transmission tree comprised 150 cases linked to nosocomial events, where each case was classified according to occupational and social exposure. It became clear relatively early on that all cases were linked to exposure in the healthcare setting: 150 cases, included 107 hospital patients (including the index patient), 28 visitors or family members, 12 healthcare workers, and 3 nonclinical staff.
The South Korean MERS outbreak comprised 3 disease generations, with the index patient representing generation 0. Estimates of the reproduction number according to disease generation can be derived by averaging the number of secondary cases in each generation [18]. The average reproduction number followed a declining trend, from 30 cases in the first generation to 3.8 in the second generation and 0.1 in the third generation (Supplementary Figure 1). Overall, this outbreak followed a similar trajectory to previous hospital clusters involving coronaviruses [18], with early super-spreading events generating a disproportionate number of secondary infections, followed by a rapid decline of the reproduction number to <1.0 in subsequent generations as infection control measures gained strength.
THE 2014–2015 EBOLA EPIDEMIC IN WEST AFRICA
Our second example of using Internet data to characterize transmission dynamics is based on the 2014–2015 Ebola outbreak in West Africa. In contrast to the MERS outbreak previously described, scarce epidemiological data were available throughout the Ebola outbreak from traditional surveillance systems. To alleviate the need for solid epidemiological information and assess Ebola transmission characteristics, we designed an approach to systematically collect information on Ebola case clusters from Internet news reports published during the outbreak [19]. Below we extend and update this work and reflect on future developments needed to generalize Internet-based approaches to study transmission dynamics more broadly.
To obtain detailed information on Ebola transmission chains, we reviewed news stories and investigative reports published between January 2014 and January 2016 and describing suspected, probable, and confirmed cases of Ebola in the 3 most affected countries (Guinea, Sierra Leone, and Liberia). We focused on reports available from the World Health Organization Web site, particularly news segments published in the section “Stories from the field on Ebola,” and Ebola situational reports, as well as online authoritative media outlets (see Supplementary Table 1 for a complete list of case clusters, their characteristics and corresponding sources).
We manually reviewed and selected articles that contained detailed stories about Ebola case clusters arising within families or via funerals or hospital exposure. Each patient with Ebola was assigned one or several types of exposure (family/household, hospital, sexual, or funeral). We also analyzed Ebola transmission dynamics for a subset of clusters for which transmission chains were explicitly described in the articles or could be inferred based on chronological information on the timing of symptoms of successive cases (Figure 1).
Figure 1.
Set of representative clusters of Ebola transmission chains extracted from Internet reports. A, Cluster 1 (Supplementary Table 1). In March 2014, the index patient traveled from his village to Conakry to be treated after visiting and infecting a physician. He stayed with family, 4 members of which became ill, and died in the hospital. His body was taken back to the village for a traditional burial, where 3 uncles washed his body and soon became sick. B, Cluster 22 (Supplementary Table 1). During June–September 2014, after the hospital from cluster 5 closed, a Monrovian patient resorted to receiving care from her church caretaker, who then went to a clinic and infected a guard, whom a healthcare worker and father treated. The guard then infected his son, whose mother denied that it was Ebola. This led to the rest of the family becoming infected. C, Cluster 47 (Supplementary Table 1). In October and November 2014, an imam developed symptoms in Guinea and then visited a family in Bamako, Mali. He went to a clinic and died there, infecting a nurse, physician, and all members of the family he stayed with. His body was returned to Guinea, where at least 1 infection occurred from his large traditional funeral. D, Cluster 62 (Supplementary Table 1). From December 2014 to February 2015, all of the cases in Liberia stemmed from one woman, who infected family members, a neighbor, and an herbalist she went to for treatment. During the third generation of secondary cases, contact tracing efforts helped stop further spreading.
Based on Internet news reports, we identified 104 Ebola virus disease (EVD) clusters between January 2014 and January 2016 originating in Guinea (18 clusters), Sierra Leone (40 clusters), and Liberia (46 clusters). The monthly number of clusters identified from news reports tracked the total number of EVD cases reported by traditional surveillance systems during the study period (Spearman rho = 0.86; P < .001; Figure 2B).
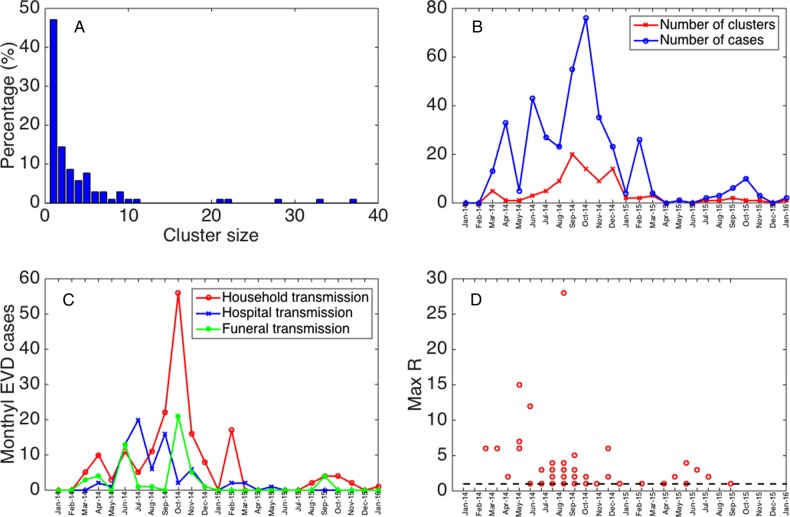
Figure 2.
Characteristics of the Ebola virus disease (EVD) case clusters derived from Internet reports, January 2014–January 2016. A, The distribution of cluster sizes of Ebola cases in our sample of clusters. B, The temporal variation in the number of clusters and total cases reported across clusters. C, The temporal changes in the distribution of Ebola exposures through the family, hospitals, and funerals. D, The temporal variation in the maximum number of secondary cases per case across all disease generations in each of the clusters with available information. The horizontal dashed line at R = 1 is shown for reference.
Of the 104 clusters, 101 (97%) were limited to a single country. The reported cluster size ranged from 1 to 37 cases (Figure 2A) and included up to 6 disease generations. The mean cluster size was estimated at 3.9 (95% confidence interval [CI], 3.1–4.7) based on fitting a negative binomial distribution. Most of the secondary cases were linked to the the index case (46.5%), while only 13.9% stemmed from first-generation cases, and 8% stemmed from second-generation cases. Overall, the mean reproduction number was estimated to be 2.4 (95% CI, 1.4–3.4) for index cases, ranging from 0 to 28. The maximum reproduction number was higher during the first few months of the epidemic, prior to November 2014 (Figure 2D), but the temporal trend in the reproduction number was not significant.
Of particular interest is the cluster of EVD cases associated with the outbreak in Nigeria, sparked by a single importation from Liberia on 20 July 2014 [2]. The transmission tree comprises 20 Ebola cases, including 11 healthcare workers, 9 of whom acquired the virus from the index case before the disease was identified in the country. The index case generated 12 secondary cases in the first generation, 5 in the second generation, and 2 in the third generation. This led to a declining reproduction number, from 12 in the first generation to <1.0 for subsequent generations, coinciding with the implementation of stringent contact tracing.
Overall, in our 104 clusters, exposure via family contacts (58.6%) was the most frequent, followed by hospital-based exposures (23.9%) and funerals (17.5%). The frequency of Ebola cases arising from funeral and hospital cases peaked during the early months of the epidemic; hospital exposure in particular declined considerably after July 2014.
DISCUSSION
Our study offers a proof of concept that publicly available online reports released in real-time by ministries of health, local surveillance systems, the WHO, and authoritative media outlets are useful to identify key information on exposure and transmission patterns during epidemic emergencies. We illustrate our findings with data from recent and well-publicized outbreaks of MERS and Ebola; EVD; our Internet-based findings on exposure patterns are in good agreement with those derived from traditional epidemiological surveillance data, which can be available after considerable delays [20, 21]. Our reproduction number analysis confirms or brings new light to important aspects of transmission characteristics, in particular amplification of the outbreak in hospital or funeral settings and a rapid clamp down in transmission rates as control interventions are strengthened.
The 2014–2015 Ebola epidemic in West Africa is a particularly interesting case study to explore the relevance of digital data streams to elucidate transmission patterns in a data-poor environment. Publicly available epidemiological data from the WHO were largely limited to aggregate weekly EVD case counts at the country level. In fact, this was the primary publicly available data set that many researchers around the world used to calibrate epidemic models. Subnational case data became available later and revealed substantial spatial heterogeneity in transmission patterns across West Africa, which could have affected epidemic forecasts and transmission potential estimates [22]. And at the time of this writing, >2.5 years after the onset of what may be one the most important outbreaks of the decade, detailed transmission chain data arising from official contact tracing efforts remain scarce and limited to a few clusters [20, 21, 23]. While our study is restricted by the amount of online information that could be processed manually, scaling-up would be possible with more-sophisticated computational tools that scour the Internet, social media, and other big-data streams to identify information on a larger set of transmission chains. These automatically sensed data sets could then be fed into modeling studies of the type we have shown here.
In our data, the main exposure to EVD was via family contacts (58.6%), which is in line with exposure patterns from prior Ebola outbreaks [24–27] and chains of transmission for the ongoing epidemic in Guinea (February–August 2014) [20]. The frequency of Ebola cases arising from funeral and hospital cases in our data peaked during the early months of the epidemic, which suggests that amplified transmission events in the healthcare setting and during funeral ceremonies facilitated the transmission of the virus across communities. Hospital exposures declined considerably after July 2014, likely as a result of the improvement in infection control measures in healthcare settings. Similarly, funeral exposures have occurred sporadically during the later containment phase of the epidemic. The decline in hospital-based transmission is in line with a decline in the proportion of healthcare workers in our data and statistics retrieved from the WHO situational reports [28].
Our analysis of the temporal variation in exposure patterns provides useful information to assess the impact of control measures and behavior changes during epidemics. Our mean estimate of the reproduction number for EVD is on the higher end of published estimates based on time series analysis of outbreaks in Central [29, 30] and West Africa [31–35] or estimates based on transmission trees in Guinea during March–August 2014 [20].
Overall, our transmission chains for EVD and MERS clusters indicate a rapidly declining trend in the reproduction number over disease generations, consistent with the presence of early super-spreading events. This pattern could result from a combination of factors, including changes in population behavior that mitigate transmission, characteristics of and spatial heterogeneity in the underlying network of contact over which the disease spreads, and control interventions. Standard compartment model theory stipulates that an outbreak should follow exponential growth before susceptible depletion or interventions set in. In contrast, a number of real epidemics have been shown to follow subexponential growth, with the effective reproduction number declining toward 1.0 in the first 3–5 disease generations, even in the absence of control interventions, depletion of susceptible individuals, or population behavioral changes [36]. The observed transmission patterns in EVD and MERS case clusters presented in this study, particularly the decline in the effective reproduction number over few disease generations, are in agreement with the subexponential growth rate behavior identified from population-level time series data [22, 37, 38] Temporal variation in exposure patterns and reproduction number should provide crucial information for the design and calibration of epidemic models particularly when these are intended to generate forecasts of the epidemic trajectory.
The analysis of case clusters from Internet news reports is not exempt of limitations. First, case clusters are subject to reporting bias. For instance, news stories could be focused on survivor or sensationalist stories or could reflect an American-centric bias, with a higher coverage of news reports for countries that are more connected to the United States. Second, larger clusters tend to be included in news reports. Finally, the amount of information in each story or news report varies, with variables such as age and sex missing in the great majority of the clusters. Reassuringly, while our sample of case clusters extracted from Internet reports corresponds to just a small fraction of the total EVD case burden in the 3 most affected countries in West Africa, the broad epidemiological features of the Internet-based data (age, timing, and exposure) were well in line with summary statistics from health authorities.
Our work is placed in the context of a growing body of digital epidemiology studies promoting the use of nontraditional online data sources to enhance detection, forecasting, and response to infectious disease threats [3–6] even before official surveillance reports are released. Other applications include HealthMap [39], a system that extracts epidemiological outbreak data from the Internet in near real time and has been used to analyze large-scale epidemics, such as the 2010 cholera outbreak in Haiti [40, 41]. Other studies have related temporal changes in the implementation of control interventions extracted from Internet reports with the trajectory of the Ebola epidemic in West Africa (eg, variation in the reproduction number) [42]. Here we have provided further evidence that systematic collection and analysis of unstructured news from authoritative sources and surveillance reports may provide a reliable mean to assess epidemiological patterns in near real time. It is worth pointing out that translational research in this field has been slow or has not been properly recorded. Careful documentation and feedback from end users and stakeholders (eg, clinicians, policymakers, and public health officials) is warranted to foster further development and refinement of digital epidemiology tools.
Our work on analysis of transmission patterns from Internet sources has relied on manual extraction and analysis of text information, a task that requires a significant amount of time that grows approximately proportionally to the amount of data in the sample. Moreover, our search focused on Internet reports and did not capture other digital data streams, including Twitter and other Web-based resources. Scaling-up the amount of information used would yield more powerful and potentially less biased analyses but would require design of novel computational tools to search, extract, analyze, interpret and visualize unstructured data from different sources. Highly flexible open source programs have been developed to scrape Internet data and could be the bedrock of such computational tools, especially in light of the rapid expansion of mining packages in R [43]. Yet the development of these tools poses several challenges related to the need for systematic and integrated search, extraction, and curation of diverse Internet data sets that contribute information about travel patterns, changes in social behavior, exposure settings, and healthcare demand and capacity. Moreover, the analysis tool kit could include the detection of critical events (eg, effects of interventions), sentiment analysis [5], and visualization of temporal and spatial epidemiological patterns. Other challenges in this process include the classification of large volumes of data with varying levels of reliability [39]. Clearly, further computational work and careful ground-truthing are needed to assess the potential of state-of-the art text-data-mining tools to compile information that can be used to model the dynamics of emerging and reemerging infections.
Supplementary Data
Supplementary materials are available at http://jid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Financial support. This work was supported by the Division of International Epidemiology and Population Studies, Fogarty International Center, National Institutes of Health (NIH; support to G. C. and C. V.); the RAPIDD Program, Science and Technology Directorate, Department of Homeland Security (support to G. C. and C. V.); the National Science Foundation (NDF; grant 1414374 to G. C.), as part of the joint NSF-NIH-US Department of Agriculture Ecology and Evolution of Infectious Diseases program; and the United Kingdom Biotechnology and Biological Sciences Research Council (grant BB/M008894/1).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Metcalf CJ, Edmunds WJ, Lessler J. Six challenges in modelling for public health policy. Epidemics 2015; 10:93–6. [DOI] [PubMed] [Google Scholar]
- 2.van Panhuis WG, Paul P, Emerson C et al. . A systematic review of barriers to data sharing in public health. BMC Public Health 2014; 14:1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salathe M, Bengtsson L, Bodnar TJ et al. . Digital epidemiology. PLoS Comput Biol 2012; 8:e1002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brownstein JS, Freifeld CC, Madoff LC. Digital disease detection—harnessing the Web for public health surveillance. N Engl J Med 2009; 360:2153–5, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salathe M, Khandelwal S. Assessing vaccination sentiments with online social media: implications for infectious disease dynamics and control. PLoS Comput Biol 2011; 7:e1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lazer D, Kennedy R, King G, Vespignani A. Big data. The parable of Google Flu: traps in big data analysis. Science 2014; 343:1203–5. [DOI] [PubMed] [Google Scholar]
- 7.Anderson RM, May RM. Infectious diseases of humans. Oxford: Oxford University Press, 1991. [Google Scholar]
- 8.Diekmann O, Heesterbeek JA, Metz JA. On the definition and the computation of the basic reproduction ratio R0 in models for infectious diseases in heterogeneous populations. J Math Biol 1990; 28:365–82. [DOI] [PubMed] [Google Scholar]
- 9.Cauchemez S, Fraser C, Van Kerkhove MD et al. . Middle East respiratory syndrome coronavirus: quantification of the extent of the epidemic, surveillance biases, and transmissibility. Lancet Infect Dis 2014; 14:50–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller MA, Meyer B, Corman VM et al. . Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: a nationwide, cross-sectional, serological study. Lancet Infect Dis 2015; 15:559–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kayali G, Peiris M. A more detailed picture of the epidemiology of Middle East respiratory syndrome coronavirus. Lancet Infect Dis 2015; 15:495–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breban R, Riou J, Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet 2013; 382:694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chowell G, Blumberg S, Simonsen L, Miller MA, Viboud C. Synthesizing data and models for the spread of MERS-CoV, 2013: key role of index cases and hospital transmission. Epidemics 2014; 9:40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization. Global Alert and Response (GAR). Coronavirus infections, 2015. http://www.who.int/csr/don/archive/disease/coronavirus_infections/en/ Accessed 15 June 2015.
- 15.Flutrackers. South Korea Coronavirus MERS Case List - including imported and exported cases. 2015. https://flutrackers.com/forum/forum/novel-coronavirus-ncov-mers-2012-2014/novel-coronavirus-who-chp-wpro-ecdc-oie-fao-moa-reports-and-updates/south-korea-coronavirus/732065-south-korea-coronavirus-mers-case-list-including-imported-and-exported-cases Accessed 15 July 2015.
- 16.Korean Centers for Disease Control. Middle East respiratory syndrome report [09 June 2015]. http://www.cdc.go.kr/CDC/notice/CdcKrIntro0201.jsp?menuIds=HOME001-MNU1154-MNU0005-MNU0011&fid=21&q_type=&q_value=&cid=63295&pageNum= Accessed 15 June 2015.
- 17.Ministry of Health and Welfare of South Korea. Information on Middle East respiratory syndrome (MERS) reported in South Korea. http://english.mw.go.kr Accessed 15 July 2015.
- 18.Chowell G, Abdirizak F, Lee S et al. . Transmission characteristics of MERS and SARS in the healthcare setting: a comparative study. BMC Med 2015; 13:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cleaton JM, Viboud C, Simonsen L, Hurtado AM, Chowell G. Characterizing Ebola transmission patterns based on Internet news reports. Clin Infect Dis 2015; 62:24–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faye O, Boelle PY, Heleze E et al. . Chains of transmission and control of Ebola virus disease in Conakry, Guinea, in 2014: an observational study. Lancet Infect Dis 2015; 15:320–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shuaib F, Gunnala R, Musa EO et al. . Ebola virus disease outbreak - Nigeria, July-September 2014. MMWR Morb Mortal Wkly Rep 2014; 63:867–72. [PMC free article] [PubMed] [Google Scholar]
- 22.Chowell G, Viboud C, Hyman JM, Simonsen L. The Western Africa Ebola virus disease epidemic exhibits both global exponential and local polynomial growth rates. PLoS Curr 2015; doi:10.1371/currents.outbreaks.8b55f4bad99ac5c5db3663e916803261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Curran KG, Gibson JJ, Marke D et al. . Cluster of Ebola virus disease linked to a single funeral—Moyamba District, Sierra Leone, 2014. MMWR Morb Mortal Wkly Rep 2016; 65:202–5. [DOI] [PubMed] [Google Scholar]
- 24.Dowell SF, Mukunu R, Ksiazek TG, Khan AS, Rollin PE, Peters CJ. Transmission of Ebola hemorrhagic fever: a study of risk factors in family members, Kikwit, Democratic Republic of the Congo, 1995. Commission de Lutte contre les Epidemies a Kikwit. J Infect Dis 1999; 179(suppl 1):S87–91. [DOI] [PubMed] [Google Scholar]
- 25.Breman JG, Piot P, Johnson KM et al. . The epidemiology of Ebola hemorrhagic fever in Zaire, 1976. Ebola Virus Haemorrhagic Fever 1978:103–24. [Google Scholar]
- 26.Francesconi P, Yoti Z, Declich S et al. . Ebola hemorrhagic fever transmission and risk factors of contacts, Uganda. Emerg Infect Dis 2003; 9:1430–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wamala JF, Lukwago L, Malimbo M et al. . Ebola hemorrhagic fever associated with novel virus strain, Uganda, 2007–2008. Emerg Infect Dis 2010; 16:1087–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.World Health Organization. Unprecedented number of medical staff infected with Ebola. http://www.who.int/mediacentre/news/ebola/25-august-2014/en/ Accessed 25 August 2014.
- 29.Chowell G, Hengartner NW, Castillo-Chavez C, Fenimore PW, Hyman JM. The basic reproductive number of Ebola and the effects of public health measures: the cases of Congo and Uganda. J Theor Biol 2004; 229:119–26. [DOI] [PubMed] [Google Scholar]
- 30.Legrand J, Grais RF, Boelle PY, Valleron AJ, Flahault A. Understanding the dynamics of Ebola epidemics. Epidemiol Infect 2007; 135:610–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chowell G, Nishiura H. Transmission dynamics and control of Ebola virus disease (EVD): a review. BMC Med 2014; 12:196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Althaus CL. Estimating the reproduction number of Zaire ebolavirus (EBOV) during the 2014 outbreak in West Africa. PLOS Curr Outbreaks 2014; 1 doi:101371/currentsoutbreaks91afb5e0f279e7f29e7056095255b288. [DOI] [PMC free article] [PubMed]
- 33.Towers S, Patterson-Lomba O, Castillo-Chavez C. Temporal variations in the effective reproduction number of the 2014 West Africa EbolaoOutbreak. PLOS Curr Outbreaks 2014; doi:10.1371/currents.outbreaks.9e4c4294ec8ce1adad283172b16bc908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiura H, Chowell G. Early transmission dynamics of Ebola virus disease (EVD), West Africa, March to August 2014. Euro Surveill 2014; 19:pii:20894. [DOI] [PubMed] [Google Scholar]
- 35.Fisman D, Khoo E, Tuite A. Early epidemic dynamics of the West African 2014 Ebola outbreak: estimates derived with a simple two-parameter model. PLoS Curr 2014; doi:10.1371/currents.outbreaks.89c0d3783f36958d96ebbae97348d571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chowell G, Viboud C, Simonsen L, Moghadas SM. Characterizing the reproduction number of epidemics with early sub-exponential growth dynamics. 2016. http://arxivorg/abs/160301216. [DOI] [PMC free article] [PubMed]
- 37.Viboud C, Simonsen L, Chowell G. A generalized-growth model to characterize the early ascending phase of infectious disease outbreaks. Epidemics 2016; 15:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chowell G, Sattenspiel L, Bansal S, Viboud C. Mathematical models to characterize early epidemic growth: A Review. Phys Life Rev 2016; doi:10.1016/j.plrev.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brownstein JS, Freifeld CC. HealthMap: the development of automated real-time internet surveillance for epidemic intelligence. Euro Surveill 2007; 12:E071129 5. [DOI] [PubMed] [Google Scholar]
- 40.Chunara R, Andrews JR, Brownstein JS. Social and news media enable estimation of epidemiological patterns early in the 2010 Haitian cholera outbreak. Am J Trop Med Hyg 2012; 86:39–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bahk CY, Scales DA, Mekaru SR, Brownstein JS, Freifeld CC. Comparing timeliness, content, and disease severity of formal and informal source outbreak reporting. BMC Infect Dis 2015; 15:135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majumder MS, Kluberg S, Santillana M, Mekaru S, Brownstein JS. 2014 Ebola outbreak: media events track changes in observed reproductive number. PLoS Curr 2015; doi:10.1371/currents.outbreaks.e6659013c1d7f11bdab6a20705d1e865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R Functions to Perform and Display Google Trends Queries. Package ‘gtrendsR’. https://cran.r-project.org/web/packages/gtrendsR/gtrendsR.pdf Accessed 26 July 2016.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.