Abstract
While big data have proven immensely useful in fields such as marketing and earth sciences, public health is still relying on more traditional surveillance systems and awaiting the fruits of a big data revolution. A new generation of big data surveillance systems is needed to achieve rapid, flexible, and local tracking of infectious diseases, especially for emerging pathogens. In this opinion piece, we reflect on the long and distinguished history of disease surveillance and discuss recent developments related to use of big data. We start with a brief review of traditional systems relying on clinical and laboratory reports. We then examine how large-volume medical claims data can, with great spatiotemporal resolution, help elucidate local disease patterns. Finally, we review efforts to develop surveillance systems based on digital and social data streams, including the recent rise and fall of Google Flu Trends. We conclude by advocating for increased use of hybrid systems combining information from traditional surveillance and big data sources, which seems the most promising option moving forward. Throughout the article, we use influenza as an exemplar of an emerging and reemerging infection which has traditionally been considered a model system for surveillance and modeling.
Keywords: big data, medical claims, Internet search queries, syndromic data, death certificates, electronic patient records, influenza, infectious diseases surveillance, real-time monitoring
A BRIEF HISTORY OF INFECTIOUS DISEASE SURVEILLANCE
Systems capturing disease incidence and mortality have been in place for centuries in high-income countries and have increased in complexity and granularity over time (see Table 1 for a timeline based on [1–12]). An ideal surveillance system is representative of the population, flexible, economic, and resilient, with timely reporting and validation of its outputs [13, 14]. Further, full situational awareness requires availability of multiple surveillance data streams that capture mild and severe clinical outcomes (death certificates, hospital admissions, and emergency department and outpatient visits), as well as laboratory-based information (confirmed cases, genetic sequences, and serologic findings).
Table 1.
A Brief History of Disease Surveillance and Beginnings of Big Data
| Year | Disease(s) | Key Thinker/Surveillance System | Institution(s) |
|---|---|---|---|
| 1662 | Plague | John Graunt | Bills of Mortality, UK [1] |
| 1817 | Smallpox | J. C. Moore | [2] |
| 1847 | Influenza | William Farr | General Registrar Office, UK [3] |
| 1854 | Cholera | John Snow | General Registrar Office, UK [4] |
| ∼1900 | All | International Classification of Diseases | WHO [5] |
| 1918 | Influenza | 121 Cities Mortality Reporting System | CDC [6] |
| 1976 | Influenza | Weekly viral surveillance | CDC [6] |
| 1984 | Influenza, viral hepatitis, acute urethritis, measles, mumps | First computerized disease surveillance network | French Sentinelles Network [7] |
| ∼2000 | All | Medical claims forms are first used | Private companies and government [8, 9] |
| 2008 | Influenza | Google Flu Trends launched | Google [10, 11] |
| 2015 | Influenza | Birth of hybrid systems | Public/private partnership [12] |
Select key events are listed. Historical beginnings coincide with the origins of the field of epidemiology.
Abbreviations: CDC, Centers for Disease Control and Prevention; UK, United Kingdom; WHO, World Health Organization.
The 19th century saw the rapid development of systematic sentinel surveillance systems in large cities of Europe and North America. Physicians systematically reported on weekly incidences of diseases and deaths with increasingly richer stratification by cause, age, and sex. Such data supported important health policy decisions, such as the introduction the smallpox vaccination programs and subsequent evaluation of intervention [15]. Meanwhile, cause-of-death coding evolved around 1900 into what is now known as the International Classification of Diseases (ICD; Table 1) [5, 13].
In the 20th century, rapid technological advances in microbiology led to laboratory surveillance systems that still form the core of disease surveillance today. Additionally, advances in computing power and information technology allowed for the development of electronic reporting of common illnesses by physicians, providing a platform for public health authorities to communicate back to clinicians and the public in a timely fashion (eg, the French Sentinelles system [7]).
In the 21st century, laboratory-based surveillance benefits from increased use of multiplex reverse transcription–polymerase chain reaction and increasingly rapid pathogen identification. Advanced detection tools can dramatically cut the time to accurate diagnosis in low-resource and crisis settings and can be deployed in the field, as illustrated by proof-of-concept studies during the 2014 West African Ebola virus outbreak and, more recently, the Zika virus epidemic [16–19]. Also, the availability of accurate antibody-based diagnostic tests has made seroepidemiologic studies feasible in near real time for emerging viral threats such as pandemic influenza [20].
Along with the increasing sophistication of these classical surveillance components, we have seen an accelerated development of novel systems that rely on big data streams. These systems include electronic death certificates, patient-level hospital discharge records, and medical claims data, in which use of ICD coding allows comparison of syndromic disease patterns over time and between locales. In parallel, novel surveillance approaches using big data streams from Internet search queries, social media, and crowdsourcing have been proposed and are in use. In the following sections, we discuss the upsides and downsides of these novel systems, highlight recent applications, and suggest opportunities for cross-fertilization.
THE BIG DATA ERA
Electronic Health Records
The use of big data in the public health surveillance arena lags decades behind that in other sectors, such as marketing, climatology, and earth sciences. Although mortality- and national hospital discharge records have been available in electronic format since the 1970s, lack of timeliness has typically been a barrier as data release is delayed by years in most countries. There is now a push from public health agencies to use electronic patient records in a more timely fashion to track important events. For example, the Food and Drug Administration launched the Sentinel Initiative, which aims to use private sector electronic health data to identify severe adverse drug events [21]. The aim of this initiative is to augment traditional surveillance that relies on passive case reporting by physicians. Another example is the Centers for Disease and Control and Prevention's (CDC's) Biosense platform, where a core aspect of the National Syndromic Surveillance Project [22] is to collect electronic health records for real-time awareness and tracking of pandemic influenza or any other novel health threat. Further, in Germany, researchers have demonstrated the utility of medical claims data to track vaccine uptake [23]. There are clearly wonderful possibilities for use of large volumes of deidentified electronic health medical records to monitor infectious diseases outcomes (physicians' visits, hospitalizations, and deaths), as well as the uptake patterns of vaccines, drugs, and their associated adverse events. However, in reality these developments have been slow due in part to bioethical concerns regarding protection of patient privacy. Indeed, even with deidentified and aggregated health data, privacy remains a concern with highly granular data sets. Other obstacles include the prohibitive cost of purchasing such data from private sector vendors and delays in the development of analytics platforms. Also, academic studies demonstrating the performances of electronic health data against ground-truth traditional surveillance systems remain relatively scarce [8]. There is continued need for proper validation of electronic health–based surveillance systems going forward, to ensure that the output of new data systems are useful and practically accurate.
The Age of the Internet and Social Media
Internet communications have opened up novel types of big data that can be harnessed for disease surveillance, including social media and search query data [24]. Such data are not directly related to medical encounters or disease outcomes and can also potentially capture health information even on those individuals who did not seek formal medical care. An example is the seminal work by Google to track influenza epidemics by using Internet search query data [10, 25, 26], as well as recent analyses of Wikipedia and Twitter feeds for near-timely studies of the 2009 influenza pandemic and other outbreaks [12, 27–30]. These initiatives offer exciting opportunities but have been shown to have important drawbacks, as we will discuss in more detail later.
Recent years have also seen the rise of participatory Internet-based surveillance systems, in which individuals report on their disease symptoms on a voluntary basis by email, text messaging, Tweets, or web interface. These systems harness the huge capacity of crowdsourcing, as many individuals actively contribute to these networks. The best established examples are for influenza [31–33], but application of similar methods would be possible for other diseases. The upsides are numerous, including volume, speed, intended utility for research, relatively minimal cost, and availability of validation against traditional surveillance systems for the longest running of these participatory systems [32]. But challenges remain, however, particularly due to differing reporting and health-seeking behaviors across time, countries, and age groups, although these issues are pervasive to most big data systems.
In parallel, online computational systems, such as Healthmap, hosted at Harvard University, or the Global Public Health Intelligence Network in Canada [34, 35], allow intelligent synthesis of multiple sources of disease outbreak information. These reactive high-volume surveillance systems scan a variety of structured and unstructured online reports to identify and track novel outbreaks and other health issues, such as drug resistance (see the article by Mc Fadden et al in this issue). Looking ahead, we can hope for entirely novel and more specific data streams; for example, technology is close to enabling an individual to self-diagnose, using immunoassays embedded on a smartphone [36]. Taken together, these innovative big data efforts offer the tantalizing opportunity to greatly increase the amount of information available in surveillance systems, echoing the satellite data revolution that boosted earth sciences decades ago. This would help shift disease surveillance beyond large administrative units and to the local level, which is often an important knowledge gap in traditional surveillance systems that still operate at the state, regional, or national level.
INFLUENZA SURVEILLANCE AS A CASE STUDY
Influenza is a particularly rich disease system for illustrate the state of the art of outbreak surveillance and make recommendations for the future. In the United States and the United Kingdom, for example, influenza surveillance relies on multicomponent systems involving a variety of laboratory and clinical elements, many of which have been in place for decades. Each component adds a piece of information regarding geographic spread, genetic and antigenic make-up of circulating viruses, and health impact, both in terms of milder outcomes (outpatient visits) and severe outcomes (hospitalizations and deaths). This information is systematically updated and disseminated in weekly reports and seasonal summaries that are publicly available online [6, 37]. In the United States, an effort has been made in the past 5 years to make surveillance data publicly available through relatively user-friendly Internet tools [6]. However, the finest spatial resolution continues to be the state or region for most components, and age breakdown is available for a subset of outcomes only. Further, timing is not ideal, as there is currently a 2-week delay in reporting of mortality and laboratory confirmed cases.
When considering augmenting or replacing these traditional surveillance components with big data elements, it is critical to validate the performance of the new systems. As a case in point, the CDC recently launched an initiative to monitor influenza-related deaths based on a real-time electronic sample of US death certificates—a new system that has just replaced the age-old 122 Cities Mortality Reporting System of manually evaluated death certificates [6].
In other countries, rapid availability of large patient-level administrative databases and detailed mortality statistics is already in place, which proved instrumental to elucidate the transmission dynamics, age patterns, and health burden of the 2009 influenza A(H1N1) pandemic in near real time [38, 39]. In Mexico, for example, timely hospital data helped identify an unusual excess of severe pneumonia among younger adults and a sharp drop among elderly in the first few weeks of the 2009 A(H1N1) pandemic [38]. In parallel, New Zealand used population-based national health data to quantify the pandemic case-fatality rate in summer 2009, a heavily debated severity indicator at the time [39]. In another study, highly timely electronic hospital and mortality records were used to document the occurrence and severity of a fourth A(H1N1) pandemic wave in Mexico as it was unfolding during February 2014 [40]. Such timely studies were particularly useful to gauge the severity of a novel influenza virus early on and helped inform national and global intervention strategies. Looking ahead, one can envision a global surveillance system for pandemic influenza that consists of a set of sentinel countries in various world regions and climatic zones, with the ability to analyze hospital and mortality records in near real-time.
THE RISE AND FALL OF GOOGLE FLU TRENDS (GFT) AND OTHER SOCIAL MEDIA
In 2008–2009, innovative studies proposed the use of Google Internet search queries to track influenza-like illnesses (ILIs) in the United States [10, 11]. The algorithm developed for GFT was very attractive because it promised a more timely system than classical influenza surveillance (lagging by approximately 1 week only), with reporting at the local level (cities) [10]. Further, once validated, Google was working to develop similar influenza surveillance components in other countries.
There were, however, some difficulties with this new approach [10, 11]. In their raw form, disease-related search queries may capture a variety of signals unrelated to the occurrence of real infectious disease outbreaks. Without filters, the data can be misleading; for example, our search for the word “cholera” in the general Google tool revealed a cholera “epidemic” in the United States in 2007, as a result of Oprah Winfrey picking Love in the Time of Cholera as book of the month in her book club [41]. Likewise, other viral or bacterial respiratory epidemics may lead to a false spike in “flu” queries, as one can search about influenza vaccination in the autumn without being ill. Furthermore, intense media coverage of pandemic threats in faraway locales, such as sporadic human A(H5N1) influenza cases in Asia, can fuel digital outbreaks in other world regions. Therefore, to tease out the true influenza signal from background noise and spurious reporting spikes, Google developed algorithms to process queries that were true to influenza and calibrated those against traditional CDC surveillance ILI data. The original GFT algorithm included 45 search terms biologically plausible with regard to their link with influenza, selected from a list of top 100 search queries that were most correlated with ILI [10].
However, GFT suffered a devastating blow by largely missing the first wave of the influenza A(H1N1) pandemic outbreak, as illustrated by New York City, in May–June 2009 [26, 42, 43]. This was likely due to severe overfitting of the GFT algorithm [25, 26]. Despite initial validation against seasonal influenza data, the GFT algorithm was not suitable for tracking of a pandemic outbreak, which, in 2009, was characterized by unusual timing (summer and early fall activity [25]), a profound change in age patterns (elderly sparing [44]), and profound geographical heterogeneity (the first wave was limited to a few Northern US cities [8]). Taken together, these signature epidemiologic features were dramatically different in the pandemic period and prepandemic GFT training set [26]. While this was not much discussed at the time, the GFT algorithm was quickly revised to better capture the first pandemic wave in retrospective analysis [42].
A few years later, GFT malfunctioned again, this time by greatly overestimating the severity of the 2012–2013 winter influenza epidemic [26]. As Google did not wish to pursue subsequent updates of the algorithm, they instead elected to release search query data to repositories maintained by the CDC and other institutions. In the meantime, other high- and middle-income countries had followed suit, validated GFT against traditional country-specific surveillance data, expanded the approach to monitor and model trends in respiratory and enteric pathogens, and reported various levels of success [45, 46].
These recent malfunctions of GFT have put a damper on the initial hopes for surveillance based on search engine data. This is unfortunate, as approaches harnessing Internet search queries could have captured disease patterns in countries where traditional surveillance data are scarce.
MEDICAL CLAIMS DATA AS THE MISSING LINK
Meanwhile, while much attention has been devoted to Internet and social media data, other sources of big data representing actual medically attended health events and outcomes have remained underused [9]. In the United States and many other developed countries, every outpatient visit results in an electronic claim form submitted for insurance purposes (form CMS-1500 in the United States). This form has information about the location, date of visit, and ICD-coded diagnoses and is therefore particularly useful to track disease outbreaks; further, in large populations, the sheer volume of this system provides unprecedented spatial and temporal resolution. Such syndromic data are deidentified and maintained in large databases in the private sector, where they form the backbone of tracking events such as cough and cold, influenza, vaccines, and drug uptake. However, such electronic databases must be validated if they are to be routinely used for influenza surveillance and other public health purposes.
To address this gap, we recently partnered with a major private sector company in this business (SDI; now IMS Health), who graciously made their surveillance data available for research. We accessed their CMS-1500 form records for multiple years and set out to develop a sensitive and specific definition of electronically sourced ILI data, by studying how physicians coded outpatient respiratory illnesses during intense influenza periods. The resulting time series were validated against the CDC's ILI and viral surveillance data from past seasons; we reported an excellent alignment with the timing and amplitude of proven influenza activity, including accurate capture of the first 2009 pandemic wave on a local level [8]. Transmission models incorporating demographic, geographic, and environmental covariates were fit to city-level onset times for the fall 2009 pandemic wave and revealed a highly structured mode of disease spread in the United States [47]. In contrast to predictions from earlier hierarchical models used for pandemic preparedness [48, 49], the bulk of the pandemic spread was highly localized and radially diffusive, with a small number of long-range jumps [47]. The intricate spatial structure of influenza dissemination would not have been apparent in coarsely grained data, such as data from traditional surveillance available at the state or regional level. This, we believe, was the first study of influenza dissemination, using quantitative data at the local level [47].
In this work we could not test the timeliness of medical claims data, as we had no access to real-time data streams to assess reporting delays (physicians may submit claims forms in bulk). But we infer that there is great promise for big data in the form of CMS-1500 ICD-coded medical claims data to be used as a component in the US influenza surveillance system [12]. Health encounters are so common that CMS-1500–based systems could yield timely and local information on influenza activity, with the possibility of further breakdown by age and comorbidities, for example.
Overall, the existing body of influenza work suggests that medical claims data can and should be harvested to generate timely local data for a variety of acute infections. In addition to tracking epidemic activity, timing, and magnitude, such systems are highly flexible and robust and could also be leveraged to assess vaccine and drug-uptake patterns, such as the use of antivirals and antibiotics. A caveat is that this type of system can only track what is delivered in the health sector and not outside (eg, in drug stores).
WHERE TO NEXT: MOVING INFECTIONS DISEASE SURVEILLANCE SYSTEMS INTO THE 21ST CENTURY
We have seen that traditional influenza surveillance systems can be revisited and improved to deliver real-time information as it was demonstrated by the use of timely US death certificates [6], or online hospital discharge data in New Zealand and Mexico [40, 50]. While these data systems have progressed from the recent era of fax machines and hand-coding, they have not quite moved into the 21st century.
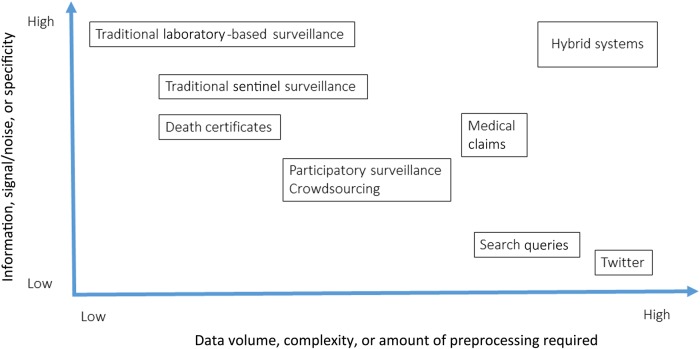
In the future, the many types of data sources described above will continue to be developed and used: traditional approaches (laboratory-based and sentinel surveillance, as well as death certificates), electronic health data (medical insurance claims from outpatient visits), and online nontraditional sources (social media, search queries, and massive participatory surveys). The relative merits of different types of source can be thought of in terms of (1) the specificity of the disease signal and (2) the volume and granularity of coverage achieved. A diagram of the relative placing of these for some of the discussed sources is given in Figure 1.
Figure 1.
Overview of the characteristics of infectious disease surveillance systems. Hybrid systems combining traditional surveillance with big data streams fall in the desirable zone associated with high information return and high data volume.
Going forward, to avoid unfortunate events as recently happened with GFT, we believe it is of the essence to insist on rigorous statistical validation of surveillance algorithms, including model fitting, leave-x-out and complete external validation, and open source access to data and code. There should also be continued demonstration that the surveillance output is useful, practically accurate, and captures profound shifts in age and spatiotemporal dynamics associated with pandemic emergence or other perturbations (eg, onset of vaccination programs).
Perhaps the most exciting direction is the development of hybrid systems, which combine data from multiple types of sources, such as laboratory components with search query, Twitter, participatory surveillance, and/or medical claims data [12, 51]. For example, the CDC has, in January 2016, launched a new website (available at: http://www.cdc.gov/flu/news/flu-forecast-website-launched.htm) to improve timeliness and near-future prediction for influenza; here, external academic teams use various digital data sources to forecast ILI activity 1 month into the future. Similarly, the FluOutlook platform (available at: http://fluoutlook.org/) has been used in real time during the 2014–2015 season to synthesize information from social media and surveillance and provide situational awareness. Hybrid tools have perhaps the most potential because they gain all the advantages of the different data sources, combining the timeliness, scale, and fine granularity of digital and social media data with a direct link to disease that is the hallmark of traditional surveillance systems, ensuring better specificity. Even though GFT suffered a severe blow in recent years, we believe that hybrid systems integrating search queries could be hugely valuable in a number of settings. With astute synthesis of information from multiple sources, one could also aim to track rare outcomes such as drug- and vaccine-related severe adverse events and severe Zika events, such as Guillain-Barré syndromes or births of infants with microcephaly.
This vision for 21st century surveillance will likely work well for data-rich countries; however, most low-income countries have little to no traditional surveillance against which to test novel systems involving digital data. In this case, validation performed in higher-income countries may lead to stand-alone algorithms, which could be extended to data-poor regions. Participatory voluntary surveillance involving big and deep data are particularly interesting systems in this respect: these can easily be extended to incorporate a broader set of diseases, particularly those most relevant to a developing country setting. From a global perspective, some level of harmonization of disease surveillance systems would be most valuable to fully capture global and regional disease trends and to elucidate the determinants of disease spread for a range of pathogens that do not respect political boundaries. From a bioethics perspective, privacy concerns must be carefully weighed against the considerable promise of improved surveillance with respect to timing, accuracy, and flexibility, particularly for emerging threats.
We envision that infectious disease surveillance will soon reap the benefits of the big data era. With more-granular epidemiological data available to academics, research in improved analytical methods will naturally follow, leading to breakthrough studies of transmission dynamics and disease burden, and more-timely and -accurate assessments of the impact of vaccines and other public health interventions.
Notes
Financial support. This work was supported by the DHS RAPIDD program, maintained by the Fogarty International Center, National Institutes of Health (NIH; support to L. S. and J. R. G.); the European Commission (Marie Curie senior fellowship to L. S.); the Lundbeck Foundation (visiting professorship grant to L. S.); and the in-house research program of the Fogarty International Center, NIH (support to C. V. and D. O.).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Graunt J. Natural and political observations made upon the bills of mortality, 1662. http://www.edstephan.org/Graunt/bills.html. Accessed 1 July 2016.
- 2.Moore J. The history and practice of vaccination. 1817. https://archive.org/details/b2135473x. Accessed 1 July 2016.
- 3.Farr W. Vital statistics: a memorial volume of selections from the reports and writings with a biographical sketch. Noel A Humphreys Editions. London Offices of the Sanitary Institute of Great Britain; 1885:166–205. [Google Scholar]
- 4.Snow J. On the mode of communication of cholera, 1854. http://collections.nlm.nih.gov/ext/cholera/PDF/0050707.pdf. Accessed 1 July 2016.
- 5.World Health Organization. International classification of diseases, 2016. http://www.who.int/classifications/icd/en/. Accessed 1 July 2016.
- 6.Centers for Disease Control and Prevention. Influenza activity in the US, 2016. http://www.cdc.gov/flu/. Accessed 1 July 2016.
- 7.Valleron AJ, Bouvet E, Garnerin P et al. A computer network for the surveillance of communicable diseases: the French experiment. Am J Public Health 1986; 76:1289–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Viboud C, Charu V, Olson D et al. Demonstrating the use of high-volume electronic medical claims data to monitor local and regional influenza activity in the US. PLoS One 2014; 9:e102429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pavlin JA, Mostashari F, Kortepeter MG et al. Innovative surveillance methods for rapid detection of disease outbreaks and bioterrorism: results of an interagency workshop on health indicator surveillance. Am J Public Health 2003; 93:1230–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ginsberg J, Mohebbi MH, Patel RS, Brammer L, Smolinski MS, Brilliant L. Detecting influenza epidemics using search engine query data. Nature 2009; 457:1012–4. [DOI] [PubMed] [Google Scholar]
- 11.Polgreen PM, Chen Y, Pennock DM, Nelson FD. Using internet searches for influenza surveillance. Clin Infect Dis 2008; 47:1443–8. [DOI] [PubMed] [Google Scholar]
- 12.Santillana M, Nguyen AT, Dredze M, Paul MJ, Nsoesie EO, Brownstein JS. Combining Search, Social Media, and Traditional Data Sources to Improve Influenza Surveillance. PLoS Comput Biol 2015; 11:e1004513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Declich S, Carter AO. Public health surveillance: historical origins, methods and evaluation. Bull World Health Organ 1994; 72:285–304. [PMC free article] [PubMed] [Google Scholar]
- 14.Thacker SB, Stroup DF. Future directions for comprehensive public health surveillance and health information systems in the United States. Am J Epidemiol 1994; 140:383–97. [DOI] [PubMed] [Google Scholar]
- 15.Newsholme A. The Epidemiology of Smallpox in the Nineteenth Century. BMJ 1902. [Google Scholar]
- 16.Quick J, Loman NJ, Duraffour S et al. Real-time, portable genome sequencing for Ebola surveillance. Nature 2016; 530:228–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woolhouse ME, Rambaut A, Kellam P. Lessons from Ebola: Improving infectious disease surveillance to inform outbreak management. Sci Transl Med 2015; 7:307rv5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Project Z. Zika in Brazil real time analysis, 2016. http://zibraproject.github.io/about/. Accessed 1 July 2016.
- 19.Kuehnert M, Basavaraju K, Moseley R et al. Screening of blood donations for Zika virus infection — Puerto Rico, April 3–June 11, 2016. MMWR Morb Mortal Wkly Rep 2016; 65:627–8. [DOI] [PubMed] [Google Scholar]
- 20.Van Kerkhove MD, Hirve S, Koukounari A, Mounts AW; H1N1pdm serology working group. Estimating age-specific cumulative incidence for the 2009 influenza pandemic: a meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influenza Other Respir Viruses 2013; 7:872–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Food and Drug Administration's Sentinel Initiative, 2016. http://www.fda.gov/Safety/FDAsSentinelInitiative/default.htm. Accessed 1 July 2016.
- 22.Centers for Disease Control and Prevention. National Syndromic Surveillance Program (NSSP), 2016. http://www.cdc.gov/nssp/biosense/. Accessed 1 July 2016.
- 23.Kalies H, Redel R, Varga R, Tauscher M, von Kries R. Vaccination coverage in children can be estimated from health insurance data. BMC Public Health 2008; 8:82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Althouse BM, Scarpino SV, Meyers LA et al. Enhancing disease surveillance with novel data streams: challenges and opportunities. EPJ Data Science 2015; 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butler D. When Google got flu wrong. Nature 2013; 494:155–6. [DOI] [PubMed] [Google Scholar]
- 26.Olson DR, Konty KJ, Paladini M, Viboud C, Simonsen L. Reassessing Google Flu Trends data for detection of seasonal and pandemic influenza: a comparative epidemiological study at three geographic scales. PLoS Comput Biol 2013; 9:e1003256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salathe M, Khandelwal S. Assessing vaccination sentiments with online social media: implications for infectious disease dynamics and control. PLoS Comput Biol 2011; 7:e1002199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nagar R, Yuan Q, Freifeld CC et al. A case study of the New York City 2012–2013 influenza season with daily geocoded Twitter data from temporal and spatiotemporal perspectives. J Medical Internet Res 2014; 16:e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McIver DJ, Brownstein JS. Wikipedia usage estimates prevalence of influenza-like illness in the United States in near real-time. PLoS Comput Biol 2014; 10:e1003581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hickmann KS, Fairchild G, Priedhorsky R et al. Forecasting the 2013–2014 influenza season using Wikipedia. PLoS Comput Biol 2015; 11:e1004239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adler AJ, Eames KT, Funk S, Edmunds WJ. Incidence and risk factors for influenza-like-illness in the UK: online surveillance using Flusurvey. BMC Infect Dis 2014; 14:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Noort SP, Codeco CT, Koppeschaar CE, van Ranst M, Paolotti D, Gomes MG. Ten-year performance of Influenzanet: ILI time series, risks, vaccine effects, and care-seeking behaviour. Epidemics 2015; 13:28–36. [DOI] [PubMed] [Google Scholar]
- 33.Smolinski MS, Crawley AW, Baltrusaitis K et al. Flu near you: crowdsourced symptom reporting spanning 2 influenza seasons. Am J Public Health 2015; 105:2124–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harvard School of Public Health. HealthMap, 2016. http://www.healthmap.org/site/about. Accessed 1 July 2016.
- 35.World Health Organization, Public Health Agency Canada. GPHIN, 2016. http://www.who.int/csr/alertresponse/epidemicintelligence/en/. Accessed 1 July 2016.
- 36.Laksanasopin T, Guo TW, Nayak S et al. A smartphone dongle for diagnosis of infectious diseases at the point of care. Sci Transl Med 2015; 7:273re1. [DOI] [PubMed] [Google Scholar]
- 37.UK National Statistics. Weekly National Flu Reports. 2016. https://www.gov.uk/government/statistics/weekly-national-flu-reports. Accessed 1 July 2016.
- 38.Chowell G, Bertozzi SM, Colchero MA et al. Severe respiratory disease concurrent with the circulation of H1N1 influenza. N Engl J Med 2009; 361:674–9. [DOI] [PubMed] [Google Scholar]
- 39.Baker MG, Wilson N, Huang QS et al. Pandemic influenza A(H1N1)v in New Zealand: the experience from April to August 2009. Euro Surveill 2009; 14:pii:19319. [DOI] [PubMed] [Google Scholar]
- 40.Davila J, Chowell G, Borja-Aburto VH, Viboud C, Grajales Muniz C, Miller M. Substantial morbidity and mortality associated with pandemic A/H1N1 influenza in Mexico, winter 2013–2014: gradual age shift and severity. PLoS Curr 2014; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oprah. Oprah book club, 2007. http://www.oprah.com/oprahsbookclub/love-in-the-time-of-cholera-by-gabriel-garcia-marquez. Accessed 1 July 2016.
- 42.Cook S, Conrad C, Fowlkes AL, Mohebbi MH. Assessing Google flu trends performance in the United States during the 2009 influenza virus A (H1N1) pandemic. PLoS One 2011; 6:e23610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lazer D, Kennedy R, King G, Vespignani A. Big data. The parable of Google Flu: traps in big data analysis. Science 2014; 343:1203–5. [DOI] [PubMed] [Google Scholar]
- 44.Miller MA, Viboud C, Balinska M, Simonsen L. The signature features of influenza pandemics--implications for policy. N Engl J Med 2009; 360:2595–8. [DOI] [PubMed] [Google Scholar]
- 45.Gluskin RT, Johansson MA, Santillana M, Brownstein JS. Evaluation of Internet-based dengue query data: Google Dengue Trends. PLoS Negl Trop Dis 2014; 8:e2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bakker KM, Martinez-Bakker ME, Helm B, Stevenson TJ. Digital epidemiology reveals global childhood disease seasonality and the effects of immunization. Proc Natl Acad Sci U S A 2016; 113:6689–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gog JR, Ballesteros S, Viboud C et al. Spatial transmission of 2009 pandemic influenza in the US. PLoS Comput Biol 2014; 10:e1003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hufnagel L, Brockmann D, Geisel T. Forecast and control of epidemics in a globalized world. Proc Natl Acad Sci U S A 2004; 101:15124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Germann TC, Kadau K, Longini IM Jr, Macken CA. Mitigation strategies for pandemic influenza in the United States. Proc Natl Acad Sci U S A 2006; 103:5935–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Centers for Disease Control and Prevention. Surveillance for the 2009 pandemic influenza A (H1N1) virus and seasonal influenza viruses - New Zealand, 2009. MMWR Morb Mortal Wkly Rep 2009; 58:918–21. [PubMed] [Google Scholar]
- 51.Shaman J, Karspeck A, Yang W, Tamerius J, Lipsitch M. Real-time influenza forecasts during the 2012–2013 season. Nat Commun 2013; 4:2837. [DOI] [PMC free article] [PubMed] [Google Scholar]