Abstract
Calorie restriction slows aging and increases life span in many organisms. In yeast, a mechanistic explanation has been proposed whereby calorie restriction slows aging by activating Sir2. Here we report the identification of a Sir2-independent pathway responsible for a majority of the longevity benefit associated with calorie restriction. Deletion of FOB1 and overexpression of SIR2 have been previously found to increase life span by reducing the levels of toxic rDNA circles in aged mother cells. We find that combining calorie restriction with either of these genetic interventions dramatically enhances longevity, resulting in the longest-lived yeast strain reported thus far. Further, calorie restriction results in a greater life span extension in cells lacking both Sir2 and Fob1 than in cells where Sir2 is present. These findings indicate that Sir2 and calorie restriction act in parallel pathways to promote longevity in yeast and, perhaps, higher eukaryotes.
This study indicates that calorie restriction and Sir2 promote longevity in yeast through distinct pathways. This undermines the accepted view, and has implications for aging in higher organisms
Introduction
The budding yeast Saccharomyces cerevisiae has served as a useful model for aging research, leading to the identification of new longevity genes and pathways whose counterparts can be examined in higher eukaryotes (Kaeberlein et al. 2001). One measure of aging in yeast is the finite replicative life span (RLS) of mother cells, defined as the number of mitotic cycles completed prior to senescence (Mortimer and Johnston 1959). Alternatively, the survival of nondividing yeast cells over time can be monitored and has been termed chronological aging (Fabrizio and Longo 2003). It has been proposed that replicative aging in yeast may be a suitable model for the aging of dividing cells in mammals, such as germ cells; whereas, chronological aging of yeast may be related to the aging of postmitotic tissues.
Replicative aging of yeast can be caused by the accumulation of extrachromosomal rDNA circles (ERCs) in the mother cell nucleus (Sinclair and Guarente 1997), and mutations that decrease ERC formation correlate with increased life span. One example of such a mutation is deletion of the gene encoding the rDNA replication fork barrier protein Fob1, which results in a dramatic decrease in ERC levels accompanied by a 30%–40% increase in mean and maximum RLS (Defossez et al. 1999).
In addition to Fob1, the Sir2 protein has also been found to affect longevity by regulating the rate at which ERCs are formed (Kaeberlein et al. 1999). Sir2 is an NAD-dependent histone deacetylase (Imai et al. 2000; Landry et al. 2000; Tanner et al. 2000) necessary for transcriptional silencing near telomeres (Aparicio et al. 1991), HM loci (Ivy et al. 1986; Rine and Herskowitz 1987), and rDNA (Bryk et al. 1997; Smith and Boeke 1997). Deletion of Sir2 increases both rDNA recombination (Gottlieb and Esposito 1989) and ERC formation, while shortening life span by approximately 50% (Kaeberlein et al. 1999). Conversely, overexpression of Sir2 increases life span by 30%–40%. Overexpression of Sir2 in the context of FOB1 deletion fails to further extend life span, consistent with the idea that Sir2 and Fob1 both impact aging by regulating ERC levels (Kaeberlein et al. 1999).
Calorie restriction (CR) of yeast cells can be accomplished by a reduction in the glucose concentration of growth media from 2% to 0.5% (or lower) and results in a 30%–40% increase in life span (Lin et al. 2000). Several genetic models of CR have also been described. In one model, deletion of the HXK2 gene, coding for hexokinase, reduces the availability of glucose for glycolysis; while in the others, deletion of other genes, including gpa2Δ and gpr1Δ, decreases cAMP-dependent protein kinase activity (Lin et al. 2000). Growth in low glucose and the various genetic models of CR have been treated as experimentally interchangeable. While they are clearly not identical, evidence to date suggests that they behave in a similar manner with respect to yeast aging (Lin et al. 2000, 2002, 2004; Kaeberlein et al. 2002, 2004).
Several reports have suggested a link between the enhanced longevity associated with CR and increased activity of Sir2 (Koubova and Guarente 2003). In one genetic model of CR, cdc25-10 is reported to decrease both rDNA recombination and ERC levels (Lin et al. 2000). In addition, deletion of Sir2 has been shown to prevent life span extension by cdc25-10 and low glucose (Lin et al. 2000, 2002). These data have been used to support a model whereby CR activates Sir2, thus causing decreased ERC accumulation and increased life span. It was initially proposed that life span extension by CR is the consequence of a metabolic shift resulting in increased cellular NAD available as a substrate for Sir2-dependent histone deacetylation (Lin et al. 2002). More recently, this theory has been supplanted by two competing models for activation of Sir2 by CR: (1) a decrease in cellular nicotinamide (a product inhibitor of Sir2) via upregulation of PNC1 (Anderson et al. 2003a), and (2) a decrease in cellular NADH (a competitive inhibitor of Sir2) (Lin et al. 2004).
We present here evidence that CR and Sir2 act in different genetic pathways to promote longevity and show that Sir2 is not required for full life span extension in response to CR. In addition, we offer data suggesting that previous experiments were misinterpreted. Finally, we propose a model that reconciles our findings with earlier reports and suggests a greater level of conservation between aging in yeast and higher eukaryotes.
Results
We recently carried out a large-scale study of more than 40 single-gene deletions reported to affect aging in yeast (unpublished data). This analysis was performed in the BY4742 genetic background, which has a mean life span significantly longer than most other yeast strains commonly used for aging research (Table 1). Included in this analysis were three genetic models of CR (hxk2Δ, gpa2Δ, and gpr1Δ) and fob1Δ. As previously reported for shorter-lived strain backgrounds (Defossez et al. 1999; Lin et al. 2000), each of these single-gene deletions resulted in a 30%–40% increase in life span in BY4742 (Figure 1A).
Table 1. BY4742 Is Long Lived Relative to Other Yeast Strains Commonly Used in Aging Research.
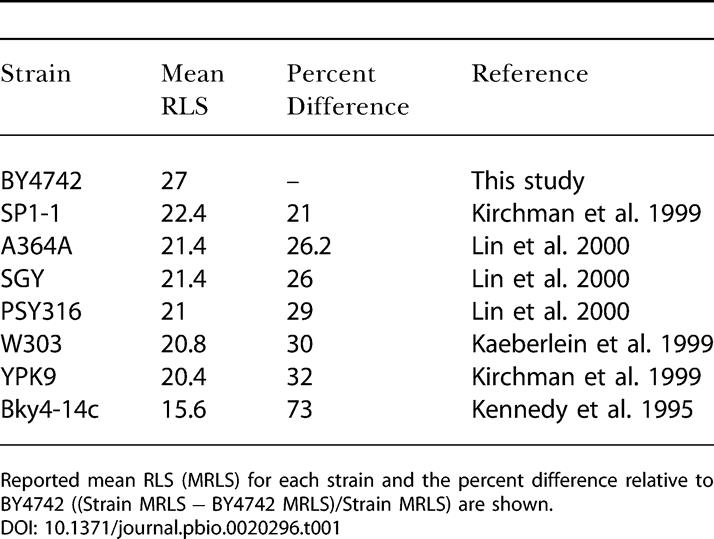
Reported mean RLS (MRLS) for each strain and the percent difference relative to BY4742 ((Strain MRLS − BY4742 MRLS)/Strain MRLS) are shown
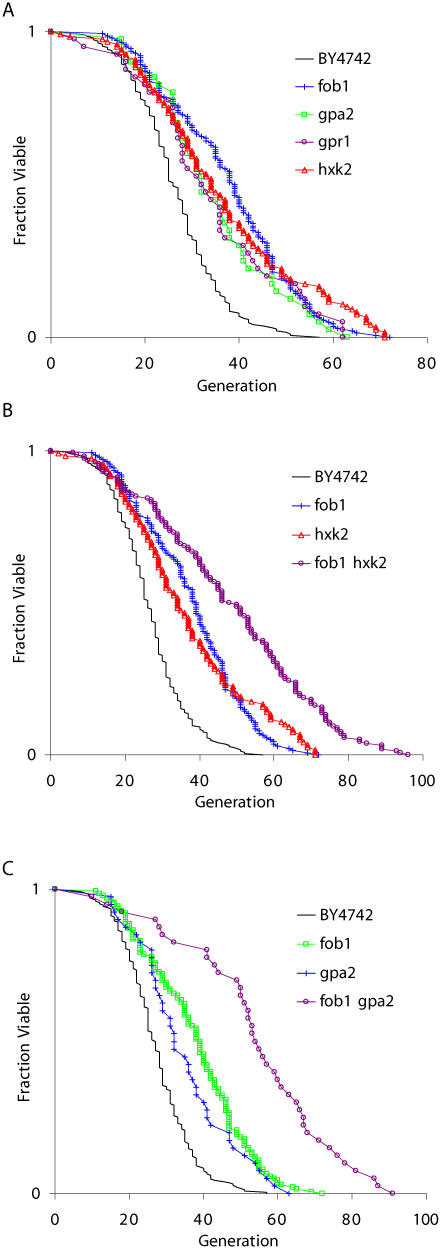
Figure 1. Regulation of Longevity by CR and Fob1.
(A) Life span analysis for three genetic models of CR and deletion of FOB1. Strains shown (and mean life spans) are BY4742 (26.7), fob1Δ (37.8), gpa2Δ (34.9), gpr1 (34.4), and hxk2Δ (36.7).
(B) fob1Δ and hxk2Δ increase life span additively. Strains shown (and mean life spans) are BY4742 (26.6), fob1Δ (37.3), hxk2Δ (36.7), and fob1Δ hxk2Δ (48.3).
(C) fob1Δ and gpa2Δ increase life span additively. Strains shown (and mean life spans) are BY4742 (27.2), fob1Δ (37.8), gpa2Δ (36.7), and fob1Δ gpa2Δ (54.5).
Since both CR and deletion of FOB1 increased life span individually in BY4742, we examined the effect of CR combined with deletion of FOB1. It is notable that this experiment has not to our knowledge been previously reported. We constructed a fob1Δ hxk2Δ double mutant and determined the replicative aging potential of this strain. As expected, both single mutants lived longer than wild-type mother cells (p < 0.001). However, the life span of the fob1Δ hxk2Δ double mutant greatly exceeded that of either single mutant (p < 0.001), suggesting an additional effect on longevity as a result of combining deletion of FOB1 with CR (Figure 1B).
In order to demonstrate that this synthetic lengthening of life span in combination with fob1Δ was not specific to the hxk2Δ model of CR, we determined the life span of a fob1Δ gpa2Δ double mutant. As observed for HXK2, deletion of GPA2 combined with deletion of FOB1 resulted in a mean life span significantly greater than was observed for either single mutant (p < 0.001), and nearly double that of wild-type cells (Figure 1C). With mean and maximum life spans of 54.5 and 94 generations, respectively, the fob1Δ gpa2Δ and fob1Δ hxk2Δ double mutants are to our knowledge the longest-lived yeast strains reported to date.
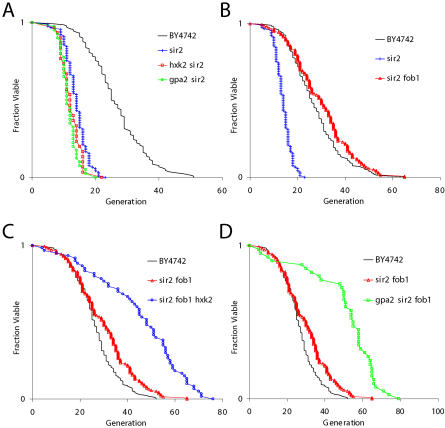
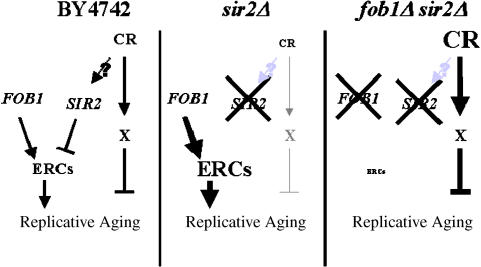
The observation that CR further increases the long life span of a fob1Δ mutant is inconsistent with the model that CR increases life span solely by activation of Sir2. Since overexpression of SIR2 is sufficient to increase the life span of wild-type cells but fails to further extend the life span of a fob1Δ mutant (Kaeberlein et al. 1999), CR (acting through Sir2) should also fail to further extend the life span of a fob1Δ strain, by this model. Our data therefore suggest the existence of a Sir2-independent pathway by which CR enhances longevity. In order to test this possibility, we determined whether CR would increase life span in the absence of Sir2. As observed in other strain backgrounds, deletion of SIR2 shortens life span by approximately 50% in BY4742 (Figure 2A), likely because of an elevated level of ERCs (Kaeberlein et al. 1999). Neither deletion of HXK2 nor deletion of GPA2 conferred increased life span to the sir2Δ mutant. As expected, deletion of FOB1 was sufficient to suppress the life span defect of cells lacking Sir2 (Figure 2B), consistent with the idea that accelerated ERC accumulation is responsible for the severe life span defect of the sir2Δ strain. Surprisingly, in the sir2Δ fob1Δ double mutant, deletion of HXK2 resulted in a robust life span extension (Figure 2C; p < 0.001). Similarly, the life span of sir2Δ fob1Δ gpa2Δ triple mutant cells was significantly longer than that of sir2Δ fob1Δ double mutant cells (Figure 2D; p < 0.001). In fact, the life spans of sir2Δ fob1Δ hxk2Δ and sir2Δ fob1Δ gpa2Δ cells did not differ significantly (p ≈ 0.4) from fob1Δ hxk2Δ and fob1Δ gpa2Δ cells, respectively. Thus, CR clearly enhances longevity in the absence of both Sir2 and Fob1, but not in the absence of Sir2 alone. While seemingly contradictory (see below), these findings demonstrate that Sir2 is dispensable for life span extension by CR, at least in the context of reduced ERC levels (as a result of fob1Δ).
Figure 2. Life Span Extension by CR Does Not Require Sir2.
(A) CR fails to increase life span of a sir2Δ mutant. Strains shown (and mean life span) are BY4742 (26.7), sir2Δ (14.0), hxk2Δ sir2Δ (12.4), and gpa2Δ sir2Δ (11.7).
(B) Deletion of FOB1 suppresses the short life span of a sir2Δ strain. Strains shown and mean life spans are: BY4742 (27.5), sir2Δ (14.0), sir2Δ fob1Δ (30.0).
(C) Deletion of HXK2 increases the life span of a sir2Δ fob1Δ double mutant. Strains shown (and mean life spans) are BY4742 (26.5), sir2Δ fob1Δ (30.0), and sir2Δ fob1Δ hxk2Δ (45.3).
(D) Deletion of GPA2 increases the life span of a sir2Δ fob1Δ double mutant. Strains shown (and mean life spans) are BY4742 (26.6), sir2Δ fob1Δ (30.0), and sir2Δ fob1Δ gpa2Δ (51.0).
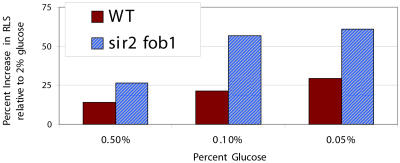
Genetic models of CR, such as hxk2Δ and gpa2Δ, have been used as convenient surrogates for CR by growth on low glucose (Lin et al. 2000, 2002); however, it is possible that these genetic models of CR may not completely recapitulate the effects of glucose deprivation. Additionally, unlike genetic models of CR, growth on low glucose provides an opportunity to control the degree of CR by manipulating the glucose concentration within a range of values (Kaeberlein et al. 2002). Taking advantage of this property, we examined the life span of wild-type and sir2Δ fob1Δ double mutant cells on 2%, 0.5%, 0.1%, and 0.05% glucose (Figure 3; Figure S1). Wild-type cells showed an increase in mean life span ranging from 15% to 25%, with maximal increases observed at 0.05% glucose (p < 0.05). The effect of growth on low glucose was even more pronounced in the sir2Δ fob1Δ double mutant, with mean life span increased by 25% on 0.5% glucose (p < 0.01) and by 60% on 0.05% glucose (p < 0.001).
Figure 3. CR Is More Effective at Enhancing Longevity in a sir2Δ fob1Δ Double Mutant than in Wild-Type Cells.
Percent increase in mean life span relative to growth on 2% glucose was determined for 20 mother cells from each strain at 0.5%, 0.1%, and 0.05% glucose.
Our data conflict with the report that CR fails to increase the life span of a sir2Δ fob1Δ double mutant (Lin et al. 2000). However, all of these prior experiments, as well as nearly all of the published life span data on CR in yeast, were carried out in the PSY316 strain background (Lin et al. 2000, 2002, 2004; Anderson et al. 2002, 2003a, 2003b; Bitterman et al. 2002). We therefore asked whether strain-specific effects might account for this apparent discrepancy. Consistent with prior reports, we observed that growth on low glucose fails to increase the life span of a sir2Δ fob1Δ double mutant derived from strain PSY316 (unpublished data). However, the previous experiments demonstrating that either deletion of FOB1 or overexpression of SIR2 increase life span were carried out in W303R (Kaeberlein et al. 1999), a genetic background apparently unrelated to PSY316. Notably, life span phenotypes for a fob1Δ mutant or SIR2-overexpressing strain in PSY316 have not been reported. Thus, we created these strains and measured their life span. Neither deletion of FOB1 (p = 0.29) nor overexpression of SIR2 (p = 0.76) was sufficient to increase life span in the PSY316 background (Figure 4A). In fact, PSY316 behaves differently from the majority of other yeast strains with respect to the roles of SIR2 and FOB1 as regulators of longevity, since overexpression of SIR2 or deletion of FOB1 has been found to increase longevity in multiple genetic backgrounds, including BY4742 (Table 2).
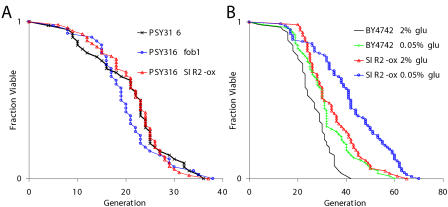
Figure 4. CR Increases the Life Span of Cells Overexpressing SIR2 .
(A) Neither deletion of FOB1 nor overexpression of SIR2 impact longevity in PSY316. Strains shown (and mean life spans) are PSY316 (21.1), PSY316 fob1Δ (20.7), and PSY316 SIR2-ox (21.7).
(B) Overexpression of SIR2 and CR increase life span additively in BY4742. Strains shown (and mean life spans) are BY4742 on 2% glucose (26.1), BY4742 on 0.05% glucose (31.8), BY4742 SIR2-ox on 2% glucose (34.6), and BY4742 SIR2-ox on 0.05% glucose (42.2).
Table 2. FOB1 Deletion or SIR2 Overexpression Increase Life Span in Multiple Genetic Backgrounds.
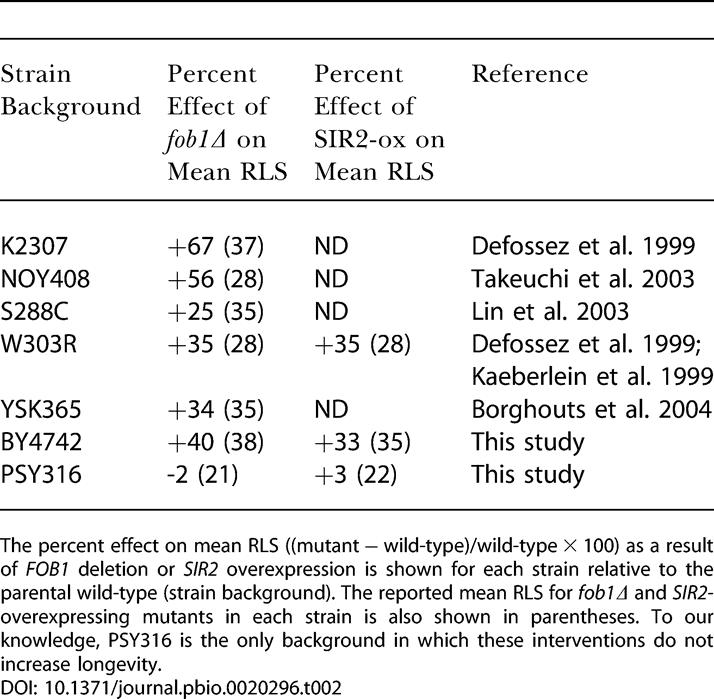
The percent effect on mean RLS ((mutant − wild-type)/wild-type × 100) as a result of FOB1 deletion or SIR2 overexpression is shown for each strain relative to the parental wild-type (strain background). The reported mean RLS for fob1Δ and SIR2-overexpressing mutants in each strain is also shown in parentheses. To our knowledge, PSY316 is the only background in which these interventions do not increase longevity
Unlike in PSY316, overexpression of SIR2 in BY4742 significantly increases life span (Figure 4B; p < 0.001). Further, growth of SIR2-overexpressing cells on low glucose results in an additional life span increase (p < 0.001), similar to that observed for sir2Δ fob1Δ double mutant cells on low glucose. The observation that CR further enhances the already long life span of cells in which SIR2 is overexpressed reinforces our model that CR and SIR2 promote longevity by influencing different pathways (Figure 5).
Figure 5. Two Pathways Determine Yeast Longevity.
The longevity of mother cells can be modified by at least two independent interventions: altered ERC levels and CR. In cells lacking Sir2 but containing Fob1, senescence due to ERCs predominates, causing an extremely short life span that cannot be increased by CR. In cells lacking FOB1, ERCs are greatly reduced and the CR pathway predominates. The presence or absence of Sir2 does not impact the longevity benefits of CR under this condition.
Discussion
We present substantial genetic evidence that CR and Sir2 act in different genetic pathways to promote longevity. The combination of CR with SIR2 overexpression results in an additive life span increase, as expected for two genetic interventions acting in parallel pathways. Further, in the context of FOB1 deletion, CR results in a larger relative increase in life span in the absence of Sir2 than in cells where Sir2 is expressed. Finally, the ability of CR to promote longevity in a strain lacking Sir2 definitively demonstrates the existence of a Sir2-independent aging pathway responsive to CR.
Experiments have previously suggested that life span extension by CR in yeast is partially Sir2-independent (Jiang et al. 2002). It is important to note, however, that the conditions employed for these experiments involved maintaining the cells on defined medium, which is known to slow growth rate and shorten life span by about 50% (Jiang et al. 2000). Under these conditions, CR is reported to modestly increase mean life span of sir2Δ mother cells from seven generations to nine generations. This differs from our results, which demonstrate that CR has no significant effect on life span in the sir2Δ background when cells are grown under standard conditions (see Figure 2A). We speculate that the apparently toxic effects of growth on defined medium (as evidenced by dramatically reduced life span and fitness) are partially mitigated by CR in a Sir2-independent manner. It is not clear whether this modest effect (two generations) is related in any way to the robust (20–30 generations) Sir2-independent life span extension caused by CR under standard growth conditions.
The seemingly disparate findings that CR fails to extend the life span of a sir2Δstrain (see Figure 2A) but dramatically extends life span in a sir2Δ fob1Δ double mutant background (see Figure 2C–2E) can be explained by a model in which there are (at least) two pathways that regulate aging in yeast: one is ERC accumulation and the other is undefined at a molecular level, but responsive to CR (see Figure 5). In our long-lived wild-type background, both processes influence longevity. To explain why CR fails to extend life span in the sir2Δ strain, we postulate that the ERC pathway predominates in this mutant. Cells lacking Sir2 exhibit elevated rDNA recombination and increased levels of ERCs (Kaeberlein et al. 1999), resulting in the premature death of nearly all mother cells prior to an age where the CR pathway becomes limiting. Thus, CR, acting through the alternative pathway, fails to impact aging in the sir2Δ mutant. In the fob1Δ mutant or sir2Δ fob1Δ double mutant, strains in which ERCs are greatly reduced (Defossez et al. 1999; Kaeberlein et al. 1999), CR slows aging through the Sir2-independent alternative pathway. This independent pathway should be more important when ERC levels are reduced, and, consistent with this model, we find that CR has a more pronounced effect on life span under these conditions (see Figure 3; Figure S1).
Our findings do not preclude the possibility that CR enhances Sir2 function through previously proposed mechanisms. However, the fact that the life spans of sir2Δ fob1Δ hxk2Δ and sir2Δ fob1Δ gpa2Δ triple mutants do not differ significantly from those of fob1Δ hxk2Δ and fob1Δ gpa2Δ double mutants, respectively, suggests that any role for Sir2 in the CR pathway is, at best, minor. Alternatively, it is possible that another protein can substitute for Sir2 as a downstream effecter of CR when Sir2 is absent. This model seems unlikely, however, given a need to postulate that the hypothetical Sir2-like protein could function as a substitute for Sir2 only in a strain lacking Fob1, since CR fails to increase life span in the sir2Δ single mutant. The most likely candidate for such a Sir2-like protein is the Sir2 homolog, Hst1. We find that deletion of HST1 has no effect on life span (unpublished data), suggesting that, at least under normal conditions, Hst1 is not an important determinant of longevity.
Nearly all of the evidence supporting a role for Sir2 in CR-mediated life span extension is derived from experiments carried out in PSY316, further weakening the case for a Sir2-dependent model. The inability of SIR2 overexpression, in particular, to increase life span in the PSY316 background supports the idea that Sir2 does not play a primary role in CR-mediated life span extension, as it is not straightforward to postulate a model whereby CR would increase life span via activation of Sir2 in a strain background that is insensitive to Sir2 dosage. Further, the inability of the fob1Δ mutation to increase life span in PSY316 provides a plausible explanation for why CR is unable to enhance longevity in the PSY316 sir2Δ fob1Δ double mutant, and suggests that either deletion of FOB1 fails to impact ERCs in this background or ERCs are not limiting for life span. While we cannot rule out the possibility that the Sir2-independent nature of CR is unique to BY4742, we note that BY4742 behaves like the majority of other strains with respect to increased life span in response to deletion of FOB1 or overexpression of SIR2, while PSY316 is the only strain (to our knowledge) that is unresponsive to these interventions (Table 2).
The observation that CR further increases the long life span of a fob1Δ strain (see Figure 1B and 1C) suggests that the mechanism of enhanced longevity by CR is unrelated to ERCs, as cells lacking Fob1 have dramatically reduced ERC levels. However, it is still possible that ERCs limit the life span of fob1Δ cells and that CR slows ERC accumulation by a second pathway that is insensitive to both Fob1 and Sir2. This seems unlikely, since CR is more effective at enhancing longevity in a sir2Δ fob1Δ double mutant than in wild-type cells. It has been observed that, while life span is comparable between wild-type and sir2Δ fob1Δ cells, ERCs are much reduced in the double mutant (Kaeberlein et al. 1999), suggesting that sir2Δ fob1Δ cells are not senescing as a result of ERCs. Thus, life span extension by CR in this context is likely to be unrelated to ERCs.
The existence of an ERC-independent aging pathway in yeast that is modulated by CR is of particular relevance to aging in higher organisms. CR is the only intervention shown to extend life span in a wide range of eukaryotes, including mammals (Weindruch and Walford 1988). In contrast, there is no evidence that ERCs affect aging in organisms other than budding yeast. Nevertheless, in Caenorhabditis elegans, increased expression of the Sir2 ortholog, Sir-2.1, has been found to extend life span in a manner dependent on the Daf-16 transcription factor (Tissenbaum and Guarente 2001). Similarly, the mammalian Sir2 ortholog, SirT1, has recently been reported to regulate the activity of murine Foxo3A (Brunet et al. 2004; Motta et al. 2004). These experiments support a role for Sir2 proteins in eukaryotic aging, linking Sirtuin activity to insulin/IGF-1 signaling. Evidence is accumulating, however, that CR and insulin/IGF-1 act in different pathways to regulate aging in complex eukaryotes. Life span extension by CR is independent of Daf-16 in C. elegans (Lakowski and Hekimi 1998; Houthoofd et al. 2003), and CR can further extend the life span of long-lived insulin/IGF-1 pathway mutants in both C. elegans and mice (Lakowski and Hekimi 1998; Bartke et al. 2001). We present similar evidence that the effects of CR and Sir2 are genetically distinct in yeast, raising the intriguing possibility that aspects of both aging pathways have been conserved.
Materials and Methods
Strains and plasmids.
All yeast strains used in this study are congenic derivatives of BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), except for PSY316AR (MATα RDN1::ADE2 his3- 200 leu2-3,112 lys2 ura3-52), PSY316AR fob1Δ::kanMX, and PSY316AR SIR2-ox. All gene disruptions were verified by PCR. In addition, sir2Δ mutants were verified by the sterility phenotype associated with this mutation. Strains overexpressing Sir2 were constructed by genomic integration of an extra copy of SIR2, as described (Kaeberlein et al. 1999), and life span was determined for four independent transformants.
RLS analysis.
Yeast strains for RLS analysis were removed from frozen stock (25% glycerol, −80 °C) and streaked onto YPD. After 2 d of growth, single colonies were selected and patched to YPD. The next evening, cells were lightly patched to the plates used for life span analysis (4–6 strains per plate). After overnight growth, cells were arrayed onto solid medium using a micromanipulator and allowed to undergo 1–2 divisions. Virgin cells were selected and subjected to life span analysis. Cells were grown at 30 °C during the day and stored at 4 °C at night. Daughter cells were removed by gentle agitation with a dissecting needle and tabulated every 1–2 cell divisions. All life span experiments were carried out on standard YPD plates (2% glucose), except for the low glucose experiments, which were performed on YEP plates supplemented with the indicated amounts of glucose. In order to prevent introduction of bias, strains were coded such that the researcher performing the life span experiment had no knowledge of the strain genotype for any particular strain. For each experiment, each strain was randomly coded at the time of removal from frozen stock. One individual was responsible for assigning codes (K. T. K.) while a different individual (M. K. or B. K. K.) performed the micromanipulation and was unaware of the genotypes of the strains being analyzed.
Statistical analysis of data
For statistical analysis, life span datasets were compared using a two-tailed Wilcoxon Rank-Sum test. Mother cell life span and p-value matrices for each figure are available in Dataset S1; life span data for individual mother cells are available in Dataset S2. Wilcoxon p-values were calculated using the MATLAB ranksum function. Data shown in each figure and used to calculate p-values were derived from pair-matched, pooled experiments where each mutant was compared to wild-type cells examined within the same experiment(s). Strains are stated to have a significant difference in life span for p < 0.05.
Supporting Information
Each matrix contains the Wilcoxon Rank-Sum p-values for a two-tailed test in which the life span data for the strain in the corresponding row were compared against the life span data for the strain in the corresponding column. Significant p-values (p < 0.05) are colored yellow. P-values were calculated using the MATLAB ranksum function.
(46 KB PDF).
(A) Life span extension by CR is maximized at 0.05% glucose in BY4742 mother cells. Mean life spans are shown for cells grown on 2% glucose (24.8), 0.5% glucose (28.3), 0.1% glucose (30.1), and 0.05% glucose (32.1).
(B) Life span extension by CR is maximized at 0.05% glucose in sir2Δ fob1Δ mother cells. Mean life spans are shown for cells grown on 2% glucose (26.0), 0.5% glucose (32.9), 0.1% glucose (40.8), and 0.05% glucose (42.0).
(88 KB PS).
Accession Numbers
The Saccharomyces Genome Database (http://www.yeastgenome.org/) accession numbers for the yeast genes and gene products discussed in this paper are CDC25 (SGDID S0004301), FOB1 (SGDID S0002517), GPA2 (SGDID S0000822), GPR1 (SGDID S0002193), HST1 (SGDID S0005429), HXK2 (SGDID S0003222), PNC1 (SGDID S0003005), and SIR2 (SGDID S0002200) The LocusLink (http://www.ncbi.nlm.nih.gov/LocusLink/) accession numbers for the non-yeast genes and gene products discussed in this paper are C. elegans Daf-16 (LocusLink 172981), C. elegans Sir-2.1 (LocusLink 177924), mouse Foxo3A (LocusLink 2309), and mouse SirT1 (LocusLink 23411).
Acknowledgments
We thank L. Guarente, S. Lin, G. Martin, and T. Powers for helpful discussion and G. Liszt for providing strains. MK is supported by National Institutes of Health training grant P30 AG013280. This work was funded by awards to BK from the University of Washington Nathan Shock Center of Excellence for the Basic Biology of Aging and the American Federation for Aging Research. SF is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- CR
calorie restriction
- ERC
extrachromosomal rDNA circle
- RLS
replicative life span
Conflicts of interest. The authors have declared that no conflicts of interest exist.
Author contributions. MK, KTK, SF, and BKK conceived and designed the experiments. MK, KTK, and BKK performed the experiments and analyzed the data. MK, KTK, SF, and BKK contributed reagents/materials/analysis tools. MK and BKK wrote the paper.
Academic Editor: Andy Dillin, Salk Institute
Citation: Kaeberlein M, Kirkland KT, Fields S, Kennedy BK (2004) Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol 2(9): e296.
References
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Cohen H, et al. Manipulation of a nuclear NAD+ salvage pathway delays aging without altering steady-state NAD+ levels. J Biol Chem. 2002;277:18881–18890. doi: 10.1074/jbc.M111773200. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA. Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae . Nature. 2003a;423:181–185. doi: 10.1038/nature01578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Latorre-Esteves M, Neves AR, Lavu S, Medvedik O, et al. Yeast life-span extension by calorie restriction is independent of NAD fluctuation. Science. 2003b;302:2124–2126. doi: 10.1126/science.1088697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae . Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Bartke A, Wright JC, Mattison JA, Ingram DK, Miller RA, et al. Extending the lifespan of long-lived mice. Nature. 2001;414:412. doi: 10.1038/35106646. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Borghouts C, Benguria A, Wawryn J, Jazwinski SM. Rtg2 protein links metabolism and genome stability in yeast longevity. Genetics. 2004;166:765–777. doi: 10.1534/genetics.166.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, et al. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Defossez PA, Prusty R, Kaeberlein M, Lin SJ, Ferrigno P, et al. Elimination of replication block protein Fob1 extends the life span of yeast mother cells. Mol Cell. 1999;3:447–455. doi: 10.1016/s1097-2765(00)80472-4. [DOI] [PubMed] [Google Scholar]
- Fabrizio P, Longo VD. The chronological life span of Saccharomyces cerevisiae . Aging Cell. 2003;2:73–81. doi: 10.1046/j.1474-9728.2003.00033.x. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Johnson TE, Vanfleteren JR. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans . Exp Gerontol. 2003;38:947–954. doi: 10.1016/s0531-5565(03)00161-x. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Ivy JM, Klar AJ, Hicks JB. Cloning and characterization of four SIR genes of Saccharomyces cerevisiae . Mol Cell Biol. 1986;6:688–702. doi: 10.1128/mcb.6.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang JC, Jaruga E, Repnevskaya MV, Jazwinski SM. An intervention resembling caloric restriction prolongs life span and retards aging in yeast. FASEB J. 2000;14:2135–2137. doi: 10.1096/fj.00-0242fje. [DOI] [PubMed] [Google Scholar]
- Jiang JC, Wawryn J, Shantha Kumara HM, Jazwinski SM. Distinct roles of processes modulated by histone deacetylases Rpd3p, Hda1p, and Sir2p in life extension by caloric restriction in yeast. Exp Gerontol. 2002;37:1023–1030. doi: 10.1016/s0531-5565(02)00064-5. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. Using yeast to discover the fountain of youth. Sci Aging Knowledge Environ. 2001:pe1. doi: 10.1126/sageke.2001.1.pe1. 2001. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, Andalis AA, Fink GR, Guarente L. High osmolarity extends life span in Saccharomyces cerevisiae by a mechanism related to calorie restriction. Mol Cell Biol. 2002;22:8056–8066. doi: 10.1128/MCB.22.22.8056-8066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Andalis AA, Liszt G, Fink GR, Guarente L. Saccharomyces cerevisiae SSD1-V confers longevity by a Sir2p-independent mechanism. Genetics. 2004;166:1661–1672. doi: 10.1534/genetics.166.4.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy BK, Austriaco NR, Zhang J, Guarente L. Mutation in the silencing gene SIR4 can delay aging in S. cerevisiae . Cell. 1995;80:485–496. doi: 10.1016/0092-8674(95)90499-9. [DOI] [PubMed] [Google Scholar]
- Kirchman PA, Kim S, Lai CY, Jazwinski SM. Interorganelle signaling is a determinant of longevity in Saccharomyces cerevisiae . Genetics. 1999;152:179–190. doi: 10.1093/genetics/152.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koubova J, Guarente L. How does calorie restriction work? Genes Dev. 2003;17:313–321. doi: 10.1101/gad.1052903. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans . Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae . Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, et al. Calorie restriction extends Saccharomyces cerevisiae life span by increasing respiration. Nature. 2002;418:344–348. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SS, Manchester JK, Gordon JI. Sip2, an N-myristoylated beta subunit of Snf1 kinase, regulates aging in Saccharomyces cerevisiae by affecting cellular histone kinase activity, recombination at rDNA loci, and silencing. J Biol Chem. 2003;278:13390–13397. doi: 10.1074/jbc.M212818200. [DOI] [PubMed] [Google Scholar]
- Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, et al. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae . Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Guarente L. Extrachromosomal rDNA circles—A cause of aging in yeast. Cell. 1997;91:1033–1042. doi: 10.1016/s0092-8674(00)80493-6. [DOI] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Horiuchi T, Kobayashi T. Transcription-dependent recombination and the role of fork collision in yeast rDNA. Genes Dev. 2003;17:1497–1506. doi: 10.1101/gad.1085403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci U S A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans . Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Weindruch RH, Walford RL. Springfield (Illinois): Charles C Thomas; 1988. The retardation of aging and disease by dietary restriction; 436 pp. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Each matrix contains the Wilcoxon Rank-Sum p-values for a two-tailed test in which the life span data for the strain in the corresponding row were compared against the life span data for the strain in the corresponding column. Significant p-values (p < 0.05) are colored yellow. P-values were calculated using the MATLAB ranksum function.
(46 KB PDF).
(A) Life span extension by CR is maximized at 0.05% glucose in BY4742 mother cells. Mean life spans are shown for cells grown on 2% glucose (24.8), 0.5% glucose (28.3), 0.1% glucose (30.1), and 0.05% glucose (32.1).
(B) Life span extension by CR is maximized at 0.05% glucose in sir2Δ fob1Δ mother cells. Mean life spans are shown for cells grown on 2% glucose (26.0), 0.5% glucose (32.9), 0.1% glucose (40.8), and 0.05% glucose (42.0).
(88 KB PS).