Abstract
To test the hypothesis that the cultivated peanut species possesses almost no molecular variability, we sequenced a diverse panel of 22 Arachis accessions representing Arachis hypogaea botanical classes, A-, B-, and K- genome diploids, a synthetic amphidiploid, and a tetraploid wild species. RNASeq was performed on pools of three tissues, and de novo assembly was performed. Realignment of individual accession reads to transcripts of the cultivar OLin identified 306,820 biallelic SNPs. Among 10 naturally occurring tetraploid accessions, 40,382 unique homozygous SNPs were identified in 14,719 contigs. In eight diploid accessions, 291,115 unique SNPs were identified in 26,320 contigs. The average SNP rate among the 10 cultivated tetraploids was 0.5, and among eight diploids was 9.2 per 1000 bp. Diversity analysis indicated grouping of diploids according to genome classification, and cultivated tetraploids by subspecies. Cluster analysis of variants indicated that sequences of B genome species were the most similar to the tetraploids, and the next closest diploid accession belonged to the A genome species. A subset of 66 SNPs selected from the dataset was validated; of 782 SNP calls, 636 (81.32%) were confirmed using an allele-specific discrimination assay. We conclude that substantial genetic variability exists among wild species. Additionally, significant but lesser variability at the molecular level occurs among accessions of the cultivated species. This survey is the first to report significant SNP level diversity among transcripts, and may explain some of the phenotypic differences observed in germplasm surveys. Understanding SNP variants in the Arachis accessions will benefit in developing markers for selection.
Keywords: groundnut, alleles, SNPs, domestication, evolution
Domesticated peanut (Arachis hypogaea L.) is one of many polyploid species belonging to the genus Arachis, family Fabaceae, and is native to South America; many of these species come from a region including Brazil, Bolivia, and Paraguay (Gregory et al. 1980). There are 80 species, including diploids and tetraploids, described in the genus, categorized into nine sections according to morphology and crossability (Krapovickas and Gregory 1994). A. hypogaea L. is classified into two subspecies, hypogaea and fastigiata, based on the presence or absence of flowers on the main axis, and spreading or erect growth habit. These two subspecies are further classified into six botanical varieties based on morphology (Krapovickas and Gregory 1994; Valls and Simpson 2005; Lavia et al. 2008).
The origin of A. hypogaea L. and identity of progenitor species have been of interest to plant taxonomists, geneticists, and breeders. However, our knowledge of the origin of cultivated peanut is limited compared with other major crops. More than eight diploid species having either the A- or B- genome have been considered to be involved in the origin of peanut (Norden 1973; Gregory and Gregory 1976; Kochert et al. 1991, 1996; Fernandez and Krapovickas 1994; Krapovickas and Gregory 1994; Lavia 1998; Raina and Mukai 1999; Raina et al. 2001; Moretzsohn et al. 2004; Seijo et al. 2007; Bertioli et al. 2011). More recently, Seijo et al. (2007) and Bertioli et al. (2011, 2016) provided stronger evidence of A. duranensis and A. ipaënsis being the progenitor species of modern cultivars.
All molecular studies, even using older types of molecular markers, of wild peanut species have identified significant molecular-level variability among these accessions (Halward et al. 1991; Lu and Pickersgill 1993; Kochert et al. 1991, 1996). Wild Arachis species possess genetic variability in pest and disease resistance traits, which could be used to improve the cultivated peanut (Stalker and Moss 1987). Alleles that confer resistance to pests and disease in some wild Arachis species have been successfully transferred into cultivated peanut (Simpson 2001; Mallikarjuna et al. 2011).
In contrast, many molecular studies have demonstrated no or little genetic variability in the cultivated species, A. hypogaea. This was first noticed with the use of allozyme and RFLP markers (Halward et al. 1991; Kochert et al. 1991, 1996; Lu and Pickersgill 1993; Burow et al. 2009), which demonstrated an almost complete lack of genetic diversity among the cultivated peanut accessions. It was concluded that a genetic bottleneck occurring as a result of the polyploidization event, coupled with a self-pollinating reproductive system, and the use of a few elite breeding lines with little exotic germplasm in breeding programs, has resulted in a narrow genetic base of peanut cultivars. Natural gene exchange between wild diploid species and cultivated peanut may have been limited due to genomic rearrangement as well as differences in ploidy levels (Soltis and Soltis 1999; Huang et al. 2012). Since then, >10,000 SSR markers have been identified in peanut, many solely among wild species, but few SSR marker maps possess 200 or more SSR markers, again suggesting low genetic variability in the cultivated species.
Despite the results of some molecular studies, phenotypic evaluation of germplasm collections, such as core collections of 1704 (ICRISAT), 831 (United States), and 582 (China) accessions (Upadhyaya 2003; Holbrook et al. 1993; Jiang et al. 2004), and minicore collections (Upadhyaya et al. 2002; Holbrook and Dong 2005) point to a different conclusion. Evaluation has demonstrated significant phenotypic diversity for numerous traits, including resistance to leaf spots, tomato spotted wilt virus, other biotic stresses, for tolerance to drought or heat stress, and for early maturity (Isleib et al. 1995; Anderson et al. 1996; Upadhyaya 2003, 2005, Upadhyaya et al. 2006a,b; Selvaraj et al. 2011; Wang et al. 2011a; Jiang et al. 2014; Pandey et al. 2014; Singh et al. 2014). To date, these have not been accompanied by molecular characterization at SNP levels.
Technology for DNA sequencing and SNP analysis has made great progress recently, both for high throughput and for low cost per sequence. Due to the ubiquity of SNPs, and the far greater power to identify polymorphisms than other types of marker analysis, sequencing is able to identify genetic diversity better than other marker types. RNASeq allows transcriptome profiling and SNP identification with high read depth at much lower cost than whole genome sequencing. Annotation of assembled RNASeq reads can help in understanding the function of a gene or a transcript. Annotations have been used in several crops for establishing genetic information where limited knowledge exists (Garg et al. 2011; Li et al. 2012). SNPs in annotated transcripts associated with traits would benefit functional characterization of genes. For example, a mutation in the fatty acid desaturase FAD2B gene in peanut is being used for selection of the high-oleic trait (Wang et al. 2011b). Use of RNASeq on elite peanut genotypes has provided evidence of large numbers of SNPs among accessions (Chen et al. 2013; Zhang et al. 2012; Chopra et al. 2015); such a large number of polymorphisms were not known using earlier molecular marker techniques (Kochert et al. 1996).
Validation of SNPs identified by sequencing is required for use of these SNPs in generation of molecular maps or association with specific traits. Validation of SNPs in crops such as soybean (Wu et al. 2011), peanut (Nagy et al. 2012; Khera et al. 2013), or chickpea (Deokar et al. 2014) has benefited in producing high-density maps extended to QTL and GWAS analysis. Assembly, annotation, and validation of the information generated using RNASeq is an integrated process needed for application of datasets. Integrating these processes is becoming routine, as it is advantageous for a program to develop its own resources to address research goals of a particular crop.
The hypothesis of the current work is that, commensurate with the phenotypic differences observed in the field, there is actually a substantial amount of unrealized genetic diversity in peanut at the nucleotide level (both among the diploid and tetraploid species accessions). Although recent studies at the genome levels were carried out in two progenitor wild species (Bertioli et al. 2016), there is no survey of SNP level variability among the broader array of wild species, nor among cultivated accessions. Therefore, in this present study, a collection of eight cultivated tetraploid accessions, eight wild diploids, a natural amphidiploid, and a synthetic amphidiploid were selected for transcriptome analysis, adding in also data from four previously sequenced cultivated accessions (Chopra et al. 2015). The objectives of the study were (a) to measure allelic and transcript diversity among the accessions of different species within the genus Arachis, (b) to estimate number of genes in peanut, (c) to compare to the expected phylogenic relationships of the Arachis accessions, (d) to highlight SNPs specific to one accession or to a group of market types or to a group of different genome affiliations, and (e) to validate the SNPs obtained from the bioinformatics calls that can be used in a breeding program.
Materials and Methods
Plant accessions
A total of 12 tetraploid accessions from A. hypogaea was used, 10 tetraploids representing the three botanical types cultivated in the United States, namely subsp. hypogaea var hypogaea, subsp. fastigiata var fastigiata, and subsp. fastigiata var vulgaris, and two tetraploids representing other botanical types namely, subsp. hypogaea var hirsuta, and subsp. fastigiata var aequatoriana (Table 1). A. monticola is the only known wild tetraploid relative in section Arachis, and was included for comparison to the cultivated peanut and potentially for understanding gene flow between A. monticola and A. hypogaea. Eight wild diploid accessions, including probable parental species A. duranensis and A. ipaënsis of A. hypogaea were also included. Use of these latter accessions provided a platform for assessing the extent of genetic variability among the wild species. A synthetic amphidiploid, TxAG-6 (Simpson et al. 1993) was also included in the study (Table 1); parents of this were A. cardenasii, A. diogoi, and A. batizocoi. TxAG-6 has donated alleles that have resulted in release of four nematode-resistant cultivars, namely COAN (Simpson and Starr 2001), NemaTAM (Simpson et al. 2003a,b), Tifguard (Holbrook et al. 2008), and Webb (Simpson et al. 2013).
Table 1. Description of Arachis accessions utilized for the sequence analysis: their origin and ploidy levels.
| Accession | Species | Genome | Origin | Botanical Type | References |
|---|---|---|---|---|---|
| GKP10017 | Arachis cardenasii | AA | Bolivia | Wild | Krapovickas et al. (1994) |
| GK10602 | Arachis diogoi | AA | Bolivia | Wild | Krapovickas et al. (1994) |
| K7988 | Arachis duranensis | AA | Argentina | Wild | Krapovickas et al. (1994) |
| KSSc38901 | Arachis duranensis | AA | Bolivia | Wild | Krapovickas et al. (1994) |
| GKPSSc30076 | Arachis ipaënsis | BB | Bolivia | Wild | Krapovickas et al. (1994) |
| GKSSc30097 | Arachis magna | BB | Bolivia | Wild | Krapovickas et al. (1994) |
| K9484 | Arachis batizocoi | KK | Bolivia | Wild | Krapovickas et al. (1994) |
| KSSc36024 | Arachis cruziana | KK | Bolivia | Wild | Krapovickas et al. (1994) |
| GKBSPSc30062 | Arachis monticola | AABB | Argentina | Wild | Krapovickas et al. (1994) |
| BSS56 | Arachis hypogaea | AABB | West Africa | Spanish | Burow et al. (2014) |
| Florunner (UF439-16-10-3-2) | Arachis hypogaea | AABB | Florida | Runner | Norden et al. (1969) |
| Jupiter | Arachis hypogaea | AABB | Oklahoma | Virginia | — |
| New Mexico Valencia C | Arachis hypogaea | AABB | New Mexico | Valencia | Hsi (1980) |
| OLin | Arachis hypogaea | AABB | Texas | Spanish | Simpson et al. (2003) |
| PI502111 (COC155B) | Arachis hypogaea | AABB | — | Virginia | Holbrook and Dong (2005) |
| PI290538 (COC224) | Arachis hypogaea | AABB | India | Runner | Holbrook and Dong (2005) |
| PI268868 (COC367) | Arachis hypogaea | AABB | Sudan | Virginia | Holbrook and Dong (2005) |
| PI158854 (COC559) | Arachis hypogaea | AABB | China | Valencia | Holbrook and Dong (2005) |
| PI648241 | Arachis hypogaea | AABB | Ecuador | Hirsuta | — |
| PI648242 | Arachis hypogaea | AABB | Peru | Aequatoriana | — |
| Tamrun OL07 | Arachis hypogaea | AABB | Texas | Runner | Baring et al. (2006) |
| TxAG-6 | — | AAKK | Texas | Synthetic tetraploid | Simpson (1993) |
Sequences, de novo assembly and annotation
Plants were grown in the greenhouse at Texas A&M AgriLife Research. Unopened leaves, roots, and pods from yellow, brown, and black pod maturity stages were selected for RNA isolation. RNA was isolated using the Trizol reagent (Invitrogen, Grand Island, NY). Accessions OLin, Tamrun OL07, Jupiter, and New Mexico Valencia C were sequenced on an Illumina GAIIx using 2 × 54 paired-end reads as reported previously (Chopra et al. 2015). The remaining 18 genotypes were sequenced on an Illumina HiSeq 2000 instrument using 2 × 50 paired end reads at the National Center for Genome Resources, Santa Fe, NM. De novo assemblies were performed for all 22 genotypes using the Trinity (Grabherr et al. 2011) assembler, with the parameters used in Chopra et al. (2014) for the peanut transcriptome. Assembly statistics from Trinity, which enumerate unique transcripts as an estimate of the number of genes in a transcriptome, and the measure of the total number of structural variants derived from genes, are reported.
A consensus assembly was built by pooling transcripts of all 22 accessions, followed by elimination of duplicate transcripts using CAP3 (Huang and Madan 1999) at a minimum match of 95%. Annotations of the consensus assembly were performed against the nr database from NCBI, and from the Swissprot and Trembl databases from UniProt (UniProt Consortium 2015). Sequences from the consensus assembly were assigned gene ontology (GO) categories using the Mercator web-based tool (Lohse et al. 2014). Brief annotations of the transcripts in the consensus assembly assigned to different biological and molecular functions by Mercator are provided in Supplemental Material, File S1. Transcripts assigned to different transcription factor families are provided in File S2.
Mapping and variant calling
Raw reads of all samples were aligned back to the OLin transcriptome sequence (Chopra et al. 2014) using the bwa.0.7.5a (Li and Durbin 2010) aligner, using the backtrack approach. These aligned reads were further processed using default values of GATK tools (DePristo et al. 2011) to improve the qualities of the alignment process. Aligned files were sorted and indexed using Picard tools (http://broadinstitute.github.io/picard/). The GATK variant calling tool was used to identify SNPs and indels among these genotypes using default parameters for UnifiedGenotyper at a ploidy level of 2. Parameters of MQ (Mapping Quality) > 40.0, AF (Allele Frequency) > 0.10, and DP (Read Depth) > 50 were used to filter the SNP calls using vcftools (Danecek et al. 2011). Variant statistics were calculated using the vcf-stats script, which reports variants among the samples used for comparisons.
Diversity and population structure
Using SNPs identified from sequences that had a read depth of at least 50, and variant allele frequency >10% across 22 genotypes, principal components analysis (PCA) was performed using the R package SNPRelate (Zheng et al. 2012). The SNP datasets obtained after using parameters to filter the SNPs were used to calculate the individual dissimilarities for each pair of genotypes using the command snpgdsHCluster[snpgdsDiss(genofile, num.thread = 2, autosome.only = FALSE)]. A dendrogram was generated for each of the paired accessions after 5000 permutations using the command snpgdsCutTree(“FileName,” n.prem = 5000).
SNP validation
Genomic DNA was prepared from seed tissue of the 12 sequenced accessions (File S3), including treatment with RNaseA during homogenization, and purified using the Qiagen DNAeasy miniprep kit (Qiagen, Valencia, CA). Sixty-six pairs of primers (File S4) were designed to perform validation of allele calls in the sequenced genotypes. For each putative SNP, two allele-specific forward primers, and one common reverse primer were designed (LGC Genomics, Hoddesdon, UK).
Genotyping reactions were performed on a LightCycler 480 (Roche, Branford, CT) in a final volume of 10 µl containing 1× KASP Reaction Mix (LGC Genomics, Hoddesdon, UK), 0.14 µl assay mix, and 10–20 ng of genomic DNA. The following cycling conditions were used: 15 min at 94°; 10 touchdown cycles of 20 sec at 94°, 60 sec at 65–57° (dropping 0.8° per cycle); and 26 cycles of 20 sec at 94°, 60 sec at 57°, and read at 37° for 5 sec. Fluorescence detection of the reaction was performed using a built-in scanner and the data were analyzed using the LightCycler 480 software (Roche, Branford, CT).
Phasing approach
To resolve homeologs in the peanut tetraploid reference sequence, we used the phasing approach. Genome-specific assemblies in tetraploid peanut using the phasing approach were generated by aligning OLin raw reads to the OLin reference transcriptome (60,798 contigs) using the BWA backtrack approach. Polymorphisms among the mapped reads were detected using the GATK software as it has been shown to perform well on RNAseq data (McCormick et al. 2015). Called SNPs and Multiple Nucleotide Polymorphisms (MNPs) were phased using the HapCUTv.0.5software (Bansal and Bafna 2008) with default parameters. To generate homeolog-specific subassemblies, we used the strategy employed by Krasileva et al. (2013), sorting the reads within each phased SNP block based on the HapCUT output, and reassembling de novo the reads for each block and phase using parallelized runs. Haplotype information along with bam files were passed through readphaser to sort the reads within each block into phases based on HapCUT tables, and reassembled.
To evaluate the resolution of the assemblies generated from each of these approaches, we mapped the raw reads back to each assembly. SNPs were called on each of these assemblies using GATK. Parameters used to evaluate assemblies were the number of heterozygous SNPs, homozygous SNPs, and number of SNPs per transcript.
Data availability
BAM files are deposited in SRA database under bioproject PRJNA248910: SRR1534396, SRR1535034, SRR1535035, SRR4039437, SRR4039359, SRR4039358, SRR4039357, SRR4039017, SRR4039016, SRR4039015, SRR4038966, SRR4038965, SRR4038285, SRR4037988, SRR4038281, SRR4038254, SRR4038252, SRR4038060, SRR4037987, SRR4037985, SRR4037986, and SRR4037959. De novo assemblies and annotations described in this manuscript have been deposited at Figshare at the following URLs: https://dx.doi.org/10.6084/m9.figshare.3580650.v2, https://dx.doi.org/10.6084/m9.figshare.3580656.v2, https://dx.doi.org/10.6084/m9.figshare.3580659.v1, and https://dx.doi.org/10.6084/m9.figshare.3580662.v1.
Results
Over 500 million reads provided a large dataset for de novo assembly and annotation
A total of 539 million filtered reads were obtained from 22 accessions including wild species plus cultivars and landraces of A. hypogaea, including different subspecies, and originating from different continents (Table 2). For diploids, the number of raw reads ranged from 13.8 to 24.5 million, and for tetraploids, the number of reads ranged from 18.4 to 43.4 million. De novo assemblies of filtered reads of diploid accessions using Trinity generated an average of 38,903 transcripts, and 27,045 unique transcripts (Table 2) of size >200 bp. Tetraploid de novo assemblies generated an average of 51,744 total transcripts and 29,234 unique transcripts. Overall the number of genes per diploid peanut genome averaged 28,321 based on the assemblies of 22 accessions.
Table 2. De novo assembly characteristics of sequences from twenty-two transcriptomes.
| Genotype | Reads | Total Transcriptsa | Unique Transcriptsb | N50 |
|---|---|---|---|---|
| GKP10017 | 16,015,713 | 37,767 | 27,745 | 1333 |
| GK10602 | 24,501,057 | 44,635 | 27,406 | 1484 |
| K7988 | 18,698,563 | 39,088 | 27,219 | 1393 |
| KSSc38901 | 16,206,929 | 37,379 | 25,530 | 1401 |
| GKPSSc30076 | 16,774,125 | 31,800 | 26,102 | 1107 |
| GKSSc30097 | 18,264,048 | 34,673 | 26,808 | 1238 |
| K9484 | 20,009,991 | 41,750 | 28,287 | 1405 |
| KSSc36024 | 13,852,235 | 37,654 | 27,261 | 1159 |
| GKBSPSc30062 | 16,366,546 | 37,944 | 26,654 | 1184 |
| BSS56 | 23,249,778 | 39,343 | 26,735 | 1294 |
| Florunner (UF439-16-10-3-2) | 18,498,900 | 39,349 | 27,226 | 1253 |
| Jupiter | 43,494,034 | 74,615 | 31,184 | 1687 |
| New Mexico Valencia C | 43,327,345 | 71,264 | 30,677 | 1655 |
| OLin | 38,335,246 | 67,098 | 30,673 | 1641 |
| PI502111(COC155B) | 22,844,455 | 41,513 | 26,932 | 1370 |
| PI290538 (COC224) | 19,126,674 | 39,916 | 27,174 | 1200 |
| PI268868 (COC367) | 25,275,018 | 38,983 | 25,079 | 1401 |
| PI158854 (COC559) | 31,318,101 | 47,007 | 27,038 | 1419 |
| PI648241 | 27,952,734 | 43,763 | 27,327 | 1355 |
| PI648242 | 23,770,118 | 44,206 | 27,146 | 1405 |
| Tamrun OL07 | 41,601,127 | 79,214 | 44,760 | 1535 |
| TxAG-6 | 19,865,766 | 46,399 | 28,089 | 1359 |
Number of total transcripts including splice variants, homeolog and paralog copies assembled using the de novo assembly approach for each of the accession.
Number of unique transcripts that were in each genotype excluding the derivatives such as splice variants/homeologs.
On merging the transcripts in diploids and tetraploids, we observed there was an increase in the total number of transcripts in each pool (Table 3). The resultant A-, B- and K- genome pooled assemblies had from 35,530 to 57,851 transcripts each. The A- genome pool had about 45% or more transcripts than the de novo assembly of a single A-genome species. The B- and K- genome pools had slightly lower ratios of pooled transcripts compared to individual species accessions (Table 4). For the tetraploid genotypes of A. hypogaea, assembled transcripts were merged, and 100,328 transcripts were obtained.
Table 3. Consensus assemblies generated from CAP3 for genome-specific pools.
| Assembly | No. of Contigs | N50 (bp) | Average Contig Length |
|---|---|---|---|
| A_Genome_Diploidsa | 57,851 | 1700 | 1145 |
| B_Genome_Diploidsa | 35,530 | 1362 | 910 |
| K_Genome_Diploidsa | 43,462 | 1518 | 993 |
| Arachis_hypogaea | 100,328 | 1816 | 1205 |
| Arachis_monticola | 37,944 | 1184 | 798 |
| TxAG-6 | 46,399 | 1359 | 903 |
| Arachis_Pooled_Consensus | 165,892 | 1874 | 1277 |
Assemblies pooled for each genome affiliates for estimating transcript diversity in each pool or in the Arachis genus.
Table 4. Variants in each accession were identified in comparison to OLin as the reference.
| Genotype | No. of Transcripts with SNPs | No. of Homozygous Variantsa | Average No. of SNPs per Transcriptb | Number of SNPs per kbc | |
|---|---|---|---|---|---|
| Diploids | GKP10017 | 22,192 | 117,812 | 5.31 | 1.57 |
| GK10602 | 22,564 | 106,882 | 4.74 | 1.43 | |
| K7988 | 21,931 | 122,370 | 5.58 | 1.64 | |
| KSSc38901 | 21,693 | 117,252 | 5.41 | 1.57 | |
| GKPSSc30076 | 18,475 | 96,241 | 5.21 | 1.28 | |
| GKSSc30097 | 21,638 | 109,797 | 5.07 | 1.47 | |
| K9484 | 24,929 | 168,289 | 6.75 | 2.25 | |
| KSSc36024 | 24,064 | 157,444 | 6.54 | 2.11 | |
| Tetraploids | GKBSPSc30062 | 6963 | 14,340 | 2.06 | 0.19 |
| BSS56 | 5326 | 9012 | 1.69 | 0.12 | |
| Florunner (UF439-16-10-3-2) | 5557 | 9454 | 1.7 | 0.12 | |
| Jupiter | 1844 | 3318 | 1.8 | 0.04 | |
| New Mexico Valencia C | 2146 | 3451 | 1.61 | 0.04 | |
| PI502111(COC155B) | 5361 | 9468 | 1.77 | 0.12 | |
| PI290538 (COC224) | 6068 | 10,832 | 1.79 | 0.14 | |
| PI268868 (COC367) | 4715 | 8491 | 1.8 | 0.11 | |
| PI158854 (COC559) | 4306 | 7209 | 1.67 | 0.09 | |
| PI648241 | 4669 | 8212 | 1.76 | 0.11 | |
| PI648242 | 4084 | 7081 | 1.73 | 0.09 | |
| Tamrun OL07 | 2263 | 3923 | 1.73 | 0.05 | |
| TxAG-6 | 22,607 | 87,574 | 3.87 | 1.17 |
Note: OLin is not reported here as the above comparisons were made relative to OLin transcriptome.
Number of homozygous SNPs present in each accession relative to OLin transcriptome.
Average number of transcripts calculated using Number of homozygous SNPs/Number of transcripts in an accession.
SNP rate calculated based on pair-wise comparison of each accession with OLin.
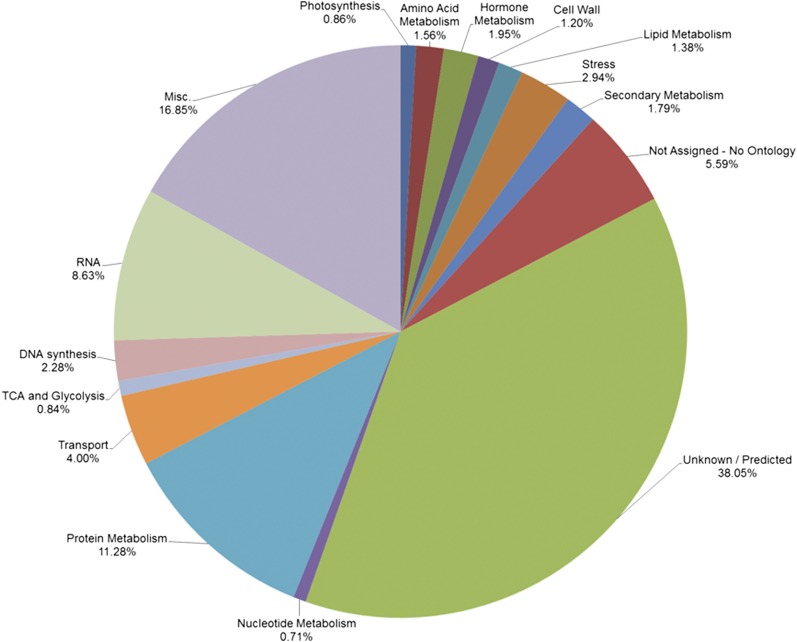
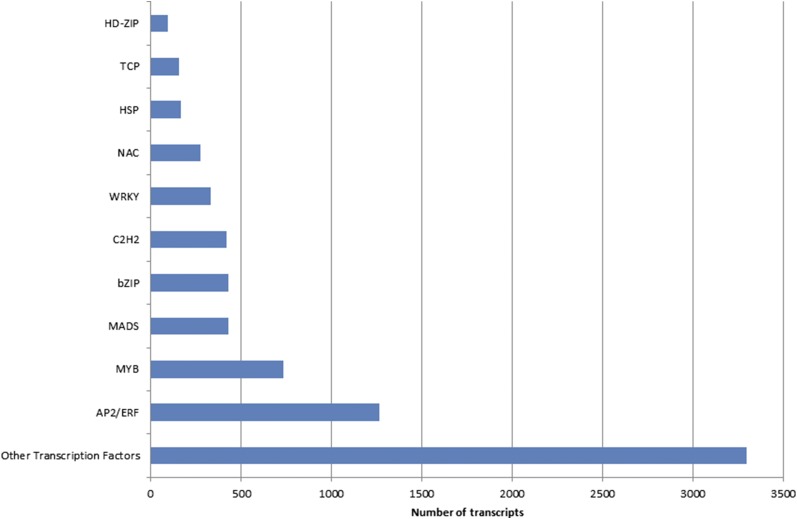
A consensus assembly resulted in 165,892 contigs built from 22 transcriptomes. About 79% of the contigs from the consensus assembly were annotated, and a description of hits from each database has been deposited in Figshare. Annotated transcripts were assigned to different GO categories and this consensus assembly of Arachis will be a valuable resource for capturing maximum number of genes (Figure 1 and File S1). Annotation of the sequences from the consensus assembly identified numerous transcription factors. Transcription factors accounted for 4.4% of the 165,892 transcripts in the consensus assembly, and these were categorized based on their DNA-binding domain (Figure 2 and File S2). Transcription factors were assigned to different groups; b-ZIP, MADS, MYB, AP2/ERF, and C2H2, were among the top five categories (Figure 2).
Figure 1.
Representation of annotations of the transcripts in the consensus assembly assigned to different biological and molecular functions. (Note: Misc. signifies the transcripts that were not assigned to a category; Unknown/Predicted signifies transcripts assigned putative categories other than those listed).
Figure 2.
Overview of transcription factor categories in the Arachis consensus assembly.
Variant analysis identified >300K SNPs
Across 22 accessions, we obtained 306,820 biallelic variants after filtering for quality parameters. SNPs were obtained by aligning raw reads of each accession to the OLin transcriptome (Chopra et al. 2014). On mapping the reads to this reference, the number of raw reads aligned ranged from 84 to 89%. These aligned files were then processed further using the GATK pipeline to improve the quality of SNP calls.
The lowest numbers of SNPs relative to OLin were found in the three cultivars Jupiter (3318), New Mexico Valencia C (3451), and Tamrun OL07 (3923). Landraces had more SNPs relative to OLin, ranging from 7081 to 10,832 SNPs (Table 4). SNP diversity averaged about three times higher among landraces and United States peanut minicore accessions, demonstrating significant genic diversity in the cultivated species. For hirsuta, and aequatoriana, the number of SNPs ranged from 7081 to 8212 relative to OLin (Table 4).
For the wild species, the number of SNPs relative to OLin was far higher: for A- genome diploid accessions, from 106,882 to 117,812 per accession; for B-genome diploids, from 96,241 to 109,797; for the K-genome diploids, 157,444 to 168,289; and for natural and synthetic amphidiploids, 14,340 and 87,574, respectively (Table 4). OLin reads were mapped back to the OLin reference for quality control of the variant calls.
A total of 40,382 unique SNPs was present among the cultivated accessions in 14,719 transcripts, and 291,115 unique SNPs were present among the diploid accessions in 26,320 transcripts (Table 4). The average number of SNPs among the tetraploids accessions was 0.5 per kilobase, and among the diploid accessions was 9.2 per kilobase (Table 4).
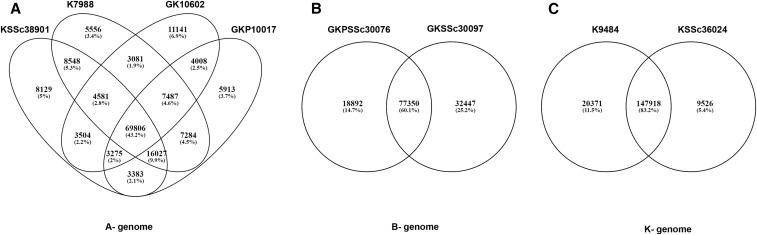
Many SNPs identified among accessions were unique to a genome or botanical class. A total of 191,723 SNPs was present when summed across four A- genome accessions compared to the OLin reference. We found that 43.2% of the 191,723 SNPs were shared by all four A- genome accessions, and the number of unique SNPs specific to each accession ranged from 3.4 to 6.9% (Figure 3A). Likewise, B- and K- genomes had 128,689 and 177,815 SNPs compared to OLin transcripts (Figure 3, B and C). The number of unique SNPs in A. magna GKSSc30097 compared to OLin was 32,430, more than the 18,092 unique SNPs between the tetraploid ancestor B- genome A. ipaënsis (GKPSSc30076) and OLin (Figure 3B). The number of shared unique SNPs among the K- genome accessions was 147,918 relative to OLin, while 20,371 unique SNPs belonged to A. batizocoi K9484, and 9526 unique SNPs to A. cruziana KSSc36024 (Figure 3C).
Figure 3.
Venn diagrams representing comparison of the number of SNPs among the diploid peanut accessions. (A) A- genome. (B) B- genome. (C) K- genome. Note: Four-way and two-way comparisons were made based on the variant sites present in the accession or a group of accessions relative to the OLin transcriptome.
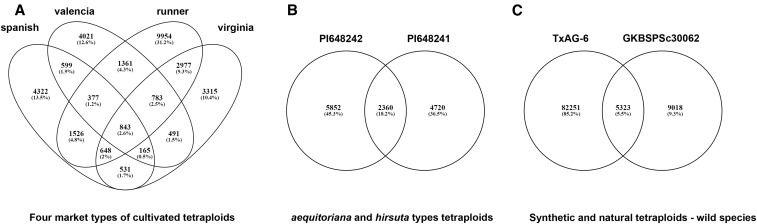
We found that 31,913 SNPs belonged to spanish, virginia, valencia and runners classes in comparison to the OLin transcripts (Figure 4A). For two botanical types hirsuta and aequatoriana, the number of unique SNPs was 45.3 and 36.5% of 12,932 SNPs (Figure 4B). The wild tetraploid group included an accession derived from A- and B- genome A. monticola (GKBSPSc30062), and the synthetic amphidiploid A- and K- genome (TxAG-6), and the differences were reflected in the number of unique SNPs (Figure 4C).
Figure 4.
Venn diagrams representing comparison of the number of SNPs among the tetraploid peanut accessions. (A) cultivated market types. (B) other botanical types. (C) tetraploid wild species and synthetic amphidiploids. Note: Four-way and two-way comparisons were made based on the variant sites present in the accession or a group of accessions relative to the OLin transcriptome.
Accuracy of SNP calls was demonstrated by validation of selected SNPs by KASP marker analysis. A subset of 66 primer pairs (File S4) was designed to validate the alleles called by the bioinformatic approach, and was tested on genomic DNA of 12 sequenced accessions (File S3). A total of 782 SNP calls (12 genotypes and 66 primers) was selected, including the calls from diploid and tetraploid accessions for validation using KASP technology. Genotyping of two diploid accessions with 66 primers confirmed 88.52% of the 122 SNP calls, while 10 tetraploid accessions confirmed 80.00% of the 660 SNP calls (Table 5 and File S3).
Table 5. Summary of total and validated bioinformatic SNP calls among the sequenced accessions that included both diploids and tetraploids using allele-specific discrimination assays.
| Genotype | Diploids | Tetraploids | Total | |
|---|---|---|---|---|
| Homozygous | 122 | 360 | 482 | Total SNP callsb |
| Heterozygousa | — | 300 | 300 | |
| Total | 122 | 660 | 782 | |
| Homozygous | 108 | 319 | 427 | Validated SNP callsb |
| Heterozygous | — | 209 | 209 | |
| Total | 108 | 528 | 636 | |
| % matched calls | 88.52 | 80.00 | 81.32 |
Heterozygous SNP calls for tetraploids are likely the result of the homeologous SNPs derived from two sub-genomes and do not indicate that genotypes are in heterozygous state.
Total SNP calls and validated SNP calls are presented in File S3.
Diversity was associated with population structure
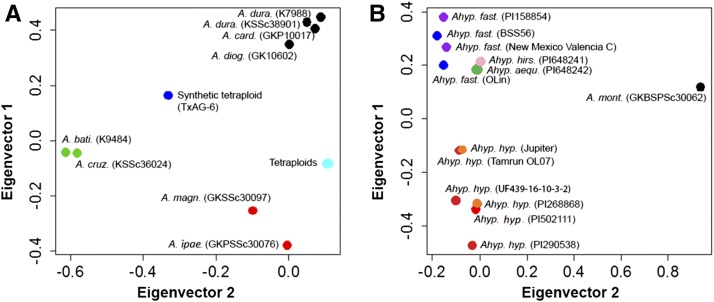
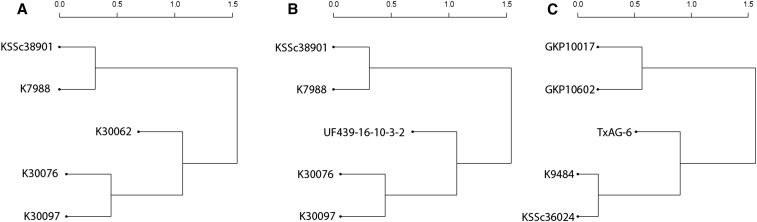
Analysis of SNP diversity demonstrated correlation with known botanical relationships or genome affinities. We assessed genetic diversity based on the above SNPs using PCA. Eigenvector 1 (Y axis) identified three groups: A-genome diploids as the first group; tetraploids, TxAG-6 (synthetic tetraploid) and K- genome as the second group; and B- genome diploids as the third group (Figure 5A). Eigenvector 2 (X axis) separated TxAG-6, K-genome diploids, and the tetraploids from each other. Clustering of genotypes for tetraploids using PCA failed to reveal evidence of subspecies of hypogaea, even after the third PCA including 22 accessions. To distinguish subspecies of tetraploids, we employed the PCA approach on the same set of SNPs from the tetraploids separately, and it was possible to distinguish var hypogaea from var fastigiata (Figure 5B). From the PCA (Figure 5A), we found that the B- or K- genome is closer to the tetraploids than the A- genome, but the number of variants for diploid accessions in Table 4 suggest that the B-genome is closer to the tetraploids than are the A- or K-genome diploids. From the dissimilarity matrix (Figure 6), the B-genome species were closer to the tetraploids, and the next closer parental species belonged to the A-genome. The hirsuta accession did not group with var hypogaea, contrary to expectation; however, validation of SNP calls was carried out using seed from the Plant Introduction station using KASP markers, and the phenotype was confirmed in the field. Surprisingly, the A. monticola accession grouped closer to the fastigiata accessions than to the hypogaea accessions, with the exception of the hirsuta accession, which was closer in the first principal component.
Figure 5.
PCA using SNP data. (A) Twenty-two genotypes analyzed, which included diploids and tetraploids. Dot colors mark genome affinities of accessions as follows: black dots, A-genome accessions; red dots, B-genome accessions; green dots, K-genome accessions; blue dot, synthetic amphidiploid; cyan dot, natural tetraploids. (B) Cultivated tetraploids along with Arachis monticola were analyzed with diploids omitted. Dot colors mark market types of accessions as follows: violet, valencias; blue, spanish; green, aequatoriana; red, runners; orange, virginias; pink, hirsutas; black A. monticola.
Figure 6.
Dendrogram based on the distance matrix obtained from the polymorphism based on biallelic sites on different groups. (A) Natural tetraploids with their possible ancestors. (B) Cultivated tetraploids with their possible ancestors. (C) Synthetic tetraploid with parents from which the amphidiploid was derived.
Phasing separated some of the merged homeologous sequences
There was evidence for significant collapse of homeologous sequences, and phasing separated a large number of merged homeologous sequences in this study. Our results among cultivars and landraces suggested that the frequency of SNPs in tetraploids was 0.5 SNP per kilobase or 0.09 per kilobase in pairwise comparisons, but we observed higher SNP rates on mapping the raw reads back to the OLin transcriptome, which could be the consequence of homeolog transcript collapse. To identify the variants in phased sequences, we aligned the sequences back to the OLin reference transcriptome. There were a total of 25,488 contigs (53.4%) that contained >1 SNP, which were therefore good candidates for polymorphism phasing (Table 6). Assuming that >1 SNP per transcript could be a result of homeolog collapse, we selected transcripts with >1 SNP to separate transcripts using the HapCUT program. We phased SNPs in 22,683 contigs containing >1 SNP into 30,672 sub-transcripts, resulting in an average of 1.35 separated transcripts per contig. During the readphaser process, only 10,815 of 22,683 phased contigs were carried forward for assembly, as the remaining contigs had either evidence of a third haplotype or low read support. Phased assemblies consisted of a total of 81,244 contigs, which were further used to evaluate the separation of homeologs.
Table 6. Summary of the assembly and SNP calls using de novo and phased approach on OLin transcriptome.
| Name | No. of Contigs | N50 | Contigs with SNPs | Contigs with >1 SNPs | Contigs with >2 SNPs | Heterozygous Callsa |
|---|---|---|---|---|---|---|
| OLin_de novo | 67,098 | 1641 | 30,918 | 25,888 | 22,071 | 241,497 |
| OLin_Phased | 87,214 | 1292 | 29,677 | 20,440 | 14,593 | 109,937 |
Heterozygous calls could be the result of homeolog collapse in the reference or alignment errors.
The phasing strategy generated higher numbers of contigs than the de novo assembly approach, but had lesser representation of the transcriptome, as only 76–78% of the reads mapped back. Variant calls from phased assemblies indicated a sharp decrease in the number of heterozygous SNPs, and the number of contigs with multiple SNPs per transcript was lower compared to the de novo assembly (Table 6). The number of contigs with >2 SNPs was reduced by ∼33%, while the number of contigs with >1 SNP was reduced by 20% in the phased assembly approach compared to the de novo approach.
Discussion
Significant genic diversity exists in peanut
SNP diversity analysis demonstrated the presence of significant genic diversity among cultivated accessions and wild species. The number of SNPs identified across all 22 genotypes suggested less genetic variation among A. hypogaea accessions compared to the wild diploids, which had substantially higher genetic variation. High phenotypic diversity observed among diploid accessions is consistent with genetic variation identified in comparison to the OLin reference. For the United States cultivars, the number of homozygous SNPs was in the range of 3000–3500, which agreed with the data previously reported by Chopra et al. (2015). Among the landraces of tetraploid peanut, homozygous SNPs ranged from 7000 to 10,000 SNPs, double to triple the number in United States cultivars. Such a high number of SNPs among the tetraploid accessions suggested high genic diversity exists compared to previous reports of low, or no, molecular variability (Kochert et al. 1996; Burow et al. 2009) using RFLP markers. Also, this genic variability among the landraces could reflect the level of phenotypic diversity observed among core and minicore collections of peanut. Indeed, four of the accessions screened in this study were chosen from the United States peanut minicore collection. Previous minicore screens have also shown that there is huge variability for traits such as water use efficiency (Singh et al. 2014), oil content (Wang et al. 2011a,b), bacterial wilt disease (Jiang et al. 2014), resistance to abiotic and biotic stresses (Upadhyaya et al. 2013), and agronomic traits (Pandey et al. 2014). Sufficient variation has been identified among tetraploid accessions through transcriptome sequencing for performing QTL analysis with high density marker linkage maps.
To call SNPs on 22 accessions, we aligned raw reads to the reference (OLin). The raw reads aligning to the reference ranged from 84 to 89%, suggesting that the assembly used in this study had a good representation of the peanut transcriptomes. A total of 520 SNPs was identified by comparing the OLin transcriptome to the reference assembly; this may suggest the presence of gene copy number variation, call errors, or artifacts in the software used. The presence of SNPs when mapping reads back to the same reference has been cited previously (Nielsen et al. 2011).
The SNP rate in tetraploid peanuts identified was 0.5 per kilobase, which is lower than other related legume species such as soybean (2.7 SNPs per kilobase) (Choi et al. 2007), field pea (2.7 SNPs per kilobase) (Leonforte et al. 2013), and Medicago truncatula (1.96 SNPs per kilobase) (Choi et al. 2004). However, in diploid species of peanut, including different genome affinities, SNP rates reported here were higher than in other legumes.
Phylogenetic analysis demonstrated that SNP diversity is associated with population structure
Previous genetic diversity analyses have indicated clear differentiation of diploid species and tetraploid subspecies (Burow et al. 2009). Based on thousands of SNPs identified in this study, we also differentiated tetraploids from the diploids (Figure 3A), and individuals of the two subspecies of A. hypogaea (Figure 3B). The number of SNPs differentiating accessions of A. hypogaea and A. monticola was also higher compared to differences among cultivars, providing evidence of additional untapped genetic diversity.
In general, the hypogaea accessions grouped together; likewise the fastigiata accessions. Two unusual results were in the first PCA results, in which the hirsuta accession and A. monticola were closer to each other and to the fastigiata accessions. This is surprising, because the spreading type of these accessions matches the hypogaea accessions. However, there are other phenotypic data to consider—for example, the pronounced reticulation of pods of hirsuta, peruviana, and aequatoriana accessions resembles each other more than the typical hypogaea or fastigiata accessions. As with the case of the greater similarity of A. monticola to the fastigiata type, inclusion of data from other accessions in a separate study would be warranted before making broader conclusions, as the present data are based on only a single accession.
Validation of SNPs demonstrated high potential utility for marker-assisted breeding using KASP markers
Another goal of this research was to identify SNP markers that could be useful in breeding programs. Of the 782 selected SNP calls tested, 81% were confirmed on the LightCycler 480, which is significantly better than results of previous studies in peanut (Khera et al. 2013; Chopra et al. 2015). The two diploid accessions used for validation were selected to demonstrate the approach used for identification of SNPs in peanut, and for development of a genetic map (R. Chopra et al., unpublished results). Tetraploid accessions were used to confirm the SNP calls that could be eventually used in population studies. The overall validation success rate of 81.32% of 782 selected SNP sites was much higher than in a previous study by Chopra et al. (2015). Validation using KASP chemistry suggested that the variant calling approach is effective in diploid and tetraploid peanut. This provides a basis for selecting SNP markers between the parents for future breeding activities.
Annotations of the transcripts in the consensus assembly and the OLin assembly by Chopra et al. (2014) provided evidence of SNPs in important trait-related transcripts. For example, SNPs were observed in oil biosynthesis genes, and in the genes of QTL regions affecting nematode resistance. Diagnostic KASP markers were designed and are being evaluated in the breeding program (R. Chopra et al., unpublished results). Collectively, these datasets have the potential for enabling SNP identification throughout the genome for any polymorphic gene of interest, thus making the tagging of specific genes with molecular markers a possibility.
Unique genes per peanut genome are in the range of 25–32K
Results of de novo assembly suggest that there are about 25,000–31,500 unique transcripts in diploid and tetraploid Arachis roots, leaves, and developing pod tissues. This number could be a minimum value, as not all tissues were sampled. The number of transcripts in tetraploids could be twice that of diploids because of homeologous copies. Given the allo-tetraploid nature of the peanut, genes from both genomes are similar, and tools used in the study in general could not distinguish homeologous genes from each other. Consensus assemblies had more contigs than any other assembly by itself, which suggested that each genotype has a certain percentage of transcripts that could not be merged at a minimum sequence similarity threshold of 95%. This increase in the number of transcripts could be due to the larger differences among species/accessions used, or differences in expression of transcripts in samples. For tetraploids, the increase in number of transcripts in the consensus assembly can also be a result of the presence of homeologs. Consensus and individual assemblies from this study could benefit annotation of the genome for rare transcripts and gene expression studies.
Phasing of homeologous copies requires longer reads
In the case of diploids, the success rate of SNP validation was greater than for tetraploid calls. Complexity of the tetraploid genome resulting from homeologous copies may have affected the outcomes of validation. Based on previous studies in complex polyploids, such as wheat (Schreiber et al. 2012) and peanut (Chopra et al. 2014), it was observed that a significant number of homeologs would be merged even after optimizing the de novo assembly parameters. Tetraploid SNP calls consisted of many apparently heterozygous state genotypes, and to reduce these heterozygous calls, we evaluated the phasing approach. We utilized the postassembly approach reported by Krasileva et al. (2013) to separate merged homeologous transcripts with assemblers. This approach gave a more complete overview of the tetraploid transcriptome with a smaller number of chimeric transcripts. Ideally, mapping the reads back to the reference derived from same accession should give fewer variant calls, but, due to collapse of some homeologs, the heterozygous variant calls were more in the de novo assembly. One way to estimate the reduction of homeolog collapse would be to observe the number of heterozygous variants upon mapping the reads back. Reduction in heterozygous SNP calls compared to the reference would be the first step for improving the assembly qualities.
After evaluating the results of resolving homeologs, we suggest that, to get a complete separation of transcripts with no collapse would require: (a) longer sequence reads, because shorter reads can affect the extension of contigs with the poor quality on the ends of reads; (b) higher read coverage to support the haplotypes, because lower read depth haplotypes could be dropped by the phasing based approach; and (c) using the genome sequence of diploid progenitor species to avoid collapse, and to obtain a better representation of transcripts.
Conclusions
We report the first survey of significant SNP level and transcript level diversity in a wide array of peanut germplasm. Highlighted untapped genetic variability among accessions may explain some of the phenotypic differences observed in germplasm surveys. Transcriptome analysis among cultivated accessions, and comparison to wild species will open many avenues for future research. Among these are relationships of cultivated tetraploid species to their diploid ancestors, evolutionary development of the cultigen, distinguishing of homologous vs. homeologous SNPs for genetic mapping and QTL analysis, study of genome rearrangements after polyploidization, and expression bias among genes and genomes. Outcomes of this research will help increase of genomic resources in peanut, and benefit QTL mapping relevant for agronomic traits.
Supplementary Material
Acknowledgments
We thank Halee Hughes, United States Department of Agriculture–Agriculture Research Service for technical support. This work was funded by grants from the Texas Peanut Producers Board award CY2008-Burow- TTU-Development to M.D.B. and C.E.S., and 2009-TTU-Burow-Genotyping to M.D.B., National Peanut Board grant #332/TX-99/1139 to M.D.B., and #332/TX-99/1213 to M.D.B. and C.E.S., Peanut Foundation grant 04-810-08 to M.D.B., Ogallala Aquifer Initiative award IPM12.06 to M.D.B., and United States Department of Agriculture/National Institute of Food and Agriculture Hatch Act award TEX08835 to M.D.B. The authors declare that they have no conflict of interest. All experiments were performed in accordance with current biomedical research ethical standards in the United States.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.115.026898/-/DC1.
Communicating editor: J. Wendel
Literature Cited
- Anderson W. F., Holbrook C. C., Culbreath A. K., 1996. Screening the peanut core collection for resistance to tomato spotted wilt virus. Peanut Sci. 23: 57–61. [Google Scholar]
- Bansal V., Bafna V., 2008. HapCUT: an efficient and accurate algorithm for the haplotype assembly problem. Bioinformatics 24: 153–159. [DOI] [PubMed] [Google Scholar]
- Baring M. R., Simpson C. E., Burow M. D., Black M. C., Cason J. M., et al. , 2006. Registration of ‘Tamrun OL07’peanut. Crop Sci. 46: 2721–2722. [Google Scholar]
- Bertioli D., Seijo G., Freitas F., Valls J., Leal-Bertioli S., et al. , 2011. An overview of peanut and its wild relatives. Plant Genet. Resour. 9: 134–149. [Google Scholar]
- Bertioli D. J., Cannon S. B., Froenicke L., Huang G., Farmer A. D., et al. , 2016. The genome sequences of Arachis duranensis and Arachis ipaënsis, the diploid ancestors of cultivated peanut. Nat. Genet. 48: 438–446. [DOI] [PubMed] [Google Scholar]
- Burow M., Simpson C., Faries W., Starr J., Paterson A., 2009. Molecular biogeography study of recently described B- and A-genome Arachis species, also providing new insights into the origins of cultivated peanut. Genome 52: 107–119. [DOI] [PubMed] [Google Scholar]
- Burow M. D., Starr J. L., Park C.-H., Simpson C. E., Paterson A. H., 2014. Introgression of homoeologous quantitative trait loci (QTLs) for resistance to the root-knot nematode [Meloidogyne arenaria (Neal) Chitwood] in an advanced backcross-QTL population of peanut (Arachis hypogaea L.). Mol. Breed. 34: 393–406. [Google Scholar]
- Chen X., Zhu W., Azam S., Li H., Zhu F., 2013. Deep sequencing analysis of the transcriptomes of peanut aerial and subterranean young pods identifies candidate genes related to early embryo abortion. Plant Biotechnol. J. 11: 115–127. [DOI] [PubMed] [Google Scholar]
- Choi H., Kim D. J., Uhm T., Limpens E., Lim H., et al. , 2004. A sequence based genetic map of Medicago truncatula and comparison of marker collinearity with M. sativa. Genetics 166: 1463–1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi I.-Y., Hyten D. L., Matukumalli L. K., Song Q., Chaky J. M., et al. , 2007. A soybean transcript map: gene distribution, haplotype and single-nucleotide polymorphism analysis. Genetics 176: 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra R., Burow G. B., Farmer A., Mudge J. A., Simpson C. E., et al. , 2014. Comparisons of de novo transcriptome assemblers in diploid and polyploid species using peanut (Arachis spp.) RNA-Seq data. PLoS One 9(12): e115055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra R., Burow G., Farmer A., Mudge J., Simpson C. E., et al. , 2015. Next-generation transcriptome sequencing, SNP discovery and validation in four market classes of peanut, Arachis hypogaea L. Mol. Genet. Genomics 290: 1169–1180. [DOI] [PubMed] [Google Scholar]
- Danecek P., Auton A., Abecasis G., Albers C. A., Banks E., et al. , 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deokar A. A., Ramsay L., Sharpe A. G., Diapari M., Sindhu A., et al. , 2014. Genome wide SNP identification in chickpea for use in development of a high density genetic map and improvement of chickpea reference genome assembly. BMC Genomics 15: 708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M. A., Banks E., Poplin R., Garimella K. V., Maguire J. R., et al. , 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43: 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A., Krapovickas A., 1994. Cromosomas y evolución en Arachis (Leguminosae). Bonplandia 8: 187–220. [Google Scholar]
- Garg R., Patel R. K., Tyagi A. K., Jain M., 2011. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 18: 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M., Haas B., Yassour M., Levin J., Thompson D., et al. , 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory W., Gregory M., 1976. Groundnut, pp. 151–154 in Evolution of Crop Plants, edited by Simmonds N. W. Longman Group, Ltd., London. [Google Scholar]
- Gregory W., Krapovickas A., Gregory M., 1980. Structure, variation, evolution, and classification in Arachis, pp. 469–481 in Advances in Legume Science, edited by Summerfield R. J., Bunting A. H. Kew: Royal Botanic Gardens, Richmond, England. [Google Scholar]
- Halward T., Stalker H. T., Larue E., Kochert G., 1991. Genetic variation detectable with molecular markers among unadapted germplasm resources of cultivated peanut and related wild species. Genome 34: 1013–1020. [Google Scholar]
- Holbrook C. C., Dong W., 2005. Development and evaluation of a minicore collection for the US peanut germplasm collection. Crop Sci. 45: 1540–1544. [Google Scholar]
- Holbrook C. C., Anderson W., Pittman R., 1993. Selection of a core collection from the US germplasm collection of peanut. Crop Sci. 33: 859–861. [Google Scholar]
- Holbrook C. C., Timper P., Culbreath A. K., Kvien C. K., 2008. Registration of ‘Tifguard’ peanut. J. of Plant Regist. 2: 2. [Google Scholar]
- Hsi D. C., 1980. Registration of ‘New Mexico Valencia C’ peanut (Reg. No. 24). Crop Sci. 20: 113–114. [Google Scholar]
- Huang L., Jiang H., Ren X., Chen Y., Xiao Y., et al. , 2012. Abundant microsatellite diversity and oil content in wild Arachis species. PLoS One 7: e50002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Madan A., 1999. CAP3: A DNA sequence assembly program. Genome Res. 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isleib T. E., Pattee H. E., Giesbrecht F. G., 1995. Ancestral Contributions to Roasted Peanut Attribute. Peanut Sci. 22: 42–48. [Google Scholar]
- Jiang H., Liao B., Duan N., Holbrook C. C., Guo B., 2004. Development of a core collection of peanut germplasm in China. Proc. Amer. Peanut Res. Educ. Soc. 36: 33. [Google Scholar]
- Jiang H., Li H., Xiaoping R., Yuning C., Xiaojing Z., et al. , 2014. Diversity characterization and association analysis of agronomic traits in a Chinese peanut (Arachis hypogaea L.) mini-core collection. J. Integr. Plant Biol. 56: 159–169. [DOI] [PubMed] [Google Scholar]
- Khera P., Upadhyaya H. D., Pandey M. K., Roorkiwal M., Sriswathi M., et al. , 2013. Single nucleotide polymorphism-based genetic diversity in the reference set of peanut (spp.) by developing and applying cost-effective kompetitive allele specific polymerase chain reaction genotyping assays. Plant Genome 6: 1–11. [Google Scholar]
- Kochert G., Halward T., Branch W., Simpson C., 1991. RFLP variability in peanut (Arachis hypogaea L.) cultivars and wild species. Theor. Appl. Genet. 81: 565–570. [DOI] [PubMed] [Google Scholar]
- Kochert G., Stalker H., Gimenes M., Galgaro L., Lopes C., et al. , 1996. RFLP and cytogenetic evidence on the origin and evolution of allotetraploid domesticated peanut Arachis hypogaea (Leguminosae). Am. J. Bot. 83: 1282–1291. [Google Scholar]
- Krapovickas A., Gregory W., 1994. Taxonomia del género Arachis (Leguminosae). Bonplandia 8: 1–186. [Google Scholar]
- Krasileva K., Buffalo V., Bailey P., Pearce S., Ayling S., et al. , 2013. Separating homoeologs by phasing in the tetraploid wheat transcriptome. Genome Biol. 14: R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavia G., 1998. Karyotypes of Arachis palustris and A. praecox (section Arachis), two species with basic chromosome number x = 9. Cytologia (Tokyo) 63: 177–181. [Google Scholar]
- Lavia G., Fernandez A., Seijo G., 2008. Cytogenetic and molecular evidences on the evolutionary relationships among Arachis species. plant genome: biodiversity and evolution. Phanerogams-Angiosperm. 1E: 101–134. [Google Scholar]
- Leonforte A., Sudheesh S., Cogan N. O. I., Salisbury P. A., Nicolas M. E., et al. , 2013. SNP marker discovery, linkage map construction and identification of QTLs for enhanced salinity tolerance in field pea (Pisum sativum L.). BMC Plant Biol. 13: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R., 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26: 589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Acharya A., Farmer A. D., Crow J. A., Bharti A. K., et al. , 2012. Prevalence of single nucleotide polymorphism among 27 diverse alfalfa genotypes as assessed by transcriptome sequencing. BMC Genomics 13: 568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse M., Nagel A., Herter T., May P., Schroda M., et al. , 2014. Mercator: a fast and simple web server for genome scale functional annotation of plant sequence data. Plant Cell Environ. 37: 1250–1258. [DOI] [PubMed] [Google Scholar]
- Lu J., Pickersgill B., 1993. Isozyme variation and species relationships in peanut and its wild relatives (Arachis L. - Leguminosae). Theor. Appl. Genet. 85: 550–560. [DOI] [PubMed] [Google Scholar]
- Mallikarjuna N., Senthilvel S., Hoisington D., 2011. Development of new sources of tetraploid Arachis to broaden the genetic base of cultivated groundnut (Arachis hypogaea L.). Genet. Resour. Crop Evol. 58: 889–907. [Google Scholar]
- McCormick R.F., Truong S. K., Mullet J. E., 2015. RIG: recalibration and interrelation of genomic sequence data with the GATK. G3 (Bethesda) 5: 655–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretzsohn M. M., Hopkins S., Mitchell S., Kresovich J., Valls J., et al. , 2004. Genetic diversity of peanut (Arachis hypogaea L.) and its wild relatives based on the analysis of hypervariable regions of the genome. BMC Plant Biol. 4: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E.D., Guo Y., Tang S., Bowers J. E., Okashah R. A., et al. , 2012. A high-density genetic map of Arachis duranensis, a diploid ancestor of cultivated peanut. BMC Genomics 13: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen R., Paul J. S., Albrechtsen A., Song Y. S., 2011. Genotype and SNP calling from next-generation sequencing data. Nat. Rev. Genet. 12: 443–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norden A. J., 1973. Breeding of the cultivated peanut (Arachis hypogaea L.), pp. 175–208 in Peanuts—Cultures and Uses, edited by Wilson C. T. Amer. Peanut Res. Educ. Soc., Stillwater. [Google Scholar]
- Norden A. J., Lipscomb R. W., Carver W. A., 1969. Registration of ‘Florunner’ peanuts (Reg No. 2). Crop Sci. 9: 850. [Google Scholar]
- Pandey M. K., Upadhyaya H. D., Rathore A., Vadez V., Sheshshayee M. S., et al. , 2014. Genomewide association studies for 50 agronomic traits in peanut using the ‘reference set’ comprising 300 genotypes from 48 countries of the semi-arid tropics of the world. PLoS One 9(8): e105228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina S., Mukai Y., 1999. Genomic in situ hybridization in Arachis (Fabaceae) identifies the diploid wild progenitors of cultivated (A. hypogaea) and related wild (A. monticola) peanut species. Plant Syst. Evol. 214: 251–262. [Google Scholar]
- Raina S., Rani V., Kojima T., Ogihara Y., Singh K., et al. , 2001. RAPD and ISSR fingerprints as useful genetic markers for analysis of genetic diversity, varietal identification, and phylogenetic relationships in peanut (Arachis hypogaea) cultivars and wild species. Genome 44: 763–772. [PubMed] [Google Scholar]
- Schreiber A. W., Hayden M. J., Forrest K. L., Kong S. L., Langridge P., et al. , 2012. Transcriptome-scale homoeolog-specific transcript assemblies of bread wheat. BMC Genomics 13: 492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seijo G., Lavia G. I., Fernández A., Krapovickas A., Ducasse D. A., et al. , 2007. Genomic relationships between the cultivated peanut (Arachis hypogaea, Leguminosae) and its close relatives revealed by double GISH. Am. J. Bot. 94: 1963–1971. [DOI] [PubMed] [Google Scholar]
- Selvaraj M. G., Burow G., Burke J. J., Belamkar V., Puppala N., et al. , 2011. Heat stress screening of peanut (Arachis hypogaea L.) seedlings for acquired thermotolerance. Plant Growth Regul. 65: 83–91. [Google Scholar]
- Simpson C., 2001. Use of wild Arachis species/introgression of genes into A. hypogaea L. Peanut Sci. 28: 114–116. [Google Scholar]
- Simpson C. E., Starr J. L., 2001. Registration of ‘COAN’ peanut. Crop Sci. 41: 918. [Google Scholar]
- Simpson C. E., Nelson S. C., Starr J. L., Woodard K. E., Smith O. D., 1993. Registration of TxAG-6 and TxAG-7 peanut germplasm lines. Crop Sci. 33: 1418. [Google Scholar]
- Simpson C. E., Baring M. R., Schubert A. M., Melouk H. A., López Y., et al. , 2003a Registration of ‘OLin’ peanut. Crop Sci. 43: 1880–1881. [Google Scholar]
- Simpson C. E., Starr J. L., Church G. T., Burow M. D., Paterson A. H., 2003b Registration of NemaTAM peanut. Crop Sci. 43: 1561. [Google Scholar]
- Simpson C. E., Starr J. L., Baring M. R., Burow M. D., Cason J. M., et al. , 2013. Registration of ‘Webb’ Peanut. J. Plant Regist. 7: 265–268. [Google Scholar]
- Singh A. L., Nakar R. N., Chakraborty K., Kalariya K. A., 2014. Physiological efficiencies in mini-core peanut germplasm accessions during summer season. Photosynthetica 52: 627–635. [Google Scholar]
- Soltis D. E., Soltis P. S., 1999. Polyploidy: recurrent formation and genome evolution. Trends Ecol. Evol. 14: 348–352. [DOI] [PubMed] [Google Scholar]
- Stalker H. T., Moss J. P., 1987. Speciation, cytogenetics, and utilization of Arachis species. Adv. Agron. 41: 1–40. [Google Scholar]
- UniProt Consortium , 2015. UniProt: a hub for protein information. Nucleic Acids Res. 43: D204–D212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya H. D., 2003. Phenotypic diversity in groundnut (Arachis hypogaea L.) core collection assessed by morphological and agronomical evaluations. Genet. Resour. Crop Evol. 50: 539–550. [Google Scholar]
- Upadhyaya H. D., 2005. Variability for drought resistance related traits in the mini core collection of peanut. Crop Sci. 45: 1432–1440. [Google Scholar]
- Upadhyaya H. D., Bramel P., Ortiz R., Singh S., 2002. Developing a mini core of peanut for utilization of genetic resources. Crop Sci. 42: 2150–2156. [Google Scholar]
- Upadhyaya H. D., Reddy L. J., Gowda C. L. L., Singh S., 2006a Identification of diverse groundnut germplasm: sources of early-maturity in a core collection. Field Crops Res. 97: 261–267. [Google Scholar]
- Upadhyaya H. D., Gowda C. L. L., Buhariwalla H. K., Crouch J. H., 2006b Efficient use of crop germplasm resources: identifying useful germplasm for crop improvement through core and mini-core collections and molecular marker approaches. Plant Gen. Res. 4: 25–35. [Google Scholar]
- Upadhyaya H. D., Sharma S., Dwivedi S. L., 2013. Genetic resources, diversity and association mapping in peanut, pp. 13–36 in Genetics, Genomics and Breeding of Peanuts, edited by Mallikarjuna N., Varshney R. K. CRC Press, Boca Raton. [Google Scholar]
- Valls J., Simpson C., 2005. New species of Arachis L. (Leguminosae) from Brazil, Paraguay and Bolivia. Bonplandia 14: 35–64. [Google Scholar]
- Wang M. L., Sukumaran S., Barkley N. A., Chen Z., Chen C. Y., et al. , 2011a Population structure and marker–trait association analysis of the US peanut (Arachis hypogaea L.) mini-core collection. Theor. Appl. Genet. 123: 1307–1317. [DOI] [PubMed] [Google Scholar]
- Wang M. L., Barkley N., Chen Z., Pittman R., 2011b FAD2 gene mutations significantly alter fatty acid profiles in cultivated peanuts (Arachis hypogaea). Biochem. Genet. 49: 748–759. [DOI] [PubMed] [Google Scholar]
- Wu X., Ren C., Joshi T., Vuong T., Xu D., et al. , 2011. SNP discovery by high-throughput sequencing in soybean. BMC Genomics 11: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Liang S., Duan J., Wang J., Chen S., et al. , 2012. De novo assembly and characterisation of the transcriptome during seed development, and generation of genic-SSR markers in peanut (Arachis hypogaea L.). BMC Genomics 13: 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Levine D., Shen J., Gogarten S. M., Laurie C., et al. , 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics 28: 3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
BAM files are deposited in SRA database under bioproject PRJNA248910: SRR1534396, SRR1535034, SRR1535035, SRR4039437, SRR4039359, SRR4039358, SRR4039357, SRR4039017, SRR4039016, SRR4039015, SRR4038966, SRR4038965, SRR4038285, SRR4037988, SRR4038281, SRR4038254, SRR4038252, SRR4038060, SRR4037987, SRR4037985, SRR4037986, and SRR4037959. De novo assemblies and annotations described in this manuscript have been deposited at Figshare at the following URLs: https://dx.doi.org/10.6084/m9.figshare.3580650.v2, https://dx.doi.org/10.6084/m9.figshare.3580656.v2, https://dx.doi.org/10.6084/m9.figshare.3580659.v1, and https://dx.doi.org/10.6084/m9.figshare.3580662.v1.