Abstract
TORC1 regulates metabolism and growth in response to a large array of upstream inputs. The evolutionarily conserved trimeric GATOR1 complex inhibits TORC1 activity in response to amino acid limitation. In humans, the GATOR1 complex has been implicated in a wide array of pathologies including cancer and hereditary forms of epilepsy. However, the precise role of GATOR1 in animal physiology remains largely undefined. Here, we characterize null mutants of the GATOR1 components nprl2, nprl3, and iml1 in Drosophila melanogaster. We demonstrate that all three mutants have inappropriately high baseline levels of TORC1 activity and decreased adult viability. Consistent with increased TORC1 activity, GATOR1 mutants exhibit a cell autonomous increase in cell growth. Notably, escaper nprl2 and nprl3 mutant adults have a profound locomotion defect. In line with a nonautonomous role in the regulation of systemic metabolism, expressing the Nprl3 protein in the fat body, a nutrient storage organ, and hemocytes but not muscles and neurons rescues the motility of nprl3 mutants. Finally, we show that nprl2 and nprl3 mutants fail to activate autophagy in response to amino acid limitation and are extremely sensitive to both amino acid and complete starvation. Thus, in Drosophila, in addition to maintaining baseline levels of TORC1 activity, the GATOR1 complex has retained a critical role in the response to nutrient stress. In summary, the TORC1 inhibitor GATOR1 contributes to multiple aspects of the development and physiology of Drosophila.
Keywords: TORC1, Nprl2, Nprl3, Iml1, metabolism
The Target of Rapamycin Complex 1 (TORC1) regulates nutrient sensing and cell metabolism from yeast to humans (Loewith and Hall 2011; Laplante and Sabatini 2012). At the heart of the TORC1 complex is the serine/threonine kinase Tor (Schmelzle and Hall 2000). In the presence of sufficient nutrients and growth signals, TORC1 is active and stimulates protein synthesis and cell growth through the phosphorylation of downstream effectors such as S6K and 4E-BP, while simultaneously inhibiting catabolic metabolism and autophagy (Hay and Sonenberg 2004; Wullschleger et al. 2006). Conversely, when nutrient or growth factors are limiting, TORC1 is inactivate resulting in the inhibition of cell growth and the promotion of catabolic metabolism (He and Klionsky 2009; Jung et al. 2010). Thus, by modulating the activity of TORC1, cells can rapidly adjust their metabolic state in response to both extracellular and intracellular stimuli.
Mutations in upstream signaling pathways that regulate TORC1 result in a wide array of human pathologies. Many of these pathologies, such as the development of benign tumors and a predisposition to cancers, are associated with increased TORC1 activity and cell growth (Laplante and Sabatini 2012). TORC1 activity also contributes to numerous age-related diseases including cancer, diabetes, and neurodegenerative disorders such as Parkinson’s (Laplante and Sabatini 2012; Johnson et al. 2013). Reducing TORC1 activity through genetic, pharmacological, or nutritional intervention extends lifespan across multiple model organisms including yeast, Caenorhabditis elegans, Drosophila, and mice, while increasing TORC1 activity results in decreased lifespan (Evans et al. 2011; Johnson et al. 2013; Fontana and Partridge 2015). Recent evidence indicates that mutations that inactivate several upstream inhibitors of TORC1 result in the development of focal epilepsies via an unknown mechanism (Dibbens et al. 2013; Tee et al. 2016). Thus, the precise regulation of TORC1 activity is critical to multiple aspects of human health.
Two GTPases, Rheb and the Rags, play a key role in the regulation of TORC1 activity. The small GTPase Rheb activates TORC1 on the surface of lysosomes (Yang et al. 2006; Sancak et al. 2008). The Rag GTPase consists of four proteins RagA, RagB, RagC, and RagD, that function as heterodimers (Kim et al. 2008; Sancak et al. 2008). While amino acids are sufficient, RagA/B binds GTP and RagC/D binds GDP. In this active configuration, the Rags function as GTPases that promote the recruitment of TORC1 to lysosomes where it encounters its activator Rheb. Thus, an important step in the activation of TORC1 is the Rag GTPase-dependent recruitment of the TORC1 complex to lysosomes.
Recently the GAP activity toward Rags (GATOR) complex, which is named the Seh1 associated (SEA) complex in yeast, was shown to regulate TORC1 activity through the Rag GTPases (Dokudovskaya et al. 2011; Wu and Tu 2011; Bar-Peled et al. 2013; Panchaud et al. 2013a). Iml1/DEPDC5, Nprl2, and Nprl3 comprise GATOR1, which functions as a GTPase activating protein (GAP) for RagA/B, and thus acts as an inhibitor of TORC1 activity. The GATOR1 complex is called the SEA Complex Inhibits TORC1 (SEACIT) in yeast (Dokudovskaya et al. 2011; Panchaud et al. 2013b). Deletion mutants of the SEACIT/GATOR1 components npr2, npr3, and iml1 have a reduced ability to grow on a poor nitrogen source or restricted methionine, but do not have proliferation defects or increased TORC1 activity under conditions of amino acid sufficiency (Neklesa and Davis 2009; Ma et al. 2013). In contrast, in mammalian and Drosophila tissue culture cells, depleting GATOR1 components results in a dramatic increase in TORC1 activity under standard culture conditions (Bar-Peled et al. 2013; Wei and Lilly 2014). Notably, recent reports have shown that GATOR1 knockouts of iml1/Depdc5 (rat), nprl2 (mouse), and nprl3 (mouse) result in late embryonic lethality, with embryos exhibiting developmental defects in the heart, liver, and brain (Kowalczyk et al. 2012; Dutchak et al. 2015; Marsan et al. 2016). Additionally, in Drosophila, the GATOR1 complex regulates entry into the meiotic cycle during oogenesis (Wei et al. 2014). Taken together, these data strongly suggest that, in metazoans, the GATOR1 complex has evolved roles beyond regulating an adaptive response to amino acid starvation.
Here, we demonstrate that the GATOR1 complex has both cell autonomous and nonautonomous effects on growth and metabolism in the model organism Drosophila melanogaster. We demonstrate that the GATOR1 complex mutants have increased baseline levels of TORC1 activity and decreased viability. Moreover, we show that GATOR1 mutants exhibit multiple phenotypes consistent with increased TORC1 activity including increased cell growth, reduced tolerance to starvation, and the inability to activate autophagy under nutrient-limiting conditions. Intriguingly, GATOR1 mutants exhibit a serious locomotion deficit that can be rescued by altering systemic metabolism. Taken together, our data demonstrate that Drosophila will provide an excellent model to study how GATOR1 influences both development and disease in multicellular animals.
Materials and Methods
Fly stocks
The stocks Df(1)BSC582, w1118/Binsinscy (BDSC#25416), w1118; Df(3L)ED4515, P{3′.RS5+3.3′}ED4515/TM6C, cu1 Sb1 (BDSC#9071,), w1118; Df(3L)ED4238, P{3′.RS5+3.3′}ED4238/TM6C, cu1 Sb1 (BDSC#8052), w*; P{GAL4-elav.L}3 (BDSC#8760), w1118; P{Cg-GAL4.A}2 (BDSC#7011), P{Ubi-mRFP.nls}1, w*, P{hsFLP}12P{neoFRT}19A (BDSC#31418), P{neoFRT}19A ry506 (BDSC#1709), w*; P{neoFRT}80B ry506 (BDSC#1988), and w*; P{w[+mW.hs]=GAL4-da.G32}2; MKRS/TM6B, Tb[1] (BDSC#55851) were obtained from Bloomington Stock Center. y,w; Tor A948V/CyO (Zhang et al. 2006) was kindly provided by Thomas P. Neufeld (University of Minnesota). All fly stocks were maintained on JAZZ-mix Drosophila food (Fisher Scientific, Waltham, MA) at 25°. For complete starvation, flies were cultured on media consisting of phosphate-buffered saline (PBS) and 0.8% agar. For amino acid starvation, flies were cultured on media consisting of PBS, 20% sucrose, and 0.8% agar.
Generation of nprl2 and iml1 deletion mutants
Guide RNAs (gRNA) that target nprl2 and iml1 were designed using the online CRISPR design tool (http://crispr.mit.edu/). To make the deletion mutants, two sets of gRNAs that target each gene were cloned into pBFv-U6.2B as previously described (Kondo and Ueda 2013). The two gRNA were separately expressed from their own U6 promoters in one plasmid. The pBFv-U6.2B-nprl2 and pBFv-U6.2B-iml1 plasmids were separately injected into w[1118]; vas-Cas9 (BDSC#51324) and y[1], vas-Cas9, w[1118] (BDSC#52669) embryos (Rainbow Transgenic Flies, Inc.). Eclosed flies were individually crossed with y, w and screened by PCR using genomic DNA. The offspring of positive screens were balanced to generate nprl2 and iml1 mutant stocks. Primers are listed in Supplemental Material, Table S1.
Generation of nprl2, nprl3, and iml1 transgenic lines
The pENTR-Nprl2 and pENTR-Nprl3 plasmids were recombined into pPFHW vectors (DGRC) to generate UASp-3×FLAG-3×HA-Nprl2 and UASp-3×FLAG-3×HA-Nprl3 plasmids using the Gateway LR Clonase II Enzyme (Invitrogen) (Wei and Lilly 2014). The fragments that contained 837 bp upstream of the iml1 transcription start, the iml1 5′UTR, the iml1 coding region, and the EGFP coding region were amplified by PCR and cloned into the attB-P[acman]-CmR-BW vector. The UASp-3×FLAG-3×HA-Nprl2, UASp-3×FLAG-3×HA-Nprl3, and attB-P[acman]-CmR-BW-Iml1 plasmids were used to generate transgenic lines (Best Gene Inc.).
Lethal phase analysis
Homozygous nprl21, nprl31/Df, and iml11/Df first-star larvae were distinguished from nprl21/FM7-GFP, nprl31/TM3-GFP, and iml11/TM3-GFP based on the absence of GFP. First-star larvae were collected and cultured in new vials at a density of 50 larvae per vial. The number of larvae that developed to the pupae and adult stage was determined for each genotype.
Geotaxis motility assay
Geotaxis motility assay was performed essentially as described by Ganetzky and Flanagan (1978). Briefly, 10–15 flies were placed in a 10 cm vial. The flies were tapped to the bottom of the vial and given 15 sec to climb from the bottom to top. The number of flies arriving at the top were counted and divided by the number of total flies in the vial. Each analysis was repeated five times to obtain an average value.
Clonal analysis
HS-FLP, UAS-RFP FRT19A/nprl21 FRT19A, HS-FLP; Ubi-GFP FRT80B/nprl31 FRT80B and HS-FLP; Ubi-GFP FRT80B/iml11 FRT80B flies were used to generate mutant clones in the germline and somatic cells of the ovary as previously reported (Wei et al. 2014). Briefly, female flies were heat-shocked for 2 hr at 37° each day for 3 d. Subsequently, flies were cultured in standard media for an additional 2 d before dissection. Homozygous mutant clones were identified by the absence of GFP or RFP.
Western blots
Drosophila third-star larvae were homogenized in RIPA buffer containing complete protease inhibitors and phosphatase inhibitors (Roche). Western blots were performed as described previously (Wei et al. 2014). Antibodies were used at the following concentrations: rabbit anti-P-S6K at 1:500 (Cell Signaling) and guinea pig anti-S6K at 1:10,000 (Hahn et al. 2010). Band intensity was quantified using Image J (NIH).
Immunofluorescence and confocal microscopy
Immunofluorescence staining and microscopy was performed as described previously (Hong et al. 2003; Iida and Lilly 2004; Senger et al. 2011), using a mouse anti-1B1 (1:30, Developmental Studies Hybridoma Bank) antibody. Anti-mouse Alexa Fluor secondary antibodies (Invitrogen) were used at a dilution of 1:1000. Nuclei were visualized by staining the DNA with 4′,6-diamidino-2-phenylindole (DAPI) (Invitrogen). Images were acquired using a Leica TCS SP5 confocal microscope. Follicle cell size was quantified using Image J (NIH) based on 1B1 staining.
Data availability
All strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results and Discussion
The GATOR1 components Nprl2, Nprl3, and Iml1 are required for viability in Drosophila
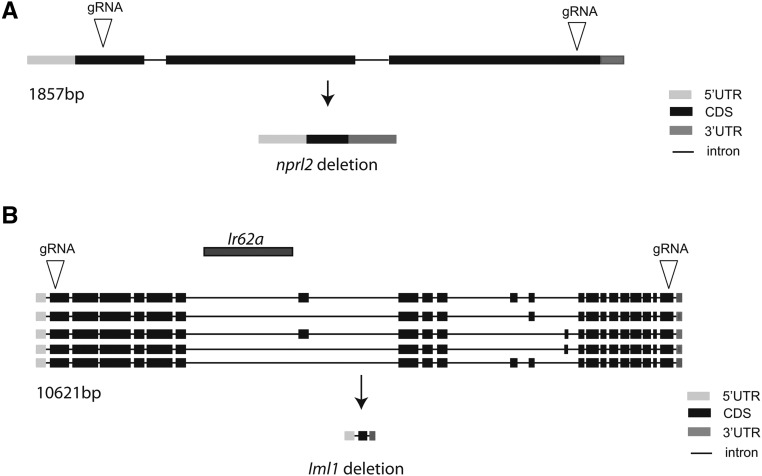
To evaluate the physiological roles of GATOR1 in vivo, we generated nprl2 and iml1 deletion alleles using CRISPR/Cas9 (Kondo and Ueda 2013). We had previously generated nprl3 deletion mutants but had not fully evaluated the mutant phenotype (Cai et al. 2016). In each gene, > 90% of the coding region was deleted (Figure 1). For each gene, multiple alleles were generated that displayed similar phenotypes. For studies here, we used the nprl21, nprl31, and iml11 alleles. In order to define the requirement for individual GATOR1 complex components, we examined the viability of individual GATOR1 deletion mutants. Notably, both nprl21 and nprl31 homozygous adults eclosed at significantly reduced rates (26 and 9% of Mendelian expectations, respectively), while no iml11 homozygous adults eclosed (Table 1). Thus, deletions of nprl2 and nprl3 are semilethal while deletions of iml1 are lethal. To rule out the possibility that the lethality of nprl21, nprl31, and iml11 were the result of the genetic background, we examined each deletion mutant in trans to the appropriate genomic deficiency (Table 1). We determined that the transheterozygotes nprl21/Df and iml11/Df eclosed at rates similar to the respective nrpl21 and iml11 homozygotes. These data are consistent with the nprl2 and iml1 mutants being null alleles. However, nrpl31/Df transheterozygotes eclosed at a significantly higher rate than nprl31 homozygous mutants (24 vs. 9%, respectively). These data suggest that at least some of the lethality associated with the nprl3 deletion may be due to genetic background.
Figure 1.
Schematic representation of GATOR1 mutants generated using CRISPR/Cas9 genome editing. To generate the nprl2 and iml1 deletion mutants, two gRNAs that target the 5′ and 3′ of each gene were used. (A) nprl2 encodes a transcript of 1707 bp and a protein of 412 amino acids (FBgn0030800). The nprl21 deletion contains 30 amino acids from the N-terminus and four frame-shifted amino acids from the C-terminus. (B) iml1 encodes five transcripts (FBgn0035227). These transcripts encode proteins of 1471, 1472, 1503, 1511, and 1544 amino acids, respectively. The iml11 deletion contains 17 amino acids from the N-terminus and 53 amino acids from the C-terminus of all iml1 transcripts. Notably, the intron of iml1 also encodes a transcript Ir62a, which is also deleted. CDS, coding sequences; CRISPR, clustered regularly interspaced short palindromic repeats; gRNA, guide RNA; UTR, untranslated region.
Table 1. Lethality of GATOR1 mutants.
| Cross | Genotype (Number of Adult Flies) | % of Expected Ratio | |
|---|---|---|---|
| nprl21 (Male) × nprl21/FM7 (Female) | nprl21/FM7 (Female) | nprl21 (Female) | 26.3a |
| 388 | 102 | ||
| nprl21 (Male) × Df/FM7 (Female) | nprl21/FM7 (Female) | nprl21/Df (Female) | 28.1a |
| 203 | 57 | ||
| nprl31/TM3 × nprl31/TM3 | nprl31/TM3 | nprl31 | 9.4b |
| 641 | 30 | ||
| nprl31/TM3 × Df/TM3 | nprl31/TM3 and Df/TM3 | nprl31/Df | 24.1b |
| 705 | 85 | ||
| nprl21/FM7; nprl31/TM3 (Male) × Df/TM3 (Female) | nprl31/TM3 (Female) and Df/TM3 (Female) | nprl21; nprl31/Df (Male) | 29.4c |
| 571 | 42 | ||
| iml11/TM3 × iml11/TM3 | iml11/TM3 | iml11 | 0b |
| 1200 | 0 | ||
| iml11/TM3 × Df/TM3 | iml11/TM3 and Df/TM3 | iml11/Df | 0.5b |
| 387 | 1 | ||
In these crosses, the FM7 (first chromosome balancer) was identified by the Bar eye. The expected Medelian ratio of non-Bar eyes to Bar eye female flies was 1:1.
In these crosses, the TM3 (third chromosome balancer) was identified by the Sb marker. The expected Medelian ratio of non-Sb to Sb flies was 1:2, since the TM3/TM3 is embryonic lethal.
In these crosses, the FM7 and TM3 was identified by the Bar and Sb markers. The expected Medelian ratio of male non-Sb and non-Bar to female Sb (both Bar and non-Bar) flies was 1:4, since the TM3/TM3 is embryonic lethal.
To demonstrate formally that the lethality of the mutants was due to the deletion of the GATOR1 genes, we overexpressed HA-tagged Nprl2 and Nprl3 using the Ubi-GAL4 driver and GFP-tagged Iml1 using the native iml1 promoter in the nprl2, nprl3, and iml1 mutant backgrounds, respectively. As shown in Table 2, the expression of each GATOR1 protein fully rescued the lethality associated with the respective GATOR1 mutant. Taken together, our data support the conclusion that null alleles of nrpl2 and nprl3 are semilethal while null mutants of iml1 are nearly fully lethal. Thus, in Drosophila, all three components of the GATOR1 complex are required for full viability.
Table 2. Rescue of GATOR1 mutants’ lethality.
| Cross | Genotype (Number of Adult Flies) | % of Expected Ratio | |
|---|---|---|---|
| nprl21; UAS-Nprl2 (male) × nprl21/FM7; Ubi-GAL4 (Female) | nprl21/FM7;Ubi > Nprl2 (Female) | nprl21; Ubi > Nprl2 (Female) | 111.6a |
| 155 | 173 | ||
| Ubi-GAL4; nprl31/TM3 × UAS-Nprl3; nprl31/TM3 | Ubi > Nprl3; nprl31/TM3 | Ubi > Nprl3; nprl31 | 109.3b |
| 302 | 165 | ||
| iml1-Iml1/SM6; iml11/TM3 × iml11/TM3 | iml1-Iml1; iml11/TM3 | iml1-Iml1; iml11 | 102.0c |
| 394 | 201 | ||
| Ubi-GAL4; nprl31/TM3 × UAS-Nprl3; Df/TM3 | Ubi > Nprl3; nprl31/TM3 and Ubi > Nprl3; Df/TM3 | Ubi > Nprl3; nprl31/Df | 113.6b |
| 405 | 230 | ||
| iml1-Iml1/SM6; iml11/TM3 × Df/TM3 | iml1-Iml1; iml11/TM3 and Iml1; Df/TM3 | iml1-Iml1; iml11/Df | 106.7c |
| 283 | 151 | ||
| nprl21; TorAV/SM6 (Male) × nprl21/FM7 (Female) | nprl21/FM7;TorAV/+ (Female) | nprl21; TorAV/+ (Female) | 89.7d |
| 290 | 260 | ||
| TorAV/SM6; nprl31/TM3 × TorAV/SM6; Df/TM3 | TorAV/+; nprl31/TM3 and TorAV/+; Df/TM3 | TorAV/+; nprl31/Df | 90.7c |
| 344 | 156 | ||
| CG-GAL4; nprl31/TM3 × UAS-Nprl3; Df/TM3 | CG > Nprl3; nprl31/TM3 and CG > Nprl3; Def/TM3 | CG > Nprl3; nprl31/Df | 125.3b |
| 241 | 151 | ||
In these crosses, the FM7 (first chromosome balancer) was identified by the Bar eye. The expected Medelian ratio of non-Bar eyes to Bar eye female flies was 1:1.
In these crosses, the TM3 (third chromosome balancer) was identified by the Sb marker. The expected Medelian ratio of non-Sb to Sb flies was 1:2, since the TM3/TM3 is embryonic lethal.
In these crosses, the SM6 (second chromosome balancer) and TM3 was identified by the Cy and Sb markers. The expected Medelian ratio of non-Cy and non-Sb to non-Cy and Sb flies was 1:2, since the TM3/TM3 is embryonic lethal.
In these crosses, the FM7 and SM6 was identified by the Bar and Cy markers. The expected Medelian ratio of non-Cy and non-Bar to non-Cy and Bar female flies was 1:1.
There are at least two possible models to explain why iml1 has a stronger phenotype than either nprl2 or nprl3. First, the Iml1 protein may retain some GAP activity toward Rags or another critical activity in the nprl2 and nprl3 mutant background. In this model, Iml1 functions, at least in part, independently of Nprl2 and Nprl3. Consistent with the first model, the overexpression of Iml1 in yeast bypasses the requirement for the Npr2 and Npr3 proteins, suggesting that the enzymatic component of the GATOR1 complex resides within the Iml1 protein (Panchaud et al. 2013a). A second possible model is that Nprl2 and Nprl3, which share a similar domain structure (Dokudovskaya et al. 2011), are functionally redundant. Both Nprl2 and Nprl3 contain a C-terminal Longin domain, which is often found in proteins that regulate membrane trafficking (Levine et al. 2013), followed by two or three N-terminal helix-turn-helix domains (Zhang et al. 2012; Levine et al. 2013). To test if nprl2 and nprl3 are redundant, we generated nprl2 and nprl3 double mutants. We reasoned that if nprl2 and nprl3 are redundant, then the double mutants should have a stronger phenotype, as measured by lethality rates, than either single mutant. However, we found that nprl2, nprl3 double-mutants eclosed at approximately the same rate as nprl2 and nprl3 single mutants (Table 1). Thus, although structurally similar, the Nprl2 and Nprl3 proteins are not functionally redundant. Moreover, the stronger phenotype observed in the iml11 mutants is consistent with the Iml1 protein of Drosophila retaining critical activities independent of the GATOR1 components Nprl2 and Nprl3.
In order to determine the specific developmental stage at which the GATOR1 mutants die, newly hatched first instar larvae were cultured and counted at both pupal and adult stages. As shown in Figure S1, nprl2, nprl3, and iml1 mutant larvae advance to the pupae stage at rates similar to those observed for wild-type larvae. However, only 37% of nprl21, 36% of nprl31/Df, and 2% of iml11/Df mutant larvae ultimately eclosed. In contrast, 82% wild-type larvae eclosed. Thus, the activity of the GATOR1 complex is required to transit from the pupal to the adult stage of development. We noticed that the dead nprl21, nprl31/Df, and iml11/Df pupae had finished metamorphosis and died beyond the pharate adult stage P13, as determined by the presence of black wings (Bainbridge and Bownes 1981). Thus, the GATOR1 complex is required for animals to transit the last stage of pupal development and eclose as adults.
GATOR1 maintains baseline levels of TORC1 activity in vivo
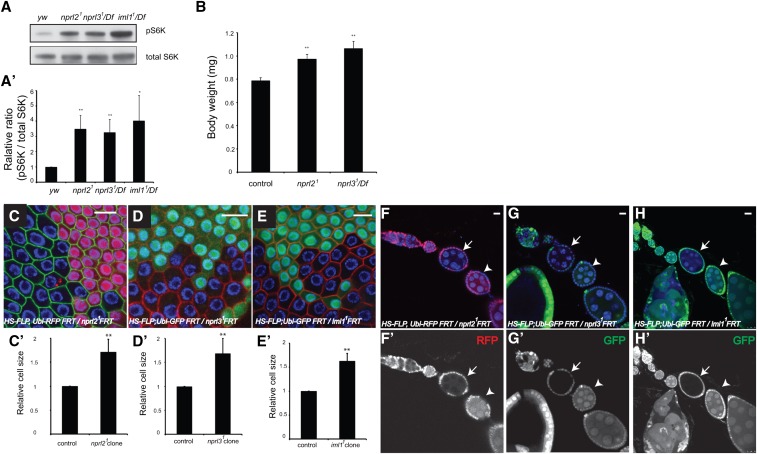
In yeast, GATOR1 downregulates TORC1 activity in response to amino acid restriction but is not required to maintain baseline levels of TORC1 activity under nutrient rich conditions (Neklesa and Davis 2009). In contrast, in mice and rats, components of the GATOR1 complex are required for viability, strongly suggesting that, in metazoans, the GATOR1 complex has evolved roles beyond the response to nutrient stress (Table 1) (Kowalczyk et al. 2012; Dutchak et al. 2015; Marsan et al. 2016). In order to determine if the GATOR1 complex is required to maintain baseline levels of TORC1 activity in vivo in Drosophila, we measured phosphorylation of S6 Kinase (S6K), a direct readout for TORC1 activity, in well-fed whole animal lysates by western blot using an anti-phosopho-Thr398 S6K antibody (Hara et al. 1998). From these experiments, we found that the levels of phospho-T398-S6K were significantly increased in lysates from nprl2, nprl3, and iml1 mutants (Figure 2A). Thus, unlike what is observed in single-celled eukaryotes, in Drosophila the GATOR1 complex is required to maintain baseline levels of TORC1 activity in vivo under nutrient-replete conditions. Similarly, rat and mouse cells cultured from nprl2, nprl3, or iml1/Depdc5 mutants have increased TORC1 activity relative to controls (Kowalczyk et al. 2012; Dutchak et al. 2015; Marsan et al. 2016). Thus, in both Drosophila and mammals, the GATOR1 complex is required to maintain baseline levels of TORC1 activity independent of nutrient status.
Figure 2.
GATOR1 is required to maintain baseline levels of TORC1 activity in Drosophila. (A) Third star larvae were lysed in RIPA buffer. The protein levels of phospho-T398-S6K and total S6K were determined by western blot. (A’) The ratio of pS6K/S6K in the y, w lysis was set as 1. Error bars indicate SD of three independent experiments. * P < 0.05, ** P < 0.01. (B) The body weight of male flies. Error bars indicate SD of five independent experiments. ** P < 0.01. (C–E) Ovaries were dissected and stained with DAPI and anti-1B1 antibody. The nprl21, nprl31, and iml1 1 homozygous follicles cell clones were identified by the absence of RFP or GFP. The 1B1 antibody stains cell membranes and was used to mark the outline of the cells (C’–E’). The area of homozygous cells and adjacent wild-type cells were quantified using Image J. The mean value of the wild-type cell size was set as 1 for each image. For each genotype, eight images were quantified. Error bars indicate SD. ** P < 0.01. (F–H) Ovaries were dissected and stained with DAPI. The nprl21, nprl31, and iml1 1 homozygous egg chamber clones were identified by the absence of RFP and GFP. Homozygous mutant cells are indicated by an arrow, while older wild-type cells are indicated by an arrowhead. Note that younger homozygous mutant cells are larger than older wild-type cells. (F’–H’) In order to clearly show the clonal boundary, RFP and GFP channels are shown separately. DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; RFP, red fluorescent protein; RIPA, radioimmunoprecipitation assay.
We reasoned that the lethality observed in GATOR1 mutants is due to the hyperactivation of TORC1. To test our hypothesis we used the mutation TorA948V, which interferes with wild-type Tor function (Zhang et al. 2006). Notably, placing one copy of the TorA948V allele in the nprl2 or nprl3 mutant backgrounds resulted in eclosion rates that were three-fold higher than nprl2 and nprl3 mutants that contained two functional copies of the Tor gene (Table 2). These data confirm that the lethality associated with nprl2 and nprl3 mutations is due to the hyperactivation of TORC1.
GATOR1 inhibits cell growth in multiple cell types
TORC1 is a potent activator of cell growth (Laplante and Sabatini 2012; Tee 2014). Consistent with the overall increase in TORC1 activity observed in whole animals, nprl2 and nprl3 escaper adults exhibit a small increase in body weight relative to wild-type controls (Figure 2B). In order to determine if the increased growth observed in GATOR1 mutants is at least in part due to a cell autonomous requirement for the GATOR1 complex, we used the FLP/FRT system to generate homozygous nprl2, nprl3, or iml1 mutant clones in multiple tissues (Theodosiou and Xu 1998). In the Drosophila ovary, developing egg chambers contain a 16-cell germline cyst that is comprised of 15 large polyploid nurse cells and a single oocyte (de Cuevas et al. 1997). During oogenesis, the polyploid nurse cells grow via endoreplication and are responsible for providing the oocyte with nutrients. The growing germline cyst is surrounded by a layer of somatically-derived follicle cells that also increase in size through the process of endoreplication (Lilly and Duronio 2005). Notably, TORC1 activity is an important factor driving endoreplication and growth in polyploid tissues (Saucedo et al. 2003). We found that homozygous mutant nprl21, nprl31, and iml11 follicle cells were larger than adjacent heterozygous cells (Figure 2, C–E). Similarly, egg chambers that contained homozygous mutant germline clones (arrow) of nprl2, nprl3, or iml1 were significantly increased in size relative to older egg chambers (arrowhead) that contained a wild-type heterozygous germline (Figure 2, F–H). Taken together, these data are consistent with the GATOR1 complex having cell autonomous effects on TORC1 activity and cell growth in vivo.
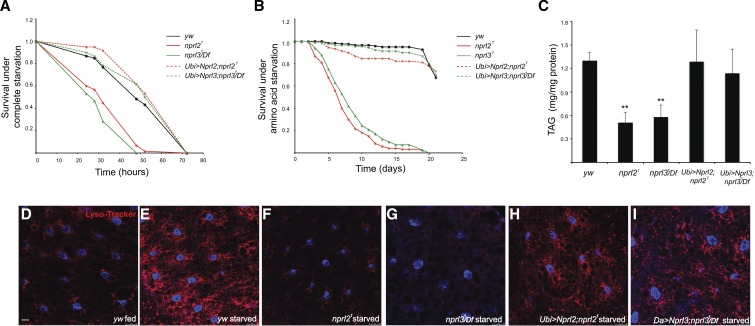
Nprl2 and Nprl3 promote starvation resistance, starvation-induced autophagy, and triglyceride storage
The GATOR1 complex downregulates TORC1 activity in response to nutrient stress triggering the activation of catabolic metabolism and autophagy (Chang et al. 2009; Neklesa and Davis 2009; Bar-Peled et al. 2013; Panchaud et al. 2013a). Previous work has shown that the GATOR1 complex promotes a cell autonomous response to nutrient stress in the female germline of Drosophila (Wei and Lilly 2014). Specifically, in the absence of GATOR1, egg chambers undergo apoptosis when females are cultured under conditions of amino acid restriction. In order to determine if nprl2 and nprl3 are required to mediate a robust response to nutrient stress in Drosophila at the organismal level, we examined the survival rates of nprl2 and nprl3 mutants under conditions of complete starvation and amino acid starvation (20% sucrose). As shown in Figure 3, A and B, the nprl2 and nprl3 mutants have decreased survival rates under both nutrient-limiting culture conditions. Thus, Nprl2 and Nprl3 promote starvation tolerance at the organismal level. In Drosophila, triglycerides (TAG) stored in the fat body are the primary energy resource and are essential to starvation resistance (Arrese et al. 2010). Consistent with increased sensitivity to starvation, nprl2 and nprl3 mutant adults contain reduced amounts of stored TAG relative to wild-type animals (Figure 3C). Importantly, the decreased survival in response to nutrient stress and the TAG storage deficit of nprl2 and nprl3 mutants were rescued by the ubiquitous expression of the corresponding transgene (Figure 3, A–C). These results are in line with previous reports that treatment with the TORC1 inhibitor Rapamycin results in increased stress resistance and elevated TAG levels (Bjedov et al. 2010).
Figure 3.
nprl2 and nprl3 mutants are sensitive to starvation. (A) Newly hatched male flies were cultured at 25° on standard food for 3 d, and then transferred to starvation media (0.8% agar in PBS) and counted at the indicated time points. The survival curves of wild-type, nprl21, nprl31/Df, Ubi-GAL4/UAS-Nprl2; nprl21, and Ubi-GAL4/UAS-Nprl3; nprl31/Df are shown. (B) Newly hatched male flies were cultured at 25° on standard food for 3 d, and then transferred to amino acid starvation media (20% sucrose, 0.8% agar in PBS) and counted each day. The survival curves of wild-type, nprl21, nprl31/Df, Ubi-GAL4/UAS-Nprl2; nprl21, and Ubi-GAL4/UAS-Nprl3; nprl31/Df are shown. (C) Newly hatched male flies were cultured at 25° on standard food for 3 d and then lysed. Total body TAG and protein were measured. Protein levels were used for normalization. The TAG to protein ratio is shown. Error bars indicate SD of three independent experiments. ** P < 0.01. (D–I) Third instar wild-type, nprl21, nprl31/Df, Ubi-GAL4/UAS-Nprl2; nprl21, and Da>-GAL4/UAS-Nprl3; nprl31/Df larvae were fed or starved for 4 hr. Subsequently, fat bodies were removed and stained with Hoechst and LysoTracker Red. PBS, phosphate-buffered saline; TAG, triglycerides.
The autophagy pathway promotes the digestion of nonessential cellular components to provide basic nutrients for cell survival during times of nutrient scarcity (Rabinowitz and White 2010). An important trigger for the activation of autophagy is the down-regulation of TORC1 activity (Kamada et al. 2010). We wanted to test the idea that the high TORC1 activity observed in the nprl2 and nprl3 mutants prevents the activation of autophagy, thus contributing to the starvation sensitivity of the mutants. LysoTracker, which highlights acidic compartments and thus stains lysosomes and autolysosomes, has been commonly used for examining autophagy in the Drosophila fat body, an organ that stores energy and is sensitive to nutrient status (Klionsky et al. 2016). In order to test this model, we stained mutant and wild-type fat bodies from third instar larvae with LysoTracker. In wild-type fat body cells, amino acid starvation results in the accumulation of lysosomes/autolysosomes as indicated by the accumulation of LysoTracker-positive puncta (Scott et al. 2004; Mauvezin et al. 2014) (Figure 3, D and E). In contrast, fat bodies from nprl2 and nprl3 mutant larvae fail to accumulate large numbers of LysoTracker-positive puncta in response to amino acid starvation (Figure 3, F and G). The failure to activate autophagy in the nprl2 and nprl3 mutants is rescued by the ubiquitous expression of the corresponding transgene. Thus, our data are consistent with the model that nprl2 and nprl3 mutants fail to activate catabolic metabolism and autophagy in response to amino acid starvation because of the failure to downregulate TORC1 activity. Notably, a similar phenotype is observed in mutants of the potent TORC1 inhibitor tuberous sclerosis complex (TSC) (Scott et al. 2004). In Drosophila, autophagy is essential for the response to nutrient deprivation (Scott et al. 2004; Lenardo et al. 2009; Neufeld 2010). Thus, we predict that the increased susceptibility to starvation observed in nprl2 and nprl3 mutants may be due to both decreased TAG storage and the inability to downregulate TORC1 activity and activate the autophagy pathway during periods of nutrient scarcity.
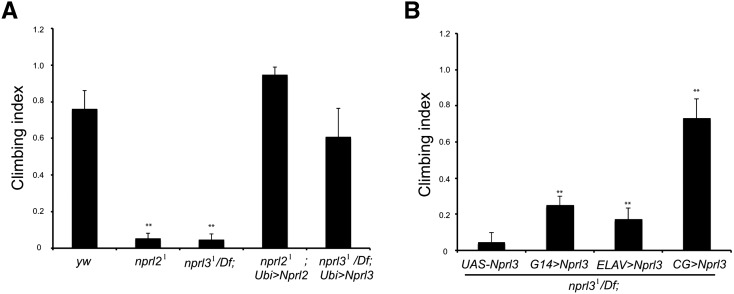
Nprl2 and Nprl3 function to promote Drosophila motility
During the course of our studies, we noted that nprl2 and nprl3 mutants had reduced motility. We quantified this motility defect using a standard geotaxis-climbing assay (Ganetzky and Flanagan 1978; Feany and Bender 2000). Consistent with our initial observations, nprl2 and nprl3 mutant adults had reduced climbing indices relative to wild-type controls (Figure 4A). The climbing deficits were rescued by the expression of the nprl2 or nprl3 transgene from a ubiquitous Ubi-GAL4 driver in the respective mutant backgrounds (Figure 4A). There are at least two possible models for how the GATOR1 complex might affect Drosophila motility. First, GATOR1 may be required cell autonomously for the development or the maintenance of metabolic homeostasis in neurons and muscles. Notably, several Drosophila neurodegenerative disease models have locomotive defects that can be rescued through genetic inhibition of TORC1 in neurons and muscle (Tain et al. 2009; Hirth 2010; Liu and Lu 2010). Alternatively, GATOR1 may act nonautonomously to control the systemic metabolism and energy availability that are required for motility. The Drosophila fat body plays a critical role in maintaining metabolic homeostasis (Colombani et al. 2003; Arrese and Soulages 2010). In the presence of amino acids, TORC1 activity in the fat body promotes global growth and anabolic metabolism through a noncell autonomous insulin-dependent pathway (Colombani et al. 2003). To distinguish between these two possibilities, we compared the climbing index of nprl3 mutant flies that expressed Nprl3 from a transgene in neurons and muscle using the neuronal and muscle drivers ELAV-GAL4 and G14-GAL4 (Aberle et al. 2002) to mutants that expressed Nprl3 in the fat body and hemocytes using the driver CG-GAL4 (Asha et al. 2003). Somewhat surprisingly, we found that while the expression of Nprl3 in neurons and muscle resulted in a small increase in the climbing index, the expression of Nprl3 in the fat body and hemocytes resulted in a near full rescue of the nprl31/Df mobility deficits (Figure 4B). These data strongly suggest that the activity of the GATOR1 complex in the fat body is required to promote adult motility. Furthermore, we found that overexpression of Nprl3 in the fat body and hemocytes can rescue the lethality of nprl3 mutants (Table 2). Taken together, our data indicate that the GATOR1 complex has both autonomous and nonautonomous functions in the regulation of growth and metabolism in Drosophila.
Figure 4.
Nprl2 and Nprl3 are required in the fat body for Drosophila locomotion. (A) The climbing indices of the indicated genotypes are shown. Overexpression of Nprl2 and Nprl3 using the ubiquitous Ubi-GAL4 driver suppressed the climbing defects of nprl21 and nprl31/Df mutant flies. Error bars indicate SD of at least three independent experiments. ** P < 0.01. (B) The climbing indices of nprl31/Df mutant flies that overexpress the Nprl3 protein using a neuronal driver ELAV-GAL4, a muscle driver G14-GAL4, or a fat body, hemocyte driver CG-GAL4 are shown. Error bars indicate SD of at least six independent experiments. ** P < 0.01.
Conclusions
In summary, we have defined the developmental and metabolic role of the newly identified TORC1 inhibitor GATOR1 in the genetically tractable animal model D. melanogaster. We have shown that the GATOR1 complex regulates multiple aspects of cell growth and metabolism, influencing processes as diverse as fat storage, growth, autophagy, and motility. Additionally, we found that the GATOR1 complex has both cell autonomous and nonautonomous functions. Taken together, our findings define Drosophila as an excellent model for the genetic dissection of the GATOR1 complex and its role in preventing the developmental defects and pathologies associated with deregulated TORC1 activity.
Supplementary Material
Acknowledgments
We thank members of the Lilly laboratory for comments on the manuscript. Multiple stocks used in this study were obtained from the Bloomington Stock Center. We thank Thomas Neufeld (University of Minnesota) for providing the TorA948V/CyO stock. This work was supported by grant ZIA HD001613 16 from the Intramural Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.035337/-/DC1.
Communicating editor: A. Bashirullah
Literature Cited
- Aberle H., Haghighi A. P., Fetter R. D., McCabe B. D., Magalhaes T. R., et al. , 2002. Wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron 33: 545–558. [DOI] [PubMed] [Google Scholar]
- Arrese E. L., Soulages J. L., 2010. Insect fat body: energy, metabolism, and regulation. Annu. Rev. Entomol. 55: 207–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrese E. L., Howard A. D., Patel R. T., Rimoldi O. J., Soulages J. L., 2010. Mobilization of lipid stores in Manduca sexta: cDNA cloning and developmental expression of fat body triglyceride lipase, TGL. Insect Biochem. Mol. Biol. 40: 91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asha H., Nagy I., Kovacs G., Stetson D., Ando I., et al. , 2003. Analysis of Ras-induced overproliferation in Drosophila hemocytes. Genetics 163: 203–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge S. P., Bownes M., 1981. Staging the metamorphosis of Drosophila melanogaster. J. Embryol. Exp. Morphol. 66: 57–80. [PubMed] [Google Scholar]
- Bar-Peled L., Chantranupong L., Cherniack A. D., Chen W. W., Ottina K. A., et al. , 2013. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340: 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I., Toivonen J. M., Kerr F., Slack C., Jacobson J., et al. , 2010. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab. 11: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W., Wei Y., Jarnik M., Reich J., Lilly M. A., 2016. The GATOR2 component Wdr24 regulates TORC1 activity and lysosome function. PLoS Genet. 12: e1006036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y. Y., Juhasz G., Goraksha-Hicks P., Arsham A. M., Mallin D. R., et al. , 2009. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem. Soc. Trans. 37: 232–236. [DOI] [PubMed] [Google Scholar]
- Colombani J., Raisin S., Pantalacci S., Radimerski T., Montagne J., et al. , 2003. A nutrient sensor mechanism controls Drosophila growth. Cell 114: 739–749. [DOI] [PubMed] [Google Scholar]
- de Cuevas M., Lilly M. A., Spradling A. C., 1997. Germline cyst formation in Drosophila. Annu. Rev. Genet. 31: 405–428. [DOI] [PubMed] [Google Scholar]
- Dibbens L. M., de Vries B., Donatello S., Heron S. E., Hodgson B. L., et al. , 2013. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat. Genet. 45: 546–551. [DOI] [PubMed] [Google Scholar]
- Dokudovskaya S., Waharte F., Schlessinger A., Pieper U., Devos D. P., et al. , 2011. A conserved coatomer-related complex containing Sec13 and Seh1 dynamically associates with the vacuole in Saccharomyces cerevisiae. Mol. Cell. Proteomics 10: M110.006478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutchak P. A., Laxman S., Estill S. J., Wang C., Wang Y., et al. , 2015. Regulation of hematopoiesis and methionine homeostasis by mTORC1 inhibitor NPRL2. Cell Reports 12: 371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D. S., Kapahi P., Hsueh W. C., Kockel L., 2011. TOR signaling never gets old: aging, longevity and TORC1 activity. Ageing Res. Rev. 10: 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feany M. B., Bender W. W., 2000. A Drosophila model of Parkinson’s disease. Nature 404: 394–398. [DOI] [PubMed] [Google Scholar]
- Fontana L., Partridge L., 2015. Promoting health and longevity through diet: from model organisms to humans. Cell 161: 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganetzky B., Flanagan J. R., 1978. On the relationship between senescence and age-related changes in two wild-type strains of Drosophila melanogaster. Exp. Gerontol. 13: 189–196. [DOI] [PubMed] [Google Scholar]
- Hahn K., Miranda M., Francis V. A., Vendrell J., Zorzano A., et al. , 2010. PP2A regulatory subunit PP2A-B’ counteracts S6K phosphorylation. Cell Metab. 11: 438–444. [DOI] [PubMed] [Google Scholar]
- Hara K., Yonezawa K., Weng Q. P., Kozlowski M. T., Belham C., et al. , 1998. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 273: 14484–14494. [DOI] [PubMed] [Google Scholar]
- Hay N., Sonenberg N., 2004. Upstream and downstream of mTOR. Genes Dev. 18: 1926–1945. [DOI] [PubMed] [Google Scholar]
- He C., Klionsky D. J., 2009. Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43: 67–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth F., 2010. Drosophila melanogaster in the study of human neurodegeneration. CNS Neurol. Disord. Drug Targets 9: 504–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong A., Lee-Kong S., Iida T., Sugimura I., Lilly M. A., 2003. The p27cip/kip ortholog dacapo maintains the Drosophila oocyte in prophase of meiosis I. Development 130: 1235–1242. [DOI] [PubMed] [Google Scholar]
- Iida T., Lilly M. A., 2004. Missing oocyte encodes a highly conserved nuclear protein required for the maintenance of the meiotic cycle and oocyte identity in Drosophila. Development 131: 1029–1039. [DOI] [PubMed] [Google Scholar]
- Johnson S. C., Rabinovitch P. S., Kaeberlein M., 2013. mTOR is a key modulator of ageing and age-related disease. Nature 493: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C. H., Ro S. H., Cao J., Otto N. M., Kim D. H., 2010. mTOR regulation of autophagy. FEBS Lett. 584: 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada Y., Yoshino K., Kondo C., Kawamata T., Oshiro N., et al. , 2010. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol. Cell. Biol. 30: 1049–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E., Goraksha-Hicks P., Li L., Neufeld T. P., Guan K. L., 2008. Regulation of TORC1 by Rag GTPases in nutrient response. Nat. Cell Biol. 10: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D. J., Abdelmohsen K., Abe A., Abedin M. J., Abeliovich H., et al. , 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12: 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S., Ueda R., 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195: 715–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk M. S., Hughes J. R., Babbs C., Sanchez-Pulido L., Szumska D., et al. , 2012. Nprl3 is required for normal development of the cardiovascular system. Mamm. Genome 23: 404–415. [DOI] [PubMed] [Google Scholar]
- Laplante M., Sabatini D. M., 2012. mTOR signaling in growth control and disease. Cell 149: 274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenardo M. J., McPhee C. K., Yu L., 2009. Autophagic cell death. Methods Enzymol. 453: 17–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T. P., Daniels R. D., Wong L. H., Gatta A. T., Gerondopoulos A., et al. , 2013. Discovery of new longin and roadblock domains that form platforms for small GTPases in Ragulator and TRAPP-II. Small GTPases 4: 62–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M. A., Duronio R. J., 2005. New insights into cell cycle control from the Drosophila endocycle. Oncogene 24: 2765–2775. [DOI] [PubMed] [Google Scholar]
- Liu S., Lu B., 2010. Reduction of protein translation and activation of autophagy protect against PINK1 pathogenesis in Drosophila melanogaster. PLoS Genet. 6: e1001237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewith R., Hall M. N., 2011. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 189: 1177–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma N., Liu Q., Zhang L., Henske E. P., Ma Y., 2013. TORC1 signaling is governed by two negative regulators in fission yeast. Genetics 195: 457–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsan E., Ishida S., Schramm A., Weckhuysen S., Muraca G., et al. , 2016. Depdc5 knockout rat: a novel model of mTORopathy. Neurobiol. Dis. 89: 180–189. [DOI] [PubMed] [Google Scholar]
- Mauvezin C., Ayala C., Braden C. R., Kim J., Neufeld T. P., 2014. Assays to monitor autophagy in Drosophila. Methods 68: 134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neklesa T. K., Davis R. W., 2009. A genome-wide screen for regulators of TORC1 in response to amino acid starvation reveals a conserved Npr2/3 complex. PLoS Genet. 5: e1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T. P., 2010. TOR-dependent control of autophagy: biting the hand that feeds. Curr. Opin. Cell Biol. 22: 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchaud N., Peli-Gulli M. P., De Virgilio C., 2013a Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Sci. Signal. 6: ra42. [DOI] [PubMed] [Google Scholar]
- Panchaud N., Peli-Gulli M. P., De Virgilio C., 2013b SEACing the GAP that nEGOCiates TORC1 activation: evolutionary conservation of Rag GTPase regulation. Cell Cycle 12: 2948–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J. D., White E., 2010. Autophagy and metabolism. Science 330: 1344–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y., Peterson T. R., Shaul Y. D., Lindquist R. A., Thoreen C. C., et al. , 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320: 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucedo L. J., Gao X., Chiarelli D. A., Li L., Pan D., et al. , 2003. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat. Cell Biol. 5: 566–571. [DOI] [PubMed] [Google Scholar]
- Schmelzle T., Hall M. N., 2000. TOR, a central controller of cell growth. Cell 103: 253–262. [DOI] [PubMed] [Google Scholar]
- Scott R. C., Schuldiner O., Neufeld T. P., 2004. Role and regulation of starvation-induced autophagy in the Drosophila fat body. Dev. Cell 7: 167–178. [DOI] [PubMed] [Google Scholar]
- Senger S., Csokmay J., Iida-Jones T., Sengupta P., Lilly M. A., 2011. The nucleoporin Seh1 forms a complex with Mio and serves an essential tissue specific function in Drosophila oogenesis. Development 138: 2133–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tain L. S., Mortiboys H., Tao R. N., Ziviani E., Bandmann O., et al. , 2009. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat. Neurosci. 12: 1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tee A. R., 2014. Fundamental for life: mTOR orchestrates developing biological systems. Semin. Cell Dev. Biol. 36: 66–67. [DOI] [PubMed] [Google Scholar]
- Tee A. R., Sampson J. R., Pal D. K., Bateman J. M., 2016. The role of mTOR signalling in neurogenesis, insights from tuberous sclerosis complex. Semin. Cell Dev. Biol. 52: 12–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodosiou N. A., Xu T., 1998. Use of FLP/FRT system to study Drosophila development. Methods 14: 355–365. [DOI] [PubMed] [Google Scholar]
- Wei Y., Lilly M. A., 2014. The TORC1 inhibitors Nprl2 and Nprl3 mediate an adaptive response to amino-acid starvation in Drosophila. Cell Death Differ. 21: 1460–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y., Reveal B., Reich J., Laursen W. J., Senger S., et al. , 2014. TORC1 regulators Iml1/GATOR1 and GATOR2 control meiotic entry and oocyte development in Drosophila. Proc. Natl. Acad. Sci. USA 111: E5670–E5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Tu B. P., 2011. Selective regulation of autophagy by the Iml1-Npr2-Npr3 complex in the absence of nitrogen starvation. Mol. Biol. Cell 22: 4124–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N., 2006. TOR signaling in growth and metabolism. Cell 124: 471–484. [DOI] [PubMed] [Google Scholar]
- Yang Q., Inoki K., Kim E., Guan K. L., 2006. TSC1/TSC2 and Rheb have different effects on TORC1 and TORC2 activity. Proc. Natl. Acad. Sci. USA 103: 6811–6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Iyer L. M., He F., Aravind L., 2012. Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front. Genet. 3: 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Billington C. J., Jr, Pan D., Neufeld T. P., 2006. Drosophila target of rapamycin kinase functions as a multimer. Genetics 172: 355–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All strains are available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.