Abstract
The unicellular green alga Chlamydomonas reinhardtii is a model organism that provides an opportunity to understand the evolution and functional biology of the lineage that includes the land plants, as well as aspects of the fundamental core biology conserved throughout the eukaryotic phylogeny. Although many tools are available to facilitate genetic, molecular biological, biochemical, and cell biological studies in Chlamydomonas, expression of unselected transgenes of interest (GOIs) has been challenging. In most methods used previously, the GOI and a selectable marker are expressed from two separate mRNAs, so that their concomitant expression is not guaranteed. In this study, we developed constructs that allow expression of an upstream GOI and downstream selectable marker from a single bicistronic mRNA. Although this approach in other systems has typically required a translation-enhancing element such as an internal ribosome entry site for the downstream marker, we found that a short stretch of unstructured junction sequence was sufficient to obtain adequate expression of the downstream gene, presumably through post-termination reinitiation. With this system, we obtained robust expression of both endogenous and heterologous GOIs, including fluorescent proteins and tagged fusion proteins, in the vast majority of transformants, thus eliminating the need for tedious secondary screening for GOI-expressing transformants. This improved efficiency should greatly facilitate a variety of genetic and cell-biological studies in Chlamydomonas and also enable new applications such as expression-based screens and large-scale production of foreign proteins.
Keywords: algae, bicistronic mRNA, IRES, transgene expression, translation reinitiation
The unicellular green alga Chlamydomonas reinhardtii is a well established model organism that has been widely used for studies of photosynthesis, the structure and function of flagella and basal bodies, the cell cycle, and other processes (Harris 2001; Marshall 2008; Ostrowski et al. 2011; Heinnickel and Grossman 2013; Cross and Umen 2015). Its advantages include its haploid vegetative cells, a sequenced genome (Merchant et al. 2007), rapid growth under both auxotrophic and heterotrophic conditions, and well developed methods and resources for genetics, biochemistry, and microscopy. Genetic transformation into the Chlamydomonas nucleus has been used in many studies, and methods and reagents including promoters, terminators, enhancers, reporter genes, and auxotrophic and drug-resistance markers are available (for review, see Jinkerson and Jonikas 2015).
Nonetheless, expression of unselected transgenes, especially of heterologous origin, has remained a challenge in Chlamydomonas. In the most commonly used “two-promoter” approach, an unselected gene of interest (GOI) and a selectable marker are transcribed under the control of two separate promoter-terminator pairs (Figure 1A); the two expression modules are introduced either as separate DNA fragments or as a single cassette (Heitzer and Zschoernig 2007) and integrate into the genome at a random site(s) by nonhomologous end-joining. However, in many cases, ≤10% of the selected transformants coexpress the unselected GOI, presumably due to a low cotransformation rate of pairs of fragments, cleavage of two-gene cassettes with integration only of the fragment with the selectable marker (Zhang et al. 2014), and/or strong transcriptional and/or post-transcriptional silencing (Cerutti et al. 1997). Thus, a secondary screen—often rather tedious—for transformants expressing the GOI has typically been necessary. For example, we needed to screen >100 transformants to acquire a single clone that expressed the F-actin probe Lifeact-Venus at a level sufficient for visualization by fluorescence microscopy (Avasthi et al. 2014). These problems have handicapped cell biological studies and impeded development of expression-based screens, medium- to high-throughput imaging analyses, and the use of Chlamydomonas as a host for expression of foreign proteins.
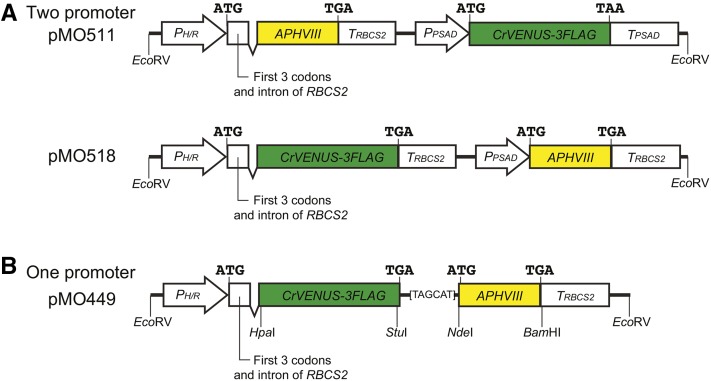
Figure 1.
Representative two-promoter and one-promoter expression constructs as used in this study. All constructs were embedded in the identical plasmid backbone (outside of the EcoRV sites shown), as described in Materials and Methods. (A) Two-promoter constructs. Translation start and stop sites for each gene are shown. PH/R, strong hybrid HSP70A/RBCS2 promoter; APHVIII, paromomycin-resistance gene; TRBCS2, RBCS2 transcription terminator; PPSAD, strong PSAD promoter; CrVENUS-3FLAG, coding sequence of Venus codon-optimized for Chlamydomonas and tagged with three copies of the FLAG epitope; TPSAD, PSAD transcription terminator; EcoRV, restriction sites that can be used to excise the construct from the vector before transformation. Other restriction sites are present [cf. (B)] but not shown. (B) General structure of the one-promoter constructs used in this study as illustrated by that of plasmid pMO449. The 6-bp linker between the two ORFs is shown by the sequence of the transcribed strand; other symbols as in (A). The restriction sites shown can be used to replace the upstream GOI, the downstream selectable marker, or the linker sequence (see text).
In an alternative “one-promoter” approach, the constructs contain a single promoter and terminator that produce a bicistronic mRNA typically containing an upstream GOI and a downstream selectable marker with separate translation start and stop sites (Figure 1B). Expression of the selectable marker then requires transcription of the entire mRNA and thus expression of the GOI. This approach has been used previously for expression of introduced genes in some animals, cultured animal cells, fungi, and plants (Martínez-Salas 1999; Hunt and Maiti 2001). Translation of the GOI is usually mediated by canonical cap-dependent initiation (Hinnebusch and Lorsch 2012), whereas that of the selectable marker is controlled by cap-independent initiation mechanisms. Most vectors have employed an internal ribosome entry site (IRES) that is inserted in the untranslated region between the ORFs to enhance translation of the selectable marker. IRES elements are sequences of different lengths and secondary and tertiary structures that provide sites for the ribosome and translation-initiation factors to assemble independent of a 5′-cap structure. Originally identified in picornaviral genomes (Jang et al. 1988; Pelletier and Sonenberg 1988), IRES-mediated translation has now been found in various other viruses, as well as in some nonviral cellular mRNAs (Hellen and Sarnow 2001; Jackson 2013). Although, to our knowledge, there is no virus known to infect Chlamydomonas, and no IRES elements have been identified in its cellular mRNAs, some viral-derived IRES elements have been shown to function in organisms evolutionarily distant from the normal host species (Iizuka et al. 1994; Urwin et al. 2000; Dorokhov et al. 2002).
In a few cases, the IRES has been omitted, and the expression vectors have simply had the two ORFs of a bicistronic mRNA connected by a short stretch of unstructured sequence; such constructs have been used successfully in both plants and mammalian cells (Levine et al. 1991; Lough et al. 1997; Hunt and Maiti 2001). Although the precise mechanism of translation initiation for the downstream ORF in these constructs is not certain, it is thought to be through post-termination reinitiation (Skabkin et al. 2013) rather than leaky scanning or ribosome shunting (Jackson et al. 2012). Because of the inefficiency of post-termination reinitiation (Hunt and Maiti 2001; Jackson et al. 2012), expression of the downstream ORF by this mechanism would normally be expected to be less efficient than that obtained with an IRES.
In this paper, we describe expression vectors for Chlamydomonas that use a single promoter and transcription terminator to express a bicistronic mRNA encoding an upstream GOI and a downstream selectable marker without an intervening IRES; they achieve very efficient expression of the GOI in the majority of transformants.
Materials and Methods
Strains, media, and growth conditions
Chlamydomonas reinhardtii wild-type strain CC-124 (also known as 137c; Chlamydomonas Resource Center) was used in most experiments. Strain CMJ030 (also known as CC-4533; Zhang et al. 2014) was also used where indicated. Cells were grown in Tris-acetate-phosphate (TAP) medium (Gorman and Levine 1965) at 21–24° under constant illumination at 75 μmol photons m−2 sec−1. In most experiments, selection of transformants used paromomycin (Sigma) at a concentration from 10 to 40 µg/ml, as indicated. Some experiments used zeocin (InvivoGen) at 5 µg/ml, spectinomycin (Sigma) at 30 µg/ml, or hygromycin B (Sigma) at 5 µg/ml.
Plasmid constructions
The plasmids used in this study are listed in Table 1 and Table 2. In construct descriptions, PXXX or TXXX indicates a promoter or terminator sequence from gene XXX; a hyphen indicates a fusion of two coding sequences or polypeptides (e.g., VENUS-3FLAG); and a colon indicates a conjoinment of two DNA sequences that does not result in such a fusion (e.g., PH/R:APHVIII). Plasmid constructions were performed using the one-step isothermal-assembly method (Gibson et al. 2009). Primer sequences are provided in Supplemental Material, File S1.
Table 1. Plasmids used in this study.
| Plasmida | Promoter for GOI | GOI | Junction | Selectable Marker |
|---|---|---|---|---|
| pMO511b | PPSAD | CrVENUS-3FLAG | PH/R | APHVIII |
| pMO424b | PPSAD | LifeAct-CrVENUS-3FLAG | PH/R | APHVIII |
| pMO448 | PH/R | N/A | N/A | APHVIII |
| pMO449 | PH/R | CrVENUS-3FLAG | TAGCAT | APHVIII |
| pMO459 | PH/R | LifeAct-CrVENUS-3FLAG | TAGCAT | APHVIII |
| pMO470 | PH/R | CrVENUS-3FLAG | TGATAGCCAT | APHVIII |
| pMO471 | PH/R | CrVENUS-3FLAG | TAGCCAT | APHVIII |
| pMO480 | PH/R | CrVENUS-3FLAG-CVIMc | TAGCAT | APHVIII |
| pMO482 | PH/R | PRO1-CrVENUS-FLAG | TAGCAT | APHVIII |
| pMO483 | PH/R | IDA5-CrVENUS-3FLAG | TAGCAT | APHVIII |
| pMO484 | PH/R | VFL2-CrVENUS-3FLAG | TAGCAT | APHVIII |
| pMO488 | PH/R | CrVENUS | TAGCAT | APHVIII |
| pMO490 | PH/R | sfGFP-3FLAG | TAGCAT | APHVIII |
| pMO507 | PPSAD | CrVENUS-3FLAG | TAGCAT | APHVIII |
| pMO508 | PTUB2 | CrVENUS-3FLAG | TAGCAT | APHVIII |
| pMO515 | PH/R | PMH1-CrVENUS-3FLAG | TAGCAT | APHVIII |
| pMO518 | PH/R | CrVENUS-3FLAG | TRBCS2-PPSAD | APHVIII |
| pMO519 | PH/R | CrmCHERRY | TAGCAT | APHVIII |
| pMO520 | PH/R | CrmCHERRY-3FLAG | TAGCAT | APHVIII |
Except for pMO511 (note b and Figure 1A), pMO424 (note b), and pMO518 (Figure 1A), all plasmids listed have the general structure shown for pMO449 in Figure 1B.
In pMO511 (Figure 1A) and pMO424 (Avasthi et al. 2014), the selectable marker is upstream under the control of the PH/R promoter, and the GOI is downstream under the control of the PPSAD promoter. pMO424 is identical to pMO511 except that the Chlamydomonas-codon-optimized Lifeact-encoding sequence is fused at the N-terminus of CrVENUS-3FLAG.
CVIM is expected to function as a “CAAX-box” for prenylation.
Table 2. Assessment of various sequences used as the junction between the GOI and the selectable marker.
| Rowa | Plasmidb | Type of Junctionc | Description or Sequence of Junction | No. of Transformants/µg DNAd | No. of Venus-Positive Clonese |
|---|---|---|---|---|---|
| 1 | pMO448 | N/A | Control (no 5′ ORF as GOI) | ∼2500 | N/A |
| 2 | pMO518 | Promoter-containing | Control (conventional two-promoter construct; see Figure 1A) | ∼400f | ∼1/16f |
| 3 | pMO455 | IRES | EMCV (porcine encephalomyocarditis virus) IRES (586 bp) | 39 | 0/16 |
| 4 | pMO467 | IRES | crTMV (crucifer-infecting tobamovirus) IRES-CP (148 bp) | 13 | 1/16 |
| 5 | pMO463 | IRES | PVY (potato virus Y) long (91 bp) | 35 | 0/16 |
| 6 | pMO464 | IRES | PVY short (66 bp) | 21 | 0/16 |
| 7 | pMO466 | IRES | PFBV (Pelargonium flower break virus) (78 bp) | 21 | 1/16 |
| 8 | pMO473 | IRES | PLRV (potato leafroll virus)g | 64 | 10/16 |
| 9 | pMO474 | IRES | PLRVmut11h | 73 | 8/16 |
| 10 | pMO449 | Non-IRES | TAGCATi | 327 | 15/16 |
| 11 | pMO471 | Non-IRES | TAGCCATi | 359 | 14/16 |
| 12 | pMO470 | Non-IRES | TGATAGCCATi | 452 | 13/16 |
Using CrVENUS-3FLAG as a sample GOI and APHVIII (conferring resistance to paromomycin) as the selectable marker.
For additional details, see Table 1 and/or Materials and Methods.
With or without an IRES.
Except for pMO518 (note f), the numbers of total and Venus-positive transformants shown here were taken from a single experiment that included all constructs; it used strain CMJ030 and the Bio-Rad electroporator with cells in in TAP medium plus 40 mM sucrose, followed by selection on TAP agar containing 20 µg/ml paromomycin. The numbers shown are consistent with those from multiple other experiments using strain CMJ030 or CC-124 with various subsets of the constructs.
In each case, 16 transformants were examined by fluorescence microscopy for expression of the Venus protein.
pMO518 was not included in the particular experiment that yielded the other data shown in this table. Thus, the numbers indicated are estimates based on several other experiments in which transformant numbers and numbers of GOI-positive transformants could be compared directly among pMO518, pMO448, and/or pMO449.
TAA C ATG ATT ATG ACT CCG ATG AGG ATT ACG GTC TGG AGA GAG AGG CTG CAA CAA ATG ATG. Additional potential start codons, in frame with that of APHVIII itself, are in bold face; the CrVENUS-3FLAG stop codon and the APHVIII start codon are indicated by italics.
TAA C ATG ATT ATG ACT CCG ATG AGG ATT ACG GTC TCC TCT CTC TCC CTG CAA CAA ATG ATG. Additional start codons, in frame with that of APHVIII itself, are in bold face; the CrVENUS-3FLAG stop codon and the APHVIII start codon are indicated by italics. The sequence altered by mut11 is underlined (Jaag et al. 2003).
Note (1) that the linkers contain one (pMO449 and pMO471) or two (pMO470) additional stop codons in frame with that of Cr-VENUS-3FLAG itself, and (2) that the APHVIII start codon is in frame with the stop codons in pMO449 but out of frame in pMO471 and pMO470.
Plasmid pMO511 (Figure 1A) was constructed by removing a HpaI-flanked fragment from pLM004-Venus (Yang et al. 2014). Plasmid pMO448 was constructed by assembling two PCR products that were amplified from pLM004-Venus. First, primers MOP651/MOP653 were used to amplify the pUC19 backbone plus the HSP70A/RBCS2 hybrid promoter (henceforth PH/R), the first three codons and first intron of RBCS2, and 29 additional base pairs of pLM004-Venus-derived sequence that include both HpaI and StuI sites. Second, primers MOP652/MOP654 were used to amplify the APHVIII coding sequence and the RBCS2 3′-UTR and TRBCS2; the fragment also includes 27 bp upstream of the APHVIII start codon that overlap with the sequence in the other fragment and provide consecutive TGA and TAG stop codons plus 3 bp of an NdeI site that is completed by the APHVIII start codon. To construct pMO449 (Figure 1B), a PCR fragment containing the codon-optimized CrVENUS-3FLAG was amplified from pLM004-Venus using primers MOP657/MOP658 and inserted into the HpaI site of pMO448, maintaining reading frame with both the RBCS2 start codon and the stop codons present in pMO448. To construct pMO470 and pMO471, preannealed oligonucleotides MOP696/MOP699 and MOP696/MOP698, respectively, were inserted between the StuI and NdeI sites of pMO449. To construct pMO518 (Figure 1A), a PCR fragment containing a stop codon, TRBCS2, and PPSAD was amplified from pLM004-Venus using primers MOP830/MOP831 and inserted between the StuI and NdeI sites of pMO449.
Plasmids like pMO449 but containing various IRES elements between the two ORFs were constructed as follows. For pMO455 (EMCV), a backbone was PCR-amplified from pMO449 using MOP658/MOP666 and assembled with an insert amplified from pIRES2-EGFP (Clontech) using MOP655/MOP665. For other plasmids, a fragment amplified from pMO449 using MOP658/MOP678 was assembled with fragments assembled from the indicated oligonucleotides: pMO467 (CrTMV cp), MOP686, MOP687, MOP688, and MOP689; pMO463 (PVY long), MOP679, MOP680, and MOP681; pMO464 (PVY short), MOP679 and MOP680; pMO466 (PFBV), MOP684 and MOP685; pMO473 (PLRV), MOP700 and MOP683; and pMO474 (PLRVmut11), MOP700, and MOP701.
To construct pMO507 and pMO508, pMO449 (except for PH/R) was PCR-amplified using MOP800/MOP801 and then assembled with a PCR fragment containing either PPSAD (amplified using MOP802/MOP803 from pLM004-Venus) or PTUB2 (amplified using MOP804/MOP805 from genomic DNA). To construct pMO490 and pMO520, a PCR fragment containing either sfGFP [amplified using MOP657/MOP766 from pGEM-sfGFP (a gift from Tomohiro Kubo and George Witman)] or Chlamydomonas-codon-optimized CrmCHERRY [amplified using MOP832/MOP833 from pBR9-mCherry (Rasala et al. 2013)] was coassembled with a fragment containing the 3FLAG coding sequence (amplified from pMO449 using MOP765/MOP663) into the HpaI site of pMO448. To construct pMO488 and pMO519, a PCR fragment containing either CrVENUS (amplified using MOP657/MOP761 from pMO449) or CrmCHERRY (amplified using MOP833/MOP834 from pBR9-mCherry) was inserted into the HpaI site of pMO448.
Plasmids containing gene fusions were constructed as follows. For pMO459, a Lifeact-CrVENUS-3FLAG sequence was amplified from pMO424 (Avasthi et al. 2014) using MOP674/MOP658 and inserted into the HpaI site of pMO448. For pMO482 (PRO1/Cre10.g427250; MOP757/MOP758), pMO483 (IDA5/Cre13.g603700; MOP741/MOP742), pMO484 (VFL2/Cre11.g468450; MOP473/MOP475), and pMO515 (PMH1/Cre03.g164600; MOP815/MOP816), fragments generated by PCR from genomic DNA were inserted into the HpaI site of pMO449; in each case, the indicated gene is fused in-frame to CrVENUS-3FLAG. For pMO480, a CVIM (“CAAX-box”)-encoding sequence assembled from oligonucleotides MOP749/MOP750 was inserted at the StuI site of pMO449.
Transformation
In most cases, plasmids were digested with EcoRV to excise the expression cassette before transformation. In a few cases in which the cassette itself contained an EcoRV site(s), ScaI was used to linearize the plasmid at a site outside the cassette. After heat-inactivation (65°, 10 min) of the enzyme used, the digestion reaction was used directly for transformation.
Three different electroporation methods were used for transformation during this study. Some early experiments used essentially the method described by Zhang et al. (2014): 240 µl of cell suspension (2 × 108 cells/ml in TAP medium plus 40 mM sucrose) at 16° were mixed with 10 µl containing 2 µg of digested plasmid and electroporated in a 4-mm-gap electroporation cuvette using a Bio-Rad Gene Pulser with Pulse Controller, with pulse parameters of 800 V, 25 µF, and infinite Ω. However, we obtained considerably higher transformation frequencies when the cells were suspended instead in CHES buffer (10 mM N-cyclohexyl-2-aminoethanesulfonic acid, pH 9.25, 40 mM sucrose, and 10 mM sorbitol) and electroporation was carried out using the Gene Pulser with parameters of 600 V, 25 µF, and 1000 Ω. Still better results were obtained with the third method, in which cells were grown to ∼8 × 106 cells/ml, resuspended at ∼8 × 108 cells/ml in CHES buffer at room temperature, and electroporated in a volume of 125 µl in a 2-mm-gap electrocuvette using a NEPA21 square-pulse electroporator (Bulldog Bio; Yamano et al. 2013), using two poring pulses of 250 and 150 V for 8 msec each, and five transfer pulses of 50 msec each starting at 20 V with a “decay rate” of 40% (i.e., successive pulses of 20, 12, 7.2, 4.3, and 2.6 V). In all three methods, electroporated cells were immediately transferred to a 15-ml centrifugation tube containing 8 ml TAP plus 40 mM sucrose. After overnight incubation at 24° under low light, cells were collected by centrifugation and spread on TAP agar (2% w/v) plates containing 10–40 µg/ml paromomycin.
Fluorescence microscopy
Venus-expressing strains were grown in TAP liquid medium without antibiotics for ≥2 d with daily dilutions. A final culture at ∼1.5 × 106 cells/ml was concentrated by centrifugation, mounted on a thin pad of TAP + 1% low-melting agarose (SeaPlaque; FMC Corporation), and sealed with a coverslip (for wide-field imaging) or placed in a glass-bottom culture dish (for confocal imaging). Wide-field imaging was performed using a Nikon Eclipse 600-FN microscope equipped with an Apochromat 100×/1.40 NA oil-immersion objective lens, an ORCA-2 cooled CCD camera (Hamamatsu Photonics), and Metamorph version 7.5 software (Molecular Devices). Confocal imaging was performed using a Leica DMI6000B microscope equipped with a Yokogawa CSU10 spinning-disk confocal-scanner unit, a Leica HCX PL Apo 63×/1.4–0.6 oil CORR lambda blue objective, an Andor iXon camera, and SlideBook 6.0 software. Images were postprocessed using ImageJ (National Institutes of Health) and Photoshop (Adobe) software.
Fluorescence-scanner and plate-reader analyses
For fluorescence-scanner analysis, cells were spotted on a TAP agar plate containing 10 µg/ml paromomycin, grown for 3–5 d, and imaged using a Typhoon Trio fluorescence scanner (GE Healthcare) with excitation and emission wavelengths of 532 and 555 nm (gain setting PMT 800), respectively, to detect Venus, and 633 and 670 nm (PMT 500), respectively, to detect the chlorophyll autofluorescence. The images were postprocessed using ImageJ and Photoshop software.
Cells for plate-reader analysis were grown without shaking in wells of a transparent 96-well microplate containing 150 µl of TAP medium without paromomycin. For measurement, 75-µl aliquots of the cultures were combined with 75 µl of fresh TAP in wells of a black-sided plate (Nunc; 265301), and fluorescence (F) and absorbance at 750 nm (A) were measured immediately using a Tecan Infinite 200 Pro microplate reader. Fluorescence readings were acquired with excitation and emission wavelengths of 515 and 550 nm for Venus, 488 and 530 nm for GFP, and 572 and 608 nm for mCherry, with bandwidths of 9 (excitation) and 20 (emission) nm and a manual gain of 150. For each well, F and A were first corrected for the average backgrounds for TAP medium as measured in a separate plate: F/A = (F – mean FTAP)/(A – mean ATAP). In figures, F/A of the wells are expressed as values relative to the average of the pMO448 control samples.
Analysis of protein expression by Western blotting
Cells growing in 5 ml TAP medium were collected by centrifugation at 2000 × g for 2 min, and the pellets were stored at −80°. For processing, samples were thawed in 100 µl of ice-cold PNE buffer (10 mM phosphate, pH 7.0, 150 mM NaCl, 2 mM EDTA) supplemented with a complete protease-inhibitor cocktail (Roche; 11697498001), and cells were disrupted by vortexing with acid-washed glass beads. Next, 100 µl of ice-cold PNE buffer + 2% NP-40 was added, followed by a 10-min incubation on ice, and the extracts were cleared by centrifugation at 12,000 × g for 10 min at 4°; 30 µg of each total cell extract were analyzed by SDS-PAGE (12%) and Western blotting, using a mouse anti-FLAG antibody (Sigma; F1804) and an HRP-conjugated rabbit anti-mouse-IgG antibody (ICN Pharmaceuticals; 55564).
Data availability
Strains, plasmids, and plasmid sequences are available upon request and will be deposited at the Chlamydomonas Resource Center. File S1 contains sequences of all DNA oligonucleotides used in this study.
Results
Comparison of two-promoter and one-promoter expression systems for Chlamydomonas
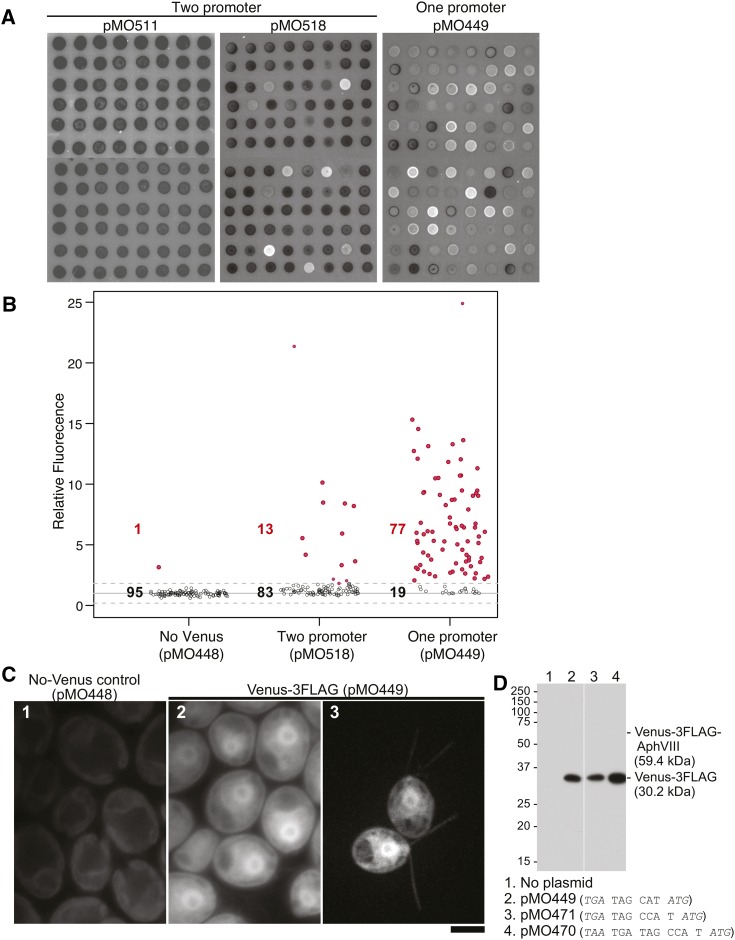
Conventional two-promoter constructs, in which the GOI and the selectable marker are expressed from separate promoters, have typically had poor efficacy in achieving expression of the unselected GOI in Chlamydomonas (see Introduction). We obtained similar results when we used two such constructs (Figure 1A) to transform cells with the gene for the fluorescent Venus protein. Although many paromomycin-resistant transformants were obtained, only a small minority of them displayed Venus fluorescence (Figure 2, A and B, and Table 2, row 2).
Figure 2.
Frequencies and levels of Venus expression after transformation of Chlamydomonas with various constructs. (A and B) Strain CC-124 was transformed with the expression construct from plasmid pMO448 (control; no VENUS gene), pMO511 or pMO518 (two-promoter constructs; see Figure 1A), or pMO449 (one-promoter construct; see Figure 1B), and transformants were selected initially on 10 µg/ml paromomycin. In each case, 96 randomly chosen clones were analyzed further. (A) The clones from the pMO511, pMO518, and pMO449 transformations were spotted on plates containing 10 µg/ml paromomycin, grown for 3 d, and imaged using a fluorescence scanner (see Materials and Methods). (B) The clones from the pMO448, pMO518, and pMO449 transformations were grown in liquid medium without paromomycin in wells of a microplate for 5 d and analyzed using a fluorescence plate reader (ex515/em550 nm; see Materials and Methods). Each individual reading was normalized to the mean value for the control (pMO448) transformants, which was set at 1.0. Solid and dashed lines show the mean ± 3 SDs for the pMO448 transformants. Open circles and black numbers denote transformants whose fluorescence values were within the ± 3 SD window; red circles and numbers denote transformants whose values were above this threshold. (C) Cells of strain CMJ030 (1 and 2) or CC-124 (3) were transformed with the expression constructs from pMO448 (control) or pMO449 and selected initially on 20 µg/ml paromomycin. The transformants were grown further and observed by fluorescence microscopy using either the Eclipse (1 and 2) or spinning-disk (3) microscope as described in Materials and Methods. Image in 3 is a maximum projection of the z-stack. Weak signal in 1 represents autofluorescence from the chloroplasts; Venus signal in 2 and 3 is seen diffusely in the cytoplasms and (for unknown reasons) more strongly in the nuclei. (D) Cells of strain CMJ030 were transformed with the expression constructs from the indicated plasmids, selected initially on 20 µg/ml paromomycin, and then grown for 2 d at 24° in liquid medium with 1 µg/ml paromomycin. From each culture, 30 µg of total cell extract was subjected to SDS-PAGE and Western blotting using an anti-FLAG antibody. The predicted molecular weights of Venus-3FLAG and of a hypothetical Venus-3FLAG-AphVIII fusion protein are indicated.
We speculated that a one-promoter system might be more effective. In this case, a single promoter upstream of the GOI drives expression of both it and a downstream selectable marker from a bicistronic mRNA (Figure 1B). This arrangement should generally prevent the isolation of transformants in which the selectable marker is integrated and expressed without the GOI, or in which the whole construct is integrated in a chromosomal location allowing only poor expression or susceptible to subsequent transcriptional silencing. Thinking that an IRES would promote translation of the selectable marker, we first tried transforming with constructs containing CrVENUS connected to the APHVIII selectable marker by various virus-derived IRES-containing junction sequences (Table 2, rows 3–8), all of which have been used successfully for one-promoter expression of nonviral proteins in other systems (see File S2). The results with five of these sequences were disappointing: the numbers of paromomycin-resistant transformants were ∼100-fold less than in a control, and very few of the selected transformants expressed detectable levels of Venus as judged by fluorescence microscopy (Table 2, rows 1 and 3–7). These results suggested that these IRES elements do not function in Chlamydomonas and that the small numbers of transformants had acquired paromomycin resistance through integration of a partially digested DNA fragment containing APHVIII downstream of an endogenous promoter.
In contrast, the construct containing the PLRV IRES yielded several-fold more transformants, and about half of these were Venus positive (Table 2, row 8). This IRES contains four potential start codons (two near its 5′ end, one in the middle, and one at the 3′ end) that are in frame with that of the selectable marker, as well as a GGAGAGAGAGG motif that is thought to be important for its IRES function (Jaag et al. 2003). However, when we replaced this motif with the sequence CCTCTCTCTCC, which dramatically reduces IRES activity in potato protoplasts (Jaag et al. 2003), there was little or no effect (Table 2, row 9). This result suggested that it was not the IRES activity of PLRV that accounted for its partial function, but rather its potential start codons, and perhaps the fact that two of them lay in close proximity to the stop codon of CrVENUS-3FLAG.
Accordingly, we next tested three constructs that contained only short linkers between CrVENUS-3FLAG and the APHVIII selectable marker; these linkers also contained one or two additional stop codons in frame with that of CrVENUS-3FLAG and positioned the APHVIII start codon just 6–10 nucleotides (nt) downstream of the CrVENUS-3FLAG stop codon, and 3 or 4 nt downstream of the most proximal stop codon (Table 2, rows 10–12). Remarkably, not only did these constructs yield 5- to 10-fold more transformants than the IRES-containing constructs, but >80% of them expressed Venus at significantly more than background levels (Figure 2, A–C, and Table 2, rows 10–12). Given the additional stop codon(s) and the fact that CrVENUS-3FLAG and APHVIII are out of frame in two of the constructs, it seemed unlikely that the translating ribosomes simply read through into APHVIII to produce a functional fusion protein that confers paromomycin resistance. Indeed, when we examined the proteins directly by Western blotting with an anti-FLAG antibody, only a protein of the predicted size of Venus-3FLAG could be detected, even after extended overexposure (Figure 2D). Thus, it appears that translation of APHVIII is mediated by post-termination ribosome reinitiation (Skabkin et al. 2013), although further investigation would be needed to be certain of the molecular mechanism (see Discussion).
It should be noted that the two two-promoter constructs examined here showed striking differences not only in the overall frequency of Venus-positive transformants but also in the expression strength observed (Figure 2A). Although some individual transformants with pMO518 displayed expression as high as those seen with the one-promoter constructs, albeit at much lower frequency (Figure 2, A and B), no such highly expressing transformants were recovered with pMO511 (Figure 2A and our unpublished data). It is not clear which of the several differences between the two constructs (Figure 1A) account(s) for these differences.
In summary, given both the higher frequency of transformants with detectable GOI expression and (in many cases) the higher levels of that expression, it appears that the use of one-promoter constructs should greatly facilitate the identification of transformants suitable for a variety of downstream applications.
Effect on expression strength of the antibiotic concentration used for selection of transformants
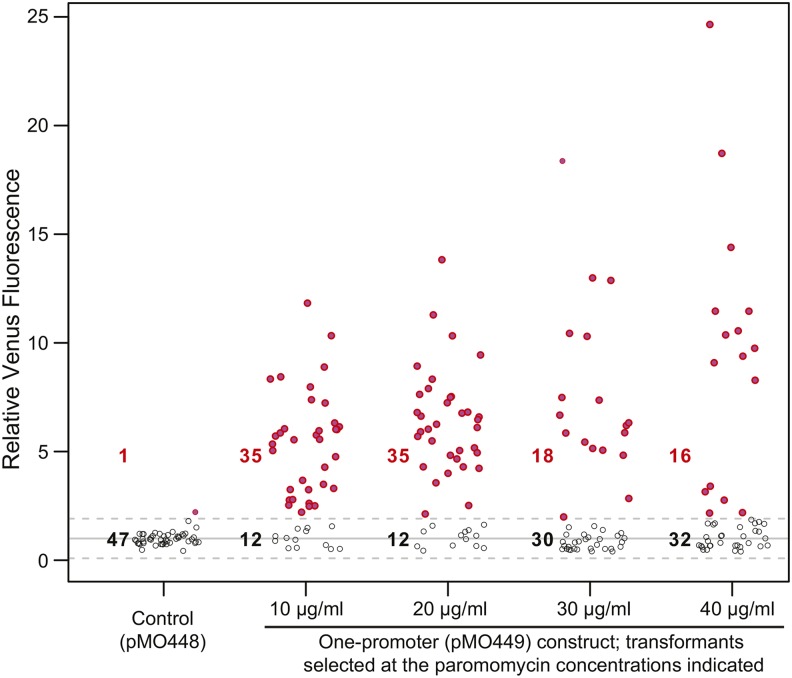
In one-promoter systems, a positive correlation would normally be expected between the expression levels of the two coexpressed proteins. With IRES-based expression vectors, this property has been utilized for screening and enrichment of cells with strong GOI expression, for example by fluorescence-activated cell sorting of transformants for high levels of expression of the marker gene downstream of the IRES (Liu et al. 2000). To ask if this would also be true for the Chlamydomonas expression constructs, we transformed wild-type cells with a one-promoter construct, a two-promoter construct, or a monocistronic APHVIII-only control, and incubated the transformants on plates containing various concentrations of paromomycin. As the antibiotic concentration was increased, a striking decrease of transformation efficiency was observed for the one-promoter construct, whereas the other constructs showed only modest decreases over the concentration range used (Table 3). These data suggest that the translation of APHVIII from the second cistron of a bicistronic mRNA is less efficient than from a monocistronic mRNA, so that relatively few transformants achieve sufficiently high expression for resistance to high paromomycin concentrations.
Table 3. Effect of paromomycin concentration on transformation efficiency.
| Plasmida | Type | Relative Transformation Efficiency at Different Paromomycin Concentrations (µg/ml) | |||
|---|---|---|---|---|---|
| 10 | 20 | 30 | 40 | ||
| pMO449 | One-promoter (see Figure 1B) | 100 | 26 | 7.1 | 3.2 |
| 100 | 34 | 11 | 3.6 | ||
| 100 | 34 | 11 | 5.0 | ||
| pMO518 | Two-promoter (see Figure 1A) | 100 | 97 | 87 | 83 |
| 100 | 103 | 95 | 90 | ||
| pMO448 | Monocistronic (APHVIII-only) control (see Materials and Methods) | 100 | 101 | 64 | 54 |
| 100 | 93 | 76 | 71 | ||
Data are shown from three (pMO449) or two (pMO518 and pMO448) separate transformation experiments using strain CC-124 and various electroporation methods (see Materials and Methods). In each experiment, the numbers of transformants per micrograms of DNA were normalized to the value obtained at 10 µg/ml paromomycin in that experiment.
Analysis of the transformants from the one-promoter construct for Venus fluorescence revealed two effects of increasing paromomycin concentration. First, at higher drug concentrations, the proportion of Venus-positive clones among the transformants decreased (Figure 3). It seems likely that this decrease occurred because relatively few of the transformants that actually expressed APHVIII via the bicistronic mRNA achieved sufficiently high levels of expression for resistance to high paromomycin concentrations, so that a larger fraction of the transformants recovered expressed APHVIII by some other mechanism such as integration of APHVIII by itself downstream of an endogenous promoter. Second, the Venus-positive clones that were recovered showed a moderate positive correlation between expression level and the antibiotic concentration that had been used to select them (Figure 3, Pearson’s r = 0.365), suggesting that the expression levels of the two proteins are indeed correlated (if somewhat weakly). This correlation probably reflects integration of the construct (containing both genes) at sites that allow different levels of transcription of the bicistronic mRNA. Taken together, the data suggest that when high levels of expression of a GOI are needed, it may be best to use a high antibiotic concentration for selection of the transformants; in this case, both a highly efficient transformation and a facile secondary screen for the transformants of interest would be desirable. When high levels of GOI expression are not required, it will probably be preferable to select transformants using a lower antibiotic concentration as determined empirically depending on the strain; we have successfully used as low as 5 µg/ml paromomycin with CC-124.
Figure 3.
Effect of the paromomycin concentration used for selection on the fraction of GOI-expressing transformants and the level of GOI expression. Strain CC-124 was transformed with the one-promoter construct from pMO449, and transformants were selected on plates containing the indicated concentrations of paromomycin. Transformants with the non-GOI control plasmid pMO448 were selected on 10 µg/ml paromomycin. For each condition, 48 randomly chosen transformants were analyzed using a fluorescence plate reader as in Figure 2B.
Effects of altering the elements in the expression constructs
We next tested the effects of replacing various elements in the expression constructs. First, we replaced PH/R in pMO449 with either of two other widely used strong promoters, PPSAD and PTUB2, yielding pMO507 and pMO508. (These constructs retain the first three codons and intron of RBCS2, whose possible contribution to expression has not been tested.) When tested in parallel, the three promoters yielded similar fractions of Venus-positive transformants and similar levels of Venus expression under the conditions used (21°; 75 μmol photons m−2 sec−1 of constant light) (Figure 4A). Because these promoters respond differently to environmental perturbations (Davies et al. 1992; Fischer and Rochaix 2001; Schroda et al. 2002; Heitzer and Zschoernig 2007), the conditions to be used may determine the best promoter to use in expressing a transgene for a particular application.
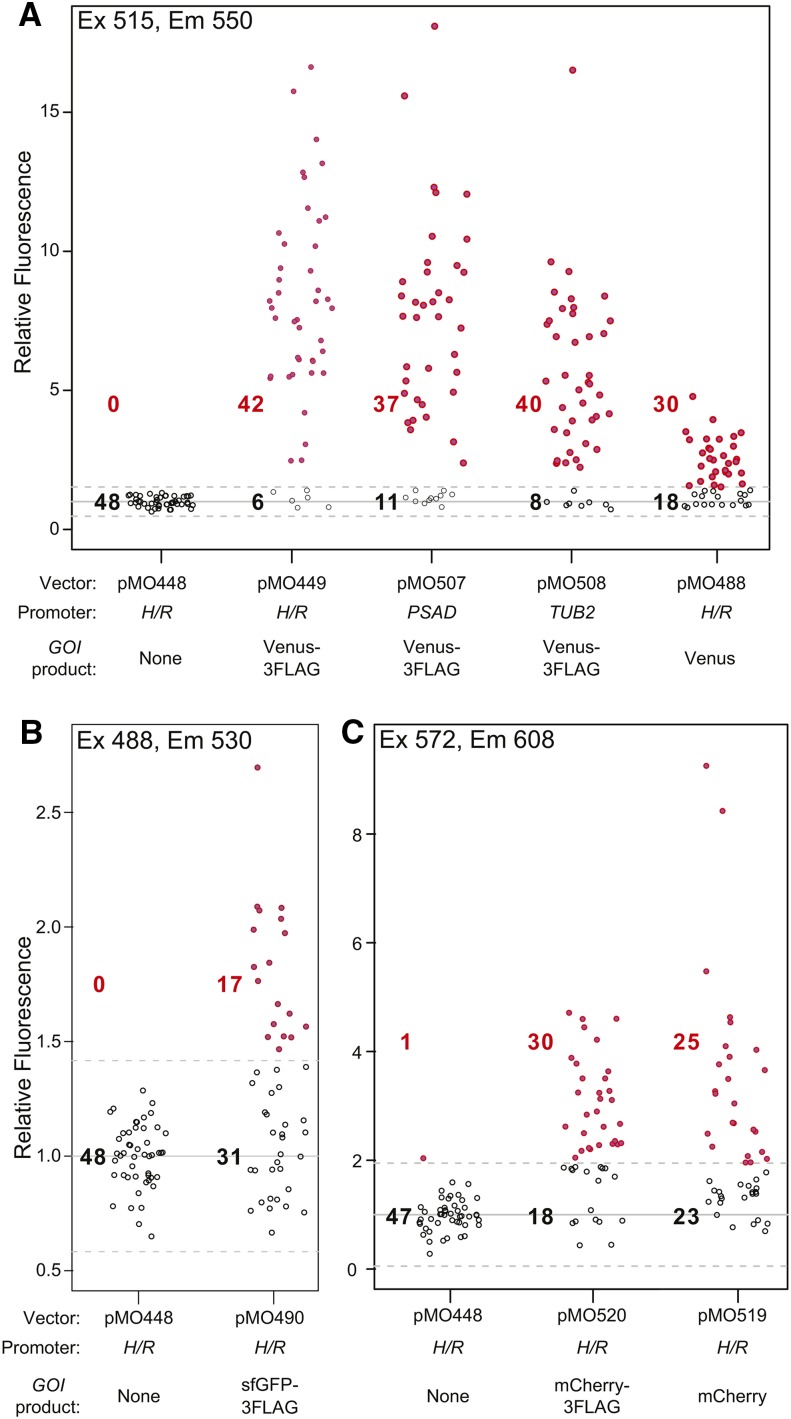
Figure 4.
Successful use of one-promoter constructs with various promoters and GOIs. Strain CC-124 was transformed with the expression constructs from the indicated plasmids. In each case, 48 randomly chosen transformants were analyzed using a fluorescence plate reader as in Figure 2B, except that the excitation and emission settings appropriate for each fluorescent protein were used, as indicated. (A) Expression of Venus constructs. (B) Expression of sfGFP constructs. Note that the expression appears weak in the plot because of the high autofluorescence of Chlamydomonas cells in this spectral range. (C) Expression of mCherry constructs.
Second, we tested the efficacy of the expression system with different GOIs. In addition to Venus-3FLAG, an untagged Venus (Figure 4A, pMO488), sfGFP-3FLAG (Figure 4B, pMO490), mCherry-3FLAG (Figure 4C, pMO520), and mCherry (Figure 4C, pMO519) were successfully expressed. Importantly, all of these genes had been codon-optimized for Chlamydomonas, and we failed to obtain GOI-expressing transformants using genes codon-optimized for other organisms (EGFP and mCHERRY, mammals; yEGFP, yeast) (data not shown). This failure probably reflects a need for efficient translation of the 5′ cistron in order to achieve efficient post-termination reinitiation for expression of APHVIII, so that the transformants actually recovered carry APHVIII alone (see also preceding section). Importantly, the recovery of transformants expressing Venus or mCherry without the 3FLAG tag indicates that nothing in the 3FLAG coding sequence functions as a cryptic IRES or an essential element for post-termination reinitiation, as is seen in some viruses (Powell et al. 2008). Interestingly, the modest reduction in the average expression of Venus (pMO488) relative to that of Venus-3FLAG (pMO449) (Figure 4A) suggests that, in the pMO449 construct, the 3FLAG sequence is actually somewhat inhibitory of APHVIII expression; thus, in the absence of this sequence, transformants can be recovered that have sufficiently high AphVIII expression for paromomycin resistance, even though expression of the bicistronic mRNA (and hence of the 5′ GOI) is at a lower level. Alternatively, it is possible that the stability and/or folding of Venus may be somewhat improved by the 3FLAG tag. Because CrVENUS and CrmCHERRY are of different origins, and have distinct nucleic-acid sequences (∼52% overall identity, ∼39% within the 120 nt upstream of the stop codons), the successful expression of both genes without a common 3FLAG tag suggests that the RNA-sequence context immediately upstream of the intergenic region is not critical for the function of the constructs described here.
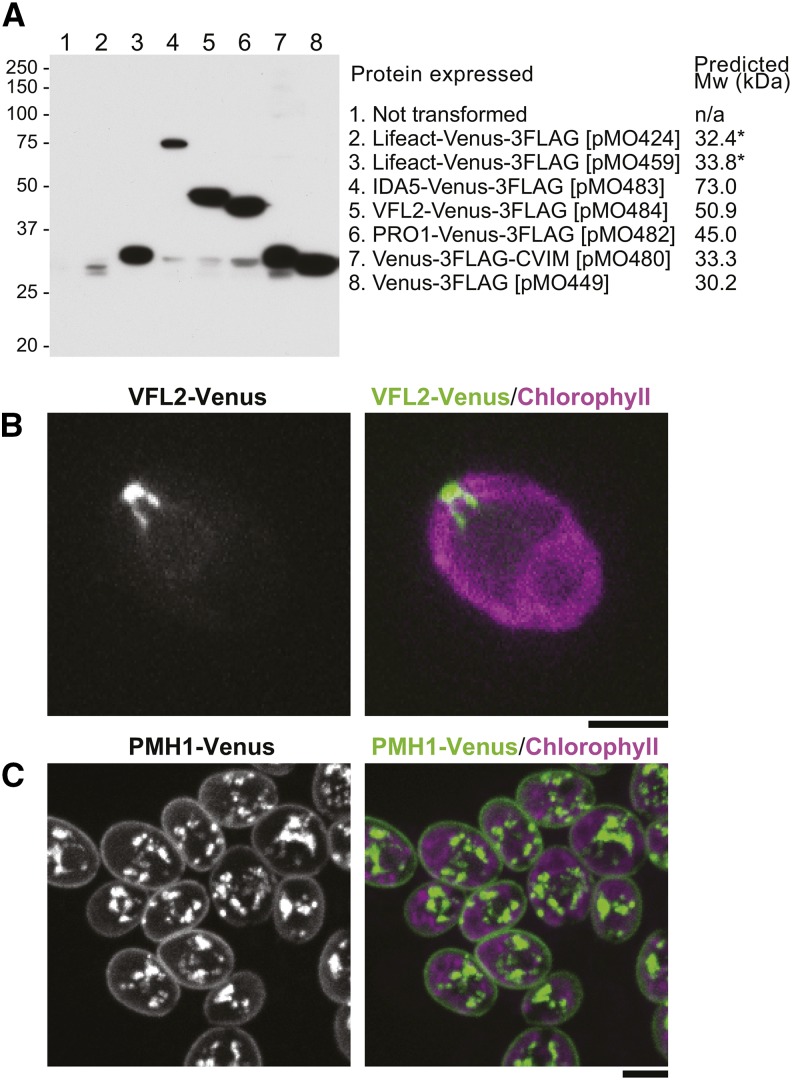
A particularly useful class of transgenes is those expressing a protein of interest tagged with a fluorescent protein, so we checked to be sure that the one-promoter constructs would give efficient recovery of transformants expressing such genes. Indeed, we were able easily to recover transformants showing good expression of either the tagged Lifeact peptide or any of several full-length proteins carrying a C-terminal Venus-3FLAG tag, as well as of Venus-3FLAG carrying a C-terminal CVIM (“CAAX-box”) peptide, as shown by Western blotting (Figure 5A), observations of in vivo fluorescence (Figure 5, B and C), or both. Note that although we screened many transformants with the two-promoter construct from pMO424 to find the one with the strongest expression of Lifeact-Venus, the expression level obtained (Figure 5A, lane 2) was considerably less than that in a randomly picked transformant obtained with the one-promoter construct from pMO459 (Figure 5A, lane 3). The intracellular localizations observed for VFL2 (Figure 5B), and PMH1 (Figure 5C), as well as for Lifeact, IDA5, and PRO1 (data not shown), were consistent with expectations (Taillon et al. 1992; Kato-Minoura et al. 1998; Kovar et al. 2001; Onishi et al. 2016), although Venus-3FLAG-CVIM failed to localize to the plasma membrane for unknown reasons. Interestingly, with both IDA5-CrVENUS-3FLAG and VFL2-CrVENUS-3FLAG, successful expression was achieved only when the full genomic coding sequences (i.e., all exons and introns) were used, whereas the corresponding cDNA sequences failed even though they were inserted at the HpaI site just downstream of the RBCS2 intron (see Figure 1B).
Figure 5.
Expression of Venus-3FLAG-tagged peptides and proteins from one-promoter constructs. Strain CC-124 was transformed with the constructs from the indicated plasmids. (A) Analysis of protein expression by SDS-PAGE and Western blotting using an anti-FLAG antibody as described in Figure 2D. In all cases but pMO424, one transformant for each construct was chosen at random for analysis. For pMO424, 192 transformants were screened by fluorescence microscopy, and the one with the strongest signal was picked for further analysis. Predicted molecular weights of the fusion proteins are shown. *, the 1.4-kDa difference in predicted molecular weight of Lifeact-Venus-3FLAG expressed from the pMO424 and pMO459 constructs is due to the seven additional amino acids (MARRFEV) in the latter encoded by sequences added during the construction. (B, C) Localization of Venus-3FLAG-tagged VFL2 [(B); the same transformant as in (A)] and PMH1 [(C); transformation with the construct from pMO515]. Cells were observed using the spinning-disk microscope. Bars, 5 µm.
Finally, we attempted to replace APHVIII in pMO449 with other antibiotic-resistance markers that have been used previously for selection of transformants in Chlamydomonas: AadA (spectinomycin resistance, codon-optimized; a gift from T. Yamasaki, Kochi University of Technology), ble (zeocin resistance; Lumbreras et al. 1998), and aph7″ (hygromycin B resistance; Berthold et al. 2002). The ble-containing construct did not yield any transformants (data not shown). The AadA- and aph7″-containing constructs yielded small numbers of transformants when low antibiotic concentrations were used (30 µg/ml spectinomycin or 5 µg/ml hygromycin B), but none of the colonies was Venus positive (data not shown). It is not clear why these constructs failed, but it seems possible that these markers simply require higher expression than AphVIII to render the cells drug resistant.
Discussion
We describe here a new method for expression of transgenes in Chlamydomonas in which a single promoter drives expression of a bicistronic mRNA containing two separate ORFs: an upstream GOI and a downstream paromomycin-resistance marker. With this system, we found that most paromomycin-resistant transformants also expressed the unselected GOI. The plasmids developed to date for applying this method contain one of three promoters, one of several cassettes for tagging proteins of interest, and convenient restriction sites for insertion of GOIs for tagging and/or overexpression (Table 4).
Table 4. Plasmids available for transgene expression using the one-promoter system.
| Plasmida | Promoterb | Upstream Genec | Selectable Markerd |
|---|---|---|---|
| pMO449e | HSP70A/RBCS2 | CrVENUS-3FLAG | APHVIII |
| pMO470e | HSP70A/RBCS2 | CrVENUS-3FLAG | APHVIII |
| pMO471e | HSP70A/RBCS2 | CrVENUS-3FLAG | APHVIII |
| pMO488 | HSP70A/RBCS2 | CrVENUS | APHVIII |
| pMO490 | HSP70A/RBCS2 | sfGFP-3FLAG | APHVIII |
| pMO519 | HSP70A/RBCS2 | CrmCHERRY | APHVIII |
| pMO520 | HSP70A/RBCS2 | CrmCHERRY-3FLAG | APHVIII |
| pMO507 | PSAD | CrVENUS-3FLAG | APHVIII |
| pMO508 | TUB2 | CrVENUS-3FLAG | APHVIII |
| pMO561f | TUB2 | CrVENUS-3FLAG | APHVIII |
Available plasmids with one-promoter expression constructs of the general structure shown in Figure 1B.
Except as indicated in note f, all promoters are followed by the first three codons and intron of RBCS2.
All genes indicated have been codon optimized for Chlamydomonas. These genes can be replaced in toto by HpaI–StuI fragments containing other GOIs whose expression/overexpression is desired. Fluorescence tagging of a gene product of interest can be achieved by inserting the coding sequence, in frame, at the HpaI site (C-terminal tagging) or the StuI site (N-terminal tagging).
Other selectable markers could be inserted as NdeI–BamHI fragments. To date, however, we have not had success with selectable markers other than APHVIII (see text).
These plasmids differ only in the short linker between CrVENUS-3FLAG and APHVIII (see Figure 2D and Table 2).
Same as pMO508 except that the RBCS2 ATG start codon has been changed to TTG. This allowed expression of CrVENUS-3FLAG, presumably from its own start codon (i.e., without the additional amino acids from RBCS2).
When Venus was used as a GOI with this system, a positive correlation was found between paromomycin resistance and Venus fluorescence, indicating that expression from the two ORFs is coupled. It initially seemed possible that a mechanism such as stop-codon read through (Jackson et al. 2012) might result in expression of the GOI and APHVIII products as a single fusion protein that provides paromomycin resistance. However, this appears not to be the case, because the method works equally well when two or three different stop codons are inserted between the ORFs, and/or the ORFs are placed out-of-frame, and Western blotting detected no such fusion products. Thus, it seems most likely that APHVIII is translated by post-termination reinitiation (Kozak 2007; Skabkin et al. 2013), in which the ribosome remains associated with the mRNA after termination, continues scanning, and reinitiates translation at a downstream (or, occasionally, upstream: Skabkin et al. 2013) AUG. Such events have been well documented for transcripts that have naturally occurring upstream ORFs (uORFs) in many organisms (Wang and Wessler 1998; Kozak 2007; Hinnebusch and Lorsch 2012), and many annotated transcripts in the Chlamydomonas genome also have uORFs, although it is not yet known how many of these are actually translated (Cross 2016). Most uORFs in these cases are very short (e.g., 3–4 codons for yeast GCN4 and 38 codons for maize R), and a negative correlation between uORF length and reinitiation efficiency at the downstream AUG has been reported (Kozak 2007). Thus, it is somewhat surprising that the long “uORFs” in the one-promoter constructs used in this study (e.g., 654 codons for IDA5-CrVENUS-3FLAG) allowed effective translation of the downstream APHVIII.
Indeed, it appears that the translation of APHVIII is less efficient than that of the upstream GOI, given that most transformants did not grow at a paromomycin concentration of 40 µg/ml (Figure 3 and Table 3), whereas most transformants obtained with constructs expressing APHVIII from a monocistronic mRNA were able grow at 40 µg/ml (Table 3), or even 80 µg/ml (our unpublished data), paromomycin. Enhancing translation of the downstream selectable marker might potentially enable the use of markers other than APHVIII. However, our attempts to date with virus-derived IRES-containing sequences failed to achieve this goal. We also considered the possibility that the 5′ portion of APHVIII contains a cryptic IRES-like translation enhancer activity. However, joining the first 120 nt of APHVIII to other markers (aph7″, ble, and AadA) still did not yield any Venus-positive clones with these markers (data not shown). It is possible that using other viral IRESs or endogenous Chlamydomonas sequences from bicistronic (Cenkci et al. 2003) or uORF-containing (Cross 2016) transcripts would be more successful.
The one-promoter system has significant advantages over traditional two-promoter strategies, of which the chief is clearly the high percentage of transformants that express the GOI. This feature should be particularly valuable in studies in which a given marker needs to be expressed in multiple strains. For example, in our initial attempts to use Lifeact-Venus to determine actin localization in vegetative cells (Avasthi et al. 2014), we needed to screen >100 transformants to isolate a clone with a barely sufficient level of expression (see also Figure 5A). In contrast, a one-promoter construct allowed us to quickly and easily observe Lifeact-Venus localization in multiple strains, and to screen mutants for altered F-actin structures (Onishi et al. 2016; our unpublished data). It should be possible to perform similar expression-based screens with other biomarkers or other genes of scientific or industrial importance. The efficient recovery of GOI-expressing transformants should also greatly facilitate mutagenesis-based functional analysis of GOIs.
Second, although some two-promoter constructs yield individual transformants with high levels of GOI expression, others do not (Figure 2, A and B, and Figure 5A), so that when high levels of expression of a GOI are desired, a one-promoter construct seems likely to provide the method of choice.
Finally, because translation of the GOI should be a prerequisite for that of APHVIII, propagation of transformants in the presence of paromomycin should circumvent the silencing of GOIs after repeated passages that has sometimes been observed (Cerutti et al. 1997). [However, we have also observed that, at least in most cases, paromomycin can be omitted from the medium for ≥2 wk without affecting the levels of GOI expression observed, an approach that may often be desirable given the known effects of paromomycin in decreasing the fidelity of translation (Davies et al. 1965; Mironova et al. 1982; Hirokawa et al. 2007).] Moreover, if a particular GOI fails to express efficiently for any reason, the number of transformants obtained would simply drop, saving the investigator from screening hundreds of colonies for a nonexistent GOI-expressing clone. These considerations also highlight the importance of optimizing the codons of heterologous genes to be expressed in Chlamydomonas. This seems likely to be even more important with the one-promoter system than it is with two-promoter constructs (Cerutti et al. 1997; Fuhrmann et al. 1999), and indeed we have failed to recover transformants with several heterologous genes that had not been codon optimized (our unpublished data).
Interestingly, we failed to express IDA5 and VFL2 when cDNAs were inserted at the HpaI site of pMO449 (Figure 1B), although insertion of the full genomic copies of the genes (with introns) yielded transformants with good levels of expression (Figure 5). As all of these constructs contained the first intron of RBCS2, which is known to enhance transcription (Lumbreras et al. 1998), it is not clear why the additional introns of IDA5 and VFL2 improved expression. We have also failed to express some other Chlamydomonas and heterologous genes using the one-promoter system, presumably either because their ectopic overexpression is toxic to cells, or because their translation efficiency is insufficient to support adequate expression of the downstream APHVIII through translation reinitiation.
The expression system described here is distinct from a previously described one-promoter system based on the viral 2A peptide (Rasala et al. 2012). In the 2A system, the transformants express a single mRNA encoding the ble selection marker, the viral 2A peptide, and a GOI. When a translating ribosome encounters the 2A sequence, no peptide bond is formed between the last two amino acids of the 2A peptide, but the ribosome continues to translate the downstream coding sequence, thereby achieving coexpression of two (or more) proteins from a single mRNA. However, the failure of peptide-bond formation at the 2A peptide is sometimes incomplete, so that a small proportion of the marker and GOI-encoded protein may be expressed as a fusion protein (Geu-Flores et al. 2008; Rasala et al. 2012; Kong et al. 2015), potentially impairing the function of the GOI product or otherwise complicating interpretation of the results obtained. Moreover, the coexpression efficiency of the GOI with the ble marker seems to be low, so that a PCR-based screen for transformants that received a complete cassette, and subsequent assays for strong expression, have been performed in studies using the 2A technique (Rasala et al. 2012, 2014; Kong et al. 2015). However, the 2A system also has an advantage over the system described here, in that it can express multiple proteins from a single transformed construct (Rasala et al. 2014), whereas this is not feasible with the system described here due to the inefficiency of translation reinitiation. It seems likely that each system will be useful in appropriate applications in the future.
In summary, we have described here a one-promoter, bicistronic-mRNA system for expression of transgenes in Chlamydomonas that should greatly facilitate a wide variety of experiments. Although only a limited set of plasmids has been constructed to date (Table 4), others (e.g., constructs containing other promoters or markers for gene tagging) could easily be constructed using the backbone provided by pMO449 (Figure 1B). A limitation of the system to date is the availability of only a single selectable marker; it should be possible to overcome that limitation in future studies.
Supplementary Material
Acknowledgments
We thank Luke Mackinder and Martin Jonikas for strains, plasmids, and general advice; Arthur Grossman, Fred Cross, Tomohito Yamasaki, and members of our laboratory for helpful discussions; and the Chlamydomonas Resource Center for strains. We also thank Heather Cartwright (Advanced Imaging Facility, Department of Plant Biology, Carnegie Institution for Science) for assistance with imaging. This work was supported by National Science Foundation Early-Concept Grants for Exploratory Research Award 1548533.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.033035/-/DC1.
Communicating editor: C. Boone
Literature Cited
- Avasthi P., Onishi M., Karpiak J., Yamamoto R., Mackinder L., et al. , 2014. Actin is required for IFT regulation in Chlamydomonas reinhardtii. Curr. Biol. 24: 2025–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthold P., Schmitt R., Mages W., 2002. An engineered Streptomyces hygroscopicus aph7″ gene mediates dominant resistance against Hygromycin B in Chlamydomonas reinhardtii. Protist 153: 401–412. [DOI] [PubMed] [Google Scholar]
- Cenkci B., Petersen J. L., Small G. D., 2003. REX1, a novel gene required for DNA repair. J. Biol. Chem. 278: 22574–22577. [DOI] [PubMed] [Google Scholar]
- Cerutti H., Johnson A. M., Gillham N. W., Boynton J. E., 1997. Epigenetic silencing of a foreign gene in nuclear transformants of Chlamydomonas. Plant Cell 9: 925–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R., 2016. Tying down loose ends in the Chlamydomonas genome: functional significance of abundant upstream open reading frames. G3 (Bethesda) 6: 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R., Umen J. G., 2015. The Chlamydomonas cell cycle. Plant J. 82: 370–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J., Gorini L., Davis B. D., 1965. Misreading of RNA codewords induced by aminoglycoside antibiotics. Mo.l Pharmacol. 1: 93–106. [PubMed] [Google Scholar]
- Davies J. P., Weeks D. P., Grossman A. R., 1992. Expression of the arylsulfatase gene from the β2-tubulin promoter in Chlamydomonas reinhardtii. Nucleic Acids Res. 20: 2959–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorokhov Y. L., Skulachev M. V., Ivanov P. A., Zvereva S. D., Tjulkina L. G., et al. , 2002. Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc. Natl. Acad. Sci. USA 99: 5301–5306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N., Rochaix J. D., 2001. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol. Genet. Genomics 265: 888–894. [DOI] [PubMed] [Google Scholar]
- Fuhrmann M., Oertel W., Hegemann P., 1999. bA synthetic gene coding for the green fluorescent protein (GFP) is a versatile reporter in Chlamydomonas reinhardtii. Plant J. 19: 353–361. [DOI] [PubMed] [Google Scholar]
- Geu-Flores F., Olsen C. E., Halkier B. A., 2008. Towards engineering glucosinolates into non-cruciferous plants. Planta 229: 261–270. [DOI] [PubMed] [Google Scholar]
- Gibson D. G., Young L., Chuang R.-Y., Venter J. C., Hutchison C. A., et al. , 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6: 343–345. [DOI] [PubMed] [Google Scholar]
- Gorman D. S., Levine R. P., 1965. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 54: 1665–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris E. H., 2001. Chlamydomonas as a model organism. Annu. Rev. Plant Biol. 52: 363–406. [DOI] [PubMed] [Google Scholar]
- Heinnickel M. L., Grossman A. R., 2013. The GreenCut: re-evaluation of physiological role of previously studied proteins and potential novel protein functions. Photosynth. Res. 116: 427–436. [DOI] [PubMed] [Google Scholar]
- Heitzer M., Zschoernig B., 2007. Construction of modular tandem expression vectors for the green alga Chlamydomonas reinhardtii using the Cre/lox-system. Biotechniques 43: 324–332. [DOI] [PubMed] [Google Scholar]
- Hellen C. U., Sarnow P., 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G., Lorsch J. R., 2012. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 4: a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa G., Kaji H., Kaji A., 2007. Inhibition of antiassociation activity of translation initiation factor 3 by paromomycin. Antimicrob. Agents Chemother. 51: 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt A. G., Maiti I. B., 2001. Strategies for expressing multiple foreign genes in plants as polycistronic constructs. In Vitro Cell. Dev. Biol. Plant 37: 313–320. [Google Scholar]
- Iizuka N., Najita L., Franzusoff A., Sarnow P., 1994. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 14: 7322–7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag H. M., Kawchuk L., Rohde W., Fischer R., Emans N., et al. , 2003. An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc. Natl. Acad. Sci. USA 100: 8939–8944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., 2013. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb. Perspect. Biol. 5: a011569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson R. J., Hellen C. U. T., Pestova T. V., 2012. Termination and post-termination events in eukaryotic translation. Adv. Protein Chem. Struct. Biol. 86: 45–93. [DOI] [PubMed] [Google Scholar]
- Jang S. K., Krausslich H. G., Nicklin M. J., Duke G. M., Palmenberg A. C., et al. , 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62: 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinkerson R. E., Jonikas M. C., 2015. Molecular techniques to interrogate and edit the Chlamydomonas nuclear genome. Plant J. 82: 393–412. [DOI] [PubMed] [Google Scholar]
- Kato-Minoura T., Uryu S., Hirono M., Kamiya R., 1998. Highly divergent actin expressed in a Chlamydomonas mutant lacking the conventional actin gene. Biochem. Biophys. Res. Commun. 251: 71–76. [DOI] [PubMed] [Google Scholar]
- Kong F., Yamasaki T., Kurniasih S. D., Hou L., Li X., et al. , 2015. Robust expression of heterologous genes by selection marker fusion system in improved Chlamydomonas strains. J. Biosci. Bioeng. 120: 239–245. [DOI] [PubMed] [Google Scholar]
- Kovar D. R., Yang P., Sale W. S., Drobak B. K., Staiger C. J., 2001. Chlamydomonas reinhardtii produces a profilin with unusual biochemical properties. J. Cell Sci. 114: 4293–4305. [DOI] [PubMed] [Google Scholar]
- Kozak M., 2007. Lessons (not) learned from mistakes about translation. Gene 403: 194–203. [DOI] [PubMed] [Google Scholar]
- Levine F., Yee J. K., Friedmann T., 1991. Efficient gene expression in mammalian cells from a dicistronic transcriptional unit in an improved retroviral vector. Gene 108: 167–174. [DOI] [PubMed] [Google Scholar]
- Liu X., Constantinescu S. N., Sun Y., Bogan J. S., Hirsch D., et al. , 2000. Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal. Biochem. 280: 20–28. [DOI] [PubMed] [Google Scholar]
- Lough T., Tourneur C., Masson J., Robaglia C., 1997. Expression of genes in transgenic plants from bicistronic transcriptional units. Plant Sci. 129: 91–99. [Google Scholar]
- Lumbreras V., Stevens D. R., Purton S., 1998. Efficient foreign gene expression in Chlamydomonas reinhardtii mediated by an endogenous intron. Plant J. 14: 441–447. [Google Scholar]
- Marshall W. F., 2008. Basal bodies: platforms for building cilia. Curr. Top. Dev. Biol. 85: 1–22. [DOI] [PubMed] [Google Scholar]
- Martínez-Salas E., 1999. Internal ribosome entry site biology and its use in expression vectors. Curr. Opin. Biotechnol. 10: 458–464. [DOI] [PubMed] [Google Scholar]
- Merchant S. S., Prochnik S. E., Vallon O., Harris E. H., Karpowicz S. J., et al. , 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironova L. N., Provorov N. A., Ter-Avanesyan M. D., Inge-Vechtomov S. G., Smirnov V. N., et al. , 1982. The effect of paromomycin on the expression of ribosomal suppressors in yeast. Curr. Genet. 5: 149–152. [DOI] [PubMed] [Google Scholar]
- Onishi M., Pringle J. R., Cross F. R., 2016. Evidence that an unconventional actin can provide essential F-actin function and that a surveillance system monitors F-actin integrity in Chlamydomonas. Genetics 202: 977–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski L. E., Dutcher S. K., Lo C. W., 2011. Cilia and models for studying structure and function. Proc. Am. Thorac. Soc. 8: 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J., Sonenberg N., 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. [DOI] [PubMed] [Google Scholar]
- Powell M. L., Brown T. D. K., Brierley I., 2008. Translational termination-re-initiation in viral systems. Biochem. Soc. Trans. 36: 717–722. [DOI] [PubMed] [Google Scholar]
- Rasala B. A., Lee P. A., Shen Z., Briggs S. P., Mendez M., et al. , 2012. Robust expression and secretion of Xylanase 1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. PLoS One 7: e43349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasala B. A., Barrera D. J., Ng J., Plucinak T. M., Rosenberg J. N., et al. , 2013. Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 74: 545–556. [DOI] [PubMed] [Google Scholar]
- Rasala B. A., Chao S.-S., Pier M., Barrera D. J., Mayfield S. P., 2014. Enhanced genetic tools for engineering multigene traits into green algae. PLoS One 9: e94028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroda M., Beck C. F., Vallon O., 2002. Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas. Plant J. 31: 445–455. [DOI] [PubMed] [Google Scholar]
- Skabkin M. A., Skabkina O. V., Hellen C. U. T., Pestova T. V., 2013. Reinitiation and other unconventional posttermination events during eukaryotic translation. Mol. Cell 51: 249–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taillon B. E., Adler S. A., Suhan J. P., Jarvik J. W., 1992. Mutational analysis of centrin: an EF-hand protein associated with three distinct contractile fibers in the basal body apparatus of Chlamydomonas. J. Cell Biol. 119: 1613–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin P., Yi L., Martin H., Atkinson H., Gilmartin P. M., 2000. Functional characterization of the EMCV IRES in plants. Plant J. 24: 583–589. [DOI] [PubMed] [Google Scholar]
- Wang L., Wessler S. R., 1998. Inefficient reinitiation is responsible for upstream open reading frame-mediated translational repression of the maize R gene. Plant Cell 10: 1733–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T., Iguchi H., Fukuzawa H., 2013. Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J. Biosci. Bioeng. 115: 691–694. [DOI] [PubMed] [Google Scholar]
- Yang W., Catalanotti C., D’Adamo S., Wittkopp T. M., Ingram-Smith C. J., et al. , 2014. Alternative acetate production pathways in Chlamydomonas reinhardtii during dark anoxia and the dominant role of chloroplasts in fermentative acetate production. Plant Cell 26: 4499–4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Patena W., Armbruster U., Gang S. S., Blum S. R., et al. , 2014. High-throughput genotyping of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell 26: 1398–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Strains, plasmids, and plasmid sequences are available upon request and will be deposited at the Chlamydomonas Resource Center. File S1 contains sequences of all DNA oligonucleotides used in this study.