Abstract
Tan spot and Septoria nodorum blotch (SNB) are important diseases of wheat caused by the necrotrophic fungi Pyrenophora tritici-repentis and Parastagonospora nodorum, respectively. The P. tritici-repentis necrotrophic effector (NE) Ptr ToxB causes tan spot when recognized by the Tsc2 gene. The NE ToxA is produced by both pathogens and has been associated with the development of both tan spot and SNB when recognized by the wheat Tsn1 gene. Most work to study these interactions has been conducted in common wheat, but little has been done in durum wheat. Here, quantitative trait loci (QTL) analysis of a segregating biparental population indicated that the Tsc2-Ptr ToxB interaction plays a prominent role in the development of tan spot in durum. However, analysis of two biparental populations indicated that the Tsn1-ToxA interaction was not associated with the development of tan spot, but was strongly associated with the development of SNB. Pa. nodorum expressed ToxA at high levels in infected Tsn1 plants, whereas ToxA expression in P. tritici-repentis was barely detectable, suggesting that the differences in disease levels associated with the Tsn1-ToxA interaction were due to differences in pathogen expression of ToxA. These and previous results together indicate that: (1) the effects of Tsn1-ToxA on tan spot in common wheat can range from nonsignificant to highly significant depending on the host genetic background; (2) Tsn1-ToxA is not a significant factor for tan spot development in durum wheat; and (3) Tsn1-ToxA plays a major role in SNB development in both common and durum wheat. Durum and common wheat breeders alike should strive to remove both Tsc2 and Tsn1 from their materials to achieve disease resistance.
Keywords: durum wheat, tan spot, Septoria nodorum, necrotrophic pathogen, disease resistance
Durum wheat [Triticum turgidum ssp. durum (Desf.) Husnot.], also known as pasta or macaroni wheat, is an allotetraploid (2n = 4x = 28, AABB genomes) of worldwide economic importance because it is used to make pasta and other semolina-based products. Durum wheat production is affected by numerous diseases. Among some of the most severe are the foliar diseases tan spot and SNB caused by the necrotrophic fungal pathogens P. tritici-repentis and Pa. nodorum, respectively. Both pathogens are members of the Pleosporales order of fungi and are known to produce NEs (Oliver et al. 2012; Faris et al. 2013 for reviews). When a specific NE is recognized by the corresponding host gene, a host “defense response” ensues, which leads to programmed cell death allowing these necrotrophs to penetrate, feed, and sporulate. The lack of NE recognition by the host leads to resistance. Therefore, these host–pathogen interactions operate in an inverse gene-for-gene manner (Wolpert et al. 2002; Friesen and Faris 2010; Oliver et al. 2012; Faris et al. 2013), and the dominant alleles of the host NE recognition genes are considered susceptibility genes.
P. tritici-repentis is known to produce at least three NEs including Ptr ToxA, Ptr ToxB, and Ptr ToxC, and these NEs are recognized by the host genes Tsn1, Tsc2, and Tsc1, which reside on wheat chromosome arms 5BL, 2BS, and 1AS, respectively (see Singh et al. 2010; Faris et al. 2013 for reviews). Of the three NEs, the molecular nature of Ptr ToxA and Ptr ToxB have been determined (Ciuffetti et al. 2010 for review) and both are small secreted proteins that induce necrosis and chlorosis in wheat lines harboring Tsn1 and Tsc2, respectively (Faris et al. 2013). Of the host genes, only Tsn1 has been cloned, and it has features resembling classic disease resistance genes include protein kinase, nucleotide binding, and leucine-rich repeat domains (Faris et al. 2010).
The Tsn1-Ptr ToxA, Tsc2-Ptr ToxB, and Tsc1-Ptr ToxC interactions have all been shown to play significant roles in the development of tan spot in common (hexaploid) wheat (T. aestivum L. ssp. aestivum, 2n = 6x = 42, AABBDD genomes) (Faris et al. 2013; Kariyawasam et al. 2016). These experiments were conducted by infiltrating leaves of wheat lines from segregating populations with cultures containing crude, partially purified, or purified cultures of the NEs and inoculating the same wheat lines with conidia produced by the fungus to evaluate the development of tan spot. Many studies showed statistically significant relationships between sensitivity to the NEs and susceptibility to P. tritici-repentis. For example, early work with Ptr ToxA showed strong correlations between sensitivity to culture filtrates containing Ptr ToxA and susceptibility to Ptr ToxA-producing isolates (Tomas and Bockus 1987; Lamari and Bernier 1989). Similarly, studies using Ptr ToxB and Ptr ToxC showed that these NEs were also strongly associated with tan spot caused by isolates that produced them (Effertz et al. 2002; Friesen and Faris 2004; Abeysekara et al. 2010). Therefore, the NEs were considered virulence factors, and it was assumed that sensitivity to an NE would lead to disease susceptibility (Anderson et al. 1999). This led to the notion that lines could be screened with NE-containing cultures to more or less predict their reaction to tan spot. However, more recent studies, particularly involving the Tsn1-Ptr ToxA interaction, indicated that NE sensitivity did not always define tan spot susceptibility, and the involvement of the Tsn1-Ptr ToxA interaction in the development of disease was dependent on the genetic background of the host (Friesen et al. 2003; Faris and Friesen 2005; Chu et al. 2008a; Faris et al. 2012; Kariyawasam et al. 2016).
The vast majority of studies involving tan spot have been conducted in common wheat and relatively few have been conducted in durum wheat. P. K. Singh et al. (2008) showed that two linked recessive genes on chromosome arm 3BL conferred resistance to race 3 (produces Ptr ToxC) and race 5 (produces Ptr ToxB) isolates in tetraploid wheat, indicating that the Tsc1-Ptr ToxC and Tsc2-Ptr ToxB interactions were not involved. In another tetraploid wheat mapping study, Chu et al. (2010a) evaluated a durum doubled haploid population that segregated for Ptr ToxA sensitivity with race 1 (produces Ptr ToxA and Ptr ToxC) and race 2 (produces Ptr ToxA) isolates, and found that neither the Tsn1-Ptr ToxA nor the Tsc1-Ptr ToxC interaction was relevant in the development of disease. Therefore, although the three host gene-NE interactions in the wheat-P. tritici-repentis system have been shown to play variable roles in disease development in hexaploid wheat, none of them have been shown to be associated with the development of tan spot in durum wheat thus far.
To date, nine interactions involving specific wheat genes and cognate NEs have been reported in the wheat-Pa. nodorum system, and all have been shown to play significant roles in the development of SNB (Liu et al. 2004a,b, 2006; Friesen et al. 2006, 2007, 2008, 2009, 2012; Abeysekara et al. 2009, 2012; Chu et al. 2010b; Zhang et al. 2011; Gao et al. 2015; Shi et al. 2015). One of these interactions is the Tsn1-SnToxA (hereafter, both Ptr ToxA and SnToxA will be referred to as ToxA) interaction. Friesen et al. (2006) showed that the ToxA gene was horizontally transferred from Pa. nodorum to P. tritici-repentis sometime prior to 1940. This event likely played a role in tan spot becoming an economically significant disease, and henceforth Tsn1 operated as a susceptibility gene for both tan spot and SNB. Liu et al. (2006) showed that the ToxA proteins derived from both P. tritici-repentis and Pa. nodorum functioned in the same way to elicit cell death when the proteins were infiltrated into wheat leaves. Numerous studies have shown that the Tsn1-ToxA interaction plays a major role in conferring susceptibility to Pa. nodorum in hexaploid wheat (Friesen et al. 2006, 2007, 2008, 2009, 2012; Chu et al. 2010b), and at least two studies have demonstrated the prominence of Tsn1 in conferring susceptibility in tetraploid wheat (Faris and Friesen 2009; Friesen et al. 2012). Therefore, unlike for tan spot, Tsn1 has been shown to be an important SNB susceptibility gene in both hexaploid and tetraploid wheat.
The primary objectives of this research were to determine the roles of the Tsn1-ToxA and Tsc2-Ptr ToxB interactions in a biparental population derived from two durum varieties. We also evaluated the role of the Tsn1-ToxA interaction in governing tan spot susceptibility in a second tetraploid wheat population, which was the same population used by Faris and Friesen (2009) to show that the interaction explained 95% of the variation in the development of SNB. Finally, we evaluated ToxA transcription in plants inoculated with Pa. nodorum or P. tritici-repentis to determine whether ToxA expression levels were correlated with the levels of disease caused by these two pathogens.
Materials and Methods
Plant materials
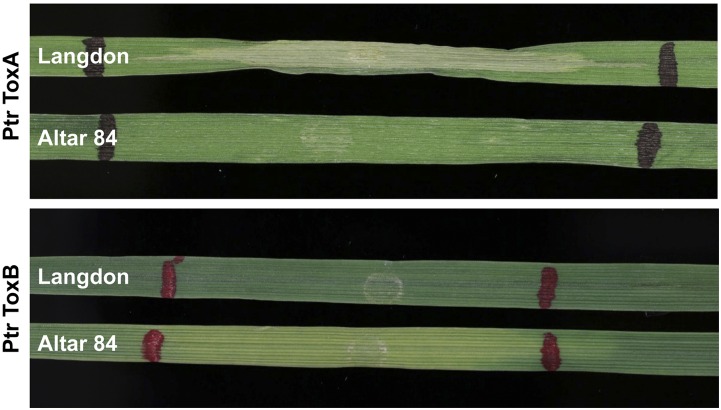
A segregating population of 127 recombinant inbred lines (RILs) was developed from a cross between the CYMMIT-bred durum variety “Altar 84” and the North Dakota durum variety “Langdon” (LDN). The RILs were developed by advancing the plants to the F7 generation by single seed descent (SSD). Preliminary experiments indicated that Altar 84 was sensitive to Ptr ToxB and insensitive to ToxA and Langdon was sensitive to ToxA and insensitive to Ptr ToxB, respectively (Figure 1). This population is hereafter referred to as the AL population.
Figure 1.
Leaves of Langdon and Altar 84 infiltrated with ToxA and Ptr ToxB. Langdon is sensitive to necrosis caused by ToxA and Altar 84 is insensitive, whereas Langdon is insensitive to the chlorosis caused by Ptr ToxB and Altar 84 is sensitive.
The second population was derived from a cross between LDN and LDN-DIC 5B, which is a genetic stock where a pair of 5B chromosomes derived from the T. turgidum ssp. dicoccoides accession Israel A was substituted for the native 5B chromosomes in the LDN background (Joppa 1993). This population, hereafter referred to as the LD5B population, consisted of 85 recombinant inbred chromosome lines (RICLs) and was used by Faris and Friesen (2009) to evaluate the role of the Tsn1-ToxA interaction in conferring SNB susceptibility. Here, it was used to evaluate the role of the Tsn1-ToxA interaction in conferring susceptibility to tan spot caused by the race 2 (ToxA producing) isolate 86–124.
All plants were grown in cones containing SB100 (Sun Gro Sunshine; Sun Gro Horticulture, Vancouver, BC) soil mix with 10–20 granules of Osmocote fertilizer (Scotts Company LLC, Marysville, OH) added to each cone. For disease evaluations and NE infiltrations, all plants were grown in the greenhouse at an average temperature of 21° with a 16 hr photoperiod.
NE production and infiltration assays
ToxA and Ptr ToxB were expressed in the yeast strain Pichia pastoris X33 and cultured as described for the NE SnTox3 in Liu et al. (2009). Harvested cultures were used to directly infiltrate the second leaf of wheat plants at the two-leaf stage. Infiltrations were conducted using a 1 ml needleless syringe on the secondary leaf until 2–3 cm of leaf was infiltrated. The boundaries of the infiltrated region were marked using a nontoxic permanent marker. The infiltrated plants were then placed in a growth chamber at 21° with a 16 hr photoperiod. Plants were evaluated 4 d after infiltration and scored as sensitive or insensitive based on the presence or absence of chlorosis for Ptr ToxB or necrosis for ToxA. The entire AL population and parents were infiltrated twice with ToxA and three times with Ptr ToxB. The infiltration scores, i.e., insensitive vs. sensitive, were converted to genotypic scores for placement of the Tsn1 and Tsc2 loci on the genetic linkage maps relative to the molecular markers. For the LD5B population, the reaction of each line to ToxA and the placement of the Tsn1 locus on the genetic linkage map were previously determined (Faris and Friesen 2009).
Disease evaluations
For P. tritici-repentis disease evaluation, the AL population and parents were screened with race 2 isolates 86–124 and L13-35, which produce ToxA, and the race 5 isolate DW5 which produces Ptr ToxB. The LD5B population was screened with 86–124. Isolates were grown on V8-potato dextrose agar (Difco PDA; Becton, Dickinson and Company, Sparks, MD) plates for 5–7 d in the dark, and inoculum was prepared as described in Lamari and Bernier (1989) and Ali et al. (2010). Parents and the RIL population were planted in a completely randomized design consisting of three replicates separated by time for conidial inoculations. Each replicate consisted of a single cone per line with three plants per cone placed in racks of 98 cones. The tan spot-susceptible wheat variety “Jerry” was planted in the borders of each rack to reduce edge effects. Plants were inoculated until runoff at the two- to three-leaf stage with 3000 spores per ml and two drops of Tween 20 (polyoxyethylene sorbitan monolaurate; J.T. Baker Chemical Co., Phillipsburg, NJ) per 100 ml of inoculum. Inoculated plants were placed in a mist chamber with 100% relative humidity at 21° for 24 hr, and then moved to a growth chamber for 6 d of incubation at 21° under a 12 hr photoperiod. Inoculated plants were rated using a 1–5 lesion type scale (Lamari and Bernier 1989) at 7 d postinoculation, where one is resistant and five is susceptible.
For Pa. nodorum disease evaluations, the AL population was screened with Pa. nodorum isolate Sn2000, which is known to produce ToxA (Friesen et al. 2006). Inoculum production and inoculation procedures were done as described in Liu et al. (2004a). After inoculation, plants were placed in a mist chamber with 100% relative humidity at 21° for 24 hr, and then moved to a growth chamber at 21° with a 12 hr photoperiod. Disease evaluation was carried out at 7 d after inoculation by scoring lesions on the second leaf using the 0–5 scale described by Liu et al. (2004).
Marker genotyping
The AL population and parents were genotyped using the iSelect array containing 9000 wheat single nucleotide polymorphism (SNP) markers (Cavanagh et al. 2013) as described in Faris et al. (2014). Simple sequence repeat (SSR) markers were also used to genotype the AL population and were selected from the following libraries: MAG (Xue et al. 2008), WMS (gwm) (Röder et al. 1998), WMC (Somers et al. 2004), HBG (Torada et al. 2006), CFD (Sourdille et al. 2004), and BARC (Song et al. 2005). SSR primer sets were used to amplify the parental DNA using polymerase chain reaction (PCR) conditions as outlined in Lu and Faris (2006). PCR amplifications were performed in 10 µl reactions consisting of 100 ng of DNA template, 1.5 mM MgCl2, 0.125 mM dNTPs, 4 pmol of primers, and 1 unit of Taq DNA polymerase. The PCR conditions were 94° for 4 min, followed by 35 cycles of 94° for 30 sec, the appropriate annealing temperature for 30 sec, and 72° for 1 min. The annealing temperature of each primer was obtained from the Graingenes website (http://wheat.pw.usda.gov/GG2/index.shtm). Fragments were electrophoresed on 6% polyacrylamide gels which were made using 46 ml of H2O, 6 ml 10 × TBE (Tris-borate-EDTA), 9 ml of 40% acrylamide/bis-acrylamide, 40 µl tetramethylethylenediamine (TEMED), and 350 µl of 10% ammonium persulfate. Gels were stained with GelRed (Biotium, Inc.) for 10 min and then visualized with a Typhoon 9410 variable mode imager (GE Healthcare, Waukesha, WI). Markers that revealed polymorphisms between the parents were then used to genotype the AL population.
In addition to the SSR and SNP markers, a cleaved amplified polymorphic sequence (CAPS) marker (Xfcp667) based on the genomic sequence of the Snn1 gene (Shi et al. 2016) was used to map the Snn1 locus. A fragment of the Snn1 gene was PCR-amplified as described for the SSR markers above using an annealing temperature of 65° and the primers FCP667F: 5ʹ-TGCGTCGATAGGAGTG-3ʹ and FCP667R: 5ʹ-ATGGCGTAGGAGCACGGGTA-3ʹ. The 898 bp amplicon was then digested with the restriction enzyme HpyCH4IV, which cleaves the Altar 84 fragment, but not the LDN fragment, thus revealing a codominant polymorphism. The digested amplicons were electrophoresed on 2% agarose gels, stained with ethidium bromide, and photographed.
Linkage, QTL, and statistical analysis
Linkage analysis was conducted using the computer program MapDisto 1.8.1 (Lorieux 2012) to generate linkage maps. First, marker grouping was done by using the command “find groups” with a logarithm of odds (LOD) > 3.0 and an Rmax value = 30.0. The “order sequence,” “check inversions,” “ripple order,” and “drop locus” commands were then used to determine the best order for each group. The Kosambi mapping function (Kosambi 1944) was used to calculate linkage distances.
In the AL population, multiple interval mapping (MIM) was used to determine the effects of the Tsn1 and Tsc2 loci in causing disease and to identify additional QTL associated with disease using the computer program QGene v.4.3 (Joehanes and Nelson 2008). An LOD of 3.6 was declared as the threshold for QTL significance based on permutation tests of 1000 iterations for each trait. The homogeneity of variances among the three replicates was determined by Bartlett’s χ2 test using SAS program version 9.4 (SAS Institute Inc., Cary, NC). Mean separation of the genotypic means were determined by Fisher’s protected LSD at an α level of 0.01. For the LD5B population, the critical LOD threshold of 1.8 declared in Faris and Friesen (2009) was used for determining significant associations for tan spot caused by 86–124. Because the LD5B population segregates for only one linkage group (5B), QTL analysis was conducted using the simple interval mapping (SIM) function in QGene v.4.3 (Joehanes and Nelson 2008).
Gene expression analysis
Plants of LDN and LDN-DIC 5B were grown and inoculated with water, P. tritici-repentis isolate 86–124, or Pa. nodorum isolate Sn2000 as described above. The youngest fully expanded leaf at the time of inoculation was harvested for RNA extraction. Samples for each treatment of both genotypes were collected at 0, 6, 24, 48, 72, and 96 hr after inoculation and immediately frozen in liquid nitrogen. Total RNA was extracted using the RNeasy Plant Mini Kit (QIAGEN, Hilden, Germany), and first-strand cDNA was synthesized from 1 µg of total RNA using Taqman Reverse Transcription Reagents including an oligo d(T)16 primer (Applied Biosystems, Foster City, CA). Relative quantitative (RQ)-PCR was performed to evaluate ToxA gene expression using primers ToxA.RT.F3 (5ʹ-AACGCCAATACAGTGCGAGT-3ʹ) and ToxA.cod.1R (5ʹ-GCTGCATTCTCCAATTTTCACG-3ʹ) in all treatments and sampled time points. Expression of the ToxA gene was compared to the expression of the endogenous wheat ubiquitin gene as described in Faris et al. (2010) using primers Ta.Ubiquitin.F: 5ʹ-GCACCTTGGCGGACTACAACATTC-3ʹ and Ta.Ubiquitin.R: 5ʹ-GACACCGAAGACGAGACTTGTGAACC-3ʹ. All RQ-PCR experiments were conducted using a 7500 Real-Time PCR System (Applied Biosystems). Each experiment was conducted using three biological replicates each consisting of a single inoculated leaf, and at least three technical replicates per biological replicate were performed. The 10 μl PCR reactions contained 1 × SYBR PCR MasterMix (Applied Biosystems), 0.25 μM each primer, and 5 μl of 10-fold diluted cDNA. The thermocycler procedure was as follows: 10 min of preincubation at 95°, followed by 40 cycles for 15 sec at 95° and for 1 min at 60°. Efficiencies of the different primer combinations were evaluated using serial dilutions of cDNA (1:5, 1:10, 1:20, and 1:40) and only primers with efficiencies higher than 95% were used for the RQ-PCR. The expression level of the Sn2000-inoculated 48 hr sample was set at 1 as a calibration point. Threshold cycles of the ToxA gene and the endogenous ubiquitin gene were used to calculate the relative expression levels using the 2−ΔΔCT method.
Data availability
Parents and mapping populations are available upon request. Mapping data for the LD5B and the AL populations is also available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Marker analysis and linkage map construction
The parental lines of the AL population were screened with 250 SSR primer pairs. Of these, 119 (47.6%) revealed polymorphisms between the parents and were used to genotype the AL population. The 9K SNP array yielded a total of 833 polymorphic SNP markers. One CAPS marker (Xfcp667) that was developed based on the Snn1 gene sequence on chromosome arm 1BS (Shi et al. 2016) was added to the marker set along with the two phenotypic markers Tsn1 and Tsc2 (see below). Therefore, the initial marker dataset consisted of a total of 955 markers. After initial linkage analysis, a total of 111 markers, including 19 SSRs and 92 SNPs, were eliminated from the dataset because they were unlinked, leaving a total of 844 markers in the dataset consisting of 100 SSRs, 741 SNPs, one CAPS, and two phenotypic (Tsn1 and Tsc2) markers.
These markers were assembled into 14 linkage groups that corresponded to the 14 durum wheat chromosomes (Supplemental Material, Table S1) and spanned a total genetic distance of 2207.15 cM with an average marker density of one marker per 2.6 cM (Table 1). The A-genome chromosomes had 414 markers and spanned 1128.22 cM with an average density of one marker per 2.7 cM, whereas the B-genome chromosomes had a total of 430 markers spanning 1078.93 cM for an average marker density of one marker per 2.5 cM (Table 1). Chromosome 7A was the longest linkage group (262.41 cM) and chromosome 4B was the shortest (107.95 cM). The number of markers per chromosome ranged from 17 (4B) to 104 (5B) (Table 1 and Table S1). Chromosome 6B had the highest marker density at one marker per 1.2 cM, whereas chromosome 4B had the lowest with one marker every 6.3 cM (Table 1). Of the 844 markers, 208 (24.6%), had segregation ratios that deviated significantly (P < 0.05) from the expected 1:1 ratio. These distorted markers were located on 10 chromosomes (1A, 2A, 2B, 3B, 4B, 5A, 5B, 6A, 6B, and 7B).
Table 1. Summary of markers mapped in each chromosome/genome in the Altar 84 × Langdon population.
| Chromosome | Markers | Phenotype | Total | Length (cM) | Marker Density (cM/Marker) | Markers with Distorted Ratios | ||
|---|---|---|---|---|---|---|---|---|
| SSR | CAPS | SNP | ||||||
| 1A | 4 | − | 65 | − | 69 | 144.49 | 2.1 | 0 |
| 1B | 20 | 1 | 53 | − | 74 | 159.84 | 2.2 | 12 |
| 2A | 4 | − | 57 | − | 61 | 178.68 | 2.9 | 2 |
| 2B | 9 | − | 36 | 1 | 46 | 196.73 | 4.2 | 22 |
| 3A | 6 | − | 47 | − | 53 | 159.97 | 3.0 | 0 |
| 3B | 3 | − | 31 | − | 34 | 167.19 | 4.9 | 9 |
| 4A | 5 | − | 50 | − | 55 | 128.57 | 2.3 | 0 |
| 4B | 2 | − | 15 | − | 17 | 107.95 | 6.3 | 8 |
| 5A | 6 | − | 52 | − | 58 | 130.76 | 2.3 | 26 |
| 5B | 23 | − | 80 | 1 | 104 | 211.72 | 2.0 | 71 |
| 6A | 3 | − | 43 | − | 46 | 123.34 | 2.7 | 2 |
| 6B | 7 | − | 88 | − | 95 | 111.10 | 1.2 | 52 |
| 7A | 6 | − | 66 | − | 72 | 262.41 | 3.6 | 0 |
| 7B | 2 | − | 58 | − | 60 | 124.40 | 2.1 | 4 |
| A genome | 34 | − | 380 | − | 414 | 1128.22 | 2.7 | 30 |
| B genome | 66 | 1 | 361 | 2 | 430 | 1078.93 | 2.5 | 178 |
| Total | 100 | 1 | 741 | 2 | 844 | 2207.15 | 2.6 | 208 |
SSR, simple sequence repeat; CAPS, cleaved amplified polymorphic sequence; SNP, single nucleotide polymorphism.
Genetic analysis of ToxA and Ptr ToxB sensitivity in the AL population
Altar 84 was insensitive and Langdon was sensitive to ToxA (Figure 1). The AL population segregated in a ratio of 59 insensitive:68 sensitive for reaction to ToxA and fitted the expected 1:1 ratio for a single host gene conferring sensitivity to ToxA (χ2df = 1 = 0.65, P = 0.42). Conversion of the ToxA reaction scores to genotypic scores allowed us to map the Tsn1 locus, which was located on the long arm of chromosome 5B as expected and flanked by SNP markers Xiwa7024 and Xiwa6915 at distances of 0.4 and 0.8 cM, respectively (Figure 2 and Table S1).
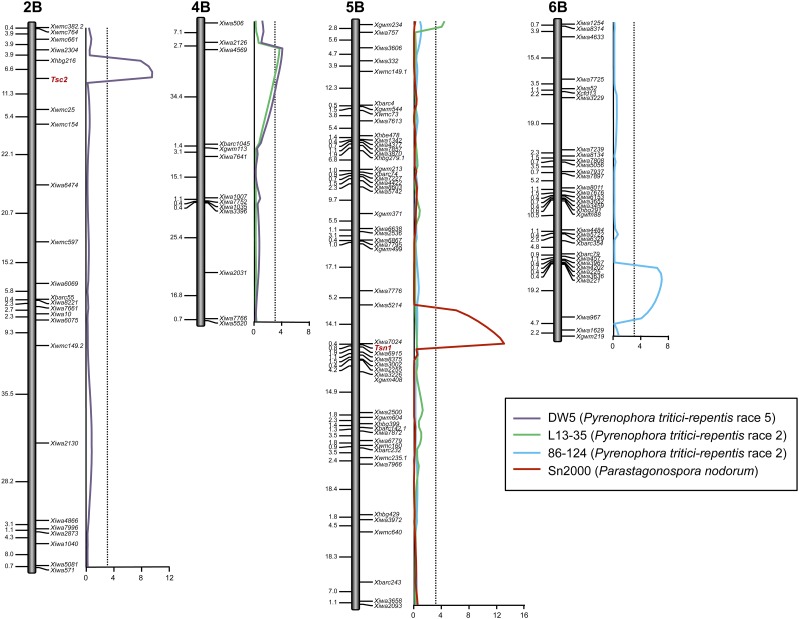
Figure 2.
Composite interval regression maps of QTL associated with the P. tritici-repentis race 5 isolate DW5, P. tritici-repentis race 2 isolates L13-25 and 86-124, and the Pa. nodorum isolate Sn2000 in the Altar 84 × Langdon recombinant inbred population. Markers are indicated to the right side of the genetic maps and cM distances are shown along the left side. The critical LOD threshold is indicated by the dotted lines and the LOD scale is indicated at the bottom along the x-axis. LOD, logarithm of odds; QTL, quantitative trait loci.
For reaction to Ptr ToxB, Altar 84 was sensitive and Langdon was insensitive (Figure 1). The AL population segregated in a ratio of 72 insensitive:55 sensitive for reaction to Ptr ToxB and fitted the expected 1:1 ratio for a single host gene conferring sensitivity to Ptr ToxB (χ2df = 1 = 3.28, P = 0.07). The reactions to Ptr ToxB were converted to genotypic scores and analyzed along with the molecular marker data. Linkage analysis showed that the Tsc2 locus mapped to the short arm of chromosome 2B flanked by SSR markers Xhbg216 and Xwmc25 at distances of 6.6 and 11.3 cM, respectively (Figure 2 and Table S1).
AL population reaction to P. tritici-repentis
Altar 84, LDN, and the AL population were screened with the ToxA-producing race 2 isolates 86–124 and L13-35, and the race 5 Ptr ToxB-producing isolate DW5. Bartlett’s Chi-squared test for homogeneity indicated that the variances among replicates for each isolate were not significantly different (86–124: χ2 df = 2 = 2.08, P = 0.35; L13-35: χ2df = 2 = 1.39, P = 0.50; DW5: χ2df = 2 = 6.01, P = 0.05); therefore, the means from the three replicates for each isolate were used for further analysis.
Altar 84 and LDN were considered resistant and moderately susceptible to 86–124 with average disease reaction types of 1.17 and 3.50, respectively (Figure 3, Figure 4, and Table 2). The average disease reaction type for the AL population was 2.31 and reaction types ranged from 1.00 to 4.16 (Figure 3 and Table 2). The mean reaction types of ToxA-insensitive and -sensitive AL lines were 2.21 and 2.31, which were not significantly different at the 0.01 level of probability (Table 2), indicating that the Tsn1-ToxA interaction was not significant in the development of tan spot caused by 86–124.
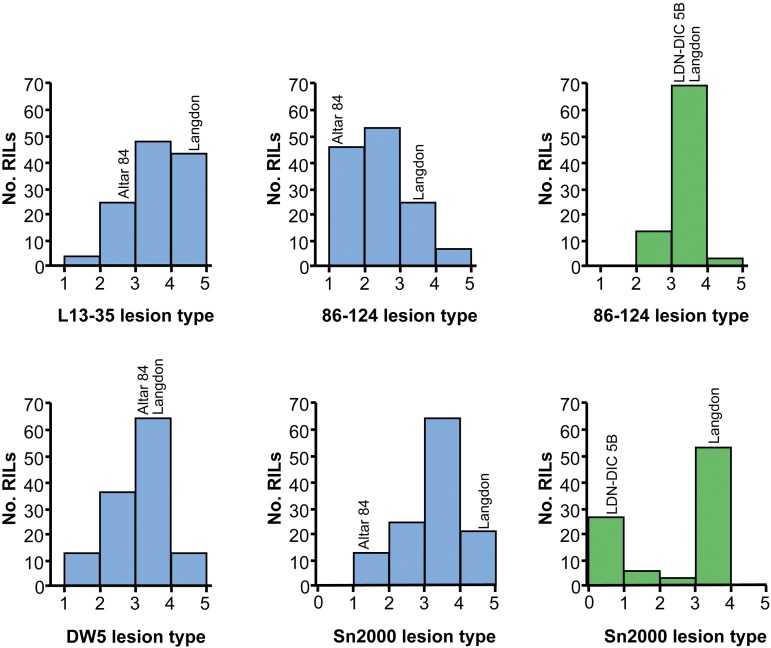
Figure 3.
Histograms of average lesion type reactions of the Altar 84 × Langdon recombinant inbred population to P. tritici-repentis race 2 isolates L13-35 and 86-124, race 5 isolate DW5, and the Pa. nodorum isolate Sn2000 (blue bars), and of average lesion type reactions of the Langdon × LDN-DIC 5B recombinant inbred chromosome line population to P. tritici-repentis race 2 isolate 86–124 and the Pa. nodorum isolate Sn2000 (Faris and Friesen 2009) (green bars). RILs, recombinant inbred lines.
Figure 4.
Leaves of Altar 84 and Langdon inoculated with P. tritici-repentis isolates 86–124 (race 2) and DW5 (race 5), and with Pa. nodorum isolate Sn2000.
Table 2. Average reaction types of Altar 84, Langdon, and the Altar 84 × Langdon population of recombinant inbred lines to tan spot and Septoria nodorum blotch.
| Isolatea | Altar 84 | Langdon | AL Population Range | AL Population Average | ToxA Sensitive Lines | ToxA Insensitive Lines | Ptr ToxB Sensitive Lines | Ptr ToxB Insensitive Lines | Difference Between Sensitive and Insensitive Lines |
|---|---|---|---|---|---|---|---|---|---|
| 86-124 (race 2) | 1.17 | 3.50 | 1.00–4.16 | 2.31 | 2.39 | 2.21 | − | − | 0.18 |
| L13-35 (race 2) | 2.75 | 4.67 | 1.33–4.83 | 3.52 | 3.64 | 3.36 | − | − | 0.28 |
| DW5 (race 5) | 3.16 | 3.50 | 1.25–4.50 | 3.02 | − | − | 2.29 | 1.33 | 0.96* |
| Sn2000 | 1.33 | 4.83 | 1.16–4.80 | 3.18 | 3.00 | 2.00 | − | − | 1.00* |
AL, Altar 84 × Langdon. *P < 0.01.
Reaction types caused by the P. tritici-repentis isolates (86–124, L13-35, and DW5) were scored using the 1–5 scale for tan spot described by Lamari and Bernier (1989), and reaction types caused by the Pa. nodorum isolate (Sn2000) were scored using the 0–5 scale for Septoria nodorum blotch described by Liu et al. (2004).
For the other P. tritici-repentis race 2 isolate, L13-35, Altar 84, and LDN were moderately susceptible and susceptible with average reaction types of 2.75 and 4.67, respectively (Figure 3 and Table 2). The AL population reaction types averaged 3.52 and ranged from 1.33 to 4.83 indicating some transgressive segregation for resistance (Figure 3 and Table 2). Average L13-35 reaction types for ToxA-sensitive and -insensitive AL lines were 3.64 and 3.36, respectively, which were not significantly different at the 0.01 level of probability (Table 2), again suggesting that the Tsn1-ToxA interaction did not play a significant role in the development of tan spot in the AL population.
For reaction to the race 5 Ptr ToxB-producing isolate DW5, Altar 84 and LDN were both moderately susceptible with average disease reactions of 3.16 and 3.50, respectively (Figure 3, Figure 4, and Table 2). The average disease reaction type for the AL population was 3.02, and reaction types ranged from 1.25 to 4.50 (Figure 3 and Table 2), indicating strong transgressive segregation and suggesting that more than one gene was involved in conditioning resistance. The mean reaction types of Ptr ToxB-sensitive and -insensitive AL lines to DW5 were 2.29 and 1.33, respectively, which were significantly different at the 0.01 level of probability (Table 2). This result indicates that the Tsc2-Ptr ToxB interaction played a significant role in the development of tan spot caused by DW5.
AL population reaction to Pa. nodorum
As with the P. tritici-repentis isolates, Bartlett’s Chi-squared test for homogeneity of variances among Pa. nodorum isolate Sn2000 replicates was not significant (χ2df = 2 = 0.78, P = 0.70), and therefore the means of the three reps were used for further analysis. Altar 84 was resistant to Sn2000 and LDN was highly susceptible with average disease reaction types of 1.33 and 4.83, respectively (Figure 3 and Table 2). The average disease reaction type for the AL population was 3.18, and reaction types ranged from 1.16 to 4.80 (Figure 3 and Table 2). The mean reaction types of ToxA-insensitive and -sensitive AL lines were 2.00 and 3.00, respectively (Table 2). These means were significantly different at the 0.01 level of probability (Table 2), indicating that the Tsn1-ToxA interaction played a significant role in the development of SNB caused by Sn2000.
QTL analysis of the AL population for reaction to P. tritici-repentis and Pa. nodorum
For the race 2 isolate 86–124, only one QTL was significantly associated with resistance (Figure 2 and Table 3). This QTL was on the long arm of 6B between markers Xiwa5148 and Xiwa967, which were at positions 84.4 and 103.4 cM, respectively. This QTL, designated QTs.fcu-6B, had an LOD of 6.9 and explained 22% of the phenotypic variation (Table 3). The resistance effects of QTs.fcu-6B were contributed by Altar 84 (Table 3).
Table 3. Composite interval mapping analysis of QTL associated with resistance to tan spot caused by P. tritici-repentis races 2 and 5 and resistance to SNB caused by Sn2000 in the Altar 84 × Langdon (AL) population.
| QTL | Marker Interval | Marker Position | 86-124 (P. tritici-repentis Race 2) | L13-35 (P. tritici-repentis Race 2) | DW5 (P. tritici-repentis Race 5) | Sn2000 (Pa. nodorum) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LOD | R2 | Add. | LOD | R2 | Add. | LOD | R2 | Add. | LOD | R2 | Add. | |||
| QTs.fcu-2B | Tsc2 | 18.7 | – | – | – | – | – | – | 12.00 | 0.26 | 0.41 | – | – | – |
| QTs.fcu-4B | Xiwa2126-Xgwm113 | 7.1-38.9 | – | – | – | 4.00 | 0.11 | −0.37 | 8.60 | 0.12 | −0.56 | – | – | – |
| QTs.fcu-5B | Xgwm234 | 0.0 | – | – | – | 4.20 | 0.12 | −0.31 | – | – | – | – | – | – |
| QTs.fcu-6B | Xiwa5148-Xiwa967 | 84.4–103.4 | 6.90 | 0.22 | −0.51 | – | – | – | – | – | – | – | – | – |
| QSnb.fcu-5B | Tsn1 | 110.2 | – | – | – | – | – | – | – | – | – | 13.00 | 0.38 | −0.59 |
A negative value for “Add.” indicates resistance effects derived from Altar 84. A dash indicates that the marker was not significantly associated with resistance. QTL, quantitative trait loci; LOD, logarithm of odds; Add., the additive effects of the QTL.
Two significant QTL were associated with the other P. tritici-repentis race 2 isolate, L13-35 (Figure 2). QTL on the short arms of chromosomes 4B and 5B, designated QTs.fcu-4B and QTs.fcu-5B, had LOD values of 4.0 and 4.2, and explained 11 and 12% of the phenotypic variation, respectively (Table 3). The resistance effects at both of these QTL were contributed to by Altar 84. The Tsn1 locus on 5BL was not significantly associated with resistance to either of the two race 2 isolates 86–124 and L13-35 (Figure 2), which both produce ToxA. In single factor regression, Tsn1 had LOD values of only 0.35 and 0.40 for 86–124 and L13-35, respectively.
For the race 5 isolate DW5, two significant QTL were identified. The QTL were located on chromosome arms 2BS and 4BL and designated QTs.fcu-2B and QTs.fcu-4B, respectively (Figure 5 and Table 3). The peak of QTs.fcu-2B was defined by the Tsc2 locus on chromosome 2BS. It had an LOD of 12.0 and explained 26% of the disease variation (Figure 2 and Table 3). Because this QTL represented the Tsc2-Ptr ToxB interaction for which Altar 84 contributed the Ptr ToxB-sensitive allele, resistance to this QTL was contributed by LDN (Table 3).
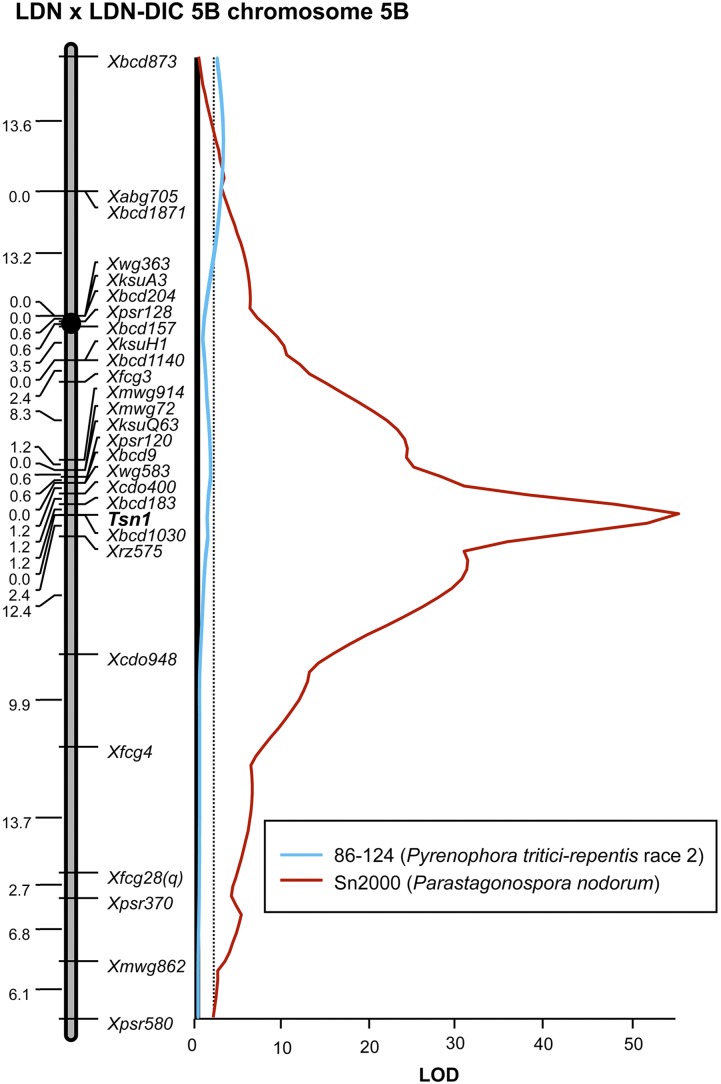
Figure 5.
Interval regression maps of QTL associated with reaction to the P. tritici-repentis race 2 isolate 86–124 and the Pa. nodorum isolate Sn2000 in the Langdon × LDN-DIC 5B recombinant inbred chromosome line population. Markers are indicated to the right side of the genetic maps and cM distances are shown along left side. The critical LOD threshold is indicated by the dotted lines and the LOD scale is indicated at the bottom along the x-axis. The linkage map and Sn2000 regression analysis were first reported in Faris and Friesen (2009). LOD, logarithm of odds; QTL, quantitative trait loci.
The QTs.fcu-4B QTL associated with reaction to tan spot caused by DW5 was that same QTL found to be associated with the P. tritici-repentis race 2 isolate L13-35 (Figure 2 and Table 3). In association with disease caused by DW5, QTs.fcu-4B had an LOD of 8.6 and explained 12% of the variation with resistance effects contributed by Altar 84 (Table 3). The QTL peaked between markers Xiwa2126 and Xgwm113, which were located at positions 7.1 and 38.9 cM, respectively (Figure 2 and Table 3).
A single QTL, designated QSnb.fcu-5B, associated with SNB caused by the ToxA-producing Pa. nodorum isolate Sn2000 was identified on the long arm of 5B (Figure 2). The Tsn1 locus, with an LOD of 13.0, defined the peak of QSnb.fcu-5B and explained 38% of the phenotypic variation (Figure 2 and Table 3). Because this QTL is due to the effects of the Tsn1-ToxA interaction, it indicates that this interaction played a significant role in the development of SNB caused by isolate Sn2000.
LD5B population reaction to P. tritici-repentis and Pa. nodorum
Bartlett’s Chi-squared test for homogeneity among the three replicates of the LD5B population inoculated with P. tritici-repentis isolate 86–124 indicated they were homogeneous (χ2df = 2 = 0.12, P = 0.94), and therefore the means of the three replicates were calculated and used for further analysis. LDN and LDN-DIC 5B had mean reaction types of 3.60 and 3.00, respectively, to 86–124 (Table 4). The LD5B population reaction types ranged from 2.25 to 4.00 with an average of 3.16 (Figure 3 and Table 4). ToxA-sensitive and -insensitive lines had average reaction types of 3.16 and 3.04 in response to 86–124, which were not significantly different (Table 4), indicating that the Tsn1-ToxA interaction played no role in the development of disease caused by P. tritici-repentis race 2 isolate 86–124.
Table 4. Average reaction types of Langdon, LDN-DIC 5B, and the Langdon × LDN-DIC 5B (LD5B) population of recombinant inbred chromosome lines to tan spot and Septoria nodorum blotch.
| Isolate | Langdon | LDN-DIC 5B | LD5B Population | LD5B Population Average | ToxA Sensitive Lines | ToxA Insensitive Lines | Difference Between Sensitive and Insensitive Lines |
|---|---|---|---|---|---|---|---|
| 86-124 | 3.60 | 3.00 | 2.25–4.00 | 3.16 | 3.16 | 3.04 | 0.12 |
| Sn2000a | 3.50 | 0.83 | 0.33–3.83 | 2.40 | 3.24 | 0.78 | 2.46* |
P < 0.01.
The data and analysis of Sn2000 in the LD5B population was taken from Faris and Friesen (2009).
As mentioned above, we previously reported the analysis of the LD5B population for reaction to Pa. nodorum isolate Sn2000 (Faris and Friesen 2009). In that study, we showed that LDN and LDN-DIC 5B had average reaction types of 3.50 and 0.83, respectively. The LD5B population had an overall average reaction type of 2.40 and ranged from 0.33 to 3.83 (Figure 3 and Table 4) (Faris and Friesen 2009). Also, ToxA-sensitive lines were highly susceptible to Sn2000 with an average reaction type of 3.24, whereas ToxA-insensitive lines were highly resistant with an average reaction type of 0.78, indicating that the Tsn1-ToxA interaction was a major factor in determining susceptibility to the Pa. nodorum isolate Sn2000 (Faris and Friesen 2009).
QTL analysis of the LD5B population
A QTL with an LOD value of 3.8, explaining 19% of the phenotypic variation, was found to be associated with tan spot caused by the P. tritici-repentis race 2 isolate 86–124 (Figure 5 and Table 5). This QTL was located in the distal region of chromosome arm 5BS and may be the same as QTs.fcu-5B, which was associated with the P. tritici-repentis race 2 isolate L13-35 in the AL population. As with QTs.fcu-5B in the AL population, the susceptible allele at this locus was contributed to by LDN (Table 5). The Tsn1 locus was not significantly associated with tan spot caused by 86–124 in this population. On the contrary, we previously showed that the Tsn1 locus was a major factor in conferring susceptibility to SNB caused by the Pa. nodorum isolate Sn2000 (Faris and Friesen 2009). In that research, the Tsn1 locus had an LOD of 54.0 and explained 95% of the phenotypic variation (Figure 5 and Table 5) (Faris and Friesen 2009).
Table 5. Simple interval mapping analysis of QTL associated with resistance to tan spot caused by P. tritici-repentis race 2 isolate 86–124 and to SNB caused by the Pa. nodorum isolate Sn2000 in the Langdon × LDN-DIC 5B (LD5B) population.
| QTL | Marker Interval | Marker Position | 86-124 (P. tritici-repentis Race 2) | Sn2000 (Pa. nodorum) | ||||
|---|---|---|---|---|---|---|---|---|
| LOD | R2 | Add. | LOD | R2 | Add. | |||
| QTs.fcu-5B | Xbcd873-Xabg705 | 0–13.6 | 3.80 | 0.19 | −0.18 | – | – | – |
| QSnb.fcu-5B | Tsn1 | 48.2 | – | – | – | 54.0 | 0.95 | −1.30 |
A dash indicates that the marker was not significantly associated with resistance. A negative value indicates resistance effects derived from LDN-DIC 5B. QTL, quantitative trait loci; LOD, logarithm of odds; Add., the additive effects of the QTL.
Expression analysis of ToxA in P. tritici-repentis- and Pa. nodorum-inoculated plants
The susceptible/ToxA-sensitive line LDN and the resistant/ToxA-insensitive line LDN-DIC 5B were inoculated with water, the P. tritici-repentis race 2 isolate 86–124, and the Pa. nodorum isolate Sn2000, and samples were collected for RNA extraction at 0, 6, 24, 48, 72, and 96 hr after inoculation. No transcriptional expression of ToxA was observed for any of the treatments at 0 and 6 hr postinoculation (Figure 6). Expression of ToxA in LDN inoculated with Sn2000 peaked at 24 hr postinoculation and from there declined through the 96 hr time point. Expression of ToxA in 86-124-treated LDN plants also peaked at 24 hr postinoculation and was detectable at the 48 and 96 hr time points as well, but the relative amounts were less than a tenth of the amount of ToxA transcription observed in the Sn2000-inoculated LDN plants (Figure 6).
Figure 6.
Relative quantitative transcriptional expression of the ToxA gene in the susceptible line Langdon inoculated with water, the P. tritici-repentis race 2 isolate 86–124, and the Pa. nodorum isolate Sn2000, and in the resistant line LDN-DIC 5B inoculated with the Pa. nodorum isolate Sn2000. The expression of the ToxA gene in each treatment was normalized to the expression of the wheat ubiquitin gene. For LDN-DIC 5B inoculated with Sn2000, ToxA transcripts were detected only at 72 and 96 hr postinoculation, and at levels that were barely visible on the graph compared to ToxA expression in Langdon inoculated with Sn2000. Expression of ToxA in the water-inoculated Langdon and LDN-DIC 5B plants and in LDN-DIC 5B plants inoculated with 86–124 was not detected, and therefore no bars representing these treatments are included on the graph. Expression of ToxA in the Langdon plants inoculated with Sn2000 was significantly greater (P < 0.01) than in 86–124 and water-inoculated Langdon plants for the 24, 48, 72, and 96 hr time points.
No expression of ToxA in 86–124-inoculated LDN-DIC 5B plants was detected. Small amounts of ToxA transcript were detected at the 72 and 96 hr time points in the Sn2000-inoculated LDN-DIC 5B plants, but the amounts were about a hundredth of the amount of ToxA transcribed in the Sn2000-inoculated LDN plants (Figure 6).
Discussion
Role of Tsc2-Ptr ToxB in tan spot susceptibility in durum wheat
One objective of this research was to determine the role of the Tsc2-Ptr ToxB interaction in conferring tan spot susceptibility in a tetraploid durum wheat population. Friesen and Faris (2004) first mapped the Ptr ToxB sensitivity gene Tsc2 on chromosome arm 2BS using the ITMI population, which was developed from a cross between the synthetic hexaploid wheat W-7984 and the hard red spring wheat variety Opata 85 (PI 591776). W-7984 was synthesized from crossing Altar 84 with the Aegilops tauschii accession CI 18 (WPI 219). Because Altar 84 donated the B-genome chromosomes to W-7984, it also donated the Tsc2 locus, thus rendering W-7984 sensitive to Ptr ToxB. QTL analysis revealed a major QTL on 2BS explaining 69% of the variation and corresponding to the Tsc2 locus, thus indicating that the Tsc2-ToxB interaction was significantly associated with development of the disease in the ITMI population. Other studies have also demonstrated that the Tsc2 gene is a major susceptibility factor in hexaploid wheat (Abeysekara et al. 2010; Singh et al. 2010).
Although the durum variety Altar 84 contributed the dominant Tsc2 allele for Ptr ToxB sensitivity to the synthetic wheat W-7984, the evaluation of the effects of the Tsc2-Ptr ToxB interaction were conducted in a hexaploid wheat background (Friesen and Faris 2004). Therefore, in this study, we chose to evaluate a tetraploid population derived from Altar 84 and LDN to determine the effects of the Tsc2-Ptr ToxB interaction in a true tetraploid wheat background. The results indicated that the Tsc2 explained up to 26% of the disease variation. This is the first study to demonstrate that the Tsc2-Ptr ToxB interaction plays a significant role in conferring susceptibility in tetraploid wheat, just as it does in hexaploid wheat.
Role of Tsn1-ToxA in tan spot
Another objective of this study was to evaluate the role of the Tsn1-ToxA interaction in conferring tan spot susceptibility in tetraploid wheat. Most of the previous tan spot studies pertaining to the Tsn1-ToxA interaction have been conducted in hexaploid wheat (Faris et al. 2013 for review), and those studies indicated that the Tsn1-ToxA interaction could play a major role (Tomas and Bockus 1987; Lamari and Bernier 1989; Cheong et al. 2004; Singh et al. 2010), a minor role (Friesen et al. 2003; Chu et al. 2008a; S. Singh et al. 2008; Faris et al. 2012), or have no effect (Faris and Friesen 2005), depending on the genetic background. Three studies prior to the current one involved the evaluation of the Tsn1-ToxA interaction in conferring tan spot susceptibility in tetraploid wheat. In one study, Chu et al. (2010a) evaluated a tetraploid wheat doubled haploid population derived from a cross between the durum variety Lebsock and accession PI 94749 of T. turgidum ssp. carthlicum (LP population) for reaction to the race 2 isolate 86-124 and the race 1 isolate Pti2, which both produce ToxA. Although the population segregated for reaction to ToxA infiltrations, sensitivity to ToxA had no effect on disease. Further QTL analysis showed no significance for the Tsn1 locus on 5BL for reaction to either 86–124 or Pti2. In the second tetraploid wheat study, Chu et al. (2008c) evaluated 172 accessions of wild emmer wheat (T. dicoccoides) for reaction to infiltrations of ToxA and inoculations with the P. tritici-repentis race 1 isolate Pti2. They reported a weak (R2 = 0.03), albeit significant, association between ToxA sensitivity and tan spot susceptibility. Similarly, in the third study, Chu et al. (2008b) evaluated 688 accessions of tetraploid wheat subspecies with Pti2 and found that the Tsn1-ToxA interaction was not associated with tan spot susceptibility.
The results of the current study agree with those of Chu et al. (2008b,c, 2010a), in that the Tsn1-ToxA interaction plays little or no role in the development of tan spot in tetraploid wheat. Consistent results were observed when evaluating two different populations (AL and LD5B) and using two different race 2 isolates (86–124 and L13-35) on the AL population. We also conducted single-replication inoculations of the AL population with the race 1 isolates Pti-2 and ASC1, both of which produce ToxA, and found that neither indicated a significant association with the Tsn1 locus (data not shown). To our knowledge, no study has demonstrated Tsn1 to play a major role in tan spot susceptibility in tetraploid wheat.
The reasons for varying levels of significance of the Tsn1-ToxA interaction in different wheat genetic backgrounds are unknown. It is unlikely that the differences are due to structural variation in the ToxA gene among isolates because first, Friesen et al. (2006) evaluated ToxA haplotypes of 54 P. tritici-repentis isolates, and found no sequence variation in the ToxA gene, and second, the same race 2 isolate (86–124) has been used in many of the above mentioned studies and has been associated with variable results including a strong role for the Tsn1-ToxA interaction (Lamari and Bernier 1989), a minor role for the interaction (Friesen et al. 2003; Chu et al. 2010b; Faris et al. 2012), and no role for the interaction such as reported in Faris and Friesen (2005), Chu et al. (2010a), and the current study.
Faris et al. (2011) showed that differential expression of the ToxA gene in different isolates of Pa. nodorum correlated with the level of SNB susceptibility. This might explain differences in the significance of the Tsn1-ToxA interaction among different P. tritici-repentis isolates. For example, Faris et al. (2012) showed that isolates ASC1, Pti2, and 86-124 explained 5, 22, and 30% of the variation, respectively, even though they all contain the same ToxA haplotype (Friesen et al. 2006). However, the same explanation for differences in Tsn1-ToxA relevance among studies that employed the same isolate, 86-124, would not be valid. In this case, differences in host genetic backgrounds must be responsible. This is in line with the results of a recent study by Manning and Ciuffetti (2015), who reported that the Tsn1-ToxA interaction may be epistatic to the production of other NEs, depending on the genetic background of the sensitive host. Perhaps some wheat genotypes possess factors that lead to altered expression levels of the ToxA gene through epistasis, or in some way inhibit the recognition of ToxA by Tsn1 in plants inoculated with fungal spores, but not in plants infiltrated with ToxA. Work is ongoing to elucidate these putative factors and their mechanisms.
Role of Tsn1-ToxA in SNB
Although this research indicated that the Tsn1-ToxA interaction played no significant role in the development of tan spot caused by ToxA-producing isolates of P. tritici-repentis in either the AL or the LD5B population, the interaction was highly significant in the development of SNB caused by Pa. nodorum in both populations. In previous research, we showed that in the hexaploid wheat BG population the Tsn1-ToxA interaction explained as much as 62% of the disease variation for resistance to Pa. nodorum, but had no significant association with tan spot caused by ToxA-producing P. tritici-repentis isolates (Faris and Friesen 2005; Liu et al. 2006). We obtained similar results using the tetraploid LP population where the Tsn1-ToxA interaction accounted for 31% of the variation in the development of SNB, but was not associated with tan spot. Therefore, the results of these studies agree with our findings in the AL and LD5B populations. On the contrary, the Tsn1-ToxA interaction was evaluated for both tan spot and SNB in one other hexaploid wheat population and found to be significantly associated with both diseases, accounting for 17% of the tan spot variation and 28% of the SNB variation (Chu et al. 2008a, 2010b).
Collectively, the currently available data indicate that the role of the Tsn1-ToxA interaction in conferring susceptibility to tan spot in hexaploid wheat can range from none to highly important (see Faris et al. 2013 for review), and for susceptibility to tan spot in tetraploid wheat the interaction is either not significant or can play a very weak role at most (Chu et al. 2008b,c, 2010a; current study). However, research to date indicates that the Tsn1-ToxA interaction always plays a highly important role in conferring susceptibility to SNB in both hexaploid and tetraploid wheat when ToxA-producing isolates are used (Abeysekara et al. 2012; Chu et al. 2008b,c, 2010b; Faris and Friesen 2009; Faris et al. 2010, 2011; Friesen et al. 2006, 2007, 2008, 2009, 2012; Liu et al. 2006).
Differential reactions to ToxA-expressing P. tritici-repentis isolates are likely due to different host genetic factors as discussed above, but the reasons for the varying levels of significance of the Tsn1-ToxA interaction in causing tan spot compared to SNB are most likely different. Including this research, there have now been four wheat populations (BG, LP, AL, and LD5B) that have shown that the Tsn1-ToxA interaction played no role in the development of tan spot, but a major role in the development of SNB. Because each of these studies evaluated disease produced by ToxA-producing isolates of both P. tritici-repentis and Pa. nodorum on the same population, the differences observed in the effects of the Tsn1-ToxA interaction must be due to differences in the biology of the pathogens, i.e., P. tritici-repentis vs. Pa. nodorum. In line with this, our expression studies indicated that Pa. nodorum expressed ToxA at much higher levels than P. tritici-repentis on Tsn1-containing plants, which is a result reminiscent of those of Faris et al. (2011) and Phan et al. (2016) who both showed that levels of NE expression in Pa. nodorum were strongly correlated with levels of disease.
The reason for the differential expression is not known, but as mentioned above, the ToxA gene has existed in Pa. nodorum for a very long time and was only recently transferred to P. tritici-repentis (Friesen et al. 2006). It is possible that ToxA functions more efficiently in Pa. nodorum compared to P. tritici-repentis, perhaps due to the presence of additional and/or different transcription factors that may enhance ToxA expression. It is perceivable that transcriptional upregulation of ToxA in Pa. nodorum-infected plants may occur due to host recognition signals. In other words, pathogen acknowledgment of host recognition, i.e., the presence of Tsn1, may lead to upregulation of ToxA expression. The fact that ToxA expression in the resistant line LDN-DIC 5B was undetectable at 24 hr postinoculation, and levels of ToxA expression in the susceptible line LDN were extremely high at the same time point, would suggest that could be the case. Therefore, it seems that Pa. nodorum might possess ToxA-regulating factors that P. tritici-repentis does not. More research is needed to test this hypothesis.
Conclusions
In conclusion, the Tsc2-Ptr ToxB interaction played a significant role in the development of tan spot caused by the Ptr ToxB-producing race 5 isolate DW5. This was the first study to demonstrate this for tetraploid wheat. Therefore, durum wheat breeders should determine whether or not their material possesses the Tsc2 gene and strive to remove it from their lines using marker-assisted selection. Diagnostic markers for Tsc2 have been developed and proven to be useful for such purposes (Abeysekara et al. 2010). Second, this research showed that the Tsn1-ToxA interaction was not associated with the development of tan spot in two tetraploid wheat populations, however, it played a significant role in the development of SNB in both populations. This is the second study to show this result in tetraploid wheat, and validates work in a different tetraploid wheat population (Chu et al. 2010a; Friesen et al. 2012). Although Tsn1 may not be relevant for susceptibility to tan spot in these two durum wheat populations, this result needs to be confirmed in other tetraploid wheat populations as well. Regardless of whether or not Tsn1 is important for tan spot susceptibility in any durum wheat genotype, breeders should still strive to remove Tsn1 from their materials in an effort to eliminate SNB susceptibility loci and render their lines more resistant. The Tsn1 gene has been cloned, and numerous diagnostic markers are available for this purpose (Zhang et al. 2009; Faris et al. 2010). More research is needed to determine why the relevance of the Tsn1-ToxA interaction in the development of tan spot is affected by different host genetic backgrounds, and why the interaction is more significant in the development of SNB than tan spot.
Supplementary Material
Acknowledgments
The authors thank Samantha Steckler for technical assistance. This research was supported by United States Department of Agriculture (USDA)-Agricultural Research Service Current Research Information System project grant 5442-22000-037-00D. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. USDA is an equal opportunity provider and employer.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3. 116.036525/-/DC1.
Communicating editor: E. Akhunov
Literature Cited
- Abeysekara N. S., Friesen T. L., Keller B., Faris J. D., 2009. Identification and characterization of a novel host-toxin interaction in the wheat-Stagonospora nodorum pathosystem. Theor. Appl. Genet. 120: 117–126. [DOI] [PubMed] [Google Scholar]
- Abeysekara N. S., Friesen T. L., Liu Z. H., McClean P. E., Faris J. D., 2010. Marker development and saturation mapping of the tan spot Ptr ToxB sensitivity locus Tsc2 in hexaploid wheat. Plant Genome 3: 179–189. [Google Scholar]
- Abeysekara N. S., Faris J. D., Chao S., McClean P. E., Friesen T. L., 2012. Whole genome QTL analysis of Stagonospora nodorum blotch resistance and validation of the SnTox4-Snn4 interaction in hexaploid wheat. Phytopathology 102: 94–104. [DOI] [PubMed] [Google Scholar]
- Ali S., Gurung S., Adhikari T. B., 2010. Identification and characterization of novel isolates of Pyrenophora tritici-repentis from Arkansas. Plant Dis. 94: 229–235. [DOI] [PubMed] [Google Scholar]
- Anderson J. A., Effertz R. J., Faris J. D., Francl L. J., Meinhardt S. W., et al. , 1999. Genetic analysis of sensitivity to a Pyrenophora tritici-repentis necrosis-inducing toxin in durum and common wheat. Phytopathology 89: 293–297. [DOI] [PubMed] [Google Scholar]
- Cavanagh C., Chao S., Wang S., Huang B. E., Stephen S., et al. , 2013. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landrace and cultivars. Proc. Natl. Acad. Sci. USA 110: 8057–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong J., Wallwork H., Williams K. J., 2004. Identification of a major QTL for yellow leaf spot resistance in the wheat varieties Brookton and Cranbook. Aust. J. Agric. Res. 55: 315–319. [Google Scholar]
- Chu C. G., Friesen T. L., Xu S. S., Faris J. D., 2008a Identification of novel tan spot resistance loci beyond the known host-selective toxin insensitivity genes in wheat. Theor. Appl. Genet. 117: 873–881. [DOI] [PubMed] [Google Scholar]
- Chu C. G., Friesen T. L., Faris J. D., Xu S. S., 2008b Evaluation of seedling resistance to tan spot and stagonospora nodorum blotch in tetraploid wheat. Crop Sci. 48: 1107–1118. [Google Scholar]
- Chu C. G., Xu S. S., Faris J. D., Nevo E., Friesen T. L., 2008c Seedling resistance to tan spot and stagonospora nodorum leaf blotch in wild emmer wheat (Triticum dicoccoides). Plant Dis. 92: 1229–1236. [DOI] [PubMed] [Google Scholar]
- Chu C. G., Chao S., Friesen T. L., Faris J. D., Zhong Z., et al. , 2010a Identification of novel tan spot resistance QTLs using an SSR-based linkage map of tetraploid wheat. Mol. Breed. 25: 327–338. [Google Scholar]
- Chu C. G., Faris J. D., Xu S. S., Friesen T. L., 2010b Genetic analysis of disease susceptibility caused by compatible Tsn1-SnToxA and Snn1-SnTox1 interactions in the wheat-Stagonospora nodorum pathosystem. Theor. Appl. Genet. 120: 1451–1459. [DOI] [PubMed] [Google Scholar]
- Ciuffetti L. M., Manning V. A., Pandelova I., Betts M. F., Martinez J. P., 2010. Host-selective toxins, Ptr ToxA and Ptr ToxB, as nectrophic effectors in the Pyrenophora tritici-repentis-wheat interaction. New Phytol. 187: 911–919. [DOI] [PubMed] [Google Scholar]
- Effertz R. J., Meinhardt S. W., Anderson J. A., Jordahl J. G., Francl L. J., 2002. Identification of a chlorosis-inducing toxin from Pyrenophora tritici-repentis and the chromosomal location of an insensitivity locus in wheat. Phytopathology 92: 527–533. [DOI] [PubMed] [Google Scholar]
- Faris J. D., Friesen T. L., 2005. Identification of quantitative trait loci for race-nonspecific resistance to tan spot of wheat. Theor. Appl. Genet. 111: 386–392. [DOI] [PubMed] [Google Scholar]
- Faris J. D., Friesen T. L., 2009. Re-evaluation of a tetraploid wheat population indicates that the Tsn1-ToxA interaction is the only factor governing susceptibility to Stagonospora nodorum blotch. Phytopathology 99: 906–912. [DOI] [PubMed] [Google Scholar]
- Faris J. D., Zhang Z., Lu H. J., Lu S. W., Reddy L., et al. , 2010. A unique wheat disease resistance-like gene governs effector-triggered susceptibility to necrotrophic pathogens. Proc. Natl. Acad. Sci. USA 107: 13544–13549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faris J. D., Zhang Z., Rasmussen J. B., Friesen T. L., 2011. Variable expression of the Stagonospora nodorum effector SnToxA among isolates is correlated with levels of disease in wheat. Mol. Plant Microbe Interact. 24: 1419–1426. [DOI] [PubMed] [Google Scholar]
- Faris J. D., Abeysekara N. S., McClean P. E., Xu S. S., Friesen T. L., 2012. Tan spot susceptibility governed by the Tsn1 locus and nonrace-specific resistance QTL in a population derived from the wheat lines Salamouni and Katepwa. Mol. Breed. 30: 1669–1678. [Google Scholar]
- Faris J. D., Liu Z. H., Xu S. S., 2013. Genetics of tan spot resistance in wheat. Theor. Appl. Genet. 126: 2197–2217. [DOI] [PubMed] [Google Scholar]
- Faris J. D., Zhang Z., Chao S., 2014. Map-based analysis of the tenacious glume gene Tg-B1 of wild emmer and its role in wheat domestication. Gene 542: 198–208. [DOI] [PubMed] [Google Scholar]
- Friesen T. L., Faris J. D., 2004. Molecular mapping of resistance to Pyrenophora tritici-repentis race 5 and sensitivity to Ptr ToxB in wheat. Theor. Appl. Genet. 109: 464–471. [DOI] [PubMed] [Google Scholar]
- Friesen T. L., Faris J. D., 2010. Characterization of the wheat-Stagonospora nodorum disease system – what is the molecular basis of this quantitative necrotrophic disease interaction? Can. J. Plant Pathol. 32: 20–28. [Google Scholar]
- Friesen T. L., Ali S., Kianian S., Francl L. J., Rasmussen J. B., 2003. Role of host sensitivity to Ptr ToxA in development of tan spot of wheat. Phytopathology 93: 397–401. [DOI] [PubMed] [Google Scholar]
- Friesen T. L., Stukenbrock E. H., Liu Z. H., Meinhardt S., Ling H., et al. , 2006. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 38: 953–956. [DOI] [PubMed] [Google Scholar]
- Friesen T. L., Meinhardt S. W., Faris J. D., 2007. The Stagonospora nodorum-wheat pathosystem involves multiple proteinaceous host-selective toxins and corresponding host sensitivity genes that interact in an inverse gene-for-gene manner. Plant J. 51: 681–692. [DOI] [PubMed] [Google Scholar]
- Friesen T. L., Zhang Z., Solomon P. S., Oliver R. P., Faris J. D., 2008. Characterization of the interaction of a novel Stagonospora nodorum host-selective toxin with a wheat susceptibility gene. Plant Physiol. 146: 682–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen T. L., Chu C.-G., Liu Z. H., Xu S. S., Halley S., et al. , 2009. Host-selective toxins produced by Stagonospora nodorum confer disease susceptibility in adult wheat plants under field conditions. Theor. Appl. Genet. 118: 1489–1497. [DOI] [PubMed] [Google Scholar]
- Friesen T. L., Chu C., Xu S. S., Faris J. D., 2012. SnTox5-Snn5: a novel Stagonospora nodorum effector-wheat gene interaction and its relationship with the SnToxA-Tsn1 and SnTox3-Snn3–B1 interactions. Mol. Plant Pathol. 13: 1101–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y., Faris J. D., Liu Z. H., Xu S. S., Friesen T. L., 2015. Identification and characterization of the SnTox6-Snn6 interaction in the wheat-Parastagonospora nodorum pathosystem. Mol. Plant Microbe Interact. 28: 615–625. [DOI] [PubMed] [Google Scholar]
- Joehanes R., Nelson J. C., 2008. QGene 4.0, an extensible Java QTL-analysis platform. Bioinformatics 24: 2788–2789. [DOI] [PubMed] [Google Scholar]
- Joppa L. R., 1993. Chromosome engineering in tetraploid wheat. Crop Sci. 33: 908–913. [Google Scholar]
- Kariyawasam G. K., Carter A. H., Rasmussen J. B., Faris J. D., Xu S. S., et al. , 2016. Genetic relationships between race-nonspecific and race specific interactions in the wheat-Pyrenophora tritici-repentis pathosystem. Theor. Appl. Genet. 129: 897–908. [DOI] [PubMed] [Google Scholar]
- Kosambi D. D., 1944. The estimation of map distances from recombination values. Ann. Eugen. 12: 172–175. [Google Scholar]
- Lamari L., Bernier C. C., 1989. Toxin of Pyrenophora tritici-repentis: host-specificity, significance in disease, and inheritance of host reaction. Phytopathology 79: 740–744. [Google Scholar]
- Liu Z. H., Friesen T. L., Meinhardt S. W., Ali S., Rasmussen J. B., et al. , 2004a QTL analysis and mapping of seedling resistance to Stagonospora nodorum leaf blotch in wheat. Phytopathology 94: 1061–1067. [DOI] [PubMed] [Google Scholar]
- Liu Z. H., Faris J. D., Meinhardt S. W., Ali S., Rasmussen J. B., et al. , 2004b Genetic and physical mapping of a gene conditioning sensitivity in wheat to a partially purified host-selective toxin produced by Stagonospora nodorum. Phytopathology 94: 1056–1060. [DOI] [PubMed] [Google Scholar]
- Liu Z. H., Friesen T. L., Ling H., Meinhardt S. W., Oliver R. P., et al. , 2006. The Tsn1-ToxA interaction in the wheat-Stagonospora nodorum pathosystem parallels that of the wheat-tan spot system. Genome 49: 1265–1273. [DOI] [PubMed] [Google Scholar]
- Liu Z. H., Faris J. D., Oliver R. P., Tan K. C., Solomon P. S., et al. , 2009. SnTox3 acts in effector-triggered susceptibility to induce disease on wheat carrying the Snn3 gene. PLoS Pathog. 5(9): e10000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorieux M., 2012. MapDisto: fast and efficient computation of genetic linkage maps. Mol. Breed. 30: 1231–1235. [Google Scholar]
- Lu H.-J., Faris J. D., 2006. Macro- and microlinearity between the genomics region of wheat chromosome 5B containing the Tsn1 gene and the rice genome. Funct. Integr. Genomics 6: 90–103. [DOI] [PubMed] [Google Scholar]
- Manning V. A., Ciuffetti L. M., 2015. Necrotrophic effector epistatis in the Pyrenophora tritici-repentis-wheat interaction. PLoS One 10: e0123548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver R. P., Friesen T. L., Faris J. D., Solomon P. S., 2012. Stagonospora nodorum: from pathology to genomics and host resistance. Annu. Rev. Phytopathol. 50: 23–43. [DOI] [PubMed] [Google Scholar]
- Phan H. T. T., Rybak K., Furuki E., Breen S., Solomon P. S., 2016. Differential effector gene expression underpins epistasis in a plant fungal disease. Plant J. 87: 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röder M. S., Korzun V., Wendehake K., Plaschke J., Tixier M.-H., et al. , 1998. A microsatellite map of wheat. Genetics 149: 2007–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi G. J., Friesen T. L., Saini S., Xu S. S., Rasmussen J. B., et al. , 2015. The wheat Snn7 gene confers susceptibility upon recognition of the Parastagonospora nodorum necrotrophic effector SnTox7. Plant Genome 8(2): 1–10. [DOI] [PubMed] [Google Scholar]
- Shi G. J., Zhang Z., Friesen T. L., Raats D., Fahima T., et al. , 2016. The hijacking of a receptor kinase-driven pathway by a wheat fungal pathogen leads to disease. Sci. Adv. 2(10): e1600822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P. K., Mergoum M., Gonzalez-Hernandez J. L., Ali S., Adhikari T. B., et al. , 2008. Genetics and molecular mapping of resistance to necrosis-inducing race 5 of Pyrenophora tritici-repentis in tetraploid wheat. Mol. Breed. 21: 293–304. [Google Scholar]
- Singh P. K., Singh R. P., Duveiller E., Mergoum M., Adhikari T. B., et al. , 2010. Genetics of wheat-Pyrenophora tritici-repentis interactions. Euphytica 171: 1–13. [Google Scholar]
- Singh S., Bockus W. W., Sharma I., Bowden R. L., 2008. A novel source of resistance in wheat to Pyrenophora tritici-repentis race 1. Plant Dis. 92: 91–95. [DOI] [PubMed] [Google Scholar]
- Somers D. J., Issac P., Edwards K., 2004. A high-density microsatellite consensus map for bread wheat. Theor. Appl. Genet. 109: 1105–1114. [DOI] [PubMed] [Google Scholar]
- Song Q. J., Shi J. R., Singh S., Fickus E. W., Costa J. M., et al. , 2005. Development and mapping of microsatellite SSR markers in wheat. Theor. Appl. Genet. 110: 550–560. [DOI] [PubMed] [Google Scholar]
- Sourdille P., Singh S., Cadalen T., Brown-Guedira G., Gay G., et al. , 2004. Microsatellite-based deletion bin system for the establishment of genetic-physical map relationship in wheat Triticum aestivum L. Funct. Integr. Genomics 4: 12–25. [DOI] [PubMed] [Google Scholar]
- Tomas A., Bockus W. W., 1987. Cultivar-specific toxicity of culture filtrate of Pyrenophora tritici-repentis. Phytopathology 77: 1337–1340. [Google Scholar]
- Torada A., Koike M., Mochida K., Ogihara Y., 2006. SSR-based linkage map with new markers using an intraspecific population of common wheat. Theor. Appl. Genet. 112: 1042–1051. [DOI] [PubMed] [Google Scholar]
- Wolpert T. J., Dunkle L. D., Ciuffetti L. M., 2002. Host-selective toxins and avirulence determinants: what’s in a name? Annu. Rev. Phytopathol. 40: 251–285. [DOI] [PubMed] [Google Scholar]
- Xue S., Zhang Z., Lin F., Kong Z., Cao Y., et al. , 2008. A high-density inter-varietal map of the wheat genome enriched with markers derived from expressed sequence tags. Theor. Appl. Genet. 117: 181–189. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Friesen T. L., Simons K. J., Xu S. S., Faris J. D., 2009. Development, identification, and validation of markers for marker-assisted selection against the Stagonospora nodorum toxin sensitivity genes Tsn1 and Snn2 in wheat. Mol. Breed. 23: 35–49. [Google Scholar]
- Zhang Z., Friesen T. L., Xu S. S., Shi G. J., Liu Z. H., et al. , 2011. Two putatively homoeologous wheat genes mediate the recognition of SnTox3 to confer effector-triggered susceptibility to Stagonospora nodorum. Plant J. 65: 27–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Parents and mapping populations are available upon request. Mapping data for the LD5B and the AL populations is also available upon request. The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.