Abstract
Enhancers physically interact with transcriptional promoters, looping over distances that can span multiple regulatory elements. Given that enhancer–promoter (EP) interactions generally occur via common protein complexes, it is unclear whether EP pairing is predominantly deterministic or proximity guided. Here, we present cross-organismic evidence suggesting that most EP pairs are compatible, largely determined by physical proximity rather than specific interactions. By reanalyzing transcriptome datasets, we find that the transcription of gene neighbors is correlated over distances that scale with genome size. We experimentally show that nonspecific EP interactions can explain such correlation, and that EP distance acts as a scaling factor for the transcriptional influence of an enhancer. We propose that enhancer sharing is commonplace among eukaryotes, and that EP distance is an important layer of information in gene regulation.
Keywords: enhancer sharing, gene coexpression, gene neighbors, enhancer specificity, transcriptional domains
Enhancers mediate the transcriptional regulation of gene expression, enabling isogenic cells to exhibit remarkable phenotypic diversity (Davidson and Peter 2015). In complex with transcription factors, they interact with promoters via chromatin looping (Marsman and Horsfield 2012), finely regulating transcription in time and space. A prevailing view is that most enhancers have a mechanism to selectively loop to a target promoter (van Arensbergen et al. 2014). Examples in this category usually require specific transcription factor binding at both enhancer and promoter sites (Davidson and Peter 2015), which could explain why some enhancers seem to influence different promoters to varying degrees (Gehrig et al. 2009). On the other hand, EP looping is generally mediated by common protein complexes (Kagey et al. 2010; Malik and Roeder 2010), conflicting with the specific molecular interactions required by such a model at a larger scale. Examples of nonspecific EP pairing also seem to be common (Butler and Kadonaga 2001). Yet given that this model could result in transcriptional crosstalk, it appears inconsistent with our current paradigm of gene regulation. The predominant EP pairing scheme, specific or nonspecific, and its determinants are thus unclear. Here, we ask to what extent are potential EP pairs compatible through a meta-analysis of the genome-wide transcription of gene neighbors in five species. We propose that enhancer sharing occurs widely across eukaryotes, test key aspects of this hypothesis in Caenorhabditis elegans, and analyze its implications in other genomic phenomena.
Materials and Methods
Computational biology
For each analyzed organism, Ensembl (Flicek et al. 2014) protein-coding genes were grouped by chromosome, sorted by position, and paired with the 100 nearest neighbors within the same chromosome. A list of duplicated gene pairs for Homo sapiens and Mus musculus was obtained from the Duplicated Genes Database (Ouedraogo et al. 2012) (http://dgd.genouest.org). A list of C. elegans genes predicted to be in operons was obtained from Allen et al. (2011), and gene pairs present in the same operon were removed from the analysis to prevent cotranscriptional bias. Processed RNA-seq data were obtained from multiple sources (Gerstein et al. 2010; Attrill et al. 2016; Ellahi et al. 2015; The ENCODE Project Consortium 2012) and converted to transcripts per million (TPM) (Li et al. 2010) when necessary. Formatted datasets are available upon request. Genes detected in < 80% of experiments were discarded. To compute the Spearman correlation coefficient, TPM values were ranked in each RNA-seq experiment and the pairwise Pearson correlation coefficient was computed on the ranked values according to the following equation:
where and are the corresponding ranks of each paired gene in a given RNA-seq experiment, their covariance and σ their SD. The list of gene pairs with intergenic distances and correlation coefficients was sorted by increasing intergenic distance, and subsequently smoothed using a sliding median with window size of 1000 gene pairs. The result was then fitted to an exponential decay function of the form:
where is the median Spearman correlation coefficient of the closest neighboring genes, d the intergenic distance, and c the baseline correlation. The mean distance at which a pair of genes remain correlated was then computed as:
To compute the background correlation, each gene was paired with 20 randomly selected genes from a different chromosome and the 95% median confidence interval was computed by bootstrapping with 10,000 samples. A list of genes annotated with RNA in situ hybridization data (Hammonds et al. 2013; Tomancak et al. 2002, 2007) was obtained from the Berkeley Drosophila Genome Project (http://insitu.fruitfly.org). Insulator Chromatin ImmunoPrecipitation coupled with microarrays (ChIP-chip) data were obtained from Negre et al. (2010) (GSE16245); the intersection of replicates was used. HiC data were obtained from Rao et al. (2014) (GSE63525, GM12878 primary replicate HiCCUPS looplist). Functional protein classification was conducted using Panther (Mi et al. 2016). Genomic manipulations were conducted using Bedtools v2.24.0 (Quinlan and Hall 2010). Data analysis was conducted using Python 2.7.9 and the Scipy library (McKinney 2010). Plots were generated using Matplotlib 1.5 (Hunter 2007).
Molecular biology
C. elegans was cultured under standard laboratory conditions (Stiernagle 2006). For enhancer additivity experiments, transgenic C. elegans lines carrying extrachromosomal arrays were generated by injecting each plasmid at 50 ng/μl into unc-119 mutant animals. The minimal Δpes-10 promoter (Fire et al. 1990) and nuclear localized GFP (Lyssenko et al. 2007) were used in all constructs. Minimal regions of the myo-2 and unc-54 enhancers (Okkema et al. 1993) able to drive tissue-specific expression were used. The BWM (body wall muscle) enhancer was obtained from the upstream region of F44B9.2; the BWM/intestine enhancer was obtained from the upstream region of rps-1. Animals were imaged at 40 × using a GFP filter on a Zeiss Axioskop microscope.
For the EP distance and ectopic enhancer experiments, we defined an EP distance of 0 to be the enhancer placed just upstream of the Δpes-10 promoter, which is ∼350 bp away from the start codon of gfp. To ensure neutrality yet maintain a similar GC content as noncoding sequences in C. elegans, we used nonoverlapping AT-rich DNA spacers obtained from the genome of Escherichia coli. Constructs were integrated in single-copy into chromosome IV via CRISPR-Cas9 using plasmids provided as gifts by Zhiping Wang and Yishi Jin (unpublished results). Briefly, plasmids containing the following expression cassettes were coinjected: reporter and hygromycin resistance genes flanked by homologous arms for recombination-directed repair (10 ng/μl), single-guide RNA (10 ng/μl), Cas9 (10 ng/μl), and an extrachromosomal array reporter for expression of either rfp or gfp outside the tissue of interest (10 ng/μl). Transformants were selected for using hygromycin at 10 μg/μl, and those not carrying extrachromosomal transgenes, which lacked gfp or rfp expression outside the tissue of interest, were subsequently isolated. Animals homozygotic for the insertion were identified by polymerase chain reaction (PCR) and Sanger sequencing.
Quantitative PCR was carried out as previously described (Ly et al. 2015) using pmp-3 as a reference gene (Zhang et al. 2012). Briefly, third-stage larval (L3) worms, when expression from the test enhancers is maximal according to RNA-seq data, were synchronized at 20° via egg-laying. Fifteen animals were lysed in 1.5 μl of Lysis Buffer [5 mM Tris pH 8.0 (MP Biomedicals), 0.5% Triton X-100, 0.5% Tween 20, 0.25 mM EDTA (Sigma-Aldrich)] with proteinase-K (Roche) at 1.5 μg/μl, and incubated at 65° for 10 min followed by 85° for 1 min. Reverse transcription was carried out using the Maxima H Minus cDNA synthesis kit (Thermo Fisher Scientific) by adding 0.3 μl H2O, 0.6 μl 5 × enzyme buffer, 0.15 μl 10 mM dNTP mix, 0.15 μl 0.5 μg/μl oligo dT primer, 0.15 μl enzyme mix, and 0.15 μl DNAse, and incubated for 2 min at 37°, followed by 30 min at 50°, and finally 2 min at 85°. The cDNA solution was diluted to 15 μl and 1 μl was used for each qPCR reaction, so that on average each well contained the amount of RNA from a single worm. All qPCR reactions were performed with three technical replicates and at least three biological replicates using the Roche LightCycler 480 SYBR Green I Master in the LightCycler 480 System. Crossing point-PCR-cycle (Cp) averages were computed for each group of three technical replicates; these values were then subtracted from the respective average Cp value of the reference gene.
Data and reagent availability
Strains are available upon request. Relevant DNA sequences, including spacers, enhancers, primers, sgRNA, and homology arms are available in Supplemental Material, Table S1. Correlation datasets are available in File S1 and File S2. qPCR data are available in Table S2. Python source code, and links to all expression datasets used in this study, are available for download on the following github repository: https://github.com/WormLabCaltech/QuinteroSternberg2016.git.
Results and Discussion
Gene neighbors are transcriptionally correlated genome-wide
We reasoned that widespread EP compatibility should result in transcriptional correlation among gene neighbors. Indeed, gene coexpression clusters have been extensively reported in eukaryotic genomes (e.g., Sémon and Duret 2006; Roy et al. 2002; Lercher et al. 2002, 2003; Lercher and Hurst 2006; Williams and Hurst 2002; Singer et al. 2005; Williams and Bowles 2004; Spellman and Rubin 2002; Purmann et al. 2007; Zhan et al. 2006; Boutanae v et al. 2002; Kalmykova et al. 2005; Caron et al. 2001; Rubin and Green 2013) in spite of order of magnitude variations in genome size (e.g., ∼12 Mb in Saccharomyces cerevisiae vs. ∼3 Gb in H. sapiens). An early informative example is the discovery of chromosomal domains of gene expression in S. cerevisiae (Cohen et al. 2000) that exhibit features that strongly support enhancer-sharing, mainly distance-dependence in transcriptional correlation that qualitatively resemble chromosome contact matrices (e.g., Rao et al. 2014), and instances in which a single enhancer seems to be responsible for the coexpression of adjacent gene pairs. The ubiquity of these features across eukaryotes would support the idea that EP interactions are largely determined by physical proximity rather than by specific interactions. Given the accumulation of transcriptome sequencing data, we decided to investigate the transcriptional correlation of gene neighbors in representative eukaryotic species as a first step to explore the average EP pairing scheme.
We paired every protein-coding gene of five organisms (S. cerevisiae, C. elegans, Drosophila melanogaster, M. musculus, and H. sapiens) with its 100 nearest neighbors within the same chromosome. This yielded lists of around 600,000 (S. cerevisiae) and 2 million (each of the rest) gene pairs. We then computed the Spearman correlation coefficient between paired genes across multiple RNA-seq datasets (Gerstein et al. 2010; Attrill et al. 2016; Ellahi et al. 2015; The ENCODE Project Consortium 2012) and the intergenic distance between the start of the 5′ untranslated region of the first gene to the start of the second gene in each pair.
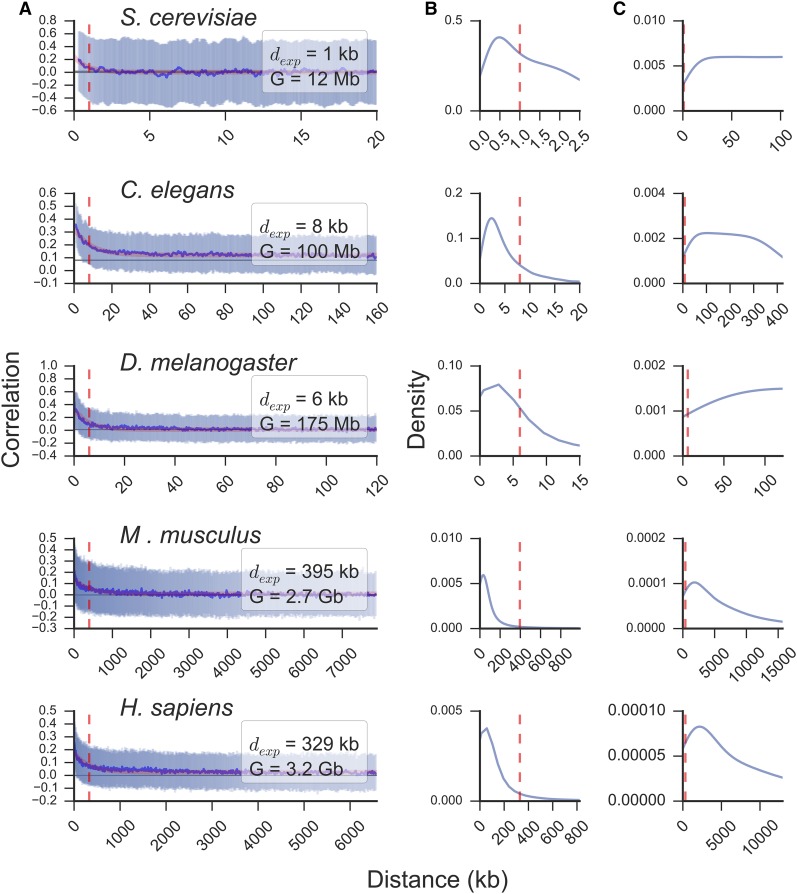
We observed that neighboring genes tend to be correlated in transcript abundance genome-wide in all analyzed organisms, and that this correlation decays exponentially with increasing intergenic distance (Figure 1A). We thus fitted the data to an exponential decay function to estimate the distance at which a pair of genes remain correlated (). Consistent with the persistence of the correlation pattern across organisms, scaled with genome size, to 1 kb in S. cerevisiae, ∼10 kb in C. elegans and D. melanogaster, and ∼350 kb in M. musculus and H. sapiens (Figure S1). This trend remained largely the same even after removing duplicated gene pairs (Figure S2). Most genes had at least one neighbor closer than in all species (Figure 1B), and the representation of gene ontology annotations remained unbiased in correlated gene pairs (Figure S3), indicating that the average gene is correlated in expression with its nearest neighbors beyond any particular gene class. In addition, sampled intergenic distances go well beyond (Figure 1C), indicating that 100 gene neighbors are a sufficient number to study this effect.
Figure 1.
Neighboring genes are transcriptionally correlated genome-wide across eukaryotes. (A) Sliding median of correlations between paired neighbors (blue line) and interquartile range (pale blue) with increasing intergenic distance. Median ± 95% C.I. of randomly paired genes is shown as a horizontal gray line. Fit to an exponential decay function (red line) was used to compute the mean distance at which gene neighbors remain correlated ( vertical red dashed line). The genome size (G) is displayed for each organism. Distribution of intergenic distances between each gene and its nearest neighbor (B) and all paired genes (C). The organism analyzed in each case is indicated for each group of three subplots.
To examine the correlation of gene expression in the spatial domain, we analyzed RNA in situ hybridization data for 6053 genes in D. melanogaster (Tomancak et al. 2002, 2007; Hammonds et al. 2013). We computed the percentage overlap in tissue expression by dividing the number of common tissues over the total number of unique tissues in which genes of any given pair are expressed (Figure S4A). This analysis revealed that close neighbors have a tendency to be expressed in the same tissues, and that this overlap also decays exponentially with intergenic distance (Figure S4B). However, the correlation extends to a longer mean distance ( = 22 compared to 6 kb), suggesting that RNA-seq analysis, which included mostly whole-organism transcriptome averages, resulted in a conservative estimate.
Given that pairing every gene with 100 proximal genes provides a complete set of distance-dependent correlations between gene pairs, we concluded that gene neighbors have a spatio-temporal correlation in expression that is highly dependent upon the spacing between them. Our meta-analysis unifies the findings of previous reports (reviewed in Michalak 2008) and highlights the distance-dependence of genome-wide and cross-organismic transcriptional correlations that transcend localized gene coexpression clusters.
Enhancer sharing explains the transcriptional correlation of gene neighbors
The pervasive nature of proximal gene coexpression supported the idea of widespread EP compatibility. This connection is, in turn, supported by several other observations in the literature: (i) enhancers regulate transcription by making contact with promoters via chromatin looping (Marsman and Horsfield 2012), whose incidence also decays exponentially as the distance between contacting sites increases (Ringrose et al. 1999; Rao et al. 2014), with the same pattern as observed here at least in some documented cases (e.g., H. sapiens, Figure S5); (ii) the average distance between a large fraction of studied EP interactions scales with genome size in ranges often consistent with < 1 kb in S. cerevisiae, (Dobi and Winston 2007), < 10 kb in C. elegans, (Araya et al. 2014), and 120 kb in H. sapiens (Sanyal et al. 2012); (iii) common protein complexes such as the mediator seem to be widely utilized bridges in EP looping (Kagey et al. 2010; Malik and Roeder 2010); (iv) a high frequency of chromatin interactions are observed within topologically-associated domains identified through high-resolution Chromosome Conformation Capture (Hi-C) (Rao et al. 2014); and (v) studied enhancers often do not show promoter specificity (Butler and Kadonaga 2001). This line of reasoning suggests a model where, as opposed to only having a specific target gene (Figure 2A), the average enhancer has a range of action in which it can influence any active promoter within its reach (Figure 2B). A concrete example consistent with this idea is the upregulation of neighboring genes upon enhancer activation by fibroblast growth factor in mammalian cells (Ebisuya et al. 2008). Transcriptome analysis could thus provide indirect evidence of genome and condition-wide EP looping that is difficult to access through Hi-C (Rao et al. 2014) due to the low signal-to-noise ratio of short-range interactions.
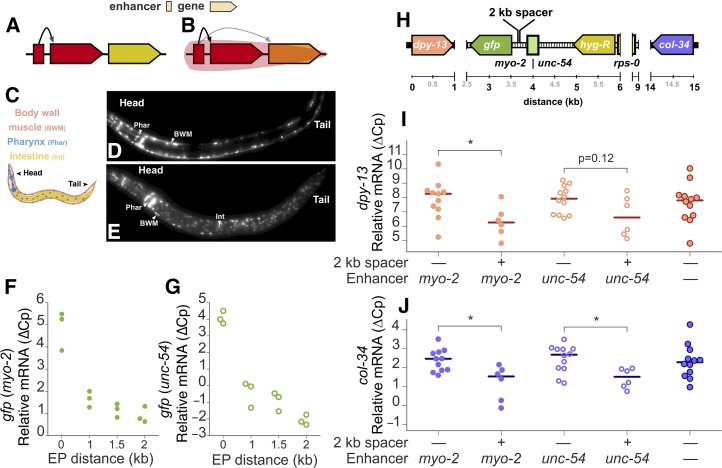
Figure 2.
Enhancer sharing explains the transcriptional correlation of gene neighbors. Two possible models for EP relationship: (A) Enhancers have specific target genes and (B) enhancers have a range of action in which they influence genes by physical proximity. Tissue-specific enhancers (C) are generally compatible. Pharynx and body wall muscle (D) and pharynx, body wall muscle, and intestine (E) enhancers driving nuclear gfp expression. mRNA levels of gfp with increasing EP distance for lines with myo-2 (filled circles) (F) and unc-54 (hollow circles) (G) enhancers. (H) Genomic context of the integration site. The inserted construct is shown over a dashed black line and includes a hygromycin resistance gene (hyg-R) regulated by a ribosomal enhancer (rps-0) and promoter in addition to the reporter (gfp) regulated by either the myo-2 or unc-54 enhancers; the native genes dpy-13 and col-34 flank the insertion site. Relative mRNA levels of dpy-13 (I) and col-34 (J) in wild-type and lines with and without the 2 kb spacer (* two tailed P-value < 0.05, Mann-Whitney U-test). The difference in crossing point-PCR-cycle (ΔCp) with the reference gene pmp-3 and the corresponding median for each group of biological replicates is shown for every qPCR experiment. EP, enhancer–promoter; mRNA, messenger RNA; PCR, polymerase chain reaction; qPCR, quantitative PCR.
Because of its compact genome, rapid development, and availability of tissue-specific enhancers (Corsi et al. 2015), we decided to use C. elegans to test the validity of a nonspecific EP pairing model. We first postulated that unrelated enhancers should generally be compatible, showing qualitative additivity when placed upstream of a single promoter. We thus paired the well-characterized myo-2 pharyngeal enhancer with a BWM and a BWM plus intestine enhancer, placed them upstream of a minimal promoter and a gfp reporter, and examined expression in transgenic animals. In both cases, we observed fluorescence in the corresponding tissues (Figure 2, C–E). This observation is consistent with typical enhancer studies in artificial constructs (Dupuy et al. 2004) and agrees with some EP compatibility studies (Butler and Kadonaga 2001).
Given that both chromatin looping and expression correlation decay exponentially, we reasoned that transcription of a given gene should also decay exponentially with increasing EP distance if the observed correlation is to be explained by enhancer sharing. To test this hypothesis, we first built a series of genetic constructs with increasing neutral EP distances (0, 1, 1.5, and 2 kb) for two different enhancers, myo-2 and unc-54 (∼400 and 300 bp, respectively). We then integrated each construct in single-copy into the genome of C. elegans and used quantitative PCR to: (i) measure the influence of EP distance on the reporter gene in native chromatin and (ii) analyze the impact of the perturbation on the two genes that natively flank the site of transgene insertion (dpy-13 and col-34, Figure 2H), which we reasoned should be affected in two counteracting ways. First, the ectopic enhancers should promote their expression. Second, the increased EP distance caused by the addition of spacers should reduce their expression by scaling down the influence of both ectopic and native enhancers (the latter of unknown identity and location) to the opposite side of the spacer.
We found that transcriptional levels of the reporter gene indeed fall rapidly with increasing EP distance with both enhancers (Figure 2, F and G); this occurred at a rate that seems congruent or faster than the genome-wide correlation decay, likely because of the dramatic separation of every regulatory element at once, as opposed to gradual separation from individual enhancers in a native environment; this dramatic effect suggests complex interactions between multiple EP loops that are disrupted with the insertion of DNA sequences devoid of regulatory elements. Transcription was still well detected even when the enhancers were placed 2 kb away, supporting the hypothesis that EP distance is a scaling factor on the enhancer’s influence. Expression of dpy-13 and col-34 was reduced with the introduction of the 2 kb spacer when compared to transgenic lines without it (Figure 2, I and J). On the other hand, spacer-free lines were comparable to wild-type, suggesting that the incorporation of ectopic enhancers compensated for the EP distance increase caused by the addition of the genetic construct itself. These observations seem to fit the corollaries of our model, even amid the complexity of a native regulatory environment. However, the distance over which we see an effect on col-34 falls outside our estimate for C. elegans (8 kb). Its expression is impacted by the presence of the 2 kb spacer outside of the interval between the myo-2/unc-54 enhancer, suggesting that enhancers >12 kb away can still influence its expression. As evidenced with the discrepancy in D. melanogaster when using in situ or RNA-seq data, this observation suggests that is only a rough estimate of the average enhancer rage of action; this is useful to gain insight into genome-wide mechanisms but not for precise individual predictions.
Chromatin modifications have been shown to have a significant impact on enhancer function (Calo and Wysocka 2013) and thus likely influence EP pairing. Thus, chromatin features and enhancer sharing might be mutually inclusive rather than stand-alone explanations for the observed correlation domains. From this perspective, transcriptionally correlated genes would have similar chromatin states, facilitated by their physical proximity, that make them accessible to enhancer action.
The existence of multiple, independent, but similar enhancers is an alternative possible explanation. However, since we are looking at genome-wide averages, this would mean that most gene neighbors have a functionally redundant set of independent enhancers that function through distinct molecular interactions. Although possible, this is a rather intricate explanation.
In agreement with the enhancer sharing hypothesis, it can be argued that the scaling of correlation domains is a result of the ability to connect EP loops over longer distances in larger genomes. Yet, in spite of having a compact genome, D. melanogaster is able to form many long-range EP interactions (>50 kb) (Ghavi-Helm et al. 2014), which is considerably different to the range of its estimated (6–22 kb). Additionally, these long-range interactions appear to be particularly specific, with enhancers selectively activating their target promoters (Ghavi-Helm et al. 2014; Kwon et al. 2009). It is, thus, possible that in compact genomes, long-range EP interactions would need to be specific, whereas nearby interactions would tend to fall in the nonspecific pairing scheme, ultimately resulting in the observed correlation domain size.
EP distance insulates neighboring genes
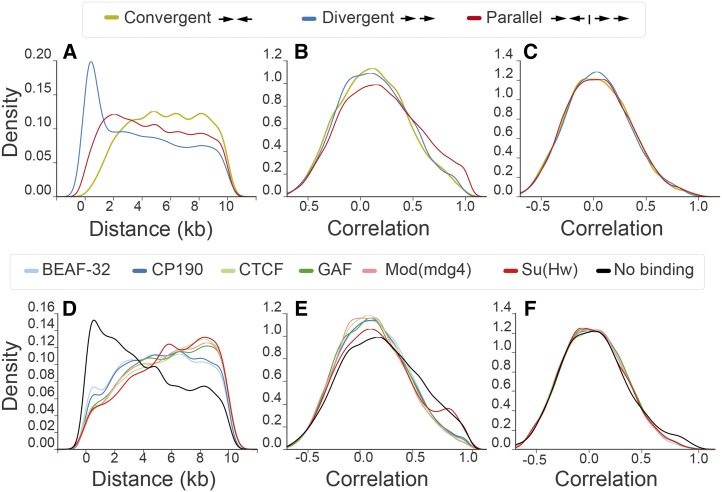
We next wished to determine the extent to which enhancer sharing could explain other genomic phenomena. Previous reports have suggested that divergent, parallel, and convergent gene pairs tend to have distinct correlation profiles (e.g., Chen and Stein 2006). To explore this hypothesis, we compared the distribution of intergenic distances of gene pairs oriented in parallel, divergent, and convergent orientations (Figure 3A and Figure S6). As expected, divergent gene pairs tend to be closest, followed by parallel, and finally convergent genes. We then confirmed that each group appears to have different distributions of correlations (Figure 3B and Figure S6). To consider the influence of EP distance, we sampled gene pairs from each orientation controlling for intergenic size. This resulted in distributions of correlations that exactly overlap (Figure 3C and Figure S6), an observation that is supported by previous reports in specific cases (Ghanbarian and Hurst 2015; Cohen et al. 2000). We thus conclude that the apparent influence of gene orientation in the transcriptional relationship of neighboring gene pairs is consistent with the enhancer sharing hypothesis. In this scenario, the effect of gene orientation can be simply explained by the different EP distance distributions for each configuration.
Figure 3.
EP distance causes gene orientation-dependent correlation and provides regulatory independence to gene neighbors. Distribution of intergenic distances < 10 kb of gene pairs in D. melanogaster by configuration (∼5–18,000 gene pairs for each group) (A) and flanking insulator binding sites identified through ChIP-chip (Negre et al. 2010) (∼5–15,000 pairs for each group) (D). The corresponding distribution of correlations is shown for the same gene pairs (B, E) and pairs with controlled distributions of intergenic distances between 30 and 40 kb (∼7–14,000 pairs for gene orientation groups, ∼10–18,000 for insulator groups) (C and F). CHIp-chip, Chromatin ImmunoPrecipitation coupled with microarrays; EP, enhancer–promoter.
From the regulatory perspective, EP distance provides independence to most gene pairs, as the vast majority have an intergenic distance that puts them in the baseline correlation regime (Figure 1C). To study the enhancer-blocking influence of insulators (Bushey et al. 2009) genome-wide, we analyzed each group of genes flanked by insulator binding sites, which were previously determined using ChIP-chip for six known insulators in D. melanogaster: BEAF-32, CP190, CTCF, GAF, Mod(mdg4), and Su(Hw) (Negre et al. 2010). We observed that gene neighbors closer than 10 kb bound by each of the insulators tend to be less correlated in gene expression than gene pairs not bound by them (Figure 3E), supporting their role in genome-wide insulation and agreeing with the observation that insulators tend to separate differentially expressed genes (Negre et al. 2010; Xie et al. 2007). Nevertheless, the same groups of gene pairs also tend to have much larger intergenic distances than genes that are not flanked by insulator binding sites (Figure 3D). After controlling for the distribution of intergenic distances, we found very similar correlation distributions between insulator and not insulator flanked gene pairs (Figure 3F). This finding agrees with previous reports suggesting that insulators do not block enhancers everywhere they bind, but rather act only on very specific genomic contexts (Schwartz et al. 2012; Liu et al. 2015; Ong and Corces 2014); it also reconciles the lack of known insulator orthologs in C. elegans (Heger et al. 2009) in the context of local enhancer-blocking. In combination, these studies strongly suggest that EP distance is the general source of transcriptional independence for close gene neighbors.
Previous EP compatibility studies have suggested that EP specificity is widespread (Gehrig et al. 2009), while others have suggested that it is restricted to a smaller subset of enhancers (Butler and Kadonaga 2001). Although our results support the latter, views arising from these studies might not be mutually exclusive, as it is likely that enhancers have specificity to promoter classes (Danino et al. 2015), whose limited number could result in general EP compatibility.
The implications from considering our observations are broadly applicable to gene regulation. Position effects, in which transgene expression levels are influenced by the insertion site (Gierman et al. 2007), are naturally expected from enhancer sharing. Chromosomal translocations and mutations involving regulatory elements likely impact genetic contexts rather than individual genes. Furthermore, enhancer sharing and distance-based scaling of enhancer influence potentially provides an additional layer of information in gene regulation, as the transcriptional output of a given gene would be the result of scaled contributions from multiple shared enhancers. Such a feature could, by itself, be under selective pressure, leading to a roughly constant size of the correlation domain in number of genes regardless of absolute physical distance, as observed in this study. Our analysis provides a clarifying perspective of gene regulation consistent with both mechanistic and genome-wide studies.
Supplementary Material
Acknowledgments
We thank Zhiping Wang and Yishi Jin for plasmids for CRISPR-Cas9 single-copy insertion; Carmie Robinson for discussions, experimental suggestions, and comments on the manuscript; Han Wang for discussions, technical advice, and comments on the manuscript; Hillel Schwartz, Mitchell Guttman, Mihoko Kato, David Angeles-Albores, Jonathan Liu, Barbara Wold, Isabelle Peter, and Angelike Stathopoulos for discussions and comments on the manuscript; the Encode and ModEncode consortiums, FlyBase, WormBase, and Ensembl databases, the Wold Lab, and the Guigo Lab for data accessibility. Our work was supported by the Howard Hughes Medical Institute, of which P.W.S. is an investigator.
Author contributions: P.Q.C. performed the experiments and analyzed the data. P.Q.C. and P.W.S. designed the experiments and wrote the paper. The authors declare no competing financial interests.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3.116.036228/-/DC1.
Communicating editor: B. J. Andrews
Literature Cited
- Allen M. A., Hillier L. W., Waterston R. H., Blumenthal T., 2011. A global analysis of C. elegans trans-splicing. Genome Res. 21: 255–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araya C. L., Kawli T., Kundaje A., Jiang L., Wu B., et al. , 2014. Regulatory analysis of the C. elegans genome with spatiotemporal resolution. Nature 512: 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attrill H., Falls K., Goodman J. L., Millburn G. H., Antonazzo G., Rey A. J., Marygold S. J., The FlyBase Consortium , 2016. Flybase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 44: D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev A. M., Kalmykova A. I., Shevelyov Y. Y., Nurminsky D. I., 2002. Large clusters of co-expressed genes in the Drosophila genome. Nature 420: 666–669. [DOI] [PubMed] [Google Scholar]
- Bushey A. M., Dorman E. R., Corces V. G., 2009. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol. Cell 32: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. E., Kadonaga J. T., 2001. Enhancer–promoter specificity mediated by DPE or TATA core promoter motifs. Genes Dev. 15: 2515–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calo E., Wysocka J., 2013. Modification of enhancer chromatin: what, how, and why? Mol. Cell 49: 825–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron H., Schaik B. v., Mee M. v. d., Baas F., Riggins G., et al. , 2001. The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science 291: 1289–1292. [DOI] [PubMed] [Google Scholar]
- Chen N., Stein L. D., 2006. Conservation and functional significance of gene topology in the genome of Caenorhabditis elegans. Genome Res. 16: 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen B. A., Mitra R. D., Hughes J. D., Church G. M., 2000. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nat. Genet. 26: 183–186. [DOI] [PubMed] [Google Scholar]
- Corsi A. K., Wightman B., Chalfie M., 2015. A transparent window into biology: a primer on Caenorhabditis elegans. Genetics 200: 387–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danino Y. M., Even D., Ideses D., Juven-Gershon T., 2015. The core promoter: at the heart of gene expression. Biochim. Biophys. Acta 1849: 1116–1131. [DOI] [PubMed] [Google Scholar]
- Davidson E. H., Peter I. S., 2015. Chapter 1 - The genome in development, pp. 1–40 in Genomic Control Process, edited by Davidson E. H., Peter I. S. Academic Press, Oxford. [Google Scholar]
- Dobi K. C., Winston F., 2007. Analysis of transcriptional activation at a distance in Saccharomyces cerevisiae. Mol. Cell. Biol. 27: 5575–5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuy D., Li Q.-R., Deplancke B., Boxem M., Hao T., et al. , 2004. A first version of the Caenorhabditis elegans promoterome. Genome Res. 14: 2169–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebisuya M., Yamamoto T., Nakajima M., Nishida E., 2008. Ripples from neighbouring transcription. Nat. Cell Biol. 10: 1106–1113. [DOI] [PubMed] [Google Scholar]
- Ellahi A., Thurtle D. M., Rine J., 2015. The chromatin and transcriptional landscape of native Saccharomyces cerevisiae telomeres and subtelomeric domains. Genetics 2: 505–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Harrison S. W., Dixon D., 1990. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene 93: 189–198. [DOI] [PubMed] [Google Scholar]
- Flicek P., Amode M. R., Barrell D., Beal K., Billis K., et al. , 2014. Ensembl 2014. Nucleic Acids Res. 42: D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig J., Reischl M., Kalmar E., Ferg M., Hadzhiev Y., et al. , 2009. Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nat. Methods 6: 911–916. [DOI] [PubMed] [Google Scholar]
- Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., et al. , 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanbarian A. T., Hurst L. D., 2015. Neighboring genes show correlated evolution in gene expression. Mol. Biol. Evol. 32: 1748–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavi-Helm Y., Klein F. A., Pakozdi T., Ciglar L., Noordermeer D., et al. , 2014. Enhancer loops appear stable during development and are associated with paused polymerase. Nature 512: 96–100. [DOI] [PubMed] [Google Scholar]
- Gierman H. J., Indemans M. H., Koster J., Goetze S., Seppen J., et al. , 2007. Domain-wide regulation of gene expression in the human genome. Genome Res. 17: 1286–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammonds A. S., Bristow C. A., Fisher W. W., Weiszmann R., Wu S., et al. , 2013. Spatial expression of transcription factors in Drosophila embryonic organ development. Genome Biol. 14: R140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger P., Marin B., Schierenberg E., 2009. Loss of the insulator protein CTCF during nematode evolution. BMC Mol. Biol. 5: 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter J. D., 2007. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9: 90–95. [Google Scholar]
- Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., et al. , 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova A. I., Nurminsky D. I., Ryzhov D. V., Shevelyov Y. Y., 2005. Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res. 33: 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon D., Mucci D., Langlais K. K., Americo J. L., DeVido S. K., et al. , 2009. Enhancer-promoter communication at the Drosophila engrailed locus. Development 136: 3067–3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher M. J., Hurst L. D., 2006. Co-expressed yeast genes cluster over a long range but are not regularly spaced. J. Mol. Biol. 359: 825–831. [DOI] [PubMed] [Google Scholar]
- Lercher M. J., Urrutia A. O., Hurst L. D., 2002. Clustering of housekeeping genes provides a unified model of gene order in the human genome. Nat. Genet. 31: 180–183. [DOI] [PubMed] [Google Scholar]
- Lercher M. J., Blumenthal T., Hurst L. D., 2003. Coexpression of neighboring genes in Caenorhabditis elegans is mostly due to operons and duplicate genes. Genome Res. 13: 238–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Ruotti V., Stewart R. M., Thomson J. A., Dewey C. N., 2010. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26: 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Maurano M. T., Wang H., Qi H., Song C.-z., et al. , 2015. Genomic discovery of potent chromatin insulators for human gene therapy. Nat. Biotechnol. 33: 198–203. [DOI] [PubMed] [Google Scholar]
- Ly K., Reid S. J., Snell R. G., 2015. Rapid RNA analysis of individual Caenorhabditis elegans. MethodsX 2: 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssenko N. N., Hanna-Rose W., Schlegel R. A., 2007. Cognate putative nuclear localization signal effects strong nuclear localization of a GFP reporter and facilitates gene expression studies in Caenorhabditis elegans. Biotechniques 43: 596–600. [DOI] [PubMed] [Google Scholar]
- Malik S., Roeder R. G., 2010. The metazoan mediator co-activator complex as an integrative hub for transcriptional regulation. Nat. Rev. Genet. 11: 761–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsman J., Horsfield J. A., 2012. Long distance relationships: enhancer–promoter communication and dynamic gene transcription. Biochim. Biophys. Acta. 1819: 1217–1227. [DOI] [PubMed] [Google Scholar]
- McKinney W., 2010. Data structures for statistical computing in Python. Proceedings of the 9th Python in Science Conference, edited by van der Walt S., Millman J. pp. 51–56. [Google Scholar]
- Mi H., Poudel S., Muruganujan A., Casagrande J. T., Thomas P. D., 2016. Panther version 10: expanded protein families and functions, and analysis tools. Nucleic Acids Res. 44: D336–D342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak P., 2008. Coexpression, coregulation, and cofunctionality of neighboring genes in eukaryotic genomes. Genomics 91: 243–248. [DOI] [PubMed] [Google Scholar]
- Negre N., Brown C. D., Shah P. K., Kheradpour P., Morrison C. A., et al. , 2010. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 6: e1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema P. G., Harrison S. W., Plunger V., Aryana A., Fire A., 1993. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics 135: 385–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong C.-T., Corces V. G., 2014. CTCF: an architectural protein bridging genome topology and function. Nat. Rev. Genet. 15: 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouedraogo M., Bettembourg C., Bretaudeau A., Sallou O., Diot C., et al. , 2012. The duplicated genes database: identification and functional annotation of co-localised duplicated genes across genomes. PLoS One 7: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purmann A., Toedling J., Schueler M., Carninci P., Lehrach H., et al. , 2007. Genomic organization of transcriptomes in mammals: coregulation and cofunctionality. Genomics 89: 580–587. [DOI] [PubMed] [Google Scholar]
- Quinlan A. R., Hall I. M., 2010. Bedtools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S. S. P., Huntley M. H., Durand N. C., Stamenova E. K., Bochkov I. D., et al. , 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L., Chabanis S., Angrand P. O., Woodroofe C., Stewart A. F., 1999. Quantitative comparison of DNA looping in vitro and in vivo: chromatin increases effective DNA flexibility at short distances. EMBO J. 18: 6630–6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy P. J., Stuart J. M., Lund J., Kim S. K., 2002. Chromosomal clustering of muscle-expressed genes in Caenorhabditis elegans. Nature 418: 975–979. [DOI] [PubMed] [Google Scholar]
- Rubin A. F., Green P., 2013. Expression-based segmentation of the Drosophila genome. BMC Genomics 14: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal A., Lajoie B. R., Jain G., Dekker J., 2012. The long-range interaction landscape of gene promoters. Nature 489: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz Y. B., Linder-basso D., Kharchenko P. V., Tolstorukov M. Y., Kim M., et al. , 2012. Nature and function of insulator protein binding sites in the Drosophila genome. Genome Res. 11: 2188–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M., Duret L., 2006. Evolutionary origin and maintenance of coexpressed gene clusters in mammals. Mol. Biol. Evol. 23: 1715–1723. [DOI] [PubMed] [Google Scholar]
- Singer G. A. C., Lloyd A. T., Huminiecki L. B., Wolfe K. H., 2005. Clusters of co-expressed genes in mammalian genomes are conserved by natural selection. Mol. Biol. Evol. 22: 767–775. [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Rubin G. M., 2002. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 1: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiernagle, T., 2006 Maintenance of C. elegans (February 11, 2006), WormBook, ed. The C. elegans Research Community WormBook, /10.1895/wormbook.1.101.1, http://www.wormbook.org.
- The ENCODE Project Consortium , 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Beaton A., Weiszmann R., Kwan E., Shu S., et al. , 2002. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 3: research0088.1–88.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomancak P., Berman B. P., Beaton A., Weiszmann R., Kwan E., et al. , 2007. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 8: R145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Arensbergen J., van Steensel B., Bussemaker H. J., 2014. In search of the determinants of enhancer–promoter interaction specificity. Trends Cell Biol. 24: 695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams E. J., Bowles D. J., 2004. Coexpression of neighboring genes in the genome of Arabidopsis thaliana. Genome Res. 14: 1060–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J. B. E., Hurst D. L., 2002. Clustering of tissue-specific genes underlies much of the similarity in rates of protein evolution of linked genes. J. Mol. Evol. 54: 511–518. [DOI] [PubMed] [Google Scholar]
- Xie X., Mikkelsen T. S., Gnirke A., Lindblad-toh K., Kellis M., et al. , 2007. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. USA 104: 7145–7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan S., Horrocks J., Lukens L. N., 2006. Islands of co-expressed neighbouring genes in Arabidopsis thaliana suggest higher-order chromosome domains. Plant J. 45: 347–357. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Chen D., Smith M. A., Zhang B., Pan X., 2012. Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PLoS One 7: e31846. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.