Abstract
Chemosensory perception allows insects to interact with the environment by perceiving odorant or tastant molecules; genes encoding chemoreceptors are the molecular interface between the environment and the insect, and play a central role in mediating its chemosensory behavior. Here, we explore how the evolution of these genes in the emerging pest Drosophila suzukii correlates with the peculiar ecology of this species. We annotated approximately 130 genes coding for gustatory receptors (GRs) and divergent ionotropic receptors (dIRs) in D. suzukii and in its close relative D. biarmipes. We then analyzed the evolution, in terms of size, of each gene family as well of the molecular evolution of the genes in a 14 Drosophila species phylogenetic framework. We show that the overall evolution of GRs parallels that of dIRs not only in D. suzukii, but also in all other analyzed Drosophila. Our results reveal an unprecedented burst of gene family size in the lineage leading to the suzukii subgroup, as well as genomic changes that characterize D. suzukii, particularly duplications and strong signs of positive selection in the putative bitter-taste receptor GR59d. Expression studies of duplicate genes in D. suzukii support a spatio-temporal subfunctionalization of the duplicate isoforms. Our results suggest that D. suzukii is not characterized by gene loss, as observed in other specialist Drosophila species, but rather by a dramatic acceleration of gene gains, compatible with a highly generalist feeding behavior. Overall, our analyses provide candidate taste receptors specific for D. suzukii that may correlate with its specific behavior, and which may be tested in functional studies to ultimately enhance its control in the field.
Keywords: gene families, Drosophilidae, gustation, gene birth-and-death, gene duplication, Spotted wing drosophila
Insects rely on stimuli detection to explore the environment for a variety of primary activities, ranging from food localization and mate choice to predator avoidance. Social and environmental chemical cues are perceived and processed through the chemosensory system, resulting in a variety of downstream responses. Thus, genes related to chemosensation are likely to be involved in the evolution of new behaviors and the adaptation to new ecological niches. Taste and smell are the two main mechanisms of chemosensation and they are mostly mediated by three receptor gene families: the highly divergent GR family, found in all arthropods and generally involved in taste; the more recently evolved olfactory receptor (OR) family (Clyne et al. 1999; Gao and Chess 1999; Vosshall et al. 1999), which is found only in insects and is mainly involved in olfaction (Missbach et al. 2014; Robertson 2015); and the ionotropic receptor (IR) family, composed of two distinct subfamilies involved in both taste and olfaction (Benton et al. 2009; Croset et al. 2010; Koh et al. 2014; Stewart et al. 2015).
Taste is used by insects for a variety of key processes, such as detection of toxins present in food, identification of secondary metabolites associated with host plants, and recognition of nonvolatile cuticle pheromones. In insects, and likely in various other arthropods, tastant compounds are detected by specialized hair-like structures, called gustatory sensilla, located in different parts of the insect body (e.g., mouthparts, legs, pharynx, and wings) (Stocker 1994). These sensilla house gustatory neurons (GRNs) that express specific transmembrane receptors, which enable the detection of external contact molecules (Freeman and Dahanukar 2015; Joseph and Carlson 2015). Initially discovered in 1999 (Clyne et al. 2000), GRs are the first and most extensively studied family of chemoreceptors to be found to be expressed in GRNs in insects. Efforts to elucidate the molecular details of their function have been steadily increasing in the past few years, but to date only a limited number of GRs have been deorphanized in a few insect species (Sato et al. 2011; Erdelyan et al. 2012; Freeman and Dahanukar 2015; French et al. 2015; Joseph and Carlson 2015). In D. melanogaster, the GR family includes 60 genes encoding 68 receptor proteins (Robertson et al. 2003; Gardiner et al. 2008; Almeida et al. 2014). Unlike ORs, some GRs are characterized by exhibiting functional plasticity beyond gustatory perception: for example, GR28bD is involved in thermosensation (Ni et al. 2013), while GR21a and GR63a are expressed in specific antennal sensilla and are associated with CO2 detection (Jones et al. 2007; Kwon et al. 2007). Such functional diversification is also revealed by the observation that, in D. melanogaster, GRs are expressed not only in GRNs, but also in other neurons scattered in different tissues (French et al. 2015).
The IRs are a divergent lineage of the ionotropic glutamate receptors (iGluRs), an ancient gene family with a synaptic role in neuronal communication (Benton et al. 2009). In D. melanogaster there are 57 genes coding for IRs, and they have been divided into two subfamilies depending on their expression, which putatively reflect their ecological role (Croset et al. 2010). The first subfamily, antennal IRs (aIRs), is highly conserved among species; it is expressed in olfactory organs and is mainly involved in the detection of air-borne molecules. The second subfamily, dIRs, comprises most the IR genes (around 40 genes in D. melanogaster); they are mainly expressed in GRNs and involved with taste, either alone or in association with GRs (Croset et al. 2010; Koh et al. 2014; Stewart et al. 2015). In contrast to aIRs, dIRs evolved under species-specific patterns, with local expansions and/or losses of family members in certain lineages, likely mirroring the natural history of species.
The evolutionary history of GRs and dIRs in Drosophila has been studied by exploiting the 12 annotated sequenced genomes (Robertson et al. 2003; McBride 2007; McBride et al. 2007; Gardiner et al. 2008; Croset et al. 2010; Almeida et al. 2014). In this work, we expanded these studies with the aim of characterizing the evolution of GRs and dIRs in D. suzukii, a pest of soft fruits characterized by a peculiar reproductive ecology. This species lays eggs inside ripening unwounded soft fruits, providing an interesting case of shift in oviposition preferences when compared to most other drosophilids, which instead oviposit on fermenting substrates (Rota-Stabelli et al. 2013; Asplen et al. 2015). We hypothesize that such behavioral change may have been accompanied by duplications and/or losses in taste receptor genes that allowed gravid females to recognize suitable oviposition sites and/or that enabled larvae to feed on fresh fruits. The availability of the genomes of D. suzukii and of its closely related species D. biarmipes, which does not share oviposition preferences with D. suzukii, enabled us to examine the evolutionary diversification of GRs and dIRs associated with this new ecological context. We previously exploited this comparison to reveal a peculiar evolution of ORs and odorant binding proteins (OBPs) in D. suzukii, and to identify various genes and ligands that likely play an active role in its attraction toward fresh rather than fermenting fruits (Ramasamy et al. 2016). The role of taste receptors in D. suzukii behavior is, on the other hand, still completely unexplored. Here, we have characterized GR and dIR families in D. suzukii and D. biarmipes, studied their evolution in a 14 Drosophila phylogenetic framework, and further analyzed expression profiles of duplicate genes in D. suzukii. We used our results to address three major questions. Is there any evidence of lineage-specific differentiation of GR or dIR repertoires in D. suzukii that may correlate with its peculiar behavior? Are the two taste perception modalities (GRs vs. dIRs) evolving with similar patterns? Finally, do recently duplicated genes have nonredundant spatio-temporal expression patterns that avoid a possible overlap in their function? Our results revealed an unusual burst of gene copies in both GR and dIR families in the branch leading to the suzukii subgroup (which includes both D. suzukii and D. biarmipes), whereas only few genomic changes uniquely characterize D. suzukii taste gene repertoire. We found evidence of a similar evolution pattern for GRs and dIRs not only in D. suzukii, but also in all the analyzed Drosophila. Lastly, reverse transcription PCR (RT-PCR) showed a tissue-specific expression pattern for some duplicate gene families which supports a role of some of them as pheromone receptors. Overall, our dataset is the first describing the complete D. suzukii taste receptor repertoire and suggests that the diversification of bitter receptors has played a central role in the evolution of the suzukii subgroup lineage.
Materials and Methods
Annotation of GRs and IRs
We used tBLASTn (cut-off values 10−5) to iteratively search against D. suzukii (Chiu et al. 2013; Ometto et al. 2013) and D. biarmipes genomes using D. melanogaster GRs and dIRs (obtained from FlyBase release FB2015_02) as query sequences. Coding sequences (CDS) were manually predicted in silico by mapping exons identified in tBLASTn searches using BioEdit (Hall 1999). Introns (following the GT-AG rule) were removed and the remaining sequences were checked for an in-frame coding sequence. D. suzukii or D. biarmipes sequences with indels leading to a premature stop codon were considered pseudogenes. In D. suzukii, GRs and IRs were retrieved from both the Italian (Ometto et al. 2013) and the American (Chiu et al. 2013) genomes. Paralogous duplications were cross-checked between the two genomes; genes that were represented by more copies located in different scaffolds and in only one genome, and which diverged for only a few SNPs, were considered allelic variants that were not well assembled during genome assembly. Genes were named based on the reconstructed phylogenetic tree (see below) and following the D. melanogaster nomenclature, while adding a two-letter prefix corresponding to the species’ names. Orthologs in D. suzukii and D. biarmipes were named with consecutive numbers: for example, IR47a has two copies in D. suzukii and D. biarmipes, which were named as DsIR47a1, DsIR47a2, DbIR47a1, and DbIR47a2. Paralogs whose orthologs could not be clearly identified in the other species were named with consecutive numbers after a point: for example, GR59d8 has three copies in D. suzukii which were named DsGR59d8.1, DsGR59d8.2, and DsGR59d8.3. We did not rename genes with previously published names (e.g., in D. ananassae IR94j or IR94k).
Phylogenetic analysis
Nucleotide sequences of GRs from D. melanogaster, D. pseudoobscura, D. ananassae, and D. erecta were downloaded from FlyBase (release FB2015_02) according to datasets used before (Gardiner et al. 2008; Almeida et al. 2014), whereas dIR sequences were obtained from Croset et al. (2010) (Supplemental Material, Table S2). These species were chosen to recreate the taxon sampling of Ramasamy et al. (2016), which proved to be useful for comparative studies. In cases of mis-annotated genes in species other than D. suzukii and D. biarmipes, sequences retrieved from databases were manually reannotated from whole-genome sequencing data to unify gene structure prediction across species (Table S2). These sequences, together with D. suzukii and D. biarmipes genes, were aligned with TranslatorX (Abascal et al. 2010) using the Muscle algorithm (Edgar 2004), and the resulting alignments were manually checked and edited. Maximum likelihood amino acid-based trees were then calculated with RAxML (Stamatakis 2014) using the PROTGAMMA+LG+F model and bootstrapping the dataset with 100 pseudoreplicates. Trees were viewed and graphically edited using iTOL (Letunic and Bork 2007). Pseudogenes were excluded from the alignments.
Gene birth and death estimation
To infer the number of GRs and dIRs duplications and losses along Drosophila phylogeny, we used Badirate (Librado et al. 2012), which reconciles gene trees onto the species tree. The species tree, which included divergence dates for 14 drosophilid species, was the one proposed by Ometto et al. (2013). The data matrix with the gene numbers for each species was taken and slightly modified from Almeida et al. (2014), except for data for D. persimilis that were added from Croset et al. (2010) and Gardiner et al. (2008) (Table S3). To obtain β (birth rate) and γ (death rate) estimates we applied the BDI-FR-CML method, which uses a full maximum-likelihood approach and assumes independent evolution for each branch along each lineage.
Analysis of selective forces
We used PAML 4.7 (Yang 2007) to infer the rate of nonsynonymous, dN, and synonymous nucleotide substitution, dS, as well as the level of selective pressure acting on a gene (ω = dN/dS). We first created multiple sequence alignments of orthologous genes from six Drosophila species (D. suzukii, D. biarmipes, D. melanogaster, D. erecta, D. pseudoobscura, and D. ananassae) with PRANK (Löytynoja and Goldman 2005) and TranslatorX. In case of paralogs in species other than D. suzukii or D. biarmipes, we used the most conserved isoform compared to the other lineages. In case of duplications in D. suzukii and D. biarmipes, the analysis was repeated for each of the paralogs using the closest ortholog from the other species. Pseudogenes were excluded from the analysis. To estimate ω values in D. suzukii and D. biarmipes, PAML was run using the “free-ratio” model, which allows branch-specific values for ω over all branches of the unrooted phylogenetic tree. Tree topology was taken from Ometto et al. (2013). To evaluate heterogeneity in the selective pressure on D. suzukii or D. biarmipes, we used a branch test that compared the likelihood of a model that assumed a single ω across branches (model = 0 and NSsites = 0) to a second that assumed a distinct ω for the focal branch (D. suzukii or D. biarmipes). To identify the occurrence of positive selected sites along D. suzukii or D. biarmipes branches, we used the branch-site test (branch-site model A, test 2; model = 2 and NSsites = 2; null model has parameters fix_ ω = 1, ω = 1; the positive selection model fix_ ω = 0, ω = 1). In both branch and branch-site tests, the value of twice the difference between the two alternative likelihoods was tested using a χ2 test with one degree of freedom. To account for multiple testing, we estimated the false discovery rate (FDR) using the Benjamini and Hochberg correction (Benjamini and Hochberg 1995).
RT-PCR
Gene expression analysis was carried on D. suzukii adults from a population collected in Trento Province (Italy) and maintained in our laboratory on a standard Drosophila semiartificial diet (Drosophila species stock center, https://stockcenter.ucsd.edu/info/food_cornmeal.php) at a temperature of 23–25°, relative humidity of 65 ± 5%, and 16L:8D photoperiod. We dissected flies from both sexes using forceps, and we pooled males and females to obtain four different samples: heads (n = 20), thoraxes (including wings and legs; n = 10), abdomens (n = 10), and antennae (n = 300 pairs). A fifth sample was composed of third instar larvae (n = 10) that were processed as whole body samples. Additionally, two more samples consisting of female forelegs and male forelegs (n = 100 legs each) were prepared. All samples were placed immediately in cold RNAlater (LifeSciences) and stored at −80° until crushed in Trizol (LifeSciences) using Tissue Lyser II (QIAGEN). Total RNA was extracted followed the Trizol manufacturer’s instructions. Extracted RNA was treated with RNAse-free DNAse (LifeSciences) and then used for first strand cDNA synthesis using Superscript RT III (LifeSciences). One µl of cDNA diluted 1:5 was amplified by PCR with GreenTaq (Promega) according to the manufacturer’s instructions using 32 amplification cycles. To control for genomic DNA contamination, each batch of total RNA underwent a parallel mock reverse transcription step in which the reverse transcriptase was omitted. The cDNA quality was checked by tubulin amplification. PCR primers (listed in Table S4) were first checked against genomic DNA. Two biological replicates were done for each sample, and each amplification was repeated at least twice.
Data availability
Sequence data are presented as File S1 and File S2, and their scaffold locations are listed in Table S1. Alignments used to build Figure 1 and Figure 2 are contained in File S3 and File S4. File S5 contains supplementary figures.
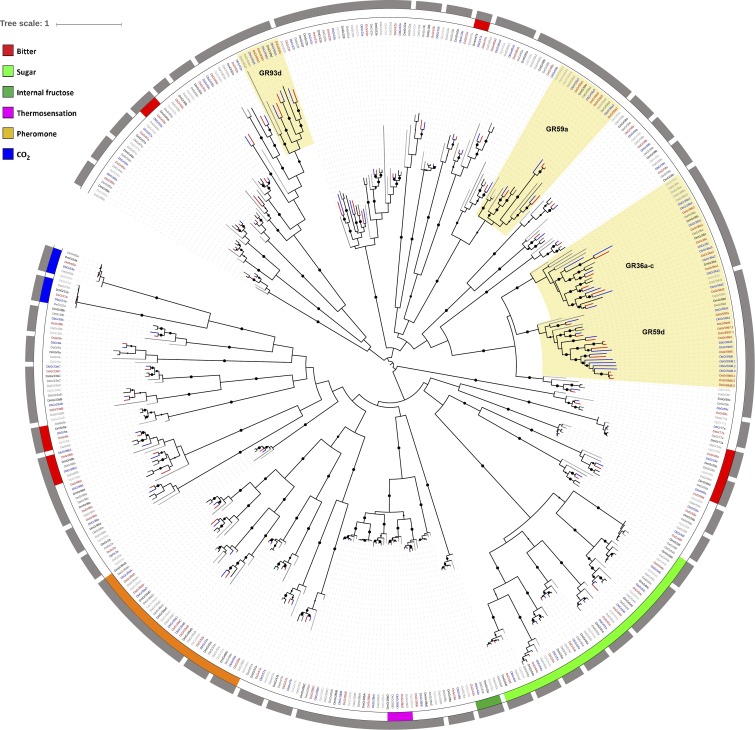
Figure 1.
Gene phylogeny of GRs from six drosophilid genomes. Maximum likelihood tree calculated with RAxML and based on Muscle nucleotide alignment implemented in TranslatorX. Bootstrap support is out of 100 replicates and support > 80 is indicated by black dots whose sizes are according to bootstrap values. Gray half circles identify the gene groups used in birth-death analysis. Branches highlighted in red show D. suzukii genes, and branches highlighted in blue show D. biarmipes genes. D. melanogaster genes are colored in black whereas other Drosophila lineages are shown with different gray shades. Color legends refer to GRs whose function has been deorphanized in D. melanogaster (Joseph and Carlson 2015). Clades highlighted in yellow have undergone specific gene expansion in the suzukii subgroup. GRs, gustatory receptors.
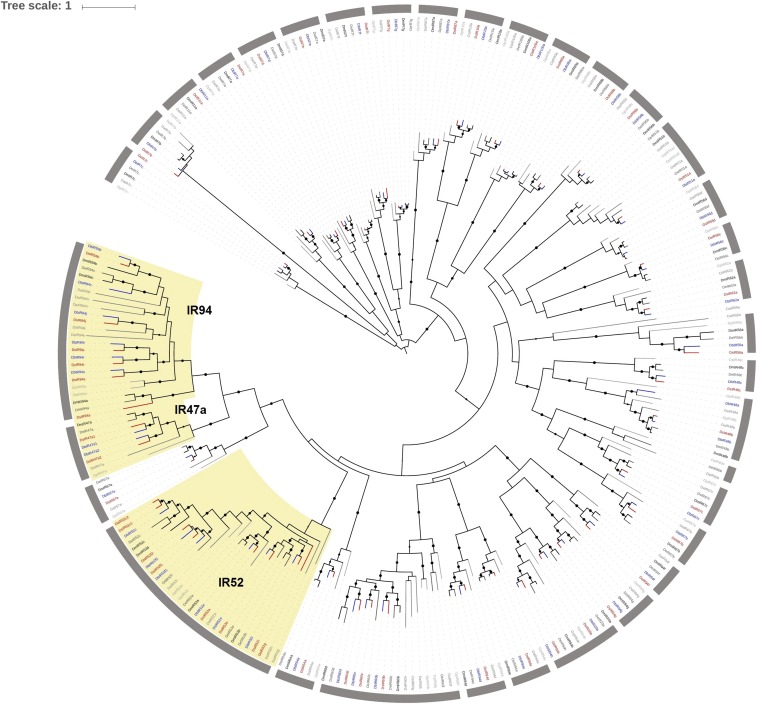
Figure 2.
Gene phylogeny of dIRs from six drosophilid genomes. Maximum likelihood tree calculated with RAxML and based on Muscle nucleotide alignment implemented in TranslatorX. Bootstrap support is out of 100 replicates and support > 80 is indicated by black dots whose sizes are according to bootstrap values. Gray half circles identify the gene groups used in birth-death analysis. Branches highlighted in red show D. suzukii genes, and branches highlighted in blue show D. biarmipes genes. D. melanogaster genes are colored in black whereas other Drosophila lineages are shown with different gray shades. Clades highlighted in yellow have undergone gene expansion in suzukii subgroup. dIRs, divergent ionotropic receptors.
Results
Annotation of GRs and dIRs in the genomes of D. suzukii and D. biarmipes
We manually annotated 77 GRs in D. suzukii and 76 in D. biarmipes. Genomic evidence of alternative splicing was found for three genes in both species (GR23a, GR28b, and GR39a), bringing the total of predicted GR proteins to 85 (File S1) and 84 (File S2), respectively; in D. biarmipes, we identified three pseudogenes (Table S1). All genes have identical intron–exon structures in the two species: exons range from 1 to 9, but most genes have one or two exons, and only two genes are intronless (GR94a and GR68a) (Table S1).
We identified 50 dIRs in the D. suzukii genome and 49 in the D. biarmipes one, and a pseudogene in D. biarmipes (File S1, File S2, and Table S1). Similar to GRs, intron–exon structures of dIRs were the same for both species. dIRs were characterized by less introns than GRs; most genes (around 70%) were intronless, and the remaining ones had one to four introns per gene (Table S1).
GR and dIR evolution in the suzukii subgroup
To obtain a detailed insight into gene duplication and loss in D. suzukii, we inferred the phylogenetic relationships of GRs and dIRs from D. suzukii to that of five other Drosophila species (D. biarmipes, D. melanogaster, D. erecta, D. pseudoobscura, and D. ananassae) (Figure 1 and Figure 2). Using the D. melanogaster orthologs as reference, 66% of GRs have a one-to-one orthologous relationship across the six species, while copy number variation occurs for the remaining genes. In particular, 15 genes are missing in at least one of the six species, whereas 10 have multiple copies in at least one species. Among dIRs, most copy number variation events are mostly species-specific (i.e., families IR52, IR60, and IR94). Considering only the 41 dIRs present in D. melanogaster genome, 68% of them have a one-to-one orthologous relationship across the six species studied.
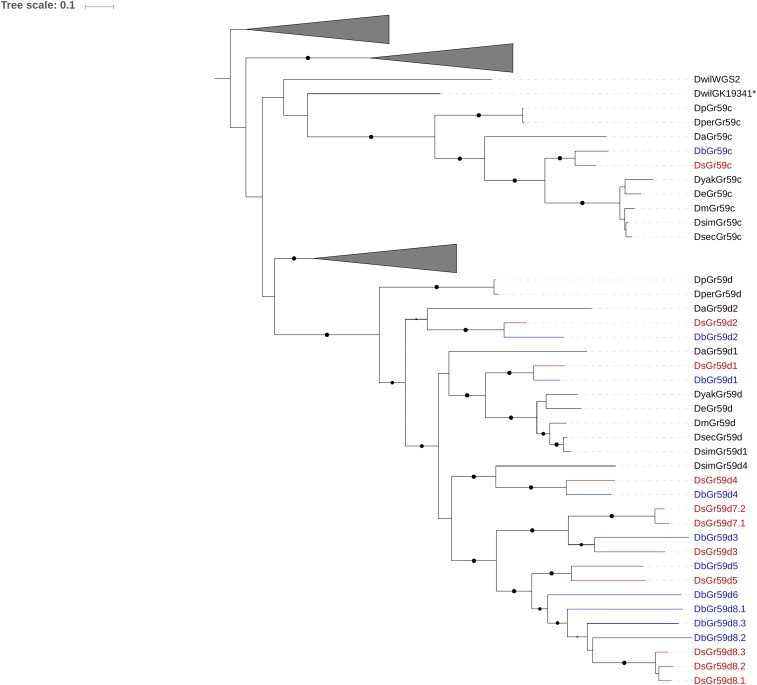
Five GR genes (GR36a, GR36b, GR59a, GR59d, and GR93d) have multiple copies in D. suzukii and D. biarmipes genomes compared to other drosophilids (a total of 17 and 16 more genes than D. melanogaster, respectively) (Figure 1 and Table 1). Based on their orthological relationships, we observed that some triplications and duplications (GR36a, GR36b, GR59a, and GR93d) are clearly shared between D. suzukii and D. biarmipes, and thus they likely originated in the common ancestor of the suzukii subgroup. Other duplications or losses are instead species-specific, such as that observed for GR59a4, which is present only in D. suzukii. The duplication pattern of GR59d is particularly complex (Figure 3). Some duplicated copies of GR59d show a one-to-one orthologous relationship, and their synteny is conserved between the two species (GR59d2, GR59d3, GR59d4, and GR59d5) (Figure S1) indicating that duplication events occurred in the common ancestor of suzukii subgroup. Within the same gene cluster, D. biarmipes possesses an additional copy (GR59d6) (Figure S1) that is not present in the D. suzukii genome. Another copy of GR59d, GR59d7, also originated from duplication in the common ancestor of the suzukii subgroup and later diverged in the two species: in D. biarmipes GR59d7 is likely a pseudogene, whereas in the D. suzukii genome it is present in two paralogous (functional) copies. GR59d8 evolution is extremely problematic; there are three distinct copies in D. suzukii and in D. biarmipes (plus a pseudogene in the latter). In both species, the three copies cluster together in a different scaffold to those containing GR59d1–6 and GR59d7 (Figure S1). Although not well resolved, their phylogenetic affinities inferred by ML tree (Figure 3) suggest various rounds of duplication in the common ancestor of D. suzukii and D. biarmipes followed by various rounds of deletion in D. suzukii, and a recent triplication in D. suzukii. Alternatively, it is possible that the three copies in D. suzukii are evolving in a concerted fashion and experienced gene conversion by homologous recombination, therefore showing high similarity among them.
Table 1. Representative GR and dIR genes characterized by duplication events and signatures of differential selective pressure in D. suzukii.
| Gene | Type of Duplication | Significant Branch Test at FDR < 0.05 | Expression in D. suzukii | Ortholog Expression in D. melanogaster |
|---|---|---|---|---|
| GR36a2 | Suzukii subgroup | ω = 1.25, FDR < 0.0001 | Head | GR36a: labellar sensillaa |
| GR36b2 | Suzukii subgroup | ω = 1.02, FDR = 0.021 | Head | GR36b: labellar sensillaa, larval sensilla innervating terminal distal groupb |
| GR59a3 | Suzukii subgroup | ω = 0.23, FDR = 0.030 | ND | GR59a: labellar sensillaa, foreleg sensillac, larval sensilla innervating terminal distal groupb |
| GR59a4 | Unique to D. suzukii | ω = 0.27, FDR = 0.021 | ND | |
| GR59d2 | Suzukii subgroup | ω = 0.48, FDR = 0.011 | Head, foreleg, larva | GR59d: labellar sensillaa, foreleg sensillac, larval sensilla innervating terminal distal groupb |
| GR59d3 | Suzukii subgroup | ω = 1.2, FDR < 0.0001 | Head, abdomen, larva | |
| GR59d5 | Suzukii subgroup | ω = 0.66, FDR < 0.0001 | Head | |
| GR59d7.1 | Unique to D. suzukii | ω = 0.41, FDR = 0.0066 | ND | |
| GR59d7.2 | Unique to D. suzukii | ω = 0.39, FDR = 0.0114 | ND | |
| GR59d8.1 | Unique to D. suzukii | ω = 1.00, FDR < 0.0001 | Head, larva | |
| GR59d8.2 | Unique to D. suzukii | ω = 1.72, FDR < 0.0001 | Head, larva | |
| GR59d8.3 | Unique to D. suzukii | ω = 1.28, FDR < 0.0001 | Head, larva | |
| IR47a2 | Suzukii subgroup | ω = 0.57, FDR = 0.0105 | Abdomen, thorax, foreleg, head, larva | IR47a: labellar sensillad, foreleg sensillad |
FDR, false discovery rate; ND not detected.
Figure 3.
Gene phylogeny of GR59c and GR59d families from 14 Drosophila species. Maximum likelihood tree calculated with RAxML and based on Muscle nucleotide alignment implemented in TranslatorX. Phylogenetic relationships of all genes included in gene family GR59c-d used in birth-death analysis are displayed in the tree. Bootstrap support is out of 100 replicates and support > 80 is indicated by black dots whose sizes are according to bootstrap values. Branches highlighted in red show D. suzukii genes, and branches highlighted in blue show D. biarmipes genes. Branches without D. suzukii or D. biarmipes genes are collapsed. GRs, gustatory receptors.
Three dIR families (IR47a, IR52, and IR94) belonging to the IR20a clade (Koh et al. 2014) experienced higher turnover in either D. suzukii and D. biarmipes genomes compared with the rest of Drosophila species (Figure 2); IR47a2 and IR52f2 are two new duplicates in both species whereas IR52i is a receptor found only in D. suzukii and D. biarmipes (Figure S2). IR52c is duplicated only in D. suzukii, while D. biarmipes does not have IR52g (Figure 2 and Figure S2). Finally, the IR94 family experienced multiple cases of gene loss and duplication; while D. biarmipes and D. suzukii possess multiple orthologs copies compared to the other Drosophila species (IR94i, IR94r, and IR94s), D. suzukii specifically lost IR94c whereas D. biarmipes lost IR94a (Figure 2 and Figure S3).
Outburst of taste receptor duplication in the branch leading to the suzukii subgroup
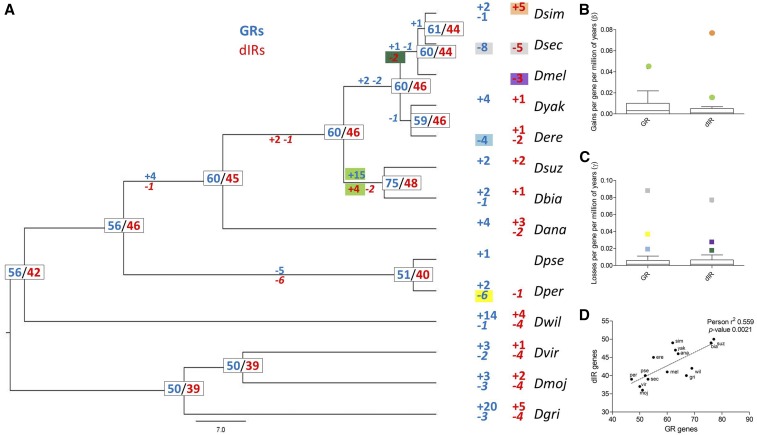
To quantify the number of GR and dIR gain and loss events, we analyzed the gene phylogeny in the context of species phylogeny using Badirate (Figure 4). Because we could not resolve the orthological relationships of GR59d7 duplicates with confidence, we have used a conservative view that implies duplications in the common ancestor of D. suzukii and D. biarmipes. Badirate estimated that along the whole Drosophila phylogeny, GRs experienced 38 losses (global death rate of 0.0022 losses per gene per millions of years, l/g/m) and 87 gains (global birth rate of 0.0045 gains per gene per millions of years, g/g/m), whereas dIRs experienced 43 losses (global death rate of 0.0029 l/g/m) and 38 gains (global birth rate of 0.0025 g/g/m) (Figure 4A). Notably, the highest birth rate for GRs occurs in the branch leading to the suzukii subgroup, where we can observe the gain of 15 GRs (birth rate 0.0439 g/g/m, tenfold higher than the average, Figure 4B). On the same branch, we can observe the second highest birth rate for dIRs (the highest is for D. simulans), characterized by four dIR gains (birth rate 0.0153 g/g/m, sixfold higher than the average, Figure 4B). Particularly high death rates occur for both gene families in the branch leading to the specialist D. sechellia (Figure 4C).
Figure 4.
Gene loss and gain in GR and dIR drosophilid repertoires. (A) Estimates of gene birth and death on internal nodes of Drosophila phylogeny [based on Ometto et al. (2013)]. Numbers on each node represent the estimated number of genes at that internal node whereas numbers on branches represent gene gains and losses. Gains and losses referred to birth rates (β) or death rates (γ), which are an outlier in Tukey boxplots, are highlighted with colors referring to the corresponding Tukey boxplot. (B) Tukey boxplot showing birth rates (β) calculated by Badirate under the BDI-FR-CML model. (C) Tukey boxplot showing death rates (γ) calculated by Badirate under the BDI-FR-CML model. (D) Correlation among number of GR and dIR gene copies along Drosophila phylogeny. dIR, divergent ionotropic receptor; GR, gustatory receptor.
Interestingly, in the 14 drosophilid genomes, the family size of GRs correlates with the family size of dIRs (Figure 4D). D. suzukii and D. biarmipes are the species with the highest number of genes in both families, whereas the two species from the obscura group (D. pseudoobscura and D. persimilis), the specialists D. sechellia and D. mojavensis, and D. virilis have the lowest number of both GRs and dIRs.
Signatures of different selective pressure on genes encoding taste receptors in D. suzukii and D. biarmipes
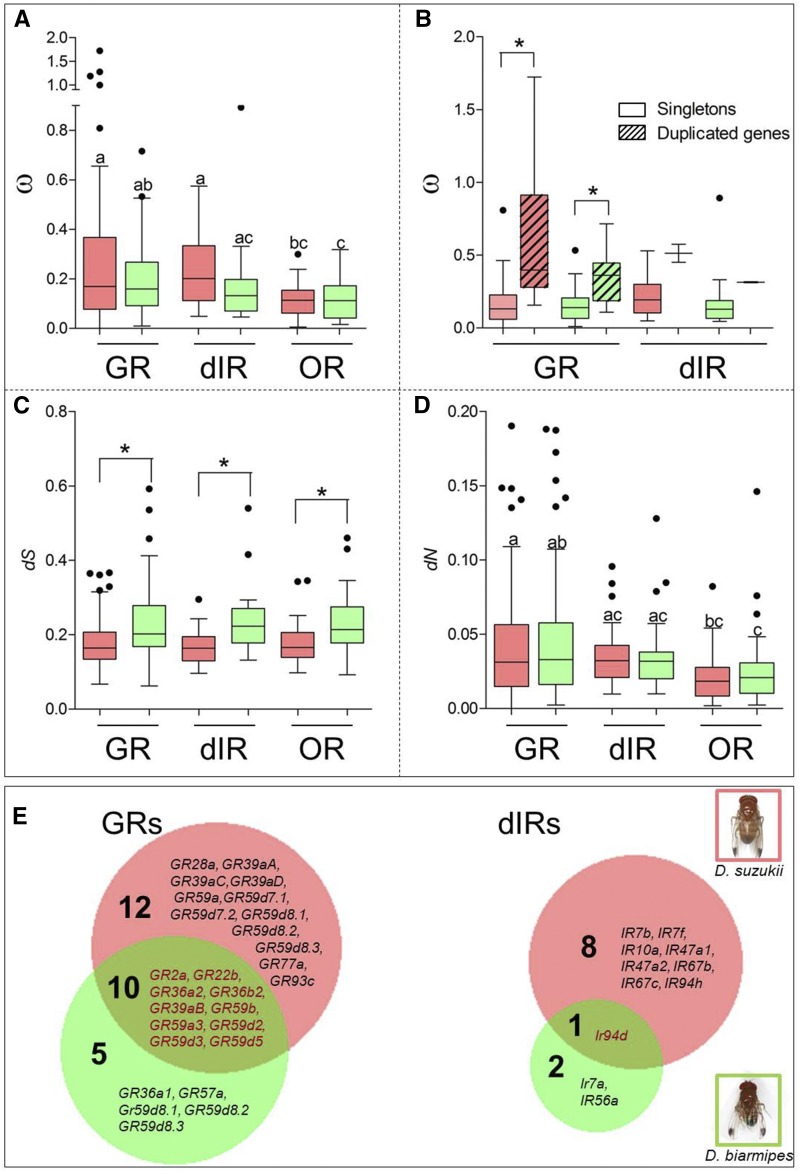
We studied the overall selective pressure acting on GRs and dIRs in D. suzukii and D. biarmipes by examining the dN/dS ratio (ω) at each locus. All tested GRs and dIRs in both species are under a gene-wide moderate to strong purifying selection regime (ω < 1), with the notable exception of six duplicate genes in D. suzukii (from the GR59d, GR36a, and GR36b families) and one (GR36b2) in D. biarmipes (Figure 5A, Table 1, and Table S5). In both species, ω ratios are always higher for duplicate genes compared to nonduplicate ones (Figure 5B). Within both species, the level of selective pressure of GRs is similar to that of dIRs (medians of 0.170 and 0.202 for DsGRs and DsdIRs, respectively; 0.159 and 0.132 for DbGR and DbdIR, respectively), and both are larger than those estimated for ORs (medians of 0.114 and 0.112 for DsOR and DbOR, respectively). Within each class of receptors, no differences between species are evident (Figure 5A). The synonymous substitution rate (dS) does not vary among GRs, dIRs, and ORs within each species (medians for D. suzukii: DsGR = 0.164, DsdIR = 0.164, and DsOR = 0.166; medians for D. biarmipes: DbGR = 0.202, DbdIR = 0.223, and DbOR = 0.214), whereas dS values in D. biarmipes are higher than D. suzukii for all receptors (Figure 5C). The nonsynonymous substitution rate (dN) does not vary among species and taste receptors (medians: DsGR = 0.031, DsdIR = 0.032, DbGR = 0.033, and DbdIR median = 0.032), whereas ORs have the lowest values in both species (medians: DsOR = 0.018 and DbOR = 0.021) (Figure 5D).
Figure 5.
Selective pressure acting on D. suzukii and D. biarmipes chemoreceptor genes. All boxplots were built using values estimated for genes present in the six lineages. OR dataset was taken from Ramasamy et al. (2016). (A) Tukey boxplots showing dN/dS (ω) for GRs, dIRs, and ORs in D. suzukii (pink) and D. biarmipes (green). Significant differences among groups were highlighted by Kruskal–Wallis test (H = 25.33, 5 d.f., and P < 0.0001) followed by Dunn’s multiple comparison post hoc test (different letters indicate significance levels at P < 0.05). (B) Tukey boxplots showing the dN/dS ratios (ω) for singleton (empty plots) or duplicate (striped plots) GRs and dIRs in D. suzukii (pink) and D. biarmipes (green). Differences between singleton and duplicate gene were tested only for GRs in both D. suzukii and D. biarmipes by Wilcoxon test using a Bonferroni-corrected P-value (P < 0.025). (C) Tukey boxplots showing synonymous substitution rate (dS) for GRs, dIRs, and ORs in D. suzukii (pink) and D. biarmipes (green). Significant differences among groups were highlighted by Kruskal–Wallis test (H = 39.61, 5 d.f., and P < 0.0001) followed by Dunn’s multiple comparison post hoc test (P < 0.05). (D) Tukey boxplots showing nonsynonymous substitution rate (dN) for GRs, dIRs, and ORs in D. suzukii (pink) and D. biarmipes (green). Significant differences among groups were highlighted by Kruskal–Wallis test (H = 23.89, 5 d.f., and P < 0.0005) followed by Dunn’s multiple comparison post hoc test (different letters indicate significance levels at P < 0.05). (E) Venn diagrams depicting the number of genes under a differential selective pressure in D. suzukii (pink) and D. biarmipes (green) identified by PAML branch-test at FDR < 0.05. dIR, divergent ionotropic receptor; FDR, false discovery rate; GR, gustatory receptor; OR, olfactory receptor.
Branch tests identified 28 GRs in D. suzukii (22 at FDR < 0.05) and 23 in D. biarmipes (15 at FDR < 0.05) that are evolving under differential selective pressure in either of these species compared to the rest of the phylogeny. Ten of these genes are shared between the two species, seven of which are duplicate genes (Figure 5E and Table S5). Among dIRs, 12 genes in D. suzukii (nine at FDR < 0.05) and five in D. biarmipes (three at FDR < 0.05) have signatures of differential selection pressure, and one of them is shared between the two species (Figure 5E). This corresponds to 38 and 29% of GRs and dIRs under differential selection pressure, respectively, suggesting a very dynamic selective regime in this class of genes. To test if few sites inside GRs or dIRs are evolving under positive selection (but are masked by purifying or relaxed selection acting on the other parts of the gene), we applied a branch-site test and obtained evidence for site-specific selection in D. suzukii and D. biarmipes. Eight GRs (one at FDR < 0.05) and two dIRs (none at FDR < 0.05) were detected as having sites under positive selection in D. suzukii, whereas in D. biarmipes five GRs (two at FDR < 0.05) and one dIR (at FDR < 0.05) were identified (Table S5).
Spatio-temporal expression of duplicate GRs and IRs in D. suzukii
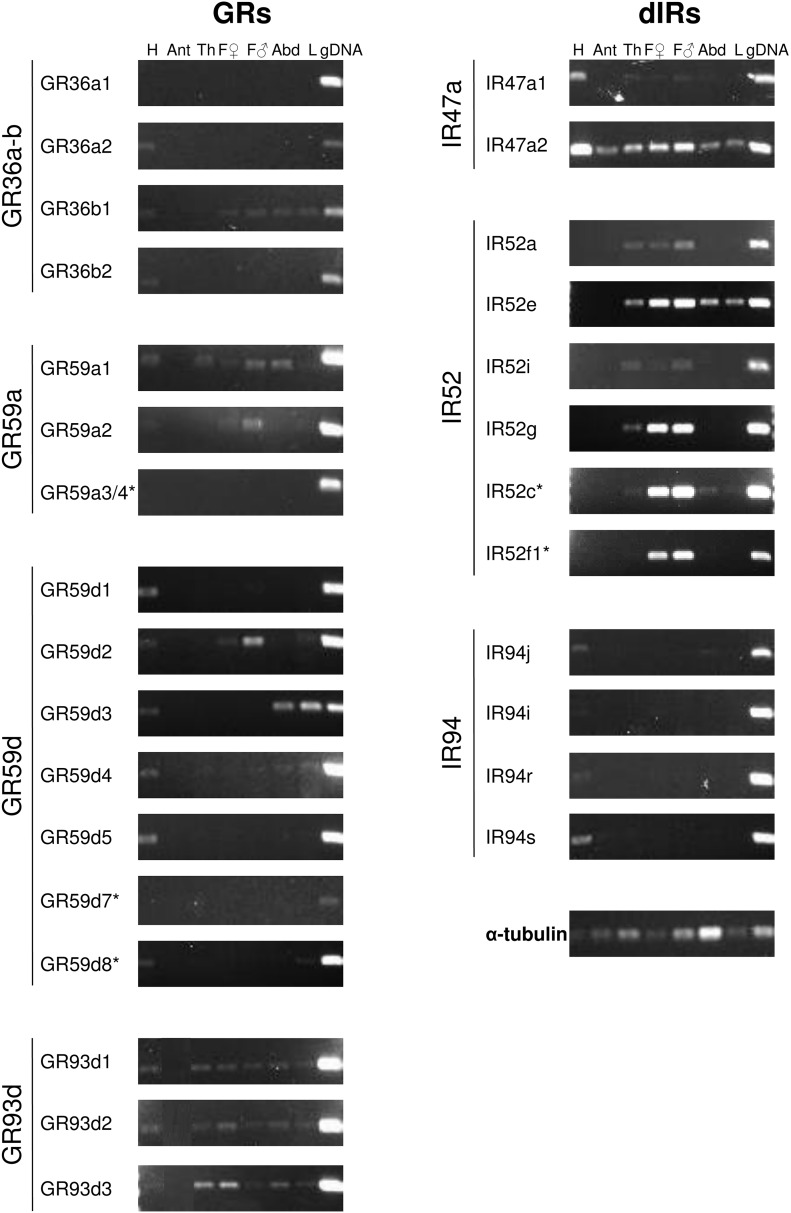
Expression patterns of the genes that underwent duplication in D. suzukii are reported in Figure 6. Family IR52 is mainly expressed in the thorax, specifically in forelegs of both females and males. One IR52 (DsIR52e) is also clearly expressed in larvae; interestingly, no members of this family are expressed in adult heads. DsIR47a2 shows the broadest expression pattern since it is expressed in all tissues and stages tested, whereas expression of DsIR47a1 was mainly detected in the head (a slight band is also observed for thorax and forelegs). Families IR94, GR36a, GR36b, GR59a, and GR59d are mainly expressed in heads, although some specific members are expressed in other tissues or during the larval stage; the three members of the GR59d family (GR59d8.1, GR59d8.2, and GR59d8.3) under positive selection are expressed in the head and in larvae (Table 1). Finally, all the three members of family GR93d are ubiquitously expressed in heads (but not antennae), thorax, and abdomen, as well as in larvae. None of the tested dIRs or GRs (except for IR47a2) were detected in antennae.
Figure 6.
Spatial and temporal expression of duplicate GRs and dIRs in D. suzukii. Gene expression analysis in different D. suzukii adult tissues and in third instar larvae was carried out by RT-PCR. The experiment was performed twice with at least two technical replicates per sample. Samples were run on 1% agarose gel stained with ethidium bromide. Asterisks indicate primer pairs that were not able to discriminate between isoforms. Gel bands for GR93d1, GR93d2, and GR93d3 are shown in a different order than gel loading. Abd, abdomen; Ant, antennae; dIR, divergent ionotropic receptor; F, forelegs; gDNA, genomic DNA; GR, gustatory receptor; H, head; L, larvae; RT-PCR, reverse transcription polymerase chain reaction; Th, thorax.
Discussion
Evolution of taste genes on the Drosophila phylogeny
In the last few years, the availability of numerous Drosophila genomes has allowed several comparative studies to be carried out that have described the evolution of chemosensory genes along the Drosophila phylogeny. Their results have highlighted lineage-specific expansions and contractions that have occasionally been associated with phenotypical differences between species (Guo and Kim 2007; McBride 2007; McBride et al. 2007; Gardiner et al. 2008; Croset et al. 2010; Almeida et al. 2014; Ramasamy et al. 2016). Some of these studies suggested that the evolutionary properties of genes encoding chemosensory receptors correlate with their putative function. For example, genes involved in taste, but belonging to phylogenetically distinct families (GRs and dIRs), evolve under evolutionary constraints that are different from those of genes involved with smell (such as ORs and aIRs) (Croset et al. 2010). Our results confirm this trend by further showing that GRs and dIRs are characterized by a similar lineage-specific turnover rate along the Drosophila phylogeny (Figure 4). Other confirmation comes from our molecular evolution studies; in D. suzukii, GRs and IRs are characterized by a more relaxed selective pressure than that observed in ORs (Figure 5A).
Overall, our average ω values are consistent with purifying selection acting in both chemosensory gene families, as reported for other Drosophila (Gardiner et al. 2008; Croset et al. 2010). Furthermore, faster molecular evolution was found for genes that underwent gene duplication, supporting the hypothesis that recently duplicate genes experience lower selective constraints and thus can be a source of genetic novelty (Lynch and Conery 2000; Kondrashov et al. 2002).
Outburst of taste receptors supports a generalist feeding behavior in the suzukii subgroup
Previous work in Drosophila and other insect species indicated that GR losses are often associated with host specialization. The specialists D. sechellia, which feeds only on Morinda fruits, and to a lesser extent D. erecta, which feeds on Pandanus spp., are losing GRs more rapidly than generalists (McBride 2007; McBride et al. 2007). In the butterflies of the genus Heliconius, whose members are specialized on different Passiflora species, there have been a number of species-specific gene losses in bitter-related GRs (Briscoe et al. 2013). In all such cases, it seems that specialization on a novel host plant is generally associated with a contraction of the GR family. D. suzukii, although being characterized by a peculiar larval feeding ecology, cannot be defined as a specialist; adults feed on fermenting substrates like other Drosophila generalists, and females lay eggs in a great variety of fruits. Accordingly, D. suzukii is not characterized by a reduced number of taste receptors as is seen in specialist species, but rather by the highest number of both GRs and IRs among the sampled Drosophila lineages. This is compatible with a generalist feeding habit, as expanded molecular taste machinery would allow perception of a large assortment of tastants from a wide variety of food sources. This is supported by experimental evidence showing that D. suzukii can oviposit in extremely different fruit species (Lee et al. 2011; Yu et al. 2013; Poyet et al. 2015). Such an increase in taste receptors is shared between D. suzukii and D. biarmipes, suggesting that an ancestral generalist feeding behavior characterized the whole suzukii subgroup, rather than D. suzukii alone.
The association between the burst in number of taste receptor genes and a change in ecology fits well with the observation that gene duplication is a major source of genetic, and phenotypic, novelty. After duplication, redundant genes may experience relaxed selection; their fate will then be defined by a combination of drift and selection, with retained duplications that will experience a distinct regime of purifying selection (Lynch and Conery 2000). Therefore, we can hypothesize that the proportion of duplicate genes in a genome is an excellent genetic proxy for adaptation to new habitats (Makino and Kawata 2012). The burst of GR and dIR duplications in the branch leading to the suzukii subgroup has a magnitude comparable only with the losses that occurred in GRs in the endemic and highly adapted D. sechellia lineage (McBride 2007; McBride et al. 2007); this suggests an adaptive role of GR and dIR duplications in expanding D. suzukii distribution to heterogeneous environments, likely promoting a generalist feeding behavior.
Do selective constraints act on D. suzukii gustative receptors?
One interesting observation that emerges from our molecular evolution analysis is that, in D. suzukii, all chemoreceptor families (GRs, dIRs, and ORs) have a higher rate of nonsynonymous site evolution (dN) compared to that of 1021 nuclear genes estimated in a previous study (Ometto et al. 2013). In contrast, this was not observed for D. biarmipes chemoreceptor genes. Results obtained by Ometto et al. (2013) showed that dN and dS rates were both higher in D. biarmipes compared to D. suzukii and no differences in ω ratio existed between the two species. In our study, we observed an increased dS rate in D. biarmipes for all the gene families tested but, unexpectedly, no differences at dN rate distributions emerged between D. biarmipes and D. suzukii. This points toward an increased evolution rate acting on D. suzukii chemosensory proteins. The increased amino acid substitution rate might be explained by the presence of fewer selective constraints acting on D. suzukii chemoreceptors or by an increase in positive selection shaping the molecular evolution of specific GRs, dIRs, and ORs. In the second case, the fixation of specific genes would lead to a high dispersion in the distribution of dN across the gene family (Betancourt and Presgraves 2002). When we compared ω variances for each gene family between D. suzukii and D. biarmipes, we observed that such a hypothesis accounts only for GRs (Levene’s test: F = 5.40; df = 1131; and P = 0.022). In particular, gene targets of positive selection that are responsible for the higher variance are four duplicate DsGRs (including the three GR59d8, see below) having a ω > 1 (Levene’s test without the four DsGRs: F = 1.33; df = 1131; and P = 0.251). In cases of dIRs and ORs, whose variance do not differ between D. suzukii and D. biarmipes, the increased rate of amino acid substitutions may be due to pervasive relaxed selection, in accordance with what has been observed at the intra specific level in D. melanogaster (Arguello et al. 2016).
Bitter taste receptors putatively relevant for the biology and control of D. suzukii
In terms of gene number, D. suzukii has only one more GR and one more dIR compared to D. biarmipes. However, differences are more pronounced because of species-specific isoforms characterizing some of the duplicate families. The most striking case is the extra duplications of GR59d8 which seems unique to D. suzukii (Figure 3); the exact affinity of these genes to their orthologs in D. biarmipes is hard to decipher and we cannot exclude an ancient origin on the common ancestor followed by different evolutionary histories in the two species. Regardless of their origin, these three genes are characterized by an ω > 1 only in D. suzukii, indicating strong positive selection acting on them (Table 1). We speculate that these paralogs may have played an active role in the adaption of D. suzukii to its fresh fruit polyphagous ovipositing behavior; we therefore consider them good candidates for functional studies and downstream practical application. Notably, these genes are expressed in heads and in the larval stage; therefore, we suggest that they may be involved in oviposition host choice and larval feeding behavior.
The limited knowledge (mostly restricted to D. melanogaster) about the function of most GRs and dIRs makes it difficult to propose a functional ecological role for duplications unique of D. suzukii, as well as for the other duplications shared with D. biarmipes. In fact, most genes whose function has been deorphanized in D. melanogaster do not exhibit copy number variation, with the notable exception of the L-canavanine receptor GR98b (Shim et al. 2015), which is duplicated only in D. biarmipes. However, most of the GRs duplicated in D. suzukii and/or in D. biarmipes (GR36b, GR59a, GR59d, GR93b, and GR98b) are expressed in bitter-sensing neurons in D. melanogaster (Weiss et al. 2011; Ling et al. 2014). This is concordant with what has been observed in Heliconius spp. and D. sechellia, where adaptive gene gain and loss appear to primarily affect GRs presumed to respond to bitter compounds (McBride 2007; McBride et al. 2007; Briscoe et al. 2013). Moreover, the proportion of putative bitter-related genes that experience a different selective pressure in both D. suzukii and D. biarmipes is particularly high (75 and 78% genes, respectively). In general, specialists tend to have a lower number of bitter receptors than generalists, since the latter enter into contact with a larger array of toxic molecules (McBride 2007; McBride et al. 2007; Briscoe et al. 2013). However, exceptions to this rule exist: Bombyx mori, which feeds exclusively on mulberry leaves, has an increased number of bitter-receptors probably used to detect the bitter compounds typical of its host plant (Wanner and Robertson 2008). We can hypothesize that the ancestor of the suzukii subgroup experienced a transition from one ecological niche to another that required the neo/subfunctionalization of newly duplicated genes to allow a wider recognition of bitter-related compounds.
Even more difficult is the prediction of the ecological function of duplicate dIRs, since less information is available compared to GRs. A family that is particularly enriched in D. suzukii (IR52), is mainly expressed in pheromone-sensing neurons located on forelegs in D. melanogaster, and DmIR52c is indeed required for normal copulation behavior (Koh et al. 2014). Duplication in DsIR52c in D. suzukii may be related to a different mating communication system specific for this species, since D. suzukii does not produce the sex pheromone cis-11-octadecenyl acetate (cVA), which is a pheromone basal to Drosophila species (Dekker et al. 2015). Expression studies also support the role of members of family IR52 as putative pheromone receptors in D. suzukii; the expression of all IR52 isoforms has been observed in thorax segments of D. suzukii adults, and more specifically in adult forelegs, but never in heads.
Expression of duplicate dIRs and GRs in different parts of the D. suzukii body (mainly in the head) is consistent with their role as taste receptors, since in the model D. melanogaster they are expressed in GRNs scattered along the body (Joseph and Carlson 2015). Within each family, each member was expressed with a different pattern, with the exception of the GR93d family, whose three members are expressed together in the three parts of the D. suzukii body. Considering the limitations of using RT-PCR to studying the expression of chemoreceptor genes (which only enabled us to examine tissue-specific and not neuron-specific patterns), our results suggest that duplicate GRs and dIRs in D. suzukii might have diverged their temporal and/or spatial expression after duplication, in response to neo-functionalization events.
Conclusions
The analysis of gene gains/losses, molecular evolution, and expression patterns of D. suzukii tastant receptors has shown that GR and dIR gene families experience rapid gene family evolution. In particular, comparison with the closely related species D. biarmipes revealed a high number of gene gains occurred on the branch leading to the suzukii subgroup, whereas few specific genomic events (for instance the GR59d duplications) characterized the D. suzukii lineage. Overall, our results bring us one step closer to understanding D. suzukii innovative ecological behavior and provide a foundation for further studies aiming to disentangle the mechanisms of oviposition preferences, for example providing candidate taste receptors specific for D. suzukii, which may be tested for their ligand affinity and their role in the oviposition behavior of this species.
Supplementary Material
Acknowledgments
We thank the editor and two anonymous reviewers for their fruitful suggestions. This work was funded by Project LExEM (Laboratory of excellence for epidemiology and modeling, http://www.lexem.eu). C.M.C. is recipient of a Marie-Curie FP7-PEOPLE-2013-IEF grant. S.R. has been funded by a PhD fellowship from the FIRS > T [FEM (Fondazione Edmund Mach) International Research School-Trentino] program at FEM, Italy.
Footnotes
Supplemental material is available online at www.g3journal.org/lookup/suppl/doi:10.1534/g3. 116.036467/-/DC1.
Communicating editor: Y. Kim
Literature Cited
- Abascal F., Zardoya R., Telford M. J., 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida F. C., Sánchez-Gracia A., Campos J. L., Rozas J., 2014. Family size evolution in Drosophila chemosensory gene families: a comparative analysis with a critical appraisal of methods. Genome Biol. Evol. 6: 1669–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello J. R., Cardoso-Moreira M., Grenier J. K., Gottipati S., Clark A. G., et al. , 2016. Extensive local adaptation within the chemosensory system following Drosophila melanogaster’s global expansion. Nat Commun. 7: ncomms11855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asplen M. K., Anfora G., Biondi A., Choi D. S., Chu D., et al. , 2015. Invasion biology of spotted wing Drosophila (Drosophila suzukii): a global perspective and future priorities. J. Pest Sci. 88: 469–494. [Google Scholar]
- Benjamini Y., Hochberg Y., 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J.R. Stat. Soc. 57: 289–300. [Google Scholar]
- Benton R., Vannice K. S., Gomez-Diaz C., Vosshall L. B., 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila. Cell 136: 149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt A. J., Presgraves D. C., 2002. Linkage limits the power of natural selection in Drosophila. Proc. Natl. Acad. Sci. USA 99: 13616–13620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briscoe A. D., Macias-Muñoz A., Kozak K. M., Walters J. R., Yuan F., et al. , 2013. Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 9: e1003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu J. C., Jiang X., Zhao L., Hamm C. A., Cridland J. M., et al. , 2013. Genome of Drosophila suzukii, the spotted wing drosophila. G3 (Bethesda) 3: 2257–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne P. J., Warr C. G., Freeman M. R., Lessing D., Kim J., et al. , 1999. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila. Neuron 22: 327–338. [DOI] [PubMed] [Google Scholar]
- Clyne P. J., Warr C. G., Carlson J. R., 2000. Candidate taste receptors in Drosophila. Science 287: 1830–1834. [DOI] [PubMed] [Google Scholar]
- Croset V., Rytz R., Cummins S. F., Budd A., Brawand D., et al. , 2010. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6: e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker T., Revadi S., Mansourian S., Ramasamy S., Lebreton S., et al. , 2015. Loss of Drosophila pheromone reverses its role in sexual communication in Drosophila suzukii. Proc. Biol. Sci. 282: 20143018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C., 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdelyan C. N. G., Mahood T. H., Bader T. S. Y., Whyard S., 2012. Functional validation of the carbon dioxide receptor genes in Aedes aegypti mosquitoes using RNA interference. Insect Mol. Biol. 21: 119–127. [DOI] [PubMed] [Google Scholar]
- Freeman E. G., Dahanukar A., 2015. Molecular neurobiology of Drosophila taste. Curr. Opin. Neurobiol. 34: 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French A., Moutaz A. A., Mitra A., Yanagawa A., Sellier M.-J., et al. , 2015. Drosophila bitter taste(s). Front. Integr. Nuerosci. 9: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Chess A., 1999. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60: 31–39. [DOI] [PubMed] [Google Scholar]
- Gardiner A., Barker D., Butlin R. K., Jordan W. C., Ritchie M. G., 2008. Drosophila chemoreceptor gene evolution: selection, specialization and genome size. Mol. Ecol. 17: 1648–1657. [DOI] [PubMed] [Google Scholar]
- Guo S., Kim J., 2007. Molecular evolution of Drosophila odorant receptor genes. Mol. Biol. Evol. 24: 1198–1207. [DOI] [PubMed] [Google Scholar]
- Hall T., 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41: 95–98. [Google Scholar]
- Jones W. D., Cayirlioglu P., Kadow I. G., Vosshall L. B., 2007. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445: 86–90. [DOI] [PubMed] [Google Scholar]
- Joseph R. M., Carlson J. R., 2015. Drosophila chemoreceptors: a molecular interface between the chemical world and the brain. Trends Genet. 31: 683–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh T. W., He Z., Gorur-Shandilya S., Menuz K., Larter N. K., et al. , 2014. The Drosophila IR20a clade of ionotropic receptors are candidate taste and pheromone receptors. Neuron 83: 850–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov F. A, Rogozin I. B., Wolf Y. I., Koonin E. V, 2002. Selection in the evolution of gene duplications. Genome Biol. 3: RESEARCH0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J. Y., Dahanukar A., Weiss L. A., Carlson J. R., 2007. The molecular basis of CO2 reception in Drosophila. Proc. Natl. Acad. Sci. USA 104: 3574–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J. Y., Dahanukar A., Weiss L. A., Carlson J. R., 2011. Molecular and cellular organization of the taste system in the Drosophila larva. J. Neurosci. 31: 15300–15309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. C., Bruck D. J., Curry H., Edwards D., Haviland D. R., et al. , 2011. The susceptibility of small fruits and cherries to the spotted-wing drosophila, Drosophila suzukii. Pest Manag. Sci. 67: 1358–1367. [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P., 2007. Interactive Tree Of Life (iTOL): an online tool for phylogenetic tree display and annotation. Bioinformatics 23: 127–128. [DOI] [PubMed] [Google Scholar]
- Librado P., Vieira F. G., Rozas J., 2012. BadiRate: estimating family turnover rates by likelihood-based methods. Bioinformatics 28: 279–281. [DOI] [PubMed] [Google Scholar]
- Ling F., Dahanukar A., Weiss L. A., Kwon J. Y., Carlson J. R., 2014. The molecular and cellular basis of taste coding in the legs of Drosophila. J. Neurosci. 34: 7148–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löytynoja A., Goldman N., 2005. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. USA 102: 10557–10562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M., Conery J. S., 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Makino T., Kawata M., 2012. Habitat variability correlates with duplicate content of Drosophila genomes. Mol. Biol. Evol. 29: 3169–3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C. S., 2007. Rapid evolution of smell and taste receptor genes during host specialization in Drosophila sechellia. Proc. Natl. Acad. Sci. USA 104: 4996–5001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride C. S., Arguello J. R., O’ Meara B. C., 2007. Five Drosophila genomes reveal nonneutral evolution and the signature of host specialization in the chemoreceptor superfamily. Genetics 177: 1395–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach C., Dweck H. K. M., Vogel H., Vilcinskas A., Stensmyr M. C., et al. , 2014. Evolution of insect olfactory receptors. eLife 3: e02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni L., Bronk P., Chang E. C., Lowell A. M., Flam J. O., et al. , 2013. A gustatory receptor paralogue controls rapid warmth avoidance in Drosophila. Nature 500: 580–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ometto L., Cestaro A., Ramasamy S., Grassi A., Revadi S., et al. , 2013. Linking genomics and ecology to investigate the complex evolution of an invasive Drosophila pest. Genome Biol. Evol. 5: 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyet M., Le Roux V., Gibert P., Meirland A., Prévost G., et al. , 2015. The wide potential trophic niche of the asiatic fruit fly Drosophila suzukii: the key of its invasion success in temperate Europe? PLoS One 10: e0142785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramasamy S., Ometto L., Crava C. M., Revadi S., Kaur R., et al. , 2016. The evolution of olfactory gene families in Drosophila and the genomic basis of chemical-ecological adaptation in Drosophila suzukii. Genome Biol. Evol. 8: 2297–2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., 2015. The insect chemoreceptor superfamily is ancient in animals. Chem. Senses 40: 609–614. [DOI] [PubMed] [Google Scholar]
- Robertson H. M., Warr C. G., Carlson J. R., 2003. Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 100: 14537–14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rota-Stabelli O., Blaxter M., Anfora G., 2013. Drosophila suzukii. Curr. Biol. 23: R8–R9. [DOI] [PubMed] [Google Scholar]
- Sato K., Tanaka K., Touhara K., 2011. Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. USA 108: 11680–11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim J., Lee Y., Jeong Y. T., Kim Y., Lee M. G., et al. , 2015. The full repertoire of Drosophila gustatory receptors for detecting an aversive compound. Nat. Commun. 6: 8867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A., 2014. RAxML version 8 a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S., Koh T., Ghosh A. C., Carlson J. R., 2015. Candidate ionotropic taste receptors in the Drosophila larva. Proc. Natl. Acad. Sci. USA 112: 4195–4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker R. F., 1994. The organization of the chemosensory system in Drosophila melanogaster: a review. Cell Tissue Res. 275: 3–26. [DOI] [PubMed] [Google Scholar]
- Vosshall L. B., Amrein H., Morozov P. S., Rzhetsky A., Axel R., 1999. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell 96: 725–736. [DOI] [PubMed] [Google Scholar]
- Wanner K. W., Robertson H. M., 2008. The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 17: 621–629. [DOI] [PubMed] [Google Scholar]
- Weiss L. A., Dahanukar A., Kwon J. Y., Banerjee D., Carlson J. R., 2011. The molecular and cellular basis of bitter taste in Drosophila. Neuron 69: 258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- Yu D., Zalom F. G., Hamby K. A., 2013. Host status and fruit odor response of Drosophila suzukii (Diptera: Drosophilidae) to figs and mulberries. J. Econ. Entomol. 106: 1932–1937. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data are presented as File S1 and File S2, and their scaffold locations are listed in Table S1. Alignments used to build Figure 1 and Figure 2 are contained in File S3 and File S4. File S5 contains supplementary figures.
Figure 1.
Gene phylogeny of GRs from six drosophilid genomes. Maximum likelihood tree calculated with RAxML and based on Muscle nucleotide alignment implemented in TranslatorX. Bootstrap support is out of 100 replicates and support > 80 is indicated by black dots whose sizes are according to bootstrap values. Gray half circles identify the gene groups used in birth-death analysis. Branches highlighted in red show D. suzukii genes, and branches highlighted in blue show D. biarmipes genes. D. melanogaster genes are colored in black whereas other Drosophila lineages are shown with different gray shades. Color legends refer to GRs whose function has been deorphanized in D. melanogaster (Joseph and Carlson 2015). Clades highlighted in yellow have undergone specific gene expansion in the suzukii subgroup. GRs, gustatory receptors.
Figure 2.
Gene phylogeny of dIRs from six drosophilid genomes. Maximum likelihood tree calculated with RAxML and based on Muscle nucleotide alignment implemented in TranslatorX. Bootstrap support is out of 100 replicates and support > 80 is indicated by black dots whose sizes are according to bootstrap values. Gray half circles identify the gene groups used in birth-death analysis. Branches highlighted in red show D. suzukii genes, and branches highlighted in blue show D. biarmipes genes. D. melanogaster genes are colored in black whereas other Drosophila lineages are shown with different gray shades. Clades highlighted in yellow have undergone gene expansion in suzukii subgroup. dIRs, divergent ionotropic receptors.