Abstract
Roles for long noncoding RNAs (lncRNAs) in gene expression are emerging, but regulation of the lncRNA itself is poorly understood. We have identified a homeodomain protein, AtNDX, that regulates COOLAIR, a set of antisense transcripts originating from the 3’ end of Arabidopsis FLOWERING LOCUS C (FLC). AtNDX associates with single-stranded DNA rather than double-stranded DNA non–sequence-specifically in vitro, and localizes to a heterochromatic region in the COOLAIR promoter in vivo. Single-stranded DNA was detected in vivo as part of an RNA-DNA hybrid, or R-loop, that covers the COOLAIR promoter. R-loop stabilization mediated by AtNDX inhibits COOLAIR transcription, which in turn modifies FLC expression. Differential stabilization of R-loops could be a general mechanism influencing gene expression in many organisms.
A major factor determining natural variation in flowering in Arabidopsis is quantitative variation in the expression and silencing of the floral repressor gene FLC (1, 2). Multiple pathways regulate FLC, and these converge on cotranscriptional mechanisms involving antisense transcripts (named COOLAIR) and different chromatin pathways (3, 4). One of these pathways is vernalization, when prolonged cold increases COOLAIR transcription and induces a Polycomb-mediated epigenetic silencing of FLC (5, 6). Another is the autonomous pathway, which involves alternative processing of COOLAIR transcripts that causes gene body histone K4 demethylation and FLC down-regulation (4, 7). Because these regulators are conserved through evolution, COOLAIR regulation may have the potential to inform long noncoding RNA (lncRNA) function generally (8, 9).
To investigate COOLAIR regulation, we undertook a forward mutagenesis screen using a luciferase reporter system ( pCOOLAIR:Luc) (fig. S1) (3). eoc1 (enhancer of COOLAIR1) was identified and mapped to a 32,000–base pair (bp) region on chromosome 4 (Fig. 1A and fig. S2, A to D). A G-A transition was detected in the ninth exon of At4g03090 that resulted in a premature stop codon (TGG to TGA). Complementation and allelic analysis confirmed that the enhanced Luc signal in eoc1-1 was due to the mutation of At4g03090 gene, previously named AtNDX (fig. S3, A to C) (10).
Fig. 1. The HD domain protein AtNDX is a regulator of FLC.
(A) A mutant showing enhanced luciferase activity was isolated and called eoc1 (enhancer of COOLAIR1). Luciferase activity is depicted with false color (blue < green < yellow < red < white). (B) eoc1 maps to AtNDX, which encodes a homeodomain-containing protein with two other conserved domains, NDX-A and NDX-B (fig. S4) (10). eoc1-1 is a point mutation; eoc1-2 and eoc1-4 are T-DNA insertion alleles in Ws and Col-0 back ground, respectively. The pink line indicates the recombinant protein used for EMSA (fig. S9A); aa, amino acids. (C) Endogenous COOLAIR RNA is up-regulated in eoc1; expression data are relative to UBC21, normalized to the wild type. Errors are SEM for three biological replicates.
AtNDX is an atypical and highly divergent plant homeodomain (HD)–containing protein conserved in moss, Selaginella, and other flowering plants (Fig. 1B and fig. S4) (10). Analysis of insertion mutants (eoc1-2, eoc1-4) showed that AtNDX represses endogenous COOLAIR expression (Fig. 1, B and C). Relative to wild-type plants, the eoc1-2 and eoc1-4 mutants flowered later (fig. S5B) and their FLC expression was up-regulated (fig. S5C). The defect in COOLAIR expression was rescued by expression of FLAG-tagged AtNDX (fig. S6). AtNDX is expressed predominantly in dividing tissues such as young leaves, root tips, flower buds, and embryos (fig. S7).
The presence of the HD domain (Fig. 1B and fig. S4) and the localization of green fluorescent protein–tagged AtNDX in the nucleus and nucleolus (fig. S7C and fig. S8) prompted us to test whether AtNDX associates with FLC chromatin. Chromatin immunoprecipitation (ChIP) experiments using the FLAG-tagged AtNDX lines showed that AtNDX is enriched at the COOLAIR promoter–FLC terminator region (Fig. 2A). This suggests that the effects of AtNDX on COOLAIR are direct. GmNDX, the homolog of AtNDX in soybean, shows DNA binding via its HD domain (11). However, HD proteins can also bind RNA (12). To test the binding properties of AtNDX, we performed electrophoretic mobility shift assay (EMSA) analysis using a glutathione S-transferase (GST) recombinant protein that included the HD and NDX-B domain (GST-AtNDX; Fig. 1B and fig. S9A). We did not see binding of GST-AtNDX to double-stranded DNA (dsDNA) probes (Fig. 2B and fig. S9), but GST-AtNDX bound single-stranded DNA (ssDNA) in a non–sequence-specific manner (Fig. 2B, fig. S9D, and table S1).No binding to ssRNA, dsRNA, or RNA-DNA hybrids was detected (Fig. 2B, fig. S9, D and E, and table S1).
Fig. 2. AtNDX associates with the COOLAIR promoter in vivo and has ssDNA binding specificity in vitro.
(A) ChIP–quantitative PCR (qPCR) analysis of AtNDX binding along FLC. ACTIN2 was used as a negative control. Errors are SEM of three biological replicates; two repeats of qPCR were tested for each ChIP replicate. The regions tested are shown in Fig. 3B. (B) Recombinant GST-AtNDX binds to ssDNA (one arrow) but not dsDNA (two arrows). Sequences of Ibc3 probes were from (11). Probes 1, 5, and 7 from the FLC locus are shown in the schematic; ssDNA and dsDNA probes were purified and labeled (fig. S9C). GST alone was used as a negative control.
Single-stranded DNA can be formed in vivo during transcription if nascent RNA transcripts invade the dsDNA and anneal to the template strand in the duplex, generating an RNA-DNA hybrid. A three-stranded nucleic acid structure formed by an RNA-DNA hybrid plus a displaced ssDNA strand is called an R-loop (13). R-loops have been considered as transcriptional by-products, but recent data suggest that R-loops may have an impact on gene expression (14–16). In Saccharomyces cerevisiae, R-loops impair RNA polymerase II (Pol II) transcription elongation (16). In mammals, an RNA/DNA helicase called senataxin resolves R-loops, and this helps transcription termination and Pol II release (15). R-loops tend to form in GC-rich genomic regions (17, 18), and recent evidence suggests that R-loop formation may maintain an unmethylated DNA state at promoters with skewed CpG islands, correlating positively with transcriptional activity in mammals (17). The COOLAIR promoter region is rich in GC nucleotides (around 60% GC; fig. S10B), is very low in DNA methylation (19), and has low nucleosome density (fig. S10C). All these features promote R-loop formation.
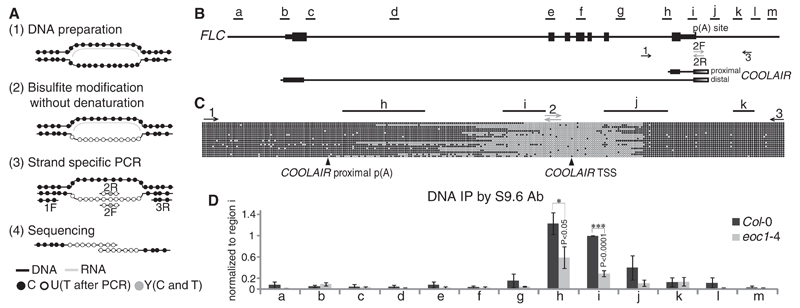
The association of AtNDX with the COOLAIR promoter–FLC terminator (Fig. 2A) and its ssDNA binding capacity led us to test whether R-loops form in this region. We used native sodium bisulfite treatment, which can convert cytosine (C) to uracil (U) if the C bases are located on the unprotected ssDNA strand (Fig. 3A and fig. S11A) (18). The mutation profile of the nontemplate ssDNA region allows definition of the position and length of the R-loop. We found C to U con version only on the noncoding DNA strand [Fig. 3C and figs. S11 and S12; thymine (T) was detected after polymerase chain reaction (PCR) amplification], indicating that R-loops are formed by antisense transcripts. The total length of the R-loop region is about 300 to 700 bp, with a well-defined 5' end starting 200 bp upstream of the multiple COOLAIR transcription start sites (3, 20). This overlaps with the FLC region that we have previously defined as a heterochromatic patch containing Histone H3 dimethyl Lys9 (H3K9me2) and homologous small RNAs (20). The 3’ end of the R-loop is more variable, terminating 100 to 500 bp downstream of the COOLAIR transcription start site window (Fig. 3C) with the longer forms terminating in the region of the COOLAIR proximal polyadenylation site (Fig. 3C). The heterogeneity at the 3’ limit of the R-loop may be caused by different rates of pausing and polymerase drop-off during transcription elongation (16, 21), or more likely cotranscriptional RNA processing (4) with factors such as SR proteins resolving the RNA-DNA hybrids (22).
Fig. 3. AtNDX stabilizes the R-loop in the endogenous FLC terminator.
(A) Schematic steps of the R-loop footprinting method. C’s (black dots) in ssDNA can be converted to U’s (white dots) by bisulfite only in nonprotected DNA (fig. S11A). The length of the R-loop is revealed after strand-specific PCR and sequencing. (B) Illustration of FLC gene structure. Amplicons from regions a to m were used in qPCR for ChIP and DIP. Primers 1 and 3 were native primers (black); primers 2F and 2R (gray) carried C to T conversions. (C) Sequence analysis of individual clones showing R-loop footprinting amplified by 1/2R and 2F/3 primer pairs. Each circle indicates a C nucleotide; empty circles indicate T’s converted from C’s, and solid circles indicate unconverted C’s. (D) DNA immunoprecipitation using RNA-DNA hybrid–specific antibody S9.6. Values of DNA enrichment were divided by input and normalized to region i shown in (B); error bars are SEM of four biological replicates. The amplicons corresponding to R-loop region are shown at the top of (C). *P < 0.05, ***P < 0.001.
We speculated that AtNDX could play a role in R-loop formation or stabilization on the basis of its ssDNA binding capacity (Fig. 2B) and its chromatin association (Fig. 2A). We therefore compared R-loop formation in Col-0 and eoc1 by DNA immunoprecipitation (DIP) using the RNA-DNA hybrid–specific antibody S9.6 (15, 17, 23). We found that the DIP signal was enriched over the COOLAIR promoter (Fig. 3D), was sensitive to ribonuclease H (fig. S13), and was reduced by a factor of ~3 in eoc1 (Fig. 3D, region i). This suggests that the R-loop naturally forms over the COOLAIR promoter, after which AtNDX binds to the displaced nontemplate ssDNA, thereby stabilizing the R-loop structure (Fig. 4D). AtNDX binding may hamper the accessibility of RNA-DNA helicases required to resolve the R-loop, which in turn could affect Pol II initiation and/or elongation (21). The presence of the R-loop and AtNDX binding would then affect initiation and/or elongation of COOLAIR transcription. Nuclear run-on data confirmed that stabilization of the R-loop reduced transcription of the endogenous COOLAIR as well as the reporter Luc fusion (Fig. 4A and fig. S14A). R-loops formed at transcriptional termination sites can affect Pol II pausing and read-through (15, 24). We therefore tested whether the R-loop and AtNDX binding also affected transcription termination of FLC. FLC read-through transcription is unchanged in eoc-1 relative to the wild type (Fig. 4B, regions j, k, and m). However, a role for R-loops in FLC sense transcription termination cannot be excluded, because the R-loop is not fully disrupted in eoc1 (Fig. 3D).
Fig. 4. Reduced R-loop formation releases transcriptional repression of COOLAIR.
(A) Strand-specific nuclear run-on (NRO) analysis (26) of transcription rate of COOLAIR. Error bars are SEM from four biological replicates. (B) Quantification of nascent and read-through transcripts of sense FLC by quantitative reverse transcription PCR. Error bars are SEM of nine biological replicates. Data were normalized to UBC21 and then to region e. (C) FLC expression in the histone K4 demethylase mutant fld (7), fld/eoc1, the RNA-binding protein mutant fca (4), and fca/eoc1 assessed by Northern blotting. (D) Model of how the R-loop influences lncRNA COOLAIR expression. AtNDX stabilizes the R-loop by binding to an ssDNA strand. This may lead to Pol II stalling and/or abortion of transcription (21), or it could affect cotranscriptional RNA metabolism (4, 22). *P < 0.05, ***P < 0.001.
We next addressed how R-loop stabilization and repression of COOLAIR might influence FLC gene expression. eoc1 did not increase FLC expression when combined with autonomous pathway mutants, which suggests that reduced stabilization of the R-loop increases FLC expression via the involvement of COOLAIR in the autonomous pathway mechanism (Fig. 4C). We also asked whether AtNDX changed FLC regulation via perturbation of the gene loop generated through physical interaction of the 5' and 3’ flanking regions of FLC (25). A robust gene loop was still detected in eoc1, suggesting independent activities (fig. S15). Previous analysis had identified a small patch of heterochromatin marked by H3K9me2 and homologous small RNAs immediately downstream of the FLC sense transcript polyadenylation site (20). The small RNAs, which are dependent on the alternative plant RNA polymerase Pol IV (20), were still detected in eoc1 mutants (fig. S16).
R-loops were initially thought to be a rare by-product of transcription but more recently have been found to cause genome instability (13). Our findings indicate that AtNDX homeodomain R-loop stabilization is an important factor regulating expression of the lncRNA COOLAIR. COOLAIR regulation is also influenced by other pathways quantitatively regulating FLC expression and flowering (8). Stabilization of the R-loop thus provides an additional regulatory layer contributing to the robustness of flowering time regulation. R-loop stabilization by ssDNA binding proteins may be a general mechanism influencing gene expression in many organisms.
Supplementary Materials
Materials and Methods
Figs. S1 to S16
Tables S1 to S4
References (27–32)
Editor’s Summary.
Making Antisense of Flowering
The recent discovery of biological roles for long noncoding RNAs raises important questions as to how they are themselves regulated. Sun et al. (p. 619) adopted a genetic approach to identify regulators of COOLAIR––a set of antisense transcripts from the locus encoding the major Arabidopsis floral repressor, FLC. Analysis of a mutant misregulating COOLAIR revealed a homeodomain protein that repressed COOLAIR expression via an R-loop covering the COOLAIR promoter. Thus, R-loop stabilization is an integral part of COOLAIR regulation, FLC expression, and flowering time.
Acknowledgments
We thank all the members of the Dean lab for useful discussions; G. Calder and S. Rosa for help with microscopy; and the Nottingham Arabidopsis Stock Centre (NASC) and Institut National de la Recherche Agronomique (INRA) for Arabidopsis lines. Supported by a Wellcome Trust programme grant (K.S.-S. and N.J.P.). The Dean lab is supported by the UK Biotechnology and Biological Sciences Research Council (BBSRC) and a European Research Council advanced investigator grant. C.D. holds stock in Mendel Biotechnology.
References and Notes
- 1.Shindo C, Lister C, Crevillen P, Nordborg M, Dean C. Genes Dev. 2006;20:3079. doi: 10.1101/gad.405306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coustham V, et al. Science. 2012;337:584. doi: 10.1126/science.1221881. [DOI] [PubMed] [Google Scholar]
- 3.Swiezewski S, Liu F, Magusin A, Dean C. Nature. 2009;462:799. doi: 10.1038/nature08618. [DOI] [PubMed] [Google Scholar]
- 4.Liu F, Marquardt S, Lister C, Swiezewski S, Dean C. Science. 2010;327:94. doi: 10.1126/science.1180278. [DOI] [PubMed] [Google Scholar]
- 5.Angel A, Song J, Dean C, Howard M. Nature. 2011;476:105. doi: 10.1038/nature10241. [DOI] [PubMed] [Google Scholar]
- 6.Song J, Angel A, Howard M, Dean C. J Cell Sci. 2012;125:3723. doi: 10.1242/jcs.084764. [DOI] [PubMed] [Google Scholar]
- 7.Liu F, et al. Mol Cell. 2007;28:398. doi: 10.1016/j.molcel.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 8.Ietswaart R, Wu Z, Dean C. Trends Genet. 2012;28:445. doi: 10.1016/j.tig.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 9.Rinn JL, Chang HY. Annu Rev Biochem. 2012;81:145. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukherjee K, Brocchieri L, Bürglin TR. Mol Biol Evol. 2009;26:2775. doi: 10.1093/molbev/msp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jørgensen JE, et al. Plant Mol Biol. 1999;40:65. doi: 10.1023/a:1026463506376. [DOI] [PubMed] [Google Scholar]
- 12.Rivera-Pomar R, Niessing D, Schmidt-Ott U, Gehring WJ, Jäckle H. Nature. 1996;379:746. doi: 10.1038/379746a0. [DOI] [PubMed] [Google Scholar]
- 13.Aguilera A, García-Muse T. Mol Cell. 2012;46:115. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 14.El Hage A, French SL, Beyer AL, Tollervey D. Genes Dev. 2010;24:1546. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Mol Cell. 2011;42:794. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huertas P, Aguilera A. Mol Cell. 2003;12:711. doi: 10.1016/j.molcel.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 17.Ginno PA, Lott PL, Christensen HC, Korf I, Chédin F. Mol Cell. 2012;45:814. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu K, Chedin F, Hsieh CL, Wilson TE, Lieber MR. Nat Immunol. 2003;4:442. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 19.Finnegan EJ, et al. Plant J. 2005;44:420. [Google Scholar]
- 20.Swiezewski S, et al. Proc Natl Acad Sci USA. 2007;104:3633. doi: 10.1073/pnas.0611459104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rondón AG, Jimeno S, Aguilera A. Biochim Biophys Acta. 2010;1799:533. doi: 10.1016/j.bbagrm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Manley JL. Cell. 2005;122:365. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Boguslawski SJ, et al. J Immunol Methods. 1986;89:123. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 24.Proudfoot NJ. Trends Biochem Sci. 1989;14:105. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- 25.Crevillén P, Sonmez C, Wu Z, Dean C. EMBO J. 2013;32:140. doi: 10.1038/emboj.2012.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu F, Bakht S, Dean C. Science. 2012;335:1621. doi: 10.1126/science.1214402. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Figs. S1 to S16
Tables S1 to S4
References (27–32)