Abstract
Terminating transcription is a highly intricate process for mammalian protein-coding genes. First, the chromatin template slows down transcription at the gene end. Then, the transcript is cleaved at the poly(A) signal to release the messenger RNA.The remaining transcript is selectively unraveled and degraded. This induces critical conformational changes in the heart of the enzyme that trigger termination. Termination can also occur at variable positions along the gene and so prevent aberrant transcript formation or intentionally make different transcripts.These may form multiple messenger RNAs with altered regulatory properties or encode different proteins. Finally, termination can be perturbed to achieve particular cellular needs or blocked in cancer or virally infected cells. In such cases, failure to terminate transcription can spell disaster for the cell.
Genes are defined as regions of the genome that correspond to a single transcription unit (TU), starting from the promoter and ending at the terminator. Although promoters are often well characterized, less is known about the mechanism and regulation of transcriptional termination.
Prokaryotes versus eukaryotes
For prokaryotic genes, protein expression units (cistrons) are usually clustered into tandem arrays transcribed as a single TU, creating a polycistronic messenger RNA (mRNA). Failure to terminate transcription results in the inclusion of extra cistrons in the extended mRNA that may cause the production of unwanted proteins with adverse biological consequences (1). The basic mechanism of termination in Escherichia coli is well defined. Formation of an RNA hairpin structure, immediately followed by an oligo(U) sequence in the nascent transcript, triggers termination (2). Alternatively, the adenosine 5´-triphosphate (ATP)–dependent translocase Rho can promote termination by recognizing a loosely defined C-rich sequence (Rho utilization transcript, RUT) (3). After initial polymerase binding, hexameric Rho translocates and unravels the nascent RNA in association with the elongating polymerase (4). Contacts between an RNA hairpin or Rho and the polymerase somehow trigger conformational changes that switch the polymerase’s enzymatic mode from elongation to termination. In prokaryotes, mRNA translation occurs on transcripts still being made by RNA polymerase (cotranscriptional). Translation elongation along the mRNA template can remove RNA hairpin structures or block access of Rho to RUT sites. Either way, translation can directly regulate termination and the consequent extent of TUs (5).
Eukaryotic gene transcription is fundamentally different from that of prokaryotes, as it occurs in the nucleus, separate from the cytoplasmic translation apparatus. Furthermore, eukaryotes employ three different classes of RNA polymerase (Pol). Pol II transcribes all protein-coding genes to generate mRNA, as well as many noncoding RNAs (ncRNAs). ncRNA can either be abundant and stable, such as small nuclear RNA (snRNA) and small nucleolar RNA (snoRNA), or be present at low levels and rapidly degraded, such as long non-coding RNA (lncRNA) that may run between or overlap with protein-coding genes (6). Pol I transcribes the highly abundant ribosomal RNA (rRNA) precursor, which is cotranscriptionally processed to mature 28S, 18S, and 5.8S rRNA, whereas Pol III transcribes transfer RNA (tRNA) and 5S rRNA. All eukaryotic mRNAs are monocistronic, with a short RNA tract before and a longer one after the coding region (5′ and 3′ untranslated regions, or UTRs). The 5′UTR begins with a 5′ terminal Cap structure, whereas the 3′ UTR ends with a polyadenylate [poly(A)] tail. Both these terminal mRNA modifications are formed as part of pre-mRNA processing that occurs cotranscriptionally and is also coordinated with removal (splicing) of introns that separate the coding exons. These complex RNA processing reactions are all required to generate translatable mRNA, which is then exported through the nuclear pore to sites of cytoplasmic translation.
Failure to terminate transcription in eukaryotic genes may have severe consequences for gene expression. For protein-coding genes arranged in tandem, readthrough transcripts from a non-terminated upstream gene will run into the promoter of the downstream gene and restrict its activity by a process called transcriptional interference (7, 8). This will in turn prevent Cap addition to the downstream gene transcript, as this can only occur on a triphosphorylated 5′ end. For genes arranged in convergent orientation, termination defects may result in the formation of overlapping transcripts that down-regulate gene expression by triggering RNA interference (RNAi) pathways (9). In severe cases, failure of convergent genes to terminate transcription will result in molecular collision between Pol II transcribing opposite DNA template strands (10, 11). Failed termination may also result in Pol II elongation complexes running into regions of the genome undergoing DNA replication. Collision with DNA polymerase complexes may disrupt DNA synthesis and trigger DNA damage and genome instability (12). The extensive lncRNA transcriptome increases the likelihood of potential interference problems between TUs. Failure of lncRNA to terminate transcription may also cause interference with adjacent protein-coding genes (13), even though our current understanding of lncRNA termination is rudimentary.
The advent of high-throughput sequencing has made it possible to visualize nascent transcription with a variety of techniques (14, 15). Consequently, it is now possible to directly map the extent of transcription past the poly(A) site (PAS) of a gene. Many Pol II genes display gradual termination profiles across multiple kilobases, whereas others terminate abruptly soon after the PAS. Here, I describe current understanding of how RNA Pol II terminates transcription, mainly focusing on mammalian protein-coding genes, but also with reference to other eukaryotic systems that exemplify specific features. I will start with a consideration of how the chromatin template signals Pol II to either slow down (pause) or completely stop (arrest). I will then consider how transcript processing and degradation can trigger Pol II termination. Finally, I will describe how termination can often be modulated to allow enhanced gene regulation or perturbed to cause genetic disease.
Transcriptional pausing
Pol II is uniquely endowed with an extra protein segment separate from the main globular enzyme that derives from the carboxyl-terminal domain (CTD) of Rpb1 (16). The CTD plays a critical role in coordinating cotranscriptional RNA processing: capping, splicing, and 3′-end cleavage and poly-adenylation. In mammals, it comprises a relatively unstructured polypeptide of 52 heptad repeats (consensus Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7) that are rich in differentially phosphorylated serine residues. In particular, phospho-Ser2 is associated with gene 3′ ends and interacts with a large complex of cleavage and polyadenylation (CPA) factors that generate mRNA 3′ ends (17–19). A further important difference between Pol II and all other types of RNA polymerase transcription is that Pol II–transcribed genes in mammals vary in length, from a few hundred nucleotides for snRNA genes through to protein-coding genes that may exceed 100 kb in length. It is evident that Pol II must have the capacity to be highly processive; thus, termination mechanisms need to be sufficiently robust to stop this molecular juggernaut (Box 1).
Box. 1.
Juggernaut was the name given for a huge wagon bearing an image of the god Krishna drawn annually in procession at Puri in Orissa, India, from the 1600s. Some devotees were crushed under its wheels in sacrifice. It now means a massive, unstoppable vehicle.
Pol II is capable of directly sensing its passage across a functional PAS. Depletion of key components of CPA, such as cleavage-polyadenylation specificity factor–73 (CPSF-73) and cleavage stimulatory factor–64 (CstF-64), reduces this Pol II pausing effect (15). Apparently, CPA recruitment to the Pol II elongation complex as it traverses the PAS induces an appreciable slow-down effect on Pol II elongation. Furthermore, in vitro transcription experiments indicate that the interaction of CPSF and CstF components with Pol II transcribing through a gene PAS can have marked pausing effects on transcription that result in the gradual release of Pol II from the DNA template (20). This termination process apparently occurs independently of PAS cleavage, arguing that Pol II conformational changes alone can induce substantial levels of transcriptional termination. Other features of the chromatin template may also induce pausing and in turn increase the dwell time of Pol II over the PAS. This will enhance CPA association with the PAS and the consequent 3′-end processing and transcriptional termination. Perhaps the most common type of transcriptional pausing for Pol II is caused by chromatin structure, especially the core nucleosome. Although it is clear that Pol II transcribes nucleosomal templates, it is also the case that nucleosome-free or nucleosome-depleted templates are more readily transcribed.
Pol II pausing can also be induced by hybridization of the nascent transcript with the antisense DNA strand outside the elongation complex. This results in the formation of RNA:DNA hybrids and displacement of the sense DNA strand, a structure referred to as an R loop (21, 22). R-loop formation is favored by the act of Pol II transcription, because the DNA template behind the elongation complex is depleted in nucleosomes that are transiently displaced during the transcription process. The DNA double helix is also torsionally underwound (negatively supercoiled). Once formed, R loops can persist, especially as the RNA:DNA hybrid is thermodynamically more stable than duplex DNA. Also, they are often associated with G-rich regions of transcribed genes because the displaced sense DNA can form stabilizing G quadruplex structures (22). R loops were originally observed in budding yeast when pre-mRNA packaging by the THO complex was inactivated by gene deletion (23). Similarly, defective splicing can lead to enhanced R-loop formation (24). In both cases, accumulation of nascent RNA in close proximity to the underwound, just transcribed DNA template leads to RNA:DNA hybrid formation.
R-loop accumulation induced by mRNA packaging or splicing defects results in increased DNA damage. This is due to the mutagenic nature of the single-strand template DNA in the R loop, which leads to single- and then double-strand breaks with elevated levels of DNA recombination (21). Helicases such as Sen1 in yeast can act to remove these potentially harmful structures. Thus, loss of Sen1 gives an equally severe DNA damage phenotype to loss of THO (25). In mammalian cells, the homolog of Sen1, called Senataxin (mutant genes cause various neurological diseases; for example, ataxia oculomotor apraxia type 2 and amyotrophic lateral sclerosis type 4), is similarly required to resolve R loops but also plays a direct role in promoting more efficient termination (26). The recruitment of Senataxin to terminator regions is likely mediated by the creation of a specific Pol II CTD mark on an arginine residue present at the 7th position of the variant 31st heptad repeat (Arg1810). Symmetric dimethylation of this residue by the methyltransferase PRMT5 recruits first SMN (survival of motor neuron disease associated protein), which then recruits Senataxin. Loss of any of these factors causes an accumulation of R loops and a defect in Pol II termination (27). An alternative Senataxin recruitment pathway involves the DNA repair factor BRCA1. This is recruited to R loops especially at Pol II terminators and in turn directly recruits Senataxin to effect the rapid resolution of R loops, promoting Pol II termination and preventing DNA damage (28).
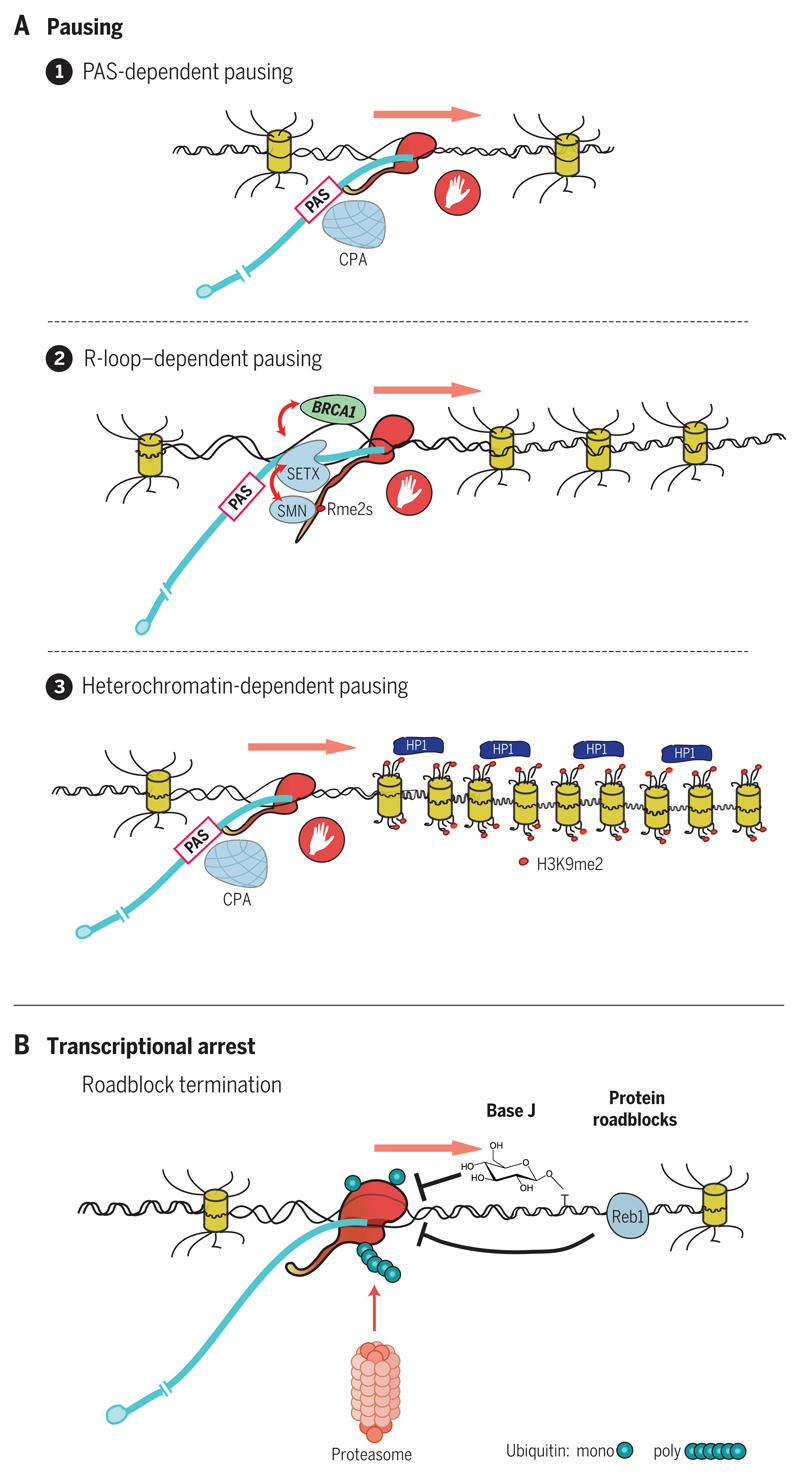
Although R-loop structures display an intrinsic slow-down effect on Pol II elongation, they have also been shown to induce low-level antisense transcription. This may result in the formation of transient double-stranded RNA (dsRNA), which will in turn trigger an RNA interference effect mediated by nuclear Dicer and Ago proteins. This leads to dimethylation of histone H3K9 by the histone methyltransferase enzyme G9a-GLP (G9a-like protein) and consequent recruitment of HP1γ (heterochromatin protein 1γ), effectively creating localized patches of repressed chromatin (29). These will act to perpetuate and enhance Pol II pausing over R-loop–associated termination regions and are a feature of relatively short and ubiquitously expressed genes (Fig. 1A depicts different types of Pol II pausing).
Fig. 1. Pausing or arresting Pol II.
(A) Three different types of Pol II pausing induced by CPA recognition of the PAS, R-loop formation, and heterochromatin patches. Elongating Pol II (red) is shown transcribing the DNA template, with extruded, capped RNA transcript (blue) indicated. Nucleosomes are depicted by yellow barrels, with histone N-terminal tails indicated. Pol II CTD is shown as an extended tail. Red dots on the CTD and histone tails denote methylation. The hand denotes Pol II pausing. (B) Pol II arrested by base J or Reb1 DNA binding protein. Pol II is then ubiquitinated and degraded by the proteasome.
Transcriptional arrest
This involves the irreversible association of Pol II with the DNA template so that it cannot be displaced by PAS-mediated termination mechanisms to allow efficient recycling. Instead, it is targeted for proteolytic degradation (Fig. 1B). PAS-dependent termination may be viewed as a productive mechanism allowing reuse of Pol II, whereas Pol II arrest can be viewed as nonproductive because Pol II is degraded on the DNA template.
The chemical modification or damage of DNA by oxidation or ultraviolet treatment results in arrested Pol II transcription complexes. Ubiquitin ligases are recruited to such complexes, resulting in Pol II degradation, effectively clearing the DNA template to allow new rounds of transcription (30). R loops may also restrict the passage of the replication fork formed in the wake of DNA replication (31). Such collisions between replication and stalled transcription complexes underlie fragile sites in the genome (32). Some trypanosome species convert the DNA base T to glucosyl hydroxymethyl uracil (base J) (33). Base J is found at the end of many TUs, especially those arranged in convergent orientation. Transcript reads terminate precisely at J sites. Deletion of genes encoding the enzymes that perform this T-J conversion result in read-through transcription and ultimately cell death (33). By analogy to DNA damage in mammals, Pol II is likely released from base J roadblocks by ubiquitin-triggered proteolysis. It is, however, unclear how trypanosomes would tolerate such a high turnover of Pol II. Also, the selection mechanism that places base J at the end of TUs has yet to be determined (34).
In Saccharomyces cerevisiae, the DNA binding protein Reb1 is well known to act as a transcription factor for ribosomal protein-coding genes (35). Reb1 also binds promiscuously to intergenic sequences, where it restricts readthrough transcription, acting to prevent global transcriptional interference between genes. Like DNA damage and T-J–mediated termination, Reb1 termination causes Pol II arrest, again requiring ubiquitin-mediated Pol II destruction (36).
Torpedo termination
The 3′ end of eukaryotic mRNA is formed by an RNA processing mechanism whereby the CPA complex assembles onto the pre-mRNA PAS as it is extruded from the RNA exit channel of Pol II. This is facilitated by prior recruitment of CPA to the nearby Pol II CTD (37). Two models are widely cited for how PAS recognition triggers termination. One, dubbed the allosteric model (indicative of Pol II conformational change), proposes that elongating Pol II somehow senses its passage through a functional PAS, as described above (20). Likely, this is caused by the association of the very large CPA complex with Pol II CTD. This in turn induces a conformational change within the Pol II active site, resulting in first pausing and then Pol II release.
The alternative model (38, 39) relates to the nascent transcript still being synthesized by Pol II after cleavage at the PAS. The nuclear 5′-3′ exonuclease Xrn2 is recruited to PAS and progressively degrades this downstream transcript in kinetic competition with ongoing Pol II elongation. When Xrn2 catches up with Pol II, this acts as a molecular trigger to release Pol II from the DNA template. Clearly, pausing of Pol II—caused, for instance, by R-loop–mediated heterochromatic marks (Fig. 1A)—will enhance Xrn2-mediated termination (29). This proposed mechanism is evocatively named the torpedo model where, in naval vernacular, Pol II is the battleship and Xrn2 the torpedo. Direct evidence came from depletion of Rat1 in S. cerevisiae or Xrn2 in mammalian cells provoking a substantial loss in termination (40, 41). Rat1 interacts with Rai1, which possesses both pyrophosphatase and some 5′-3′ exonuclease activity (42), so that both together promote more efficient RNA degradation. A third member of this torpedo complex, called Rtt103, possesses a CTD interaction domain (CID), possibly accounting for Rat1 recruitment to Pol II (40). Other CPA factors may also aid Rat1 recruitment, including Pcf11, which also possesses a CID (43). In general, the depletion of Xrn2 by RNAi technology in mammalian cells produced varying degrees of termination defect, leading to the view that other factors might cooperate with Xrn2 or Rat1 to achieve more efficient termination (15). One such factor is the RNA:DNA helicase Sen1 in yeast or Senataxin in mammals, which may act to expose the cleaved downstream RNA product to Xrn2 degradation. This may be particularly important if RNA is entwined with the DNA template in an R-loop structure (RNA:DNA hybrid) (25, 26). Interestingly, Sen1 displays termination activity independently of RNA degradation (44), indicating that the simple act of unraveling RNA structure is enough to destabilize Pol II and promote termination. Such a mechanism is very similar to the termination activity of E. coli Rho. Although depletion of Xrn2 by RNAi often causes only a marginal termination defect, combining RNAi treatment with expression of dominant negative Xrn2 (active-site mutant) gives a satisfyingly large termination defect for most protein-coding genes (45). It appears that depletion of Xrn2 cellular levels (by RNAi) may not adequately reduce levels of Xrn2 actively engaged in termination.
An underlying feature of productive termination is that 3′-end cleavage of the nascent transcript at the PAS facilitates termination by allowing Xrn2 “torpedo” action. The S. cerevisiae RNAse III Rnt1 recognizes specific hairpin structures and carries out a double endonuclease cut across the hairpin. Several yeast genes use Rnt1 cleavage as alternative 3′-end processing events that allow PAS-independent Xrn2-mediated termination (46, 47). A similar mechanism operates on lncRNA-derived primary microRNAs (lnc-pri-miRNAs), which are independently transcribed rather than more usually residing in the introns of protein-coding genes. lnc-pri-miRNAs are cotranscriptionally cleaved by Drosha, a distant relative of Rnt1. RNA cleavage releases pre-miRNAs, which are then exported from the nucleus and converted into miRNAs by another Rnt1 relative called Dicer. Notably, Drosha cleavage not only generates pre-miRNA but also promotes lncRNA gene termination further downstream, again likely involving the action of Xrn2 degradation (13, 48). To underline the effectiveness of Rnt1 cleavage as a termination mechanism, Pol I transcription is also terminated by Rnt1 cleavage of a hairpin at the 3′ end of rRNA genes. This again elicits termination by a combination of Xrn2 and Sen1 action (49).
A further category of termination has been uncovered wherein Pol II continues to generate an extended transcript multiple kilobases into the gene 3′-flanking region after passage of the PAS. Termination eventually occurs, coincident with a terminal cotranscriptionally cleaved transcript (CoTC sequence), which generates cleavage products closely associated with Pol II. These are degraded by Xrn2 and so promote termination of Pol II in their vicinity. CoTC termination still requires the presence of an upstream PAS. This suggests that the conformational change induced by CPA recognition of the PAS is required for downstream CoTC-mediated termination (50). Cleavage at the PAS to finally release polyadenylated mRNA may occur following CoTC-mediated termination, effectively after the Pol II complex, with its still associated pre-mRNA, is released into the nucleoplasm (51).
Transcriptional backtracking to promote termination
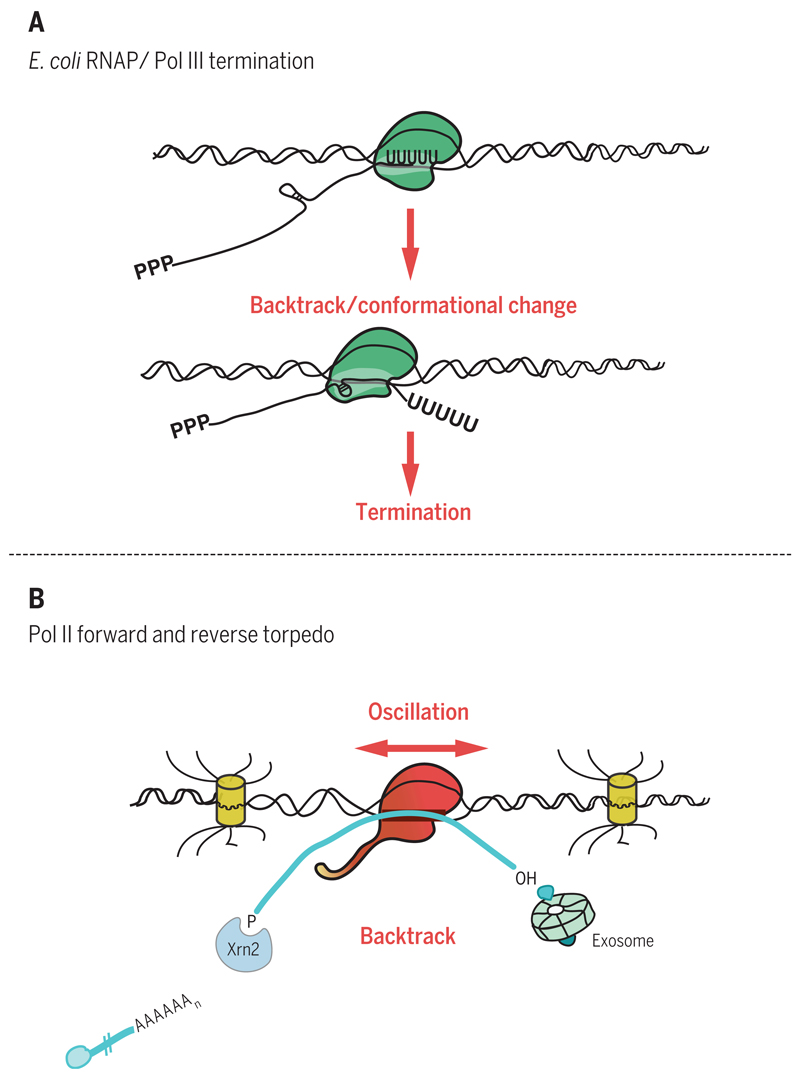
All RNA polymerases can transcribe in both forward and reverse directions on the DNA template. Forward movement results in template-dependent RNA synthesis, with the nascent transcript emerging from the RNA exit channel. Backward movement (backtracking) results in extrusion of the already synthesized nascent transcript out of the secondary channel (also referred to as the nucleotide entry channel) (52). During transcriptional elongation, such backtracking is widely used by Pol II as a proofreading mechanism. The general transcription factor TFIIS enhances an intrinsic endonuclease activity of Pol II that promotes cleavage of the mismatched extruded RNA. This allows transcription to resume, reinstating the correct nucleotide into the nascent RNA (53, 54). For both E. coli RNA polymerase and eukaryotic Pol III, backtracking has been directly implicated in termination. For intrinsic termination in E. coli, a model is envisaged wherein oligo(U) sequences promote polymerase pausing, which then favors backtracking. If an RNA hairpin forms on the upstream transcript, it can be forced into the RNA exit channel, which in turn triggers a conformational change in the polymerase that promotes its release from the DNA template (2). Pol III appears to adopt a similar strategy, as transcript 3′ ends are normally oligo(U) sequences, which pause the polymerase and so encourage backtracking. If backward polymerase movement encounters a hairpin structure, then termination ensues (55) (Fig. 2A).
Fig. 2. Pol II backtracking.
(A) Bacterial RNA polymerase or Pol III terminates at an oligo(U) transcript, which pauses polymerase and promotes backtracking. An upstream RNA hairpin is forced into polymerase active site, inducing a conformational change that results in termination. (B) Pol II moves forward to synthesize or backward to extrude transcript (oscillation). Forward transcript, once cleaved at PAS to release mRNA, is then degraded by Xrn2. Backtracked transcript is degraded by the exosome. Removal of RNA up to Pol II (forward or reverse torpedo) induces termination.
Pol II may employ a related backtracking mechanism to promote termination. In the yeast Schizosaccharomyces pombe, inactivation of the RNA exosome displays a clear general termination defect, which is counteracted by simultaneous loss of TFIIS (56). Because the multisubunit exosome possesses two separate 3′-5′exonucleases, one of these may act on the extruded RNA formed by backtracking. This may push the polymerase further backward and by so doing, induce conformational changes in the Pol II active site that promote termination. Further evidence for such a mechanism comes from in vitro termination experiments using purified yeast Pol II, Rat1, and Rai1 together with immobilized DNA templates, where transcription is artificially blocked by omitting specific nucleotides (57). Although earlier in vitro experiments failed to observe Pol II termination with these minimal components (58), substantial termination is observed if the wrong nucleotide is forced onto the transcript 3′ end, so arresting Pol II at this mismatch position. This has the effect of inducing Pol II backtracking and also, remarkably, promotes termination when coupled with degradation of the upstream transcript by Xrn2 up to the arrested Pol II (57). In effect, these experiments argue that the act of removing RNA up to or into the Pol II active site by either degradation of backtracked extruded transcript (reverse torpedo) or degradation of upstream RNA up to backtracked Pol II (forward torpedo) induces conformational changes to Pol II that promote termination (Fig. 2B).
Premature termination versus transcriptional elongation
Saccharomyces cerevisiae possesses a secondary Pol II termination mechanism that operates on short Pol II transcripts. This involves the NRD complex (Nrd1, Nab3, and Sen1) (59), which promotes termination of ncRNA, particularly that derived from antisense promoter activity associated with promoters of protein-coding genes. It also functions on genes encoding small stable RNA, snRNA, and snoRNA and plays a major role in regulating a subset of protein-coding genes by promoting their premature termination. NRD promotes termination in a sequence-specific manner (through RNA recognition domains on Nrd1 and Nab3) and recruits the exosome to rapidly degrade these transcripts. This degradation will occur unless the transcripts are protected by RNA binding proteins that package snRNA and snoRNA into functional splicing or RNA modification complexes, respectively.
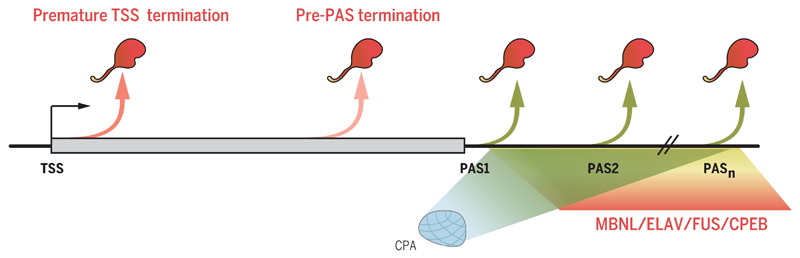
Although mammalian genes have no clear counterpart to NRD, most protein-coding genes display substantial promotor proximal pausing. This is manifested by an accumulation of actively transcribing Pol II localized to the first few hundred nucleotides of the gene (60). In contrast, S. cerevisiae genes show little Pol II pausing at the promoter. It appears that the mammalian transcriptome, possibly because of its greater complexity, has evolved mechanisms of transcriptional regulation more focused on postinitiation events. However, if these early transcripts are not intermediates waiting to be converted into full-length gene transcripts and are therefore abortive, they need to be terminated. Bona fide termination clearly operates on these TSS (transcription start site) transcripts. First, decapping of the transcript can occur by Dcp1 action, followed by Xrn2 degradation (61). Second, mis-spliced transcripts are somehow detected by nuclear surveillance and are similarly degraded by decapping and Xrn2 degradation (62). Promoter-proximal PASs are also thought to be actively recognized by CPA and Xrn2 to promote early termination. Thus, depletion of either CPA components or Xrn2 increases levels of TSS-associated transcripts (15). Some of these early terminated TSS transcripts form hairpin structures, which directly act as functional pre-microRNA without the involvement of the microprocessor (63). It is apparent that TSS-associated termination may be regulated by 5′ splice sites, which block promoter proximal PASs and thereby favor continued elongation into the gene body. This phenomenon was first shown in viruses such as HIV-1 (64), but has also been revealed as a general mechanism that acts to block cryptic PAS recognition and consequent premature termination. In particular, depletion of U1 snRNA activates TSS-proximal PASs as well as numerous PASs present across genes, often within their extensive intronic regions (65).
Transcription elongation is tightly controlled across genes (66). Specific check points operate to enforce premature termination that acts to prevent inappropriate transcription (Fig. 3). Early transcription elongation is restricted by two negative elongation factors, DRB-sensitivity–inducing factor (DSIF) and negative elongation factor (NELF), which are regulated by the major Pol II CTD Ser2 kinase Cdk9 (cyclin-dependent kinase 9). As well as acting on the CTD, this kinase further phosphorylates DSIF and NELF. Thus, when Cdk9 is experimentally inhibited by drugs such as 5,6-dichloro-1-B-d-ribofuranosylbenzimidazole (DRB) or KM05382 (KM), a substantial increase in TSS-proximal transcripts is observed, with greatly reduced transcription downstream into the gene body (67). Once Pol II escapes from TSS-proximal checkpoints and elongation is fully under way, then numerous elongation factors come into play. These promote efficient transcription across the TU, be it a modest 1-kb or much longer 1-Mb gene. Many of these elongation factors act to remodel nucleosomes encountered by elongating Pol II, as well as to coordinate efficient and often regulated (alternative) intron splicing (66). Recently, a further example of regulated premature termination has been observed near the end of gene TUs (3′-end checkpoint) that may act as a final control mechanism to prevent the production of a translatable but potentially flawed mRNA. Like the TSS checkpoint, the 3′-end checkpoint is controlled by Cdk9 activity. In this case, the substrate may be Xrn2, which is substantially activated by Cdk9-mediated phosphorylation (68). Inhibition of Cdk9 causes a nonproductive termination mechanism that still appears to use component parts of CPA, but apparently does not allow the formation of functional polyadenylated mRNA (67).
Fig. 3. Pol II alternative 3′-end formation.
Diagram depicting early release of Pol II prior to PAS or alternative PAS selection at gene 3′ ends. This later process is mediated by competitive association of CPA versus other RNA binding factors with alternative PAS.
Alternative polyadenylation and termination
Termination at gene 3′ ends is also subject to intense regulation. Many mRNAs possess variable lengths of 3′-untranslated sequence defined by the selective usage of different PASs (69). Because mRNA 3′UTRs define mRNA cytoplasmic functions, including RNA stability, translatability, and localization, the use of alternative poly(A) sites (APA) can constitute a key regulatory process in gene expression. However, the differential stability of different mRNA 3′UTR isoforms in the cytoplasm must be distinguished from actual PAS selection during pre-mRNA synthesis. Analyzing total cellular mRNA isoform levels will mainly show up differential stability, whereas analysis of nuclear mRNA isoform levels more closely reflects PAS selection. Indeed, nuclear APA shows rather less variation and consequently gene regulation than initially envisaged (70). An issue often overlooked is whether APA constitutes alternative RNA processing alone or whether selective termination defines which PAS is used. Furthermore, APA may also occur at more internal positions within a gene where it will result in truncated mRNA, which in some cases can be translated into shorter protein isoforms with important biological functions. A classic example of this phenomenon is the alternative use of an internal PAS in immunoglobulin M (IgM) that generates secreted antibody versus a distal PAS that produces membrane-bound antibody. This APA switch is regulated by levels of CstF-64, which is depleted in early B cells and favors distal PAS selection (71). In most examples of APA (as in IgM), the first (proximal) PAS will have a weaker match to the consensus sequence features than the downstream (distal) PAS (72). As mentioned above, Pol II pausing will also have notable effects here, as pausing between proximal and distal PAS favors the proximal PAS. Once a PAS is selected, this will trigger Pol II termination downstream and so preclude distal PAS usage (72).
Several specific conditions are known to affect APA. The kinetics of transcription can have a major effect. Also, rapidly proliferating cells favor proximal PAS usage (69). How a gene promoter recruits elongation factors such as the PAF complex can further influence downstream Pol II elongation and consequent PAS selection (73). Finally, directly slowing down Pol II elongation by specific Pol II mutation can favor proximal PAS usage (74). Either slowing down or speeding up Pol II elongation by specific Pol II mutation can be seen to globally shorten or extend Pol II TUs, implying effects on both APA and coupled termination (45). The availability of CPA factors can also affect APA. Systematic depletion of individual CPA components by RNAi treatment can influence APA (75, 76). For example, depletion of CFI components and PABPN1 reduces, whereas Pcf11 and Fip1 depletion enhances, distal PAS usage. However, it is unclear whether modulation of CPA factor concentration occurs naturally. CF1m25 is overexpressed in glioblastoma cells, resulting in global proximal PAS selection (77). Also, PABN1 can be lost in a rare form of muscular dystrophy called OPMD (oculopharyngeal muscular dystrophy), where a triplet expansion in the gene generates an inactive protein containing N-terminal polyalanine. Loss of PABN1 causes a clear APA shift to distal PAS usage (78, 79). U1snRNA, already mentioned as a means to block premature PAS usage at upstream gene positions, can also affect APA. Thus. lowering U1snRNA levels favors proximal PAS usage. Possibly in activated neurons, U1snRNA may be sufficiently depleted to favor proximal PAS usage (80). A series of non-CPA RNA binding factors are known to be tightly regulated in amount and/or cellular location. These include CPEB, FUS, ELAV, and MBNL (81–84). All of these factors display some degree of RNA binding specificity, and where their binding sites are near PAS, may directly influence CPA binding and recruitment (Fig. 3).
The actual mechanism by which APA may be regulated in these diverse situations remains largely unknown. However, the consequences of an mRNA having a more or less extended 3′UTR clearly relates to key functions of these mRNA regulatory regions, including microRNA binding sites and RNA stability elements. Notably, the actual selection of different PAS can also result in the recruitment of protein factors to the 3′UTR, which, when the mRNA is translated, can directly associate with the nascent protein to modulate its function (85).
Misregulated termination
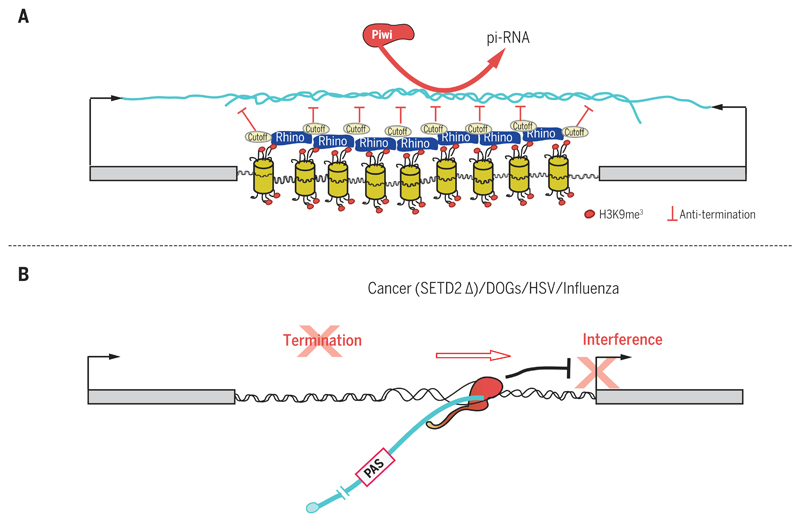
Transcriptional termination is a highly robust process capable of stopping the Pol II juggernaut at gene 3′ ends. It is therefore paradoxical that this basic mechanism appears to be readily subverted for particular cellular or viral needs. First, a notable feature of repressed termination can be seen in the primary Piwi-interacting RNA (piRNA) clusters of Drosophila. These clusters harbor a wide range of transposon-related sequences and in Drosophila are often transcribed on both DNA strands. Primary piRNAs are processed into small, 20-nucleotide piRNAs by dedicated RNA processing enzymes, including specific argonaute proteins. piRNAs act to block the spread of transposons and retroposons in a wide range of eukaryotes by targeting transposon-derived mRNA, using a mechanism analogous to that of miRNA (86). Notably, piRNA clusters lack independent promoters, but are positioned between convergent genes so that they are transcribed by readthrough transcription (87–89). Owing to dsRNA formation in these clusters, the primary piRNA chromatin is marked by H3K9me3, normally a feature of repressed, heterochromatin protein 1 (HP1)–associated heterochromatin. However, these TUs are bound by a distinct HP1 protein called Rhino, which is associated with other proteins including a Rai1 homolog called Cutoff. Rhino enhances rather than represses Pol II transcription across the piRNA TUs, and Cutoff further aids this read-through transcription process by blocking Pol II termination. Cutoff may act as a dominant negative regulator of Xrn2 (45). Not only are the normal termination sites of the flanking convergent genes blocked, thereby promoting efficient Pol II read-through transcription across both strands of the primary piRNA TUs, but terminators prevalent in transposon termini are similarly restricted (Fig. 4A).
Fig. 4. Regulated or misregulated Pol II termination.
(A) Primary piRNA clusters in Drosophila are often positioned between convergent genes. dsRNA induces heterochromatic histone tail modification (H3K9me3). This in turn recruits the HP1-like factor Rhino together with Cutoff, an anti-terminator that promotes readthrough transcription. (B) Inactivation of termination by cancer mutation (SETD2 mutation), osmotic stress, or viral infection all induce Pol II read-through and interference with downstream gene expression.
Further examples of blocked termination come from cells responding to stress. The artificial induction of osmotic stress in cultured cells results in a large number of genes failing to terminate efficiently at the normal gene 3′ end. Instead, readthrough transcripts are detectable that may extend through intergenic regions and invade downstream genes. These “downstream of gene” transcripts (DOGs), though potentially deleterious owing to interference effects, have been postulated to possess protective features for the overall integrity of the nucleus under cellular stress (90). Similarly, cancer cells display complex readthrough transcription profiles that may be related to DOGs (91–93). In renal cancer, mutations in the methyltransferase gene SETD2 correlate with readthrough transcription profiles (94). Setd2 adds the histone H3K36me3 mark to genic nucleosomes, which is required for Pol II elongation and termination. Setd2 also cooperates with Pol II elongation factors and facilitates Pol II CTD phospho-Ser2 formation, which will ultimately lead to CPA recruitment and termination (95).
Viral infection may be considered an extreme form of cellular stress. Remarkably, at least two viruses are known to drastically perturb the termination efficiency of their host genomes. Thus, influenza virus essentially blocks host transcription termination, genome-wide. In detail, the viral protein NS1 has high affinity for CPSF-30 and through this interaction destroys CPA complex integrity. This leads to a general loss in 3′-end processing with commensurate readthrough transcription (96). Herpes simplex virus, upon infecting host cells, similarly causes a massive misregulation of host gene transcription. Extensive readthrough transcription occurs with all the associated interference and mis-splicing of these extended transcripts (97) (Fig. 4B). The destruction of normal regulated Pol II termination in host cells infected with these common human viruses likely explains their pathogenicity.
Conclusions
This Review charts our increasing understanding of how transcriptional termination affects many aspects of eukaryotic gene expression. Far from acting as a constitutive mechanism to separate TUs across the genome, termination can be seen as an intricate process that displays remarkable flexibility and regulatory potential. At the beginning of the gene, termination regulates transcript release into productive elongation. It also acts as a checkpoint to prevent the synthesis of defective mRNA, which could be translated into a toxic (dominant negative) protein. At the end of the gene, termination dictates which mRNA isoform is formed by APA, thereby conferring selective expression properties on the mRNA. Finally, termination can be overridden to adjust cells to stress conditions or to adapt cells into a more pliable host for viral replication. It is likely that future analysis of the termination process has yet more surprises in store.
An end to gene transcription.
Much attention has been focused on regulating the start of gene transcription. But transcription must also be terminated, and the mechanisms are only now being defined in detail in eukaryotes. Proudfoot reviews how termination happens for RNA polymerase II genes, mainly in mammals, covering the various steps that can lead to messenger RNA (mRNA) 3 ′ end formation and how they can be regulated. Termination can occur at various positions throughout the gene, forming the wild-type mRNA, preventing the synthesis of aberrant mRNAs, or generating alternative mRNAs with different regulatory or coding properties.
Acknowledgments
I thank my lab colleagues, especially M. Dye and H. Mischo, for advice on the text. I am indebted to H. Mischo for the illustrations. The expanding field of Pol II termination has generated a large literature. I apologize that space restricts its wider citation. My laboratory is supported by a Wellcome Trust Investigator Award (107928/Z/15/Z) and European Research Council Advanced Grant (339270).
References and Notes
- 1.Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 2.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 3.Richardson JP. Preventing the synthesis of unused transcripts by Rho factor. Cell. 1991;64:1047–1049. doi: 10.1016/0092-8674(91)90257-Y. [DOI] [PubMed] [Google Scholar]
- 4.Epshtein V, Dutta D, Wade J, Nudler E. An allosteric mechanism of Rho-dependent transcription termination. Nature. 2010;463:245–249. doi: 10.1038/nature08669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henkin TM, Yanofsky C. Regulation by transcription attenuation in bacteria: How RNA provides instructions for transcription termination/antitermination decisions. BioEssays. 2002;24:700–707. doi: 10.1002/bies.10125. [DOI] [PubMed] [Google Scholar]
- 6.Gloss BS, Dinger ME. The specificity of long noncoding RNA expression. Biochim Biophys Acta. 2015;1859:16–22. doi: 10.1016/j.bbagrm.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Greger IH, Proudfoot NJ. Poly(A) signals control both transcriptional termination and initiation between the tandem GAL10 and GAL7 genes of Saccharomyces cerevisiae. EMBO J. 1998;17:4771–4779. doi: 10.1093/emboj/17.16.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shearwin KE, Callen BP, Egan JB. Transcriptional interference—a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gullerova M, Proudfoot NJ. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell. 2008;132:983–995. doi: 10.1016/j.cell.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 10.Prescott EM, Proudfoot NJ. Transcriptional collision between convergent genes in budding yeast. Proc Natl Acad Sci USA. 2002;99:8796–8801. doi: 10.1073/pnas.132270899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hobson DJ, Wei W, Steinmetz LM, Svejstrup JQ. RNA polymerase II collision interrupts convergent transcription. Mol Cell. 2012;48:365–374. doi: 10.1016/j.molcel.2012.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaillard H, García-Muse T, Aguilera A. Replication stress and cancer. Nat Rev Cancer. 2015;15:276–289. doi: 10.1038/nrc3916. [DOI] [PubMed] [Google Scholar]
- 13.Dhir A, Dhir S, Proudfoot NJ, Jopling CL. Microprocessor mediates transcriptional termination of long noncoding RNA transcripts hosting microRNAs. Nat Struct Mol Biol. 2015;22:319–327. doi: 10.1038/nsmb.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Core LJ, Waterfall JJ, Lis JT. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science. 2008;322:1845–1848. doi: 10.1126/science.1162228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nojima T, et al. Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell. 2015;161:526–540. doi: 10.1016/j.cell.2015.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vannini A, Cramer P. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 17.McCracken S, et al. The C-terminal domain of RNA polymerase II couples mRNA processing to transcription. Nature. 1997;385:357–361. doi: 10.1038/385357a0. [DOI] [PubMed] [Google Scholar]
- 18.Heidemann M, Hintermair C, Voß K, Eick D. Dynamic phosphorylation patterns of RNA polymerase II CTD during transcription. Biochim Biophys Acta. 2013;1829:55–62. doi: 10.1016/j.bbagrm.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 19.Hsin JP, Manley JL. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang H, Rigo F, Martinson HG. Poly(A) signal-dependent transcription termination occurs through a conformational change mechanism that does not require cleavage at the poly(A) site. Mol Cell. 2015;59:437–448. doi: 10.1016/j.molcel.2015.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Aguilera A, García-Muse T. R loops: From transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression. Genes Dev. 2014;28:1384–1396. doi: 10.1101/gad.242990.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jimeno S, Rondón AG, Luna R, Aguilera A. The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J. 2002;21:3526–3535. doi: 10.1093/emboj/cdf335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005;122:365–378. doi: 10.1016/j.cell.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 25.Mischo HE, et al. Yeast Sen1 helicase protects the genome from transcription-associated instability. Mol Cell. 2011;41:21–32. doi: 10.1016/j.molcel.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011;42:794–805. doi: 10.1016/j.molcel.2011.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao DY, et al. SMN and symmetric arginine dimethylation of RNA polymerase II C-terminal domain control termination. Nature. 2016;529:48–53. doi: 10.1038/nature16469. [DOI] [PubMed] [Google Scholar]
- 28.Hatchi E, et al. BRCA1 recruitment to transcriptional pause sites is required for R-loop-driven DNA damage repair. Mol Cell. 2015;57:636–647. doi: 10.1016/j.molcel.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014;516:436–439. doi: 10.1038/nature13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Svejstrup JQ. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem Sci. 2010;35:333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 31.Santos-Pereira JM, Aguilera A. R loops: New modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 32.Helmrich A, Ballarino M, Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol Cell. 2011;44:966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 33.van Luenen HG, et al. Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania. Cell. 2012;150:909–921. doi: 10.1016/j.cell.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Genest PA, et al. Defining the sequence requirements for the positioning of base J in DNA using SMRT sequencing. Nucleic Acids Res. 2015;43:2102–2115. doi: 10.1093/nar/gkv095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hartley PD, Madhani HD. Mechanisms that specify promoter nucleosome location and identity. Cell. 2009;137:445–458. doi: 10.1016/j.cell.2009.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Colin J, et al. Roadblock termination by reb1p restricts cryptic and readthrough transcription. Mol Cell. 2014;56:667–680. doi: 10.1016/j.molcel.2014.10.026. [DOI] [PubMed] [Google Scholar]
- 37.Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr Opin Cell Biol. 2004;16:272–278. doi: 10.1016/j.ceb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Connelly S, Manley JL. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 39.Proudfoot NJ. How RNA polymerase II terminates transcription in higher eukaryotes. Trends Biochem Sci. 1989;14:105–110. doi: 10.1016/0968-0004(89)90132-1. [DOI] [PubMed] [Google Scholar]
- 40.Kim M, et al. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature. 2004;432:517–522. doi: 10.1038/nature03041. [DOI] [PubMed] [Google Scholar]
- 41.West S, Gromak N, Proudfoot NJ. Human 5′ —> 3′ exonuclease Xrn2 promotes transcription termination at co-transcriptional cleavage sites. Nature. 2004;432:522–525. doi: 10.1038/nature03035. [DOI] [PubMed] [Google Scholar]
- 42.Xiang S, et al. Structure and function of the 5′—>3′ exoribonuclease Rat1 and its activating partner Rai1. Nature. 2009;458:784–788. doi: 10.1038/nature07731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Licatalosi DD, et al. Functional interaction of yeast pre-mRNA 3′ end processing factors with RNA polymerase II. Mol Cell. 2002;9:1101–1111. doi: 10.1016/S1097-2765(02)00518-X. [DOI] [PubMed] [Google Scholar]
- 44.Porrua O, Libri D. A bacterial-like mechanism for transcription termination by the Sen1p helicase in budding yeast. Nat Struct Mol Biol. 2013;20:884–891. doi: 10.1038/nsmb.2592. [DOI] [PubMed] [Google Scholar]
- 45.Fong N, et al. Effects of transcription elongation rate and Xrn2 exonuclease activity on RNA polymerase II termination suggest widespread kinetic competition. Mol Cell. 2015;60:256–267. doi: 10.1016/j.molcel.2015.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rondón AG, Mischo HE, Kawauchi J, Proudfoot NJ. Fail-safe transcriptional termination for protein-coding genes in S. cerevisiae. Mol Cell. 2009;36:88–98. doi: 10.1016/j.molcel.2009.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ghazal G, et al. Yeast RNase III triggers polyadenylation-independent transcription termination. Mol Cell. 2009;36:99–109. doi: 10.1016/j.molcel.2009.07.029. [DOI] [PubMed] [Google Scholar]
- 48.Ballarino M, et al. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol Cell Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kawauchi J, Mischo H, Braglia P, Rondon A, Proudfoot NJ. Budding yeast RNA polymerases I and II employ parallel mechanisms of transcriptional termination. Genes Dev. 2008;22:1082–1092. doi: 10.1101/gad.463408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.West S, Proudfoot NJ, Dye MJ. Molecular dissection of mammalian RNA polymerase II transcriptional termination. Mol Cell. 2008;29:600–610. doi: 10.1016/j.molcel.2007.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nojima T, Dienstbier M, Murphy S, Proudfoot NJ, Dye MJ. Definition of RNA polymerase II CoTC terminator elements in the human genome. Cell Reports. 2013;3:1080–1092. doi: 10.1016/j.celrep.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nudler E. RNA polymerase backtracking in gene regulation and genome instability. Cell. 2012;149:1438–1445. doi: 10.1016/j.cell.2012.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sigurdsson S, Dirac-Svejstrup AB, Svejstrup JQ. Evidence that transcript cleavage is essential for RNA polymerase II transcription and cell viability. Mol Cell. 2010;38:202–210. doi: 10.1016/j.molcel.2010.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Izban MG, Luse DS. The RNA polymerase II ternary complex cleaves the nascent transcript in a 3′—— 5′ direction in the presence of elongation factor SII. Genes Dev. 1992;6:1342–1356. doi: 10.1101/gad.6.7.1342. [DOI] [PubMed] [Google Scholar]
- 55.Nielsen S, Yuzenkova Y, Zenkin N. Mechanism of eukaryotic RNA polymerase III transcription termination. Science. 2013;340:1577–1580. doi: 10.1126/science.1237934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lemay JF, et al. The RNA exosome promotes transcription termination of backtracked RNA polymerase II. Nat Struct Mol Biol. 2014;21:919–926. doi: 10.1038/nsmb.2893. [DOI] [PubMed] [Google Scholar]
- 57.Park J, Kang M, Kim M. Unraveling the mechanistic features of RNA polymerase II termination by the 5′-3′ exoribonuclease Rat1. Nucleic Acids Res. 2015;43:2625–2637. doi: 10.1093/nar/gkv133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dengl S, Cramer P. Torpedo nuclease Rat1 is insufficient to terminate RNA polymerase II in vitro. J Biol Chem. 2009;284:21270–21279. doi: 10.1074/jbc.M109.013847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mischo HE, Proudfoot NJ. Disengaging polymerase: Terminating RNA polymerase II transcription in budding yeast. Biochim Biophys Acta. 2013;1829:174–185. doi: 10.1016/j.bbagrm.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Adelman K, Lis JT. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat Rev Genet. 2012;13:720–731. doi: 10.1038/nrg3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brannan K, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Davidson L, Kerr A, West S. Co-transcriptional degradation of aberrant pre-mRNA by Xrn2. EMBO J. 2012;31:2566–2578. doi: 10.1038/emboj.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xie M, et al. Mammalian 5′-capped microRNA precursors that generate a single microRNA. Cell. 2013;155:1568–1580. doi: 10.1016/j.cell.2013.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ashe MP, Griffin P, James W, Proudfoot NJ. Poly(A) site selection in the HIV-1 provirus: Inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev. 1995;9:3008–3025. doi: 10.1101/gad.9.23.3008. [DOI] [PubMed] [Google Scholar]
- 65.Kaida D, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature. 2010;468:664–668. doi: 10.1038/nature09479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kwak H, Lis JT. Control of transcriptional elongation. Annu Rev Genet. 2013;47:483–508. doi: 10.1146/annurev-genet-110711-155440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laitem C, et al. CDK9 inhibitors define elongation checkpoints at both ends of RNA polymerase II-transcribed genes. Nat Struct Mol Biol. 2015;22:396–403. doi: 10.1038/nsmb.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sansó M, et al. P-TEFb regulation of transcription termination factor Xrn2 revealed by a chemical genetic screen for Cdk9 substrates. Genes Dev. 2016;30:117–131. doi: 10.1101/gad.269589.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tian B, Manley JL. Alternative cleavage and polyadenylation: The long and short of it. Trends Biochem Sci. 2013;38:312–320. doi: 10.1016/j.tibs.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Neve J, et al. Subcellular RNA profiling links splicing and nuclear DICER1 to alternative cleavage and polyadenylation. Genome Res. 2016;26:24–35. doi: 10.1101/gr.193995.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell. 1996;87:941–952. doi: 10.1016/S0092-8674(00)82000-0. [DOI] [PubMed] [Google Scholar]
- 72.Proudfoot NJ. Ending the message: Poly(A) signals then and now. Genes Dev. 2011;25:1770–1782. doi: 10.1101/gad.17268411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nagaike T, et al. Transcriptional activators enhance polyadenylation of mRNA precursors. Mol Cell. 2011;41:409–418. doi: 10.1016/j.molcel.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pinto PA, et al. RNA polymerase II kinetics in polo polyadenylation signal selection. EMBO J. 2011;30:2431–2444. doi: 10.1038/emboj.2011.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li W, et al. Systematic profiling of poly(A)+ transcripts modulated by core 3′ end processing and splicing factors reveals regulatory rules of alternative cleavage and polyadenylation. PLOS Genet. 2015;11:e1005166. doi: 10.1371/journal.pgen.1005166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Martin G, Gruber AR, Keller W, Zavolan M. Genome-wide analysis of pre-mRNA 3′ end processing reveals a decisive role of human cleavage factor I in the regulation of 3′ UTR length. Cell Reports. 2012;1:753–763. doi: 10.1016/j.celrep.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 77.Masamha CP, et al. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jenal M, et al. The poly(A)-binding protein nuclear 1 suppresses alternative cleavage and polyadenylation sites. Cell. 2012;149:538–553. doi: 10.1016/j.cell.2012.03.022. [DOI] [PubMed] [Google Scholar]
- 79.de Klerk E, et al. Poly(A) binding protein nuclear 1 levels affect alternative polyadenylation. Nucleic Acids Res. 2012;40:9089–9101. doi: 10.1093/nar/gks655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berg MG, et al. U1 snRNP determines mRNA length and regulates isoform expression. Cell. 2012;150:53–64. doi: 10.1016/j.cell.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bava FA, et al. CPEB1 coordinates alternative 3′-UTR formation with translational regulation. Nature. 2013;495:121–125. doi: 10.1038/nature11901. [DOI] [PubMed] [Google Scholar]
- 82.Masuda A, et al. Position-specific binding of FUS to nascent RNA regulates mRNA length. Genes Dev. 2015;29:1045–1057. doi: 10.1101/gad.255737.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hilgers V, Lemke SB, Levine M. ELAV mediates 3′ UTR extension in the Drosophila nervous system. Genes Dev. 2012;26:2259–2264. doi: 10.1101/gad.199653.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Batra R, et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol Cell. 2014;56:311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berkovits BD, Mayr C. Alternative 3′ UTRs act as scaffolds to regulate membrane protein localization. Nature. 2015;522:363–367. doi: 10.1038/nature14321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Weick EM, Miska EA. piRNAs: From biogenesis to function. Development. 2014;141:3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- 87.Mohn F, Sienski G, Handler D, Brennecke J. The rhino-deadlock-cutoff complex licenses noncanonical transcription of dual-strand piRNA clusters in Drosophila. Cell. 2014;157:1364–1379. doi: 10.1016/j.cell.2014.04.031. [DOI] [PubMed] [Google Scholar]
- 88.Zhang Z, et al. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell. 2014;157:1353–1363. doi: 10.1016/j.cell.2014.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Le Thomas A, et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014;28:1667–1680. doi: 10.1101/gad.245514.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vilborg A, Passarelli MC, Yario TA, Tycowski KT, Steitz JA. Widespread Inducible Transcription Downstream of Human Genes. Mol Cell. 2015;59:449–461. doi: 10.1016/j.molcel.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maher CA, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kannan K, et al. Recurrent chimeric RNAs enriched in human prostate cancer identified by deep sequencing. Proc Natl Acad Sci USA. 2011;108:9172–9177. doi: 10.1073/pnas.1100489108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Varley KE, et al. Recurrent read-through fusion transcripts in breast cancer. Breast Cancer Res Treat. 2014;146:287–297. doi: 10.1007/s10549-014-3019-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grosso AR, et al. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. eLife. 2015;4:e09214. doi: 10.7554/eLife.09214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.de Almeida SF, Carmo-Fonseca M. Reciprocal regulatory links between cotranscriptional splicing and chromatin. Semin Cell Dev Biol. 2014;32:2–10. doi: 10.1016/j.semcdb.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 96.Nemeroff ME, Barabino SM, Li Y, Keller W, Krug RM. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/S1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 97.Rutkowski AJ, et al. Widespread disruption of host transcription termination in HSV-1 infection. Nat Commun. 2015;6:7126. doi: 10.1038/ncomms8126. [DOI] [PMC free article] [PubMed] [Google Scholar]