Abstract
Eukaryotic (+)-strand RNA viruses utilize a wide variety of gene expression strategies to achieve regulated production of their viral proteins. A common mechanism used by many is to transcribe viral subgenomic (sg) mRNAs. Transcription of sg mRNA2 in tombusviruses allows for expression of the p19 suppressor of gene silencing and p22 movement proteins. We have investigated the mechanism of transcription of this sg mRNA in Tomato bushy stunt virus and have determined that this process is facilitated by no less than three different RNA modules that are located throughout the viral genome. These RNA units perform distinct tasks and function via long-distance RNA–RNA interactions. Systematic deconstruction of the RNA network and analysis of related RNA promoter elements allowed us to identify fundamental properties necessary for productive sg mRNA2 transcription. Collectively, our results (i) establish specific roles for the different RNA components of a multipartite RNA-based control system, (ii) support a premature termination mechanism for tombusvirus sg mRNA transcription and (iii) reveal a close mechanistic relationship between sg mRNA transcription, viral RNA replication and RNA recombination.
Keywords: gene expression, gene regulation, RNA recombination, RNA structure, RNA virus
Introduction
Eukaryotic (+)-strand RNA viruses form the largest group of medically and agriculturally important viruses. Their single-stranded messenger-sensed RNA genomes encode an assortment of proteins involved in various aspects of virus reproduction. Many of these proteins function at specific stages during infections and accordingly require regulated expression. Viruses that contain polycistronic RNA genomes regulate their gene expression primarily at the transcriptional level. They do so by transcribing genome-derived subgenomic (sg) mRNAs that act as messages for the translation of viral proteins encoded either internally or 3′-proximally in their genomes (Miller and Koev, 2000). This strategy allows for the expression of downstream open reading frames (ORFs) that are normally translationally silent within a polycistronic context and is a mechanism utilized by a variety of (+)-strand RNA viruses (Miller and Koev, 2000).
Positive-strand RNA viruses replicate their genomes via a (−)-strand RNA intermediate using their virally encoded RNA-dependent RNA polymerases (vRdRps) (Buck, 1996) (Figure 1A). Sg mRNA synthesis is also carried out by vRdRps in a process referred to as transcription. Most sg mRNAs have 5′ termini that map to internal genomic regions and 3′ ends that are coterminal with the genome (Figure 1A). Three basic models have been proposed for sg mRNA transcription and two of these, internal initiation (Miller et al, 1985) and discontinuous template synthesis (DTS) (Sawicki and Sawicki, 1998), have been established convincingly (Miller et al, 1985; van Marle et al, 1999; Pasternak et al, 2001). The third model, premature termination (PT), is less well supported by experimental data, but has been implicated in a number of diverse virus groups (Zhong and Rueckert, 1993; Sit et al, 1998; Zhang et al, 1999; Price et al, 2000; Gowda et al, 2001).
Figure 1.
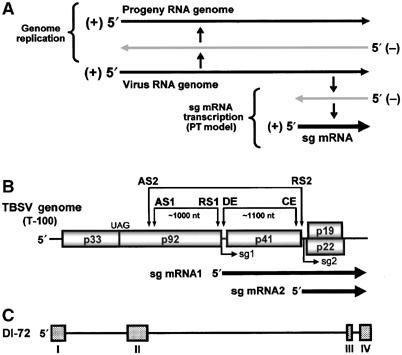
Genome replication, PT transcription model, and relevant TBSV RNAs. (A) Simplified scheme for viral RNA genome replication and sg mRNA transcription via a PT mechanism. (B) Linear representation of the TBSV RNA genome and coding organization. P92 is expressed by translational readthrough of the p33 stop codon (UAG). The relative positions (arrowheads) of interacting RNA elements involved in sg mRNA transcription are shown above the genome. Initiation sites for sg mRNA transcription are labeled sg1 and sg2 and corresponding structures of the two sg mRNAs are represented by bold arrows below the genome. (C) Schematic representation of a prototypical TBSV DI RNA, DI-72. Boxes correspond to regions (I–IV) of the TBSV genome (directly above) that are present in the DI RNA, while the lines represent genomic regions that are absent.
In the PT model, a vRdRp terminates internally during (−)-strand RNA synthesis on a genome template (Figure 1A). This leads to the generation of a smaller (−)-strand RNA that is complementary to the sg mRNA. This sg mRNA-sized (−)-strand (hereafter termed sg mRNA(−)) is then used as a template for transcription of the sg mRNA (White, 2002) (Figure 1A). The ‘primary' sg mRNAs produced by this mechanism could potentially be amplified further via replication (White, 2002). Different types of RNA–RNA interactions involving sequences located just upstream of sg mRNA start sites have been identified in viral genomes in which the PT model is proposed to operate (Sit et al, 1998; Choi et al, 2001; Gowda et al, 2001; Choi and White, 2002). In some cases, these RNA interactions were found to be essential for efficient sg mRNA transcription and, as proposed originally for Red clover necrotic mosaic virus (RCNMV) (Sit et al, 1998), could potentially act as roadblocks to mediate the vRdRp stalling/dissociation that generates the sg mRNA(−) templates (Zhang et al, 1999; Choi et al, 2001; Choi and White, 2002; Lindenbach et al, 2002).
The PT mechanism has been proposed to function in the transcription of two sg mRNAs in the (+)-strand RNA tombusvirus, Tomato bushy stunt virus (White, 2002). This virus encodes five proteins (Hearne et al, 1990) that are involved in viral RNA synthesis (p92, a vRdRp, and p33, an essential accessory protein) (Oster et al, 1998), particle assembly (p41, the capsid protein) (Hillman et al, 1989), cell-to-cell movement (p22) (Scholthof et al, 1995) and suppression of virus-induced gene silencing (p19) (Voinnet et al, 1999; Ye et al, 2003) (Figure 1B). P33 and p92 are translated directly from the genome, while the 3′-proximal ORFs are translated from either sg mRNA1 (p41) or 2 (p19 and p22) (Figure 1B).
Several observations suggest that TBSV sg mRNAs are transcribed via a PT mechanism (Zhang et al, 1999; Choi et al, 2001; Choi and White, 2002). (i) In TBSV infections, both sg mRNA (+)- and (−)-strands are detectable in total RNA extracts from infected cells. (ii) The formation of two different sets of long-distance RNA–RNA interactions, AS1/RS1 and DE/CE, involving sequences just 5′ to the two sg mRNA start sites, is required for efficient transcription of sg mRNA1 and 2, respectively (Figure 1B). (iii) These interactions function in the (+)-strand of the genome, consistent with their proposed functions as RNA-based terminators of the vRdRp during (−)-strand synthesis. (iv) Substitution of the initiating nucleotides for sg mRNA1 or 2 transcription results in inhibition of sg mRNA accumulation but not of corresponding sg mRNA(−) templates, in agreement with a PT model where the sg mRNA(−) templates are generated prior to, and independently of, their sg mRNA counterparts. (v) The autonomous synthesis of sg mRNA1(−) (as described in (iv)) is dependent on the AS1/RS1 interaction, indicating that this interaction is involved specifically in the generation of sg mRNA1(−) templates.
The previously characterized AS1 RNA element is required for efficient sg mRNA1 transcription and maps to the terminal loop of a predicted stem–loop structure (Choi and White, 2002) (Figure 2). Its base-pairing partner RS1 is positioned just three nucleotides (nt) 5′ to the sg mRNA1 start site and is directly adjacent to a conserved stem–loop structure, SL1sg1. In contrast, the previously identified DE-A/CE-A interaction that is necessary for efficient sg mRNA2 transcription occurs some 11 nt away from the start site for sg mRNA2 (Zhang et al, 1999) (Figure 2). This difference in spatial arrangement (11 versus 3 intervening nucleotides) led us to propose that sg mRNA2 transcription requires a third RNA element that base pairs within the 11 nt segment, designated CE-C, in a manner similar to the AS1/RS1 interaction (Choi and White, 2002) (Figure 2). Positionally, the sequence between RS1 and DE-A, termed DE-C, represented a good candidate (Figure 2). However, in TBSV, this sequence exhibits no significant complementarity to CE-C. Furthermore, DE-Cs are highly variable in different tombusviruses, but in all cases they are not complementary to their corresponding CE-Cs (Choi et al, 2001). Consequently, we hypothesized that this sequence maintains noncomplementarity in order to allow the missing RNA element unfettered access to CE-C.
Figure 2.
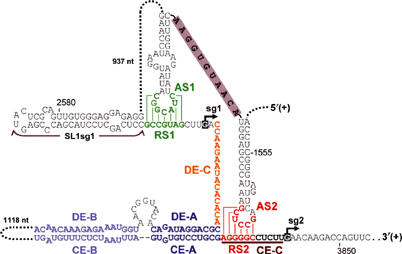
RNA elements that regulate sg mRNA transcription in TBSV. Relevant sequences of the TBSV genome are shown with corresponding coordinates provided. RNA elements involved in sg mRNA transcription are labeled and color-coded. The DE-B/CE-B interaction (light blue) is thought to stabilize the DE-A/CE-A interaction (deep blue) that is essential for sg mRNA2 transcription. The AS2/RS2 interaction (red), identified in this report, is critical for efficient transcription of sg mRNA2. Stem–loop structures containing AS1 and AS2 are connected by an 11 nt long sequence (gray bar). DE-C (gold) and CE-C (maroon and underlined) are noncomplementary elements defined previously. The AS1/RS1 interaction (green) is critical for efficient transcription of sg mRNA1. Initiation sites for the two sg mRNAs are highlighted in black and indicated by small arrows.
In the present study, we have identified the missing RNA element involved in sg mRNA2 transcription and have investigated its role within a complex multipartite RNA regulatory system. Our results provide important new evidence that a PT model of transcription operates in TBSV and suggest that this process shares mechanistic properties with the DTS model for transcription, viral genome replication, and RNA recombination.
Results
A third RNA element is required for activation of sg mRNA2 transcription
The notably different spacing of the AS1/RS1 and DE-A/CE-A interactions relative to their respective sg mRNA start sites led to the proposal that an additional interaction was involved in sg mRNA2 transcription (Choi and White, 2002) (Figure 2). A search of the TBSV genome revealed a candidate sequence, designated AS2, that was complementary to a subsection of CE-C, defined as RS2 (Figure 2). Interestingly, AS2 was located in the p92-coding region just upstream from AS1 and, similar to AS1, was situated in the terminal loop of a predicted stem–loop structure. The DE-A/CE-A interaction shown previously to be essential for efficient sg mRNA2 transcription (Zhang et al, 1999) was also of relevance, since it positions AS2 and RS2 in close proximity (Figure 2).
The proposed AS2/RS2 interaction would generate a 6-bp-long segment that is predicted to contain two non-Watson/Crick base pairs, GU and GA (Figure 2). The inclusion of the GA base pair in the interaction is based on the finding that, next to GU base pairs, sheared GA base pairs are the most common non-Watson/Crick pairs in known RNA structures (Nagaswamy et al, 2002). Also, sequence analysis of different tombusvirus genomes revealed a monovariation that maintains the predicted base pair (i.e. GA to GC) (unpublished data).
To test whether the proposed interaction was important for sg mRNA2 transcription, we carried out compensatory mutational analysis on the complementary sequences. Modifications that would disrupt and then restore AS2/RS2 base pairing were introduced into the TBSV genome and their effect on sg mRNA2 accumulation was assayed in protoplast infections and quantified by Northern blot analysis (Figure 3A and B). Substitutions in either AS2 (at degenerate codon positions) or RS2 that decreased their complementarity led to notably reduced sg mRNA2 levels (to ∼5 and 0%, respectively, relative to T100), while restoration of base pairing by combining the two substitutions resulted in partial recovery (∼33%) (Figure 3B). The relative levels of accumulation of sg mRNA2(−) strands also corresponded closely with those of their (+)-strand counterparts (Figure 3B). The reduced level of sg mRNA2 recovery observed for mutant A/RS2m1 could have been due to (i) a weaker AS2/RS2 interaction, (ii) the terminal loop modifications interfering with presentation of the AS2 residues and/or (iii) the substitutions causing either AS2 or RS2 to interact unproductively with other RNA sequences. Regardless, the six-fold or greater increase in the activity of A/RS2m1 over either AS2m1 or RS2m1 supports the importance of the AS2/RS2 interaction for sg mRNA2 transcription.
Figure 3.
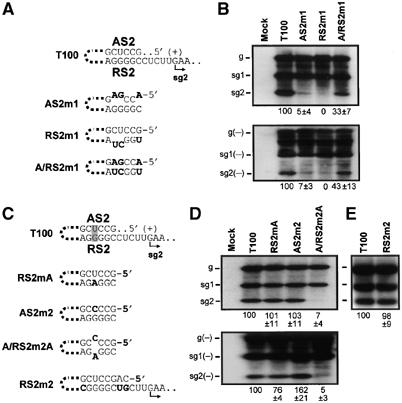
Analysis of the AS2/RS2 interaction. (A, C) AS2 and RS2 sequences in wt T100 and modified viral genomes. Substituted nucleotides are in bold. The UG base pair of interest is shaded in (C). The thick dotted lines represent a large intervening sequence between DE-A and DE-C. (B, D, E) Northern blot analysis and quantification of (+)- and (−)-strand viral RNA accumulation in protoplast infections. Following denaturation with glyoxal, total RNA extracts were separated in 1.4% agarose gels, transferred to a nylon membrane and hybridized with 32P-labeled viral strand-specific probes. Note that, although the (+)- and (−)-strands are presented with similar intensities, (−)-strand viral RNAs actually accumulate in vivo to significantly lower levels than (+)-strands. The values correspond to means from three independent experiments and represent the ratios of sg mRNA2 levels to their corresponding genomic RNA levels, all normalized to that for T100.
The presence of a naturally occurring UG base pair in the AS2/RS2 interaction is consistent with it functioning in the (+)-strand, as these residues correspond to a disruptive AC mismatch in the (−)-strand (Figure 3C). Although this inference is reasonable, additional evidence was sought to reinforce the concept. Changing the UG pair to either UA or CG did not notably affect sg mRNA2 accumulation, whereas a CA substitution, which corresponds to a GU base pair in the (−)-strand, nearly abolished accumulation (Figure 3C and D). Since the two substitutions in A/RS2m2A were tolerated well when present individually in RS2mA and AS2m2 (Figure 3C and D), these results are best explained by the CA mismatch disrupting the AS2/RS2 interaction in the (+)-strand. These results, in turn, support the proposal that this helix functions in the (+)-strand.
The AS2/RS2 interaction is distinct from the AS1/RS1 interaction in that the former is composed of different nucleotides and is positioned two residues further away from the transcriptional initiation site (Figure 2). To determine if the length and/or relative position of the interaction was relevant to sg mRNA2 transcription, we extended the complementary segment in AS2/RS2 to 8 bp by making appropriate substitutions in RS2 (Figure 3C). Interestingly, these substitutions did not lead to any notable increase in sg mRNA2 levels (Figure 3E), underscoring the effectiveness of the shorter wildtype (wt) interaction.
DE-C can be engineered to competitively inhibit AS2 binding and can functionally substitute for AS2
We hypothesized previously that DE-C plays a passive role in sg mRNA2 transcription by not base pairing with CE-C (Choi et al, 2001). This would leave CE-C free to interact with its authentic pairing partner, now identified as AS2. If correct, this hypothesis predicts that modifying DE-C to be complementary to the RS2 portion of CE-C should competitively inhibit AS2 binding and, in turn, lead to downregulation of sg mRNA2 transcription. To test this idea, the 3′ portion of DE-C was substituted with different nucleotides that were either noncomplementary or complementary to RS2 (Figure 4A). PY2, containing the noncomplementary sequence, was still fully functional for sg mRNA2 transcription (∼104%) in comparison to its genomic control Psg20/26 (a modified TBSV genome containing an engineered restriction enzyme site). Unexpectedly, PY9, containing the complementary sequence, was also fully functional (∼126%) (Figure 4A). This latter result could have been due to either AS2 maintaining its ability to bind to RS2 or the complementary segment in DE-C functionally replacing AS2. To distinguish between these two possibilities, an additional modification was introduced into both PY2 and PY9 that inactivated AS2 (Figure 4A). Analysis of the double mutants, PY2-AS2m1 and PY9-AS2m1, respectively, revealed that PY2-AS2m1 was still AS2-dependent, whereas PY9-AS2m1 was not (Figure 4A). Thus, DE-C can be modified to act as a competitive inhibitor of AS2 and can, concurrently, functionally replace AS2. Compensatory mutational analysis of the engineered DE-C/RS2 interaction confirmed its role in mediating sg mRNA2 transcription (Figure 4B).
Figure 4.
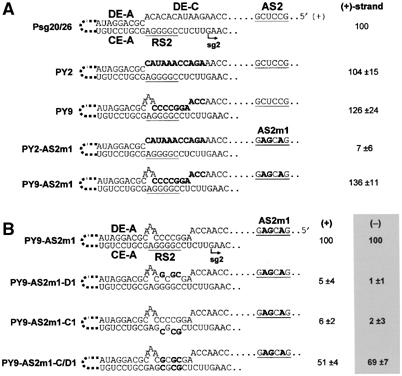
Characterization of a DE-C/RS2-based interaction. (A, B) Relevant sequences of modified viral genomes and corresponding relative levels of (+)- and/or (−)-strand sg mRNA2 accumulation in protoplasts. The values were calculated as described in the legend to Figure 3, except that all were normalized to that for Psg20/26 on PY9-AS2m1.
A local RNA hairpin structure can functionally replace the AS2/RS2 interaction
The surprising result that the AS2/RS2 interaction was dispensable for sg mRNA2 transcription prompted us to wonder whether a more radical structural replacement would also be functional. To investigate this possibility, we introduced different-sized RNA hairpins 3 nt upstream of the sg mRNA2 start site in a genomic context that had AS2 inactivated by substitutions (Figure 5A). The modified genome context used, Psg2-27, also had CE-A and -B deleted; thus any transcriptional activity detected would be independent of the DE/CE interactions formed by these subelements (refer to Figure 2). No detectable sg mRNA2 was observed for the genome containing the 4 bp stem (HL56); however, those with longer stems of 8 (Lsg2), 12 (HL45) and 16 bp (HL87) directed increasing levels of sg mRNA2 accumulation, underscoring the importance of helix length and/or stability (Figure 5A). The critical role of stem formation was confirmed through compensatory mutational analysis of the 8 bp hairpin (Figure 5B), while an important role for helix stability was supported by the increased activity observed when the 12 bp hairpin was converted to all GC base pairs in mutant HL78 (Figure 5A). The identity of the residues in the helix did not appear to be essential since HL57, with the lower 8 bp in its stem transposed, exhibited only moderately reduced levels of sg mRNA2 accumulation, compared with HL45 (Figure 5A). Strand-preferential destabilization of the helix in HL78 revealed that, similar to the wt AS2/RS2 interaction, it was far more sensitive to (+)- than to (−)-strand disruption, supporting a (+)-strand function (Figure 5C).
Figure 5.
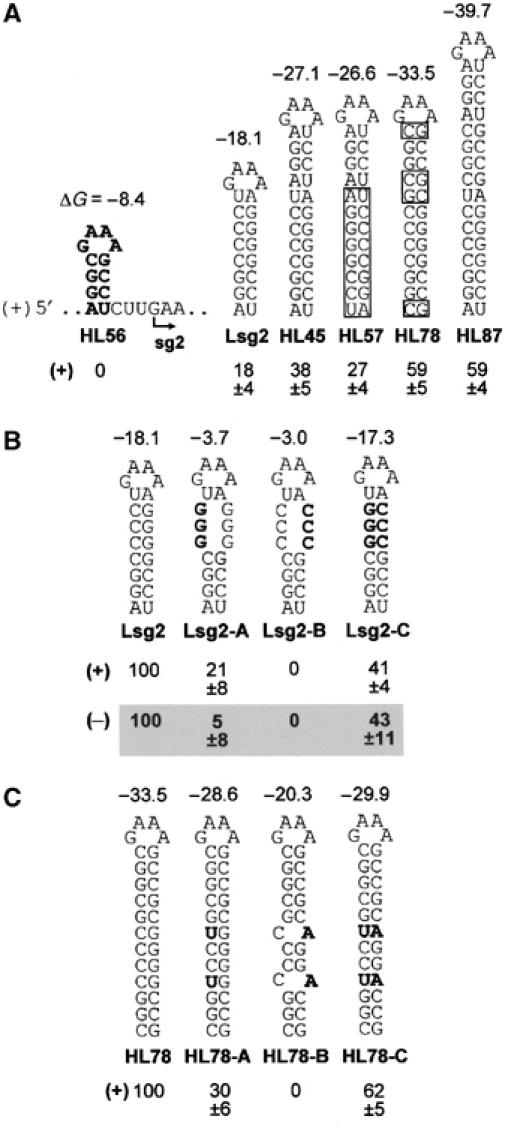
Effect of hairpin size, stability, and polarity of sg mRNA2 transcription. (A–C) Depiction of hairpin sequences introduced into modified viral genomes and corresponding levels of (+)- and/or (−)-strand sg mRNA2 accumulation in protoplasts. The boxed nucleotides in (A) are substitutions. Free energy changes (ΔG, at 37°) for formation of each RNA structure are indicated in kcal/mol (calculated using mfold 3.0). The values in (A) were normalized to that for T100, whereas those in (B, C) were normalized to those for Lsg2 and HL78, respectively.
Viral genomes containing the RNA hairpin element show delays in genomic and sg mRNA accumulation
Thus far our data have demonstrated that both wt AS2/RS2 and DE/CE interactions are not essential for productive sg mRNA2 transcription. One possible reason for the virus maintaining these wt long-distance interactions could be related to transcriptional regulation, either temporal or quantitative. Earlier studies have already ruled out any major role for the proteins encoded by the sg mRNAs in transcriptional control (Zhang et al, 1999).
To investigate whether the wt long-distance interactions conferred any distinct type of transcriptional regulation, we compared the viral RNA accumulation profiles for the wt T100 genome with PY9-AS2m1 (in which the wt AS2/RS2 interaction was replaced with DE-C/RS2), Psg20/26 (the modified genome used to generate PY9-AS2m1, that is, the control for PY9-AS2m1), HL78 (in which both wt DE/CE and AS2/RS2 interactions were replaced with a 12 bp hairpin) and Psg2-27 (the modified genome used to generate HL78, that is, the control for HL78) (Figure 6). In wt T100, sg mRNA2 accumulated to detectable levels before sg mRNA1 and the appearance of sg mRNA2 coincided with that of the genome (Figure 6). The early (2–11 h) accumulation rates of genomic and sg mRNAs for T100, Psg20/26 and PY9-AS2m1 were very similar; however, viral RNA levels were clearly higher at later time points (15–24 h) for PY9-AS2m1 (Figure 6). For HL78, a lower relative level of sg mRNA2 was evident and the initial appearance of all viral RNAs was delayed by ∼2 h (Figure 6). This delay was not related to its Psg2-27-based genomic context, since the timing of early accumulation of genomic and sg mRNA1 for Psg2-27 was similar to that for T100 (Figure 6). The very low levels of sg mRNA2 for Psg2-27 is expected due to the absence of the critical CE-A and important CE-B subelements in this modified genome (Zhang et al, 1999). Overall, this time-course analysis suggests that (i) the engineered DE-C/RS2 interaction functions similarly to the wt AS2/RS2 interaction at early time points, but causes increased accumulation of all viral RNAs later in the infection, and (ii) genomes containing the engineered 12 bp hairpin show delayed accumulation of all viral RNAs and a lower relative level of sg mRNA2; however, sg mRNA2 still appears earlier than sg mRNA1.
Figure 6.
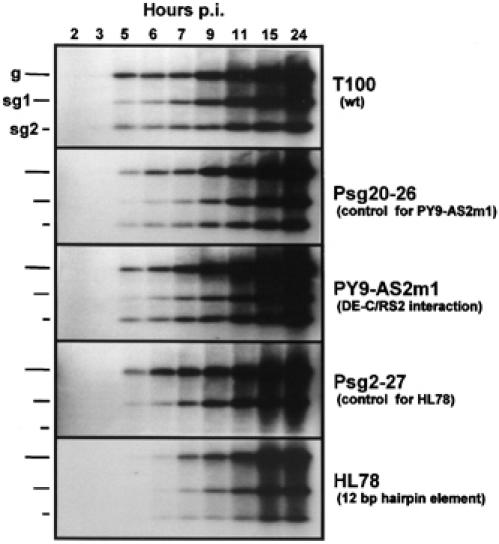
Accumulation profile of TBSV viral RNAs over time in infected protoplasts. Total RNA preparations were isolated at various times (in hours) postinfection (p.i.), as indicated above the lanes, and analyzed by Northern blotting. The positions of genomic and sg mRNAs are indicated to the left, while the genomes used in the infections are listed to the right.
An engineered hairpin-based element is able to function in a different viral context
We employed a TBSV defective interfering (DI) RNA as a surrogate genome to investigate whether sequence context was important to the activity of hairpin-based elements (Figure 1C). DI RNAs are noncoding deletion mutants of the TBSV genome that are efficiently amplified when p33 and p92 are provided in trans (e.g. in coinfections with T100) (White, 1996). These molecules have served as excellent model replicons for studying genome replication in a context that is independent of translation (White and Nagy, 2004).
A prototypical TBSV DI RNA, such as DI-72, contains four noncontiguous regions of the viral genome termed regions I–IV (Figure 1C). Interestingly, RNA B, a 5′-truncated derivative of DI-72 lacking region I, is still able to replicate at low levels (∼10% that of DI-72) in coinoculations with the wt TBSV genome (Wu and White, 1998). Structurally, the 5′ end of RNA B corresponds to an internal region of DI-72, whereas its 3′ end is coterminal with DI-72 (Figure 7A). This basic structural correspondence is analogous to that of a sg mRNA relative to its cognate viral genome. However, in contrast to sg mRNAs, RNA B is not detectable in coinfections of DI-72 and TBSV genome (Figure 7B, lane 3); thus, RNA B is not normally produced during DI-72 replication. To test whether RNA B could be ‘launched' from a DI RNA molecule in a manner comparable to sg mRNA transcription, the active hairpin-based cassette from Lsg2 (Figure 5A) was inserted between regions I and II in DI-72, thereby generating HL65 (Figure 7A). As a control, another DI 72-based molecule was constructed, HL69, which contained an unstructured sequence of similar length inserted at the same position. RNA B was not detected when HL69 was coinoculated with T100; however, it was clearly present in coinoculations with HL65 (Figure 7B). The importance of the small helix for the production of RNA B was confirmed by compensatory mutational analysis (Figure 7A and B). This ability of the 8 bp hairpin cassette to mediate RNA B production from a DI RNA suggests that this cassette contains all of the structural properties required for local context-independent activity. Additionally, we have found that substitution of the initiating nucleotide in this cassette leads to cessation of (+)- but not (−)-strand RNA B synthesis (unpublished data), which is consistent with the concept that the hairpin structure specifically mediates production of (−)-strand templates.
Figure 7.
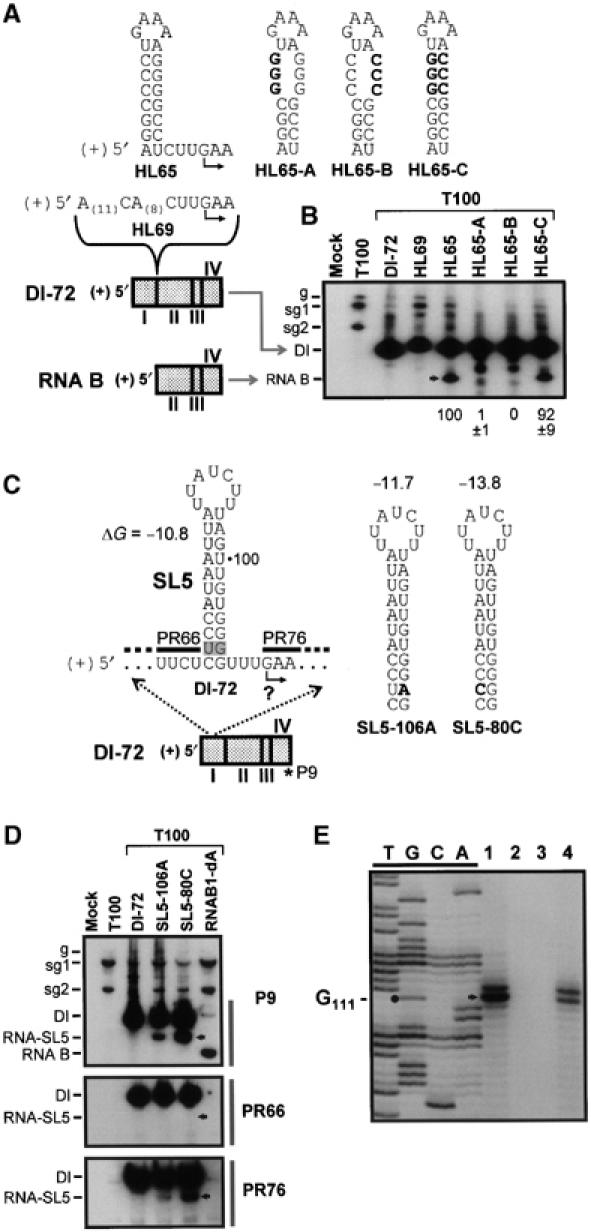
Transcriptional activity of artificial and natural hairpin elements. (A) Schematic representation of DI-72 and a DI-72-derived RNA, RNA B. Different sequences inserted between regions I and II in DI-72 are indicated above it. (B) Northern blot analysis of wt DI-72 and modified versions containing sequence insertions. Nucleic acids in this and subsequent Northern blot experiments were separated in denaturing 4.5% polyacrylamide–8% urea gels. The position of RNA B is indicated on the left and also by an arrowhead. The relative levels of accumulation of RNA B are indicated below each lane. The origin and structure of the bands in lanes 6 and 7 (with mobility between those of the DI RNA and RNA B) are unknown. (C) Schematic representation of DI-72, with SL5 and adjacent sequences expanded above it. The UG base pair of interest in SL5 is shaded and the modified versions analyzed are shown to the right. The relative positions of (−)-sensed oligonucleotide probes, PR66, PR76 and P9, used in (D) are indicated by thick lines and an asterisk, respectively. (D) Northern blot analysis of RNA-SL5 using the different oligonucleotide probes. RNAB1-dA is a replicable RNA B molecule that acts as a marker for the position of RNA B. The probes used are indicated on the right, with the relevant portion of the blot examined with the different probes indicated by thick vertical bars. The position of RNA-SL5 is indicated by the arrowheads. (E) Primer-extension (PE) analysis of RNA-SL5. Products from PE reactions using total RNA preparations from protoplasts infected with either T100+SL5-80C (lane 1), T100 only (lane 2) or T100+DI-72 (lane 3), or gel-purified RNA-SL5 (lane 4). The sequencing ladder was separated in the four lanes on the left, which are labeled with the appropriate bases. The major PE product is indicated by an arrowhead, while the nucleotide it corresponds to, G111, is denoted by a circle.
Downregulation of a naturally occurring RNA hairpin-type transcriptional element by helix destabilization
The apparent simplicity and context independence of the hairpin cassette led us to question whether other similar, and possibly functional, elements were present in the viral genome. Analysis of the TBSV genomic sequence led to the identification of a sequence in the 5′ UTR that forms a structure that very closely resembles that of our functional hairpin cassette (compare HL65 in Figure 7A with the expanded region of DI-72 in Figure 7C). Since the 5′ UTR of the genome is also involved in translational regulation (Fabian and White, 2004), we chose to investigate this sequence in the context of a nontranslated DI RNA, DI-72 (Figure 7C). Interestingly, previous analysis of the SL5 structure in a DI-72 context revealed that strengthening the UG base pair at the bottom of its stem via substitution results in the accumulation of an additional small viral RNA, which was not characterized (Ray et al, 2003). Considering its structural similarity to our functional hairpin element, we wondered whether the stabilizing substitutions were causing it to become ‘transcriptionally' active. To investigate this possibility, we characterized the smaller viral RNA generated. Northern blot analysis revealed that this additional RNA, termed RNA-SL5, was clearly larger than the previously characterized RNA B and contained the 3′-terminal end of the DI RNA and sequences just downstream, but not upstream, of SL5 (Figure 7D). Primer-extension analysis mapped the 5′ terminus of RNA-SL5 to position G111, which is the predicted initiation site if the SL5 element was functioning analogously to our transcriptional hairpin cassettes (Figure 7E). These results suggest that at least one cryptic transcriptional element is present within the TBSV genome and that its activity is downregulated by helix destabilization, probably to avoid production of unnecessary and possibly deleterious viral RNAs.
The 5′ terminus of sg mRNA2 can function as a (+)-strand promoter for viral RNA replication
In the framework of a PT model, the stalling of the vRdRp by the AS2/RS2 helix should produce a sg mRNA2(−) template containing the initiation site for sg mRNA2 transcription. Indeed, cloning and sequencing of the 3′ ends of sg mRNA2(−) templates by oligonucleotide-anchored RT–PCR revealed that all templates included the cytidylate corresponding to the initiating guanylate in sg mRNA2, and some contained an extra 3′-terminal adenylate corresponding to a uridylate in the genomic sequence (Figure 8A, T100). When this uridylate in the genome was substituted with an adenylate, thereby creating HL-UP5, one of the 10 sg mRNA2(−) analyzed contained a corresponding uridylate (Figure 8A). These results suggest that (−)-strand templates containing the extra 3′-terminal nucleotides were derived from the genomic template.
Figure 8.
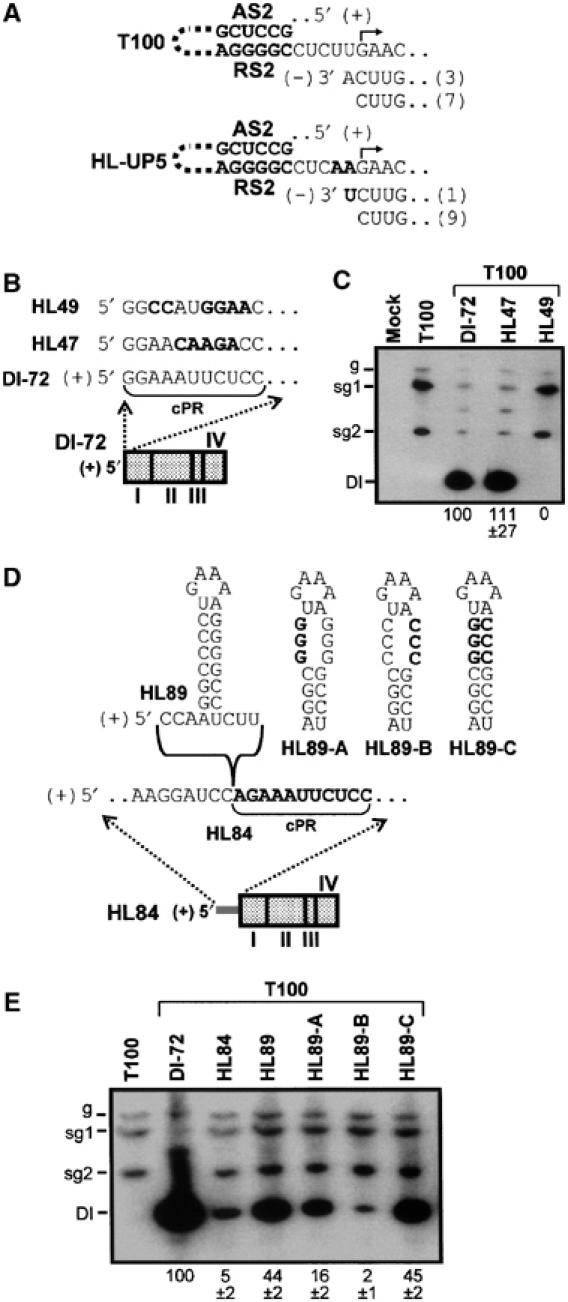
Analysis of sg mRNA2(−) strands and 5′-terminally modified DI RNAs. (A) Analysis of the 3′ terminus of sg mRNA2(−) strands. The structure of 3′-terminal regions of sg mRNA2(−) strands is shown below relevant corresponding (+)-strand sequence. The number in parentheses indicates the number of times each sequence was observed. The substituted nucleotides in HL-UP5 are in bold. (B) Schematic representation of DI-72 with wt and modified 5′-terminal sequences shown above it. The sequence corresponding to the promoter for (+)-strand RNA synthesis (cPR), which is actually located in (−)-strand (not shown), is denoted by a bracket. (C) Northern blot analysis and quantification of DI RNA accumulation levels. (D) Schematic representation of DI-72 containing a 5′-terminal sequence extension. The sequence corresponding to the junction between the 5′ extension and the normal 5′ terminus of DI-72 is expanded above the DI RNA and sequences inserted at this junction site are presented at the top. (E) Northern blot analysis and quantification of DI RNA accumulation levels.
The PT model also predicts that the 3′ end of the (−)-strand generated acts as promoter for sg mRNA transcription. Consistent with this notion, the 5′ terminus of sg mRNA2 is similar in sequence to the 5′ terminus of the viral genome, which is also the 5′ terminus of DI-72 (compare HL47 with DI-72 in Figure 8B). The complement of the 5′-terminal 11 nt in DI-72 corresponds to the core genomic (+)-strand promoter, cPR (Panavas et al, 2002); thus the similarity observed between the 5′-terminal genomic and sg mRNA sequences suggests that the complement of this sg mRNA segment may also function as a (+)-strand promoter. To test this idea, we replaced the 5′-terminal 11 nt of the DI RNA with the corresponding 5′-terminal sequence of sg mRNA2 or a random sequence, thereby generating HL47 and HL49, respectively (Figure 8B). HL47 was able to replicate and accumulate to wt DI RNA levels, whereas HL49 did not accumulate to detectable levels (Figure 8C). Collectively, these results demonstrate that the 3′ termini of sg mRNA2(−) templates contain the initiating site for sg mRNA transcription and that this terminal sequence possesses promoter activity that is capable of mediating efficient (+)-strand viral RNA synthesis.
The inhibitory effect of a 5′-terminal sequence extension on viral RNA replication is attenuated by an insertion of an RNA hairpin-based element
The functional equivalence of (+)-strand RNA synthesis in viral sg mRNA transcription and DI RNA replication suggested that these two discrete processes could share additional properties. Specifically, we wondered whether repositioning the (+)-strand promoter for replication internally in an RNA template would be inhibitory to its activity and, if so, could the defect be rescued by insertion of an active hairpin-based cassette. As before, these studies were carried out in the context of a DI RNA (Figure 8D). In HL84, a 164 nt long 5′ extension was added to DI-72 and this greatly reduced its accumulation to ∼5% that of the wt DI-72 (Figure 8D and E). However, when the 8 bp hairpin cassette was added just upstream of the internally located 5′ terminus, the accumulation level of the resulting mutant, HL89, increased by ∼10-fold compared with that of HL84 (Figure 8D and E). The importance of the hairpin component of the cassette for mediating efficient DI RNA accumulation was confirmed by compensatory mutagenesis (Figure 8D and E). These results indicate that a hairpin-based cassette can substantially rescue a replication defect caused by an inhibitory 5′-terminal extension.
Discussion
In this study, we investigated RNA elements that contribute to efficient transcription of TBSV sg mRNA2. Below we discuss our results in relation to the PT model and its apparent mechanistic parallels with genome replication, the DTS model, and RNA recombination.
The sg mRNA2 regulatory RNA network and the PT model
A third RNA element, AS2, was identified and shown to functionally base pair with the RS2 subsection of CE-C. Interestingly, AS2, like AS1, is present in the terminal loop of a predicted stem–loop structure, which likely facilitates its presentation and pairing with RS2 (Figure 2). A similarly structured stem–loop is also involved in transactivation of transcription in RCNMV (Sit et al, 1998). In TBSV, the AS2 and AS1 stem–loops are positioned tandemly and, thus, could potentially operate as a unit. However, inactivation of either AS1 or AS2 does not appreciably affect the ability of the other to mediate transcription (Choi and White, 2002) (Figure 3). Thus, although coupled positionally, these elements and their corresponding interactions do not appear to be coupled functionally. Also worth noting is the fact that both AS1 and AS2 reside in the p92 readthrough ORF, thus their functions would be negatively affected by translating ribosomes. Accordingly, efficient transcription may require downregulation of translation.
A surprising discovery of this investigation was that the wt AS2 sequence (and its distal positioning) was not required for transcription and could be functionally replaced by a local hairpin structure. This result raises the intriguing possibility that some viruses may normally use localized hairpin-type structures in their genomes to mediate this type of sg mRNA transcription. Our active hairpin elements were determined to function in the (+)-strand in a primarily identity-independent, stability-dependent manner. This is consistent with the helix acting as an RNA secondary structure-dependent terminator of (−)-strand synthesis, as proposed for the wt AS2/RS2 interaction. The sequence independence of these base-paired elements is also supported by the lack of sequence identity between the AS1/RS1 and AS2/RS2 helices (Figure 2) and the activation of an unrelated cryptic hairpin element (SL5) by strengthening the base of its helix.
The ability of the hairpin elements to function independently of the DE-A/CE-A and DE-B/CE-B interactions (which are essential and important respectively, for AS2/RS2 activity) suggests that these interactions play important auxiliary roles in positioning RS2 close to AS2 in the wt context, as predicted by our experimentally supported structural model (Figure 2). In addition, the DE-A/CE-A interaction could act to stabilize the critical AS2/RS2 helix via coaxial stacking (Figure 2). This concept is in line with our results showing that activity of both the wt AS2/RS2 interaction and local hairpin elements depends on helix stability. Additionally, the coaxial stacking-mediated stabilization is supported indirectly by a similar potential interaction between the AS1/RS1 helix and that of the adjacent SL1sg1 (Figure 2).
Our finding that the complement of the 5′-terminal sequence of sg mRNA2 is able to function as a (+)-strand promoter for DI RNA replication indicates that it could also act in a similar capacity as a transcriptional promoter in sg mRNA(−) templates to generate primary sg mRNA2 transcripts (and possibly as a replication promoter for their further amplification). Importantly, this sg mRNA2-derived promoter was able to function without any (−)-strand sequences 3′-proximal to it, a finding consistent with the AS2/RS2 interaction (i) not constituting part of the transcriptional promoter in the (−)-strand and (ii) acting as an RdRp terminator in the (+)-strand. RNA secondary structure-based termination is also supported by the significant rescue of an internally positioned replication promoter via insertion of a hairpin-based cassette. Interestingly, poor activity from internal sites seems to be a property shared by both transcriptional and (+)-strand replication promoters. The reason for this could be that in an internal context the promoters are less accessible to the TBSV vRdRp due to confinement in a putative double-stranded RNA replication intermediate and/or that the vRdRp has a strong 3′-end dependence. Regardless, these results point to a close mechanistic link between sg mRNA transcription and the (+)-strand synthesis step of genome replication.
Collectively, these and previous results can be readily integrated into a PT model for sg mRNA2 transcription (Figure 9A). In this process, formation of the AS2-containing stem–loop positions AS2 within its terminal loop and facilitates its binding to RS2. The DE-A/CE-A interaction aids in this process by bringing the AS2 and RS2 sequences into close proximity. The DE-B/CE-B interaction helps to stabilize the DE-A/CE-A interaction and DE-C assists passively by not competing for RS2. Following formation of the AS2/RS2 helix, it may be stabilized further by coaxial stacking with the DE-A/CE-A helix. The AS2/RS2 helix serves as the major secondary structure-dependent RNA termination signal for the vRdRp, and the sg mRNA2(−) template generated is used subsequently for sg mRNA2 transcription via initiation at the 3′-terminal promoter (PSG). Sg mRNA1 transcription is also thought to occur via a similar mechanism that involves the essential AS1/RS1 interaction (Figure 9A).
Figure 9.
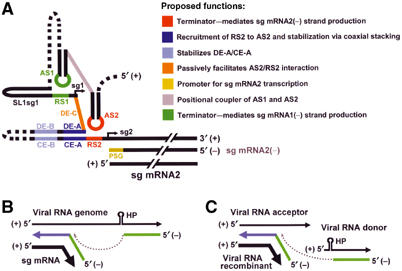
Models for sg mRNA transcription and viral RNA recombination. (A) A PT model for TBSV sg mRNA2 transcription. A list of proposed functions for the color-coded RNA elements is provided (and apply only to panel A). (B) A DTS model for BEV sg mRNA 2 transcription (van Vliet et al, 2002). (C) Hairpin-mediated RNA recombination model for TBSV (White and Morris, 1995). See text for details.
Possible roles for the wt long-distance RNA–RNA interactions
A fascinating aspect of the regulatory RNA elements involved is their location throughout the TBSV genome. Although not essential for function, this dispersed organization has been selected over time by nature based on performance. A network of long-distance interactions could offer certain advantages, such as (i) providing a more efficient system for sg mRNA transcription, (ii) allowing for better regulation of transcription and/or (iii) facilitating and/or coordinating additional viral processes. For TBSV sg mRNA2, the hairpin-based elements mediated transcription less efficiently than the wt long-distance interactions. Also, the hairpin elements caused a delay in early expression of sg mRNA2 (and also of other viral mRNAs). These highly reproducible differences, although subtle, could have significant effects on virus fitness in nature. Indeed, the early and efficient transcription of sg mRNA2 is probably beneficial to infections by facilitating rapid suppression of gene silencing by the encoded p19 and swift invasion of neighboring cells by p22.
It is also interesting to note that the more 5′-proximal location of AS1 and AS2 places the initial production of sg mRNA(−) templates strictly under genomic control (as neither sg mRNAs contain AS1 or AS2) (Figure 1B). Thus, even if the sg mRNAs are amplified further in a manner independent of the genomic template (i.e. sg mRNA replication, as shown for Flock house virus; Eckerle et al, 2003), the genome would still maintain absolute control of the launching of primary sg mRNA transcripts. This may be an important feature for proper regulation of the viral proteins expressed from these sg mRNAs.
The PT model in comparison with other transcriptional mechanisms and RNA recombination
The PT model is distinct from the internal initiation model in which a full-length genomic (−)-strand is used as the template for sg mRNA transcription (Miller et al, 1985). In the latter case, the vRdRp initiates transcription internally in a genomic (−)-strand template and, in some cases, the promoters utilized contain a functionally important RNA hairpin (Haasnoot et al, 2000). Interestingly, the hairpin-containing sg mRNA promoters of Brome mosaic virus and Alfalfa mosaic virus are similar in structure to their respective genomic promoters for (−)-strand RNA synthesis, and these two different types of promoter are functionally interchangeable (Joost Haasnoot et al, 2002; Olsthoorn et al, 2004; Sivakumaran et al, 2004). In contrast, we have shown that the TBSV sg mRNA2 promoter is structurally similar and functionally equivalent to its respective genomic promoter for (+)-strand synthesis. Additionally, the TBSV promoter lacks a hairpin constituent and does not operate well from internal positions.
The PT model proposed for sg mRNA2 is instead more mechanistically similar to the DTS model, shown to operate in arteriviruses (van Marle et al, 1999). In the DTS model, the vRdRp stalls during (−)-strand synthesis on a genome template and then transposes itself (along with its nascent RNA strand) to a more 5′-proximal site, where it proceeds to copy a section of the 5′ UTR (Sawicki and Sawicki, 1998). The hybrid (−)-strand formed by covalent joining of noncontiguous 5′- and 3′-terminal segments is then used for transcription of sg mRNAs (Figure 9B). The stalling step in the DTS model (i.e. just prior to vRdRp transposition) is comparable to the termination step in the PT model (compare Figure 9A and B).
Intriguingly, an RNA hairpin has been implicated in the stalling step of a proposed DTS mechanism of transcription for sg mRNA 2 of the torovirus Berne virus (BEV) (van Vliet et al, 2002) (Figure 9B). The RNA hairpin in the BEV genome maps immediately upstream of the 3′ junction site in BEV sg mRNA 2; thus, it has been suggested that this hairpin could facilitate the transposition step in the proposed DTS model (van Vliet et al, 2002) (Figure 9B). The BEV hairpin may function in a manner similar to the AS2/RS2 helix in the TBSV genome; therefore, local and long-distance RNA secondary structures may act in DTS and PT mechanisms, respectively, to mediate vRdRp stalling that leads to either transposition or termination. The functional equivalence of local and long-distance RNA secondary structures in mediating vRdRp stalling/termination is well supported by the data in this article. Additionally, the proposed transposition-promoting activity of RNA secondary structure is corroborated by previous studies that showed that stable RNA hairpins can promote vRdRp-mediated recombination events in TBSV (White and Morris, 1995) (Figure 9C). It is interesting to note that, mechanistically, the hairpin-mediated recombination event reported previously for TBSV is the intermolecular equivalent of the hairpin-mediated DTS transcriptional mechanism proposed for BEV (compare Figure 9B and C). The inter-relatedness and mechanistic similarities of these processes point to a common role for RNA secondary structure in defining RNA termini and junction sites in transcriptional and recombinational events through modulation of vRdRp activity.
Materials and methods
Plasmid construction
All clones used in this study were generated using the previously described constructs: T100, the wt TBSV genome construct (Hearne et al, 1990), Psg20/26 and Psg2-27, modified genomic clones (Zhang et al, 1999), and DI-72SXP (for simplicity, termed DI-72 in this work), a prototypical TBSV DI RNA (Wu et al, 2001). The detailed structures of the modifications introduced into these different base constructs are presented in the respective figures, an exception being HL47 and HL49, where additional substitutions were made more 3′ in these DI RNAs to maintain an essential helix (stem 1) in the 5′-terminal T-shaped RNA domain (Wu et al, 2001). All modifications were introduced using PCR-based mutagenesis and standard cloning techniques (Sambrook et al, 1989). PCR-derived regions introduced into constructs were sequenced completely to ensure that only the intended modifications were present.
In vitro transcription, protoplast inoculation and RNA isolation
In vitro RNA transcripts of genomic and DI RNAs were generated using T7 RNA polymerase as described previously (Wu and White, 1998). Preparation and inoculation of cucumber protoplasts and extraction of total nucleic acids were carried out as before (Choi and White, 2002). Briefly, isolated cucumber protoplasts (∼300 000) were inoculated with RNA transcripts (3 μg for genomic RNA and 1 μg for DI RNA). Inoculated protoplasts were incubated at 22°C, except for HL69, HL65, HL65-A, -B and -C (Figure 7A), HL84, HL89 and HL89-A, -B and -C (Figure 8E), which were incubated at 19°C. Total nucleic acids or gel-purified viral RNA were isolated and analyzed as described below.
Viral RNA analysis
Total nucleic acid preparations isolated from virus-inoculated protoplasts were subjected to Northern blot analysis for detection of (+)- and (−)-strand viral RNAs as described previously (Choi and White, 2002). Nucleic acids were either treated with glyoxal and separated in 1.4% agarose gels or denatured in formamide-containing buffer and separated in 4.5% polyacrylamide–8% urea gels. Equal loading of lanes was confirmed prior to transfer via staining the gels with ethidium bromide. Following electrophoretic transfer to nylon membranes, viral RNAs were detected using 32P-labeled DNA oligonucleotide probes or RNA riboprobe and their relative levels were determined by radioanalytical scanning of blots (Choi and White, 2002).
Primer-extension analysis of RNA-SL5 in total nucleic acid preparations or from gel-purified RNA was performed as described previously (Wu and White, 1998). The products of the reaction and sequencing ladder were separated in an 8% polyacrylamide–8% urea gel.
Cloning and sequencing of the 3′ termini of sg mRNA2(−) strands were accomplished by ligation of phosphorylated oligonucleotides with RNA ligase, reverse-transcription–PCR amplification of the cDNA, and subsequent cloning and sequencing (Liu and Gorovsky, 1993).
RNA structures were predicted and free energy changes calculated using mfold version 3.0 (Mathews et al, 1999; Zuker et al, 1999).
Acknowledgments
We thank members of our laboratory for reviewing the manuscript. This work was supported by NSERC and CRC.
References
- Buck KW (1996) Comparison of the replication of positive-stranded RNA viruses of plants and animals. Adv Virus Res 47: 159–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi IR, Ostrovsky M, Zhang G, White KA (2001) Regulatory activity of distal and core RNA elements in tombusvirus subgenomic mRNA2 transcription. J Biol Chem 276: 41761–41768 [DOI] [PubMed] [Google Scholar]
- Choi IR, White KA (2002) An RNA activator of subgenomic mRNA1 transcription in tomato bushy stunt virus. J Biol Chem 277: 3760–3766 [DOI] [PubMed] [Google Scholar]
- Eckerle LD, Albarino CG, Ball LA (2003) Flock house virus subgenomic RNA3 is replicated and its replication correlates with transactivation of RNA2. Virology 317: 95–108 [DOI] [PubMed] [Google Scholar]
- Fabian MR, White KA (2004) 5′–3′ RNA–RNA interaction facilitates cap- and poly(A) tail-independent translation of tomato bushy stunt virus mRNA: a potential common mechanism for tombusviridae. J Biol Chem 279: 28862–28872 [DOI] [PubMed] [Google Scholar]
- Gowda S, Satyanarayana T, Ayllon MA, Albiach-Marti MR, Mawassi M, Rabindran S, Garnsey SM, Dawson WO (2001) Characterization of the cis-acting elements controlling subgenomic mRNAs of citrus tristeza virus: production of positive- and negative-stranded 3′-terminal and positive-stranded 5′-terminal RNAs. Virology 286: 134–151 [DOI] [PubMed] [Google Scholar]
- Haasnoot PCJ, Brederode FT, Olsthoorn RCL, Bol JF (2000) A conserved hairpin structure in Alfamovirus and Bromovirus subgenomic promoters is required for efficient RNA synthesis in vitro. RNA 6: 708–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hearne PQ, Knorr DA, Hillman BI, Morris TJ (1990) The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology 177: 141–151 [DOI] [PubMed] [Google Scholar]
- Hillman BI, Hearne P, Rochon D, Morris TJ (1989) Organization of tomato bushy stunt virus genome: characterization of the coat protein gene and the 3′ terminus. Virology 169: 42–50 [DOI] [PubMed] [Google Scholar]
- Joost Haasnoot PC, Olsthoorn RC, Bol JF (2002) The Brome mosaic virus subgenomic promoter hairpin is structurally similar to the iron-responsive element and functionally equivalent to the minus-strand core promoter stem–loop C. RNA 8: 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Sgro JY, Ahlquist P (2002) Long-distance base pairing in Flock House virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J Virol 76: 3905–3919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gorovsky MA (1993) Mapping the 5′ and 3′ ends of Tetrahymena thermophila mRNAs using RNA ligase mediated amplification of cDNA ends (RLM-RACE). Nucleic Acids Res 21: 4954–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Sabina J, Zuker M, Turner DH (1999) Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J Mol Biol 288: 911–940 [DOI] [PubMed] [Google Scholar]
- Miller WA, Dreher TW, Hall TC (1985) Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genome RNA. Nature 313: 68–70 [DOI] [PubMed] [Google Scholar]
- Miller WA, Koev G (2000) Synthesis of subgenomic RNAs by positive-strand RNA viruses. Virology 273: 1–8 [DOI] [PubMed] [Google Scholar]
- Nagaswamy U, Larios-Sanz M, Hury J, Collins S, Zhang Z, Zhao Q, Fox GE (2002) NCIR: a database of non-canonical interactions in known RNA structures. Nucleic Acids Res 30: 395–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsthoorn RC, Haasnoot PC, Bol JF (2004) Similarities and differences between the subgenomic and minus-strand promoters of an RNA plant virus. J Virol 78: 4048–4053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oster SK, Wu B, White KA (1998) Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J Virol 72: 5845–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T, Pogany J, Nagy PD (2002) Analysis of minimal promoter sequences for plus-strand synthesis by the Cucumber necrosis virus RNA-dependent RNA polymerase. Virology 296: 263–274 [DOI] [PubMed] [Google Scholar]
- Pasternak AO, van den Born E, Spaan WJ, Snijder EJ (2001) Sequence requirements for RNA strand transfer during nidovirus discontinuous subgenomic RNA synthesis. EMBO J 20: 7220–7228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price BD, Roeder M, Ahlquist P (2000) DNA-directed expression of functional flock house virus RNA1 derivatives in Saccharomyces cerevisiae, heterologous gene expression, and selective effects on subgenomic mRNA synthesis. J Virol 74: 11724–11733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Wu B, White KA (2003) A second functional RNA domain in the 5′ UTR of the Tomato bushy stunt virus genome: intra- and interdomain interactions mediate viral RNA replication. RNA 9: 1232–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, 2nd edn Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Sawicki SG, Sawicki DL (1998) A new model for coronavirus transcription. Adv Exp Med Biol 440: 215–219 [DOI] [PubMed] [Google Scholar]
- Scholthof HB, Scholthof KB, Kikkert M, Jackson AO (1995) Tomato bushy stunt virus spread is regulated by two nested genes that function in cell-to-cell movement and host-dependent systemic invasion. Virology 213: 425–438 [DOI] [PubMed] [Google Scholar]
- Sit TL, Vaewhongs AA, Lommel SA (1998) RNA-mediated trans-activation of transcription from a viral RNA. Science 281: 829–832 [DOI] [PubMed] [Google Scholar]
- Sivakumaran K, Choi SK, Hema M, Kao CC (2004) Requirements for brome mosaic virus subgenomic RNA synthesis in vivo and replicase-core promoter interactions in vitro. J Virol 78: 6091–6101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Marle G, Dobbe JC, Gultyaev AP, Luytjes W, Spaan WJ, Snijder EJ (1999) Arterivirus discontinuous mRNA transcription is guided by base pairing between sense and antisense transcription-regulating sequences. Proc Natl Acad Sci USA 96: 12056–12061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vliet AL, Smits SL, Rottier PJ, de Groot RJ (2002) Discontinuous and non-discontinuous subgenomic RNA transcription in a nidovirus. EMBO J 21: 6571–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Pinto YM, Baulcombe DC (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA 96: 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA (1996) Formation and evolution of tombusvirus defective interfering RNAs. Semin Virol 7: 409–416 [Google Scholar]
- White KA (2002) The premature termination model: a possible third mechanism for subgenomic mRNA transcription in (+)-strand RNA viruses. Virology 304: 147–154 [DOI] [PubMed] [Google Scholar]
- White KA, Morris TJ (1995) RNA determinants of junction site selection in RNA virus recombinants and defective interfering RNAs. RNA 1: 1029–1040 [PMC free article] [PubMed] [Google Scholar]
- White KA, Nagy PD (2004) Advances in the molecular biology of tombusviruses: gene expression, genome replication and recombination. Prog Nucleic Acids Res Mol Biol 78: 187–226 [DOI] [PubMed] [Google Scholar]
- Wu B, Vanti WB, White KA (2001) An RNA domain within the 5′ untranslated region of the tomato bushy stunt virus genome modulates viral RNA replication. J Mol Biol 305: 741–756 [DOI] [PubMed] [Google Scholar]
- Wu B, White KA (1998) Formation and amplification of a novel tombusvirus defective RNA which lacks the 5′ nontranslated region of the viral genome. J Virol 72: 9897–9905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Malinina L, Patel DJ (2003) Recognition of small interfering RNA by a viral suppressor of RNA silencing. Nature 426: 874–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Slowinski V, White KA (1999) Subgenomic mRNA regulation by a distal RNA element in a (+)-strand RNA virus. RNA 5: 550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Rueckert RR (1993) Flock house virus: down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand. J Virol 67: 2716–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M, Mathews DH, Turner DH (1999) Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. In RNA Biochemistry and Bio/Technology, Barciszewski J, Clark BFC (eds) pp 11–43. Dordrecht/Norwell, MA: Kluwer Academic Publishers [Google Scholar]


