Abstract
Hormones are important regulators of plant growth and development. In Arabidopsis, perception of the phytohormones ethylene and cytokinin is accomplished by a family of sensor histidine kinases including ethylene-resistant (ETR) 1 and cytokinin-response (CRE) 1. We identified the Arabidopsis response regulator 2 (ARR2) as a signalling component functioning downstream of ETR1 in ethylene signal transduction. Analyses of loss-of-function and ARR2-overexpressing lines as well as functional assays in protoplasts indicate an important role of ARR2 in mediating ethylene responses. Additional investigations indicate that an ETR1-initiated phosphorelay regulates the transcription factor activity of ARR2. This mechanism may create a novel signal transfer from endoplasmic reticulum-associated ETR1 to the nucleus for the regulation of ethylene-response genes. Furthermore, global expression profiling revealed a complex ARR2-involving two-component network that interferes with a multitude of different signalling pathways and thereby contributes to the highly integrated signal processing machinery in higher plants.
Keywords: expression profiling, phosphorelay, phytohormone signalling, response regulator, two-component system
Introduction
Hormones mediate the communication between cells and organs in plants and animals. In recent years, the molecular action of the phytohormones like ethylene and cytokinin has been intensively studied in plants (Kakimoto, 2003; Guo and Ecker, 2004). The gaseous molecule ethylene affects many aspects of the plant life cycle, including seed germination, abscission and fruit ripening (Guo and Ecker, 2004). Moreover, reactions of plants to biotic and abiotic stresses involve ethylene. Insights into the ethylene-response pathway have mainly arisen from molecular studies in the model plant Arabidopsis thaliana using genetic screens based on the ‘triple response'. Several etr, ers and ein mutants have been identified in Arabidopsis, which exhibit a reduced or no triple response in the presence of ethylene or its biochemical precursor 1-aminocyclopropane-1-carboxylic acid (ACC). Cloning of the genes and characterization of mutant alleles have defined the framework of the signalling pathway leading from ethylene perception to changes in gene expression (Guo and Ecker, 2004). The first gene cloned was ETR1 (ethylene-resistant 1), which represents an endoplasmic reticulum (ER)-associated ethylene receptor with similarity to receptor histidine kinases of two-component signalling systems. Ethylene perception in Arabidopsis involves four additional ETR-related proteins. For all of them, binding of ethylene with similar affinity has been shown (Wang et al, 2003). The next known downstream signalling component interacting with ETR1 and ethylene response sensor (ERS) 1 is CTR1. CTR1 encodes a Raf mitogen-activated protein (MAP) kinase kinase kinase and functions as a negative regulator of ethylene responses. Recent data indicate that CTR1 regulates a protein kinase cascade consisting of the MAP kinase kinase SIMKK and the MAP kinases MPK6 and MPK13 (Ouaked et al, 2003). Therefore, sensor histidine kinase-like ethylene receptors link the perception of the hormone to the activity of a MAP kinase phosphorylation cascade. The integral membrane protein EIN2, which has similarity to Nramp metal ion transporters, is thought to function downstream of the CTR1-regulated protein kinase cascade (Ouaked et al, 2003; Guo and Ecker, 2004). EIN2 regulates the activity of specific transcription factors by a yet unknown mechanism. Two families of ethylene-responsive transcription factors have been described: EIN3 and EIN3-like proteins and ethylene response element-binding proteins (EREBPs; Guo and Ecker, 2004). EIN3 and other members of the EIN3-like family bind to a conserved cis-regulatory element in the promoter of the EREBP-encoding gene ethylene response factor (ERF) 1 (Solano et al, 1998). As recently shown, ethylene regulates EIN3 activity by SCFEBF1/EBF2-dependent proteolysis (Guo and Ecker, 2003; Potuschak et al, 2003; Yanagisawa et al, 2003). EIN3 activates transcription of the ERF1 gene, and ERF1 in turn binds to a conserved cis-acting sequence within promoters of secondary target genes, which eventually mediate ethylene responses (Solano et al, 1998). However, although CTR1 and the other above-described proteins are central components in ethylene signal transduction, there are several indications for the existence of additional ethylene signalling pathways (reviewed in Hass et al, 2004).
The hormone cytokinin regulates physiological and developmental processes overlapping with those of ethylene. The cloning of the cytokinin receptor CRE1 (cytokinin-response 1) and its relatives the Arabidopsis histidine kinases 2 and 3 revealed that again a sensor histidine kinase family is involved in hormone perception (Kakimoto, 2003). Although the signal transduction cascade downstream of the cytokinin receptors awaits further elucidation, recent observations suggest that a two-component signalling circuit may mediate cytokinin signal transmission from the plasma membrane to the nucleus (Kakimoto, 2003; Hass et al, 2004). In addition to the receptors, this cytokinin-regulated circuit appears to comprise several response regulators including Arabidopsis response regulator (ARR) 1 and ARR2 (Hwang and Sheen, 2001; Sakai et al, 2001; Tajima et al, 2004).
A number of additional two-component elements have been discovered in plants within recent years. These include additional histidine kinases, histidine phosphotransfer (HPt) proteins and response regulators (Hwang et al, 2002). However, although the histidine kinase activity of ETR1 and of the cytokinin receptor CRE1 has been experimentally verified (Kakimoto, 2003; Hass et al, 2004), evidence for the precise mode of action of multistep phosphorelays in plant hormone responses remains elusive.
We have shown that ARR2 represents a transcriptional regulator of the GARP transcription factor family and binds to the 5′-GAT-3′ sequence motif conserved in the promoters of nuclear genes coding for components of the Arabidopsis mitochondrial respiratory chain complex I (nCI; Lohrmann et al, 2001). Although preferentially found in pollen grains, ARR2 is expressed in all organs of the adult Arabidopsis plant as well as in seedlings (Lohrmann et al, 2001; Tajima et al, 2004). Whereas the C-terminal output domain of ARR2 mediates DNA binding and transactivation of target genes, the receiver module interacts with the Arabidopsis histidine phosphotransfer protein (AHP) 1 and AHP2 and contains the conserved aspartate residue (Asp80) predicted to be phosphorylated (Supplementary material; Sakai et al, 2000; Lohrmann et al, 2001). Hence, it was proposed that ARR2 constitutes a part of a two-component signalling pathway that in addition to the response regulator consists of at least one HPt protein and a hybrid sensor kinase (Lohrmann and Harter, 2002).
Here we describe the phenotypes of Arabidopsis arr2 loss-of-function and ARR2-overexpressing lines. These data as well as results from functional analyses in protoplasts reveal that ARR2 contributes not only to cytokinin but also to ethylene signalling. Further analyses including biochemical approaches and genome-wide expression profiling suggest that an ETR1-dependent phosphorelay regulates the transactivation capacity of ARR2 thereby establishing a novel branch in ethylene signalling. Moreover, our data suggest that ARR2-including two-component signalling systems are responsible for fine-tuning and crosstalk of a multitude of signalling pathways in higher plants.
Results
Characterization of an arr2 loss-of-function mutant from Arabidopsis
To elucidate the function of ARR2, we identified a dissociation (DS) transposon-tagged Arabidopsis line (SGT4387) in the IMA collection (Parinov et al, 1999). Characterization of this line revealed that the DS element is integrated in the ARR2 coding region at codon 19 in the first exon (Supplementary material). Southern blot and cosegregation analyses demonstrated the presence of a single transposon insertion in the genome of line SGT4387 and its linkage to the ARR2 locus. RT–PCR-based expression analysis revealed that the transposon insertion caused a complete absence of the ARR2 transcript defining this line as an arr2 null mutant (Supplementary material).
Growth and development of the arr2 mutant were monitored in comparison to the wild type. At early seedling stage, we did not observe major phenotypic differences. However, 20 days after sowing, the arr2 mutant displayed retarded growth and development and early flowering (Figure 1A and data not shown). The arr2 mutant phenotype was partially complemented by expression of the wild-type ARR2 cDNA under the control of the constitutive viral 35S promoter, indicating that the response regulator is responsible for the observed phenotype (Figure 1A). The incomplete complementation is very likely due to the 35S promoter, which may not perfectly establish the expression pattern of the native ARR2 promoter. The arr2 mutant phenotype was only observed in homozygous arr2 lines, indicating the recessive nature of the mutant allele. These analyses revealed that the arr2 loss-of-function allele causes aberrations in growth and development of Arabidopsis plants, which could be due to an altered hormone homeostasis.
Figure 1.
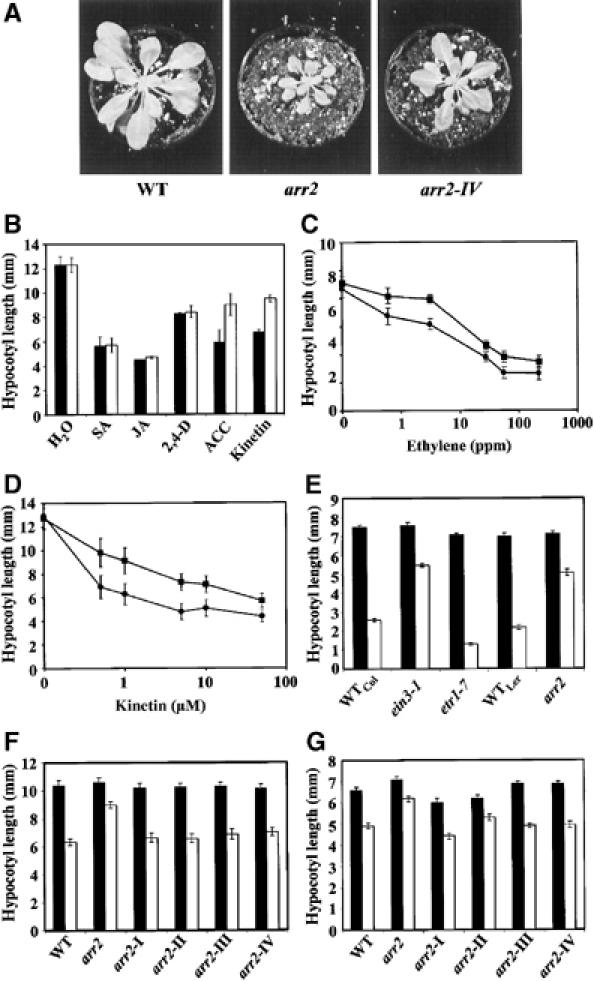
ARR2 is involved in ethylene and cytokinin signalling in Arabidopsis. (A) Wild-type and representative plants of homozygous arr2 loss-of-function mutant (arr2) and ARR2 overexpressors (arr2.IV) 30 days after sowing. Bars, 2.5 cm. (B) Hypocotyl elongation of wild-type (black) and arr2 (white) seedlings exposed to different plant hormones. Seedlings were grown on filter papers containing no hormone (control) or 1 mM salicylic acid (SA), 1 μM jasmonic acid (JA), 1 μM auxin (2,4-D), 10 μM ACC or 0.5 μM cytokinin (kinetin). (C) Dose dependence of ethylene-induced hypocotyl growth inhibition in wild-type (circles) and arr2 (squares) seedlings. A 0 ppm concentration of ethylene means no gas supplied. (D) Dose dependence of cytokinin-induced hypocotyl growth inhibition in wild-type (circles) and arr2 (squares) seedlings. Seedlings were grown in darkness on filter papers containing the indicated concentrations of kinetin. (E) Hypocotyl elongation of wild type (WTCol, WTLer), the ein3 mutant (ein3-1), the etr1-7 mutant (etr1-7) and the arr2 loss-of-function (arr2) in the absence (black) or presence of 10 μM ACC (white). (F) Hypocotyl elongation of wild-type (WT), the arr2 null mutant (arr2) and four independent ARR2-overexpressing lines in the arr2 genetic background (arr2-I to arr2-IV) in the absence (black) or presence of 10 μM ACC (white). (G) Hypocotyl elongation of wild-type (WT), the arr2 null mutant (arr2) and four independent ARR2-overexpressing lines in the arr2 loss-of-function background (arr2-I to arr2-IV) in the absence (black) or presence of 0.5 μM kinetin (white).
ARR2 also functions in ethylene signalling
We investigated in detail whether the arr2 knockout mutant affects hormone signalling by analyzing the hypocotyl growth response in seedlings. As shown in Figure 1B, ACC- and cytokinin-treated arr2 seedlings displayed a reduced response compared to wild type, whereas the application of other hormones did not affect growth. Dose–response analyses indicated that the observed altered hypocotyl elongation response of arr2 seedlings is caused by a reduced sensitivity to ethylene and cytokinin (Figure 1C and D). The hyposensitive response of arr2 seedlings to ethylene is comparable to that of the ethylene signalling mutant ein3-1 and opposite to the weak hypersensitive reaction of the etr1-7 loss-of-function receptor mutant (Figure 1E; Cancel and Larsen, 2002). To further elucidate the role of ARR2 in hormone signalling, we studied arr2 loss-of-function seedlings that express the ARR2 cDNA under the control of the 35S promoter. As shown in Figure 1F and G, expression of ARR2 in the arr2 mutant complemented the ethylene and cytokinin hyposensitive phenotype. These results demonstrate that the loss of the ARR2 gene product is responsible for the observed hormone hyposensitive phenotype and, furthermore, that ARR2 acts not only in cytokinin signal transduction (Hwang and Sheen, 2001) but also in ethylene signalling.
The primary ethylene-responsive (PER) element of the ERF1 promoter harbors two GAT boxes that are essential for ethylene regulation of ERF1 expression (Solano et al, 1998). As depicted in Figure 2A, ARR2 binds not only to the GAT box in the promoter of the PSST gene—a representative member of the nCI gene family serving as positive control—but also to the PER element of the ERF1 promoter. To elucidate the participation of ARR2 in ERF1 gene regulation, we used a leaf cell assay on the basis of ethylene-regulated transcription in Arabidopsis mesophyll protoplasts (Hwang and Sheen, 2001). In transfected protoplasts, the activity of an ERF1∷LUC reporter gene was very low and doubled by ethylene treatment (Figure 2B, control). Expression of ARR2 enhanced the activity of ERF1∷LUC 20-fold. Treatment of protoplasts with ethylene in the presence of ARR2 reduced ERF1∷LUC activity (Figure 2B). These data indicate that the response regulator ARR2 contributes to the regulation of ERF1 gene expression in Arabidopsis protoplasts.
Figure 2.
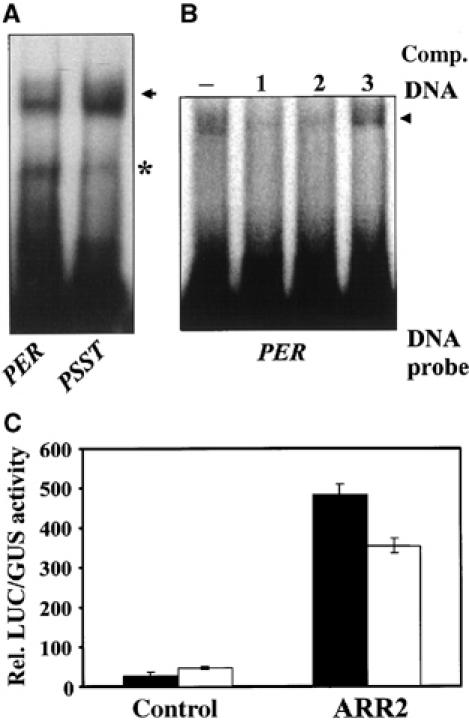
Transcriptional regulation of the ethylene-regulated ERF1 gene by ARR2. (A) ARR2 binds to the conserved PER element of the ERF1 promoter and to the GAT box of the PSST gene promoter in vitro. Gel-shift experiments were performed with the indicated radioactively labelled probes and recombinant ARR2. The arrow indicates the position of the ARR2:DNA complex. *, unspecific band. (B) Binding of ARR2 to the PER element is sequence specific. For competition experiments, a 50 × molar excess of nonlabelled PER (lane 1), PSST (lane 2) element or DNA without a GAT-box motif (lane 3) was added to the binding reaction. (−), no competitor DNA. The arrow indicates the position of the ARR2:DNA complex. (C) Transcriptional regulation of ERF1∷LUC by ARR2. Arabidopsis mesophyll protoplasts were cotransfected with UBQ10∷GUS (internal standard), the ERF1∷LUC reporter gene and an effector plasmid expressing HA-tagged ARR2. Nonfunctional GFP was used as a control. The transfected protoplasts were incubated without (black bars) or with 10 μl/l ethylene (white bars).
To further examine the role of ARR2, we carried out genome-wide expression profiling of the arr2 null mutant compared to wild type. Total RNA from rosette tissue of four independently grown 30-day-old plants was combined, extracted and subjected to microarray analysis using the Affimetrix Genome Genechip representing about 26 370 Arabidopsis genes (Zhu, 2003, ArrayExpress accession numbers are A-MEXP-79 and E-MEXP-125). Around 370 of these genes showed a significant alteration of expression levels in the arr2 null mutant compared to wild type (Table I and Supplementary material). Among them, a major fraction is known to play a crucial role in responses of plants to phytohormones or is related to pathogen defense or reactions to stress (Table I and Supplementary material; Cheong et al, 2002; Lorenzo et al, 2003). The majority of these genes are downregulated in the arr2 null mutant compared to wild type (Table I). However, we also observed a parallel up- and downregulation of genes within the same category (e.g. defense- and stress-related category; Table I). The microarray data were verified for a few selected genes by semiquantitative RT–PCR (Supplementary material). These results implicate a crucial role of ARR2 in the integration of plant signal transduction pathways.
Table 1.
Differentially regulated genes in the arr2 null mutant compared to wild type (selection)
| AGI no. | Putative function | Fold changea |
|---|---|---|
| ABA (abscisic acid)-related | ||
| AT4G24960 | Abscisic acid-induced-like protein | −2.1 |
| AT4G01600 | ABA-responsive protein | 2.4 |
| AT5G15960 | Cold- and ABA-inducible protein kin1 | −2.2 |
| Abiotic stress-related | ||
| AT2G42540 | Cold-regulated protein cor15a precursor | −3.0 |
| AT3G12580 | Heat shock protein 70 | −5.8 |
| AT3G47440 | Aquaporin protein | −2.5 |
| AT3G25760 | ERD12 | −8.5 |
| AT3G46230 | Heat shock protein 17 | −2.3 |
| AT4G30650 | Low-temperature- and salt-responsive protein | −3.3 |
| AT4G11650 | Osmotin precursor | 7.4 |
| AT4G37220 | Cold acclimation protein | 2.7 |
| AT4G21100 | UV-damaged DNA-binding protein | −2.1 |
| AT5G56010 | Heat shock protein 90 | −2.8 |
| AT1G16410 | Cytochrome P450 | −3.2 |
| AT2G22330 | Cytochrome P450 | −4.7 |
| Auxin-related | ||
| AT1G51760 | IAA-Ala hydrolase (IAR3) | −3.2 |
| AT1G78370 | 2,4-D-inducible glutathione S-transferase | −3.0 |
| AT1G05670 | Indole-3-acetate beta-glucosyltransferase | −3.0 |
| AT4G15490 | Indole-3-acetate beta-glucosyltransferase-like protein | −2.1 |
| AT5G13370 | Auxin-responsive-like protein | −3.9 |
| Ethylene-related | ||
| AT1G28370 | Ethylene-responsive element-binding protein (EREBP) | −2.5 |
| AT1G05010 | ACC oxidase (ACO) | −2.8 |
| AT2G44840 | Ethylene-responsive element-binding protein (EREBP) | −3.4 |
| AT2G31230 | Ethylene-responsive element-binding protein (EREBP) | −2.7 |
| AT3G24500 | Ethylene-responsive transcriptional coactivator | −2.2 |
| AT3G23240 | Ethylene-responsive element-binding factor 1 (ERF1) | −2.0 |
| AT3G23150 | Ethylene receptor (ETR2) | −2.2 |
| AT4G11280 | ACC synthase (AtACS-6) | −2.8 |
| Defense-related | ||
| AT1G72900 | Virus resistance protein | −5.8 |
| AT1G80840 | WRKY transcription factor | −5.1 |
| AT1G61560 | Mlo protein | −2.3 |
| AT1G75380 | Wound-responsive protein | 2.2 |
| AT1G73260 | Trypsin inhibitor | 6.8 |
| AT2G46400 | WRKY transcription factor | −5.8 |
| AT2G29470 | Glutathione S-transferase | 3.0 |
| AT2G14610 | Pathogenesis-related PR-1-like protein | 19.8 |
| AT2G38470 | WRKY transcription factor | −3.6 |
| AT3G57240 | beta-1,3-Glucanase | 3.4 |
| AT3G57260 | beta-1,3-Glucanase 2 (PR-2) | 4.7 |
| AT3G04670 | Elicitor response element-binding protein WRKY3 isolog | −2.5 |
| AT3G04720 | Hevein-like protein precursor (PR-4) | 2.8 |
| AT4G39410 | WRKY transcription factor | −2.0 |
| AT4G12880 | Blue copper-binding protein | −2.7 |
| AT5G13080 | WRKY transcription factor | 3.8 |
| AT1G60270 | beta-Glucosidase | −3.5 |
| AT5G44420 | Antifungal protein-like (PDF1.2) | 3.5 |
| Jasmonate-related | ||
| AT1G17420 | Lipoxygenase | −4.1 |
| AT3G16470 | Lectin, similar to jasmonate-inducible protein | −4.3 |
| AT3G45140 | Lipoxygenase AtLOX2 | −2.8 |
| Phenylpropane biosynthesis-related | ||
| AT1G74100 | Flavonol sulfotransferase | −3.7 |
| AT1G74710 | Isochorismate synthase (icsI) | −2.7 |
| AT2G37040 | Phenylalanine ammonia lyase (PAL1) | −3.6 |
| AT2G38240 | Anthocyanidin synthase | −6.5 |
| AT3G21230 | 4-coumarate:CoA ligase 2 | −2.8 |
| AT3G53260 | Phenylalanine ammonia-lyase | −5.5 |
| aAverage channel intensity ratio of arr2 null mutant sample over wild-type sample. | ||
ARR2 function depends on aspartate phosphorylation
Because ARR2 represents a response regulator, we considered that ARR2 activity may be regulated by phosphorylation. To investigate whether the conserved aspartate residue (Asp80) within the receiver domain of ARR2 may become phosphorylated in plants, a cell-free phosphorelay system on the basis of evacuolated protoplasts (AtEP) from Arabidopsis mesophyll tissue was established (Figure 3A). These cells enabled the isolation of total extracts as well as of microsomal and cytosolic fractions as demonstrated by Western blot analyses using compartment-specific antisera (Figure 3B). For in vitro phosphorelay experiments, Strep-tagged ARR2 was incubated in the presence of [γ-32P]ATP either with microsomes, cytosol or both fractions. Then, ARR2 was recovered from the reaction mixture by affinity purification and analyzed for incorporation of radioactive phosphate. Phosphorylation of ARR2 was only detected when both the microsomal and cytosolic fractions were present in the reaction mixture (Figure 3C).
Figure 3.
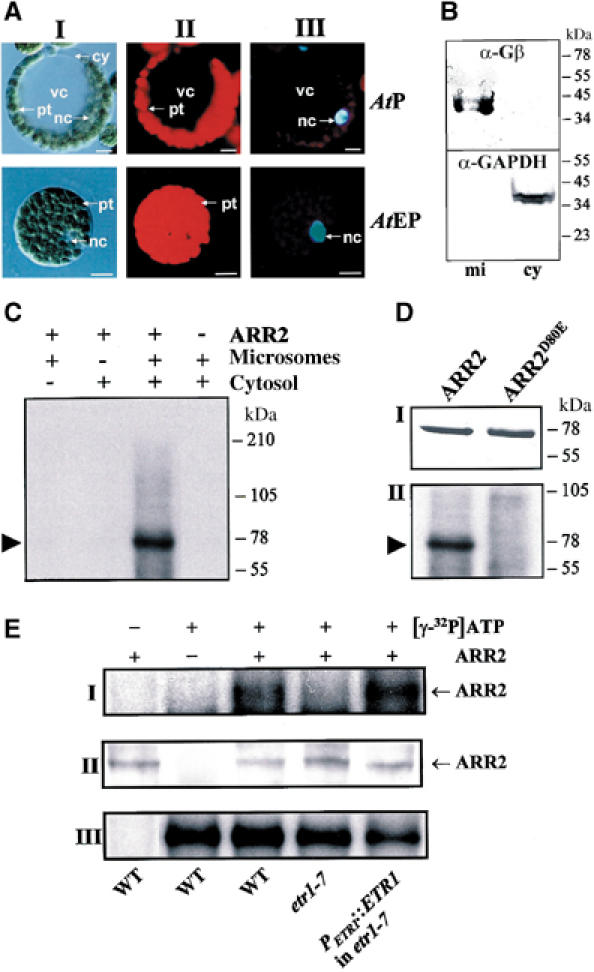
Phosphorylation of ARR2 by a plant cell-free phosphorelay system depends on Asp80 and the presence of ETR1. (A) Generation of evacuolated protoplasts from mesophyll tissue of Arabidopsis. AtEP were generated from protoplasts (AtP) by high-speed centrifugation on Percoll gradients (I). Chlorophyll fluorescence of the chloroplasts (II) and DAPI staining of the nucleus (III) were analyzed by epifluorescence microscopy. cy, cytoplasm; nc, nucleus; vc, vacuole. Bars, 10 μM. (B) Western blot analysis of microsomal (mi) and cytosolic (cy) fractions prepared from AtEP using compartment-specific antisera. α-Gβ, antiserum against membrane-associated β subunit of Arabidopsis G protein; α-GAPDH, antiserum against cytosolic glyceraldehyde-3-phosphate dehydrogenase. (C) Phosphorylation of ARR2 is mediated by microsomal and cytosolic activities. Strep-tagged ARR2 was incubated in a reaction mix containing [γ-32P]ATP and the indicated subcellular fractions. ARR2 was recovered from the mix by affinity purification and analyzed for incorporation of radioactively labelled phosphate by autoradiography. The arrow indicates the position of ARR2. (D) Phosphorylation of ARR2 targets Asp80. Equal amounts of Strep-tagged ARR2 and Strep-tagged ARR2D80E (I) were analyzed for incorporation of radioactively labelled phosphate in the presence of microsomal and cytosolic fractions (II) as outlined above. The arrow indicates the position of ARR2. (E) ETR1 contributes to Asp80 phosphorylation of ARR2. Protein extracts from AtEP of wild type (WT), the etr1-7 loss-of-function mutant (etr1-7) and the etr1-7 mutant overexpressing ETR1 under the control of the ETR1 promoter (PETR1∷ETR1 in etr1-7) were coincubated with Strep-tagged ARR2 (ARR2) and [γ-32P]ATP as indicated. After purification, phosphorylation of ARR2 was analyzed by autoradiography (panel I). An equal amount of ARR2 in the reaction mixture was tested by Western blot using an ARR2 antiserum (panel II). Nearly identical total phosphorylation activity is indicated in panel III, which shows a representative labelled protein in the extracts of evacuolated protoplasts. Phosphorylation experiments were conducted at least twice with similar results.
Mutation of the highly conserved phosphorylatable Asp to Glu within receiver domains of response regulators usually abolishes phosphorylation (Stock et al, 2000). To verify this for ARR2, the conserved Asp80 in the receiver domain was mutated to Glu generating ARR2D80E. Whereas wild-type ARR2 was efficiently phosphorylated, a modification of ARR2D80E was not observed (Figure 3D). These data show that the cell-free phosphorelay system from plant cells transfers a phosphoryl group from ATP to the receiver domain of ARR2. Furthermore, this system comprises at least two additional activities besides ARR2, which reside in two different subcellular compartments.
The membrane-associated phosphotransfer activity observed in the plant cell-free phosphorelay system very likely originates from sensor histidine kinases like ETR1. To investigate the linkage between ethylene perception and ARR2 function, we prepared protein fractions from AtEP of the ethylene receptor mutant etr1-7 and of a transgenic Arabidopsis line expressing ETR1 under its own promoter in the etr1-7 background (PETR1∷ETR1/etr1-7; Gamble et al, 2002). Etr1-7 is a loss-of-function allele of ETR1 that arises from a stop codon at Trp74 (Hua and Meyerowitz, 1998). The protein fractions from evacuolated protoplasts of etr1-7, PETR1∷ETR1/etr1-7 and wild-type plants were supplemented with Strep-tagged ARR2 and [γ-32P]ATP, and phosphotransfer assays were performed as described above. Phosphorylation of ARR2 was strongly reduced in the presence of the etr1-7 fraction as compared to wild type (Figure 3D). In contrast, if the extract from AtEP of PETR1∷ETR1/etr1-7 plants was used, ARR2 phosphorylation was restored (Figure 3D). These results indicate that ETR1 is an upstream regulator of ARR2 phosphorylation and is biochemically linked to the response regulator.
Asp80 phosphorylation is crucial for ARR2 function in vivo
To analyze the functional relevance of the Asp80-to-Glu mutation in planta, we generated wild-type and arr2 null mutant plants expressing ARR2 or ARR2D80E under the control of the constitutive viral 35S promoter (Figure 4A). Whereas plants carrying the ARR2 construct were almost indistinguishable from the corresponding wild type, expression of ARR2D80E caused severe pleiotropic aberrations in growth and development (Figure 4B). These alterations included apparent disturbances in the primary vegetative meristem manifested in a disturbed rosette and an extremely aberrant leaf shape (Figure 4B and C). Moreover, around 50% of the primary ARR2D80E-expressing transformants in wild-type background died before flowering. The ARR2D80E-induced phenotype was even more severe in the arr2 mutant background (Figure 4B and C), and surviving plants were infertile. Most remarkably, ARR2D80E-overexpressing seedlings showed a triple response phenotype in the absence of ACC (Figure 4D). Further treatment with ACC could not enhance this reaction in ARR2D80E-overexpressing plants, whereas wild-type seedlings displayed the expected ethylene-dependent phenotype (Figure 4D). In contrast to wild-type and the ethylene overproduction 1 (eto1) mutant (Chae et al, 2003) but comparable to ctr1, inhibition of ethylene biosynthesis by aminoethoxyvinyl glycine (AVG) could not significantly suppress the constitutive triple response, indicating that the phenotype seen in the ARR2D80E overexpressor is not due to a strongly enhanced ethylene production (Figure 4D). These results implicate that ARR2D80E (and ARR2) functions downstream of ethylene perception.
Figure 4.
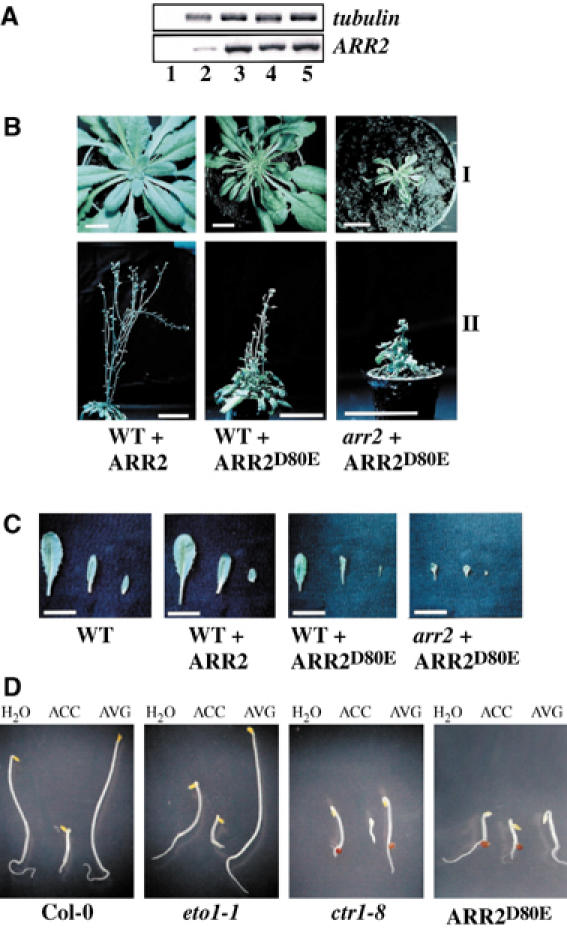
Phosphorylation of Asp80 is essential for appropriate function of ARR2 in plants. (A) ARR2, ARR2D80E and tubulin transcript levels in seedlings from wild type (lane 2), ARR2 overexpressors in a wild-type background (lane 3) and ARR2D80E overexpressors in a wild-type (lane 4) and arr2 null mutant background (lane 5) as determined by RT–PCR. Lane 1, control PCR without reverse transcription. (B) Phenotypes of representative wild-type plants (WT) and the arr2 null mutant (arr2) expressing either ARR2 (+ARR2) or ARR2D80E (+ARR2D80E) 50 days (I) and 70 days (II) after sowing. Bars, 1 cm in I and 5 cm in II. (C) Phenotypic comparison of representative leaves of different age from wild-type (WT) and arr2 null mutant plants (arr2) expressing either ARR2 (+ARR2) or ARR2D80E (+ARR2D80E). Bars, 2 cm. (D) Phenotypes of 4-day-old dark-incubated wild-type (WT), ctr1-8, eto1-1 and ARR2D80E-overexpressing seedlings (WT+ARR2D80E) grown in the absence (H2O) or presence (+) of 50 μM ACC (ACC) or 10 μM AVG (AVG). Bars, 0.25 cm.
Subsequently, we performed an additional transcriptome analysis to elucidate the gene expression pattern giving rise to the phenotype induced by ARR2D80E. Total RNA from four independently grown 30-day-old transgenic plants overexpressing to a similar extent either ARR2 or ARR2D80E (Figure 4A) was hybridized to the whole genome exon GeneChip array. This analysis revealed a significant expression change in around 1.400 genes in the ARR2D80E overexpressors compared to plants ectopically expressing wild-type ARR2. These included genes related to signal transduction of various phytohormones and light, responses to biotic and abiotic stress, protein degradation and folding, and development (Table II and Supplementary material). Remarkably, mRNA levels of several genes known to be induced by treatment of plants with auxin (e.g. Aux/IAA genes), cytokinin (e.g. type A ARR genes) and ethylene (e.g. EREBP genes) were up- or downregulated in ARR2D80E-overexpressing plants (Table II and Supplementary material). The microarray data were verified for a few selected genes by semiquantitative RT–PCR (Supplementary material). These data indicate that the substitution of the conserved Asp80 to Glu creates a dominant-active form of ARR2 that causes plants to become partially independent of exogenously applied phytohormones. Taken together, our findings indicate that Asp80 phosphorylation of ARR2 apparently occurs in plants and plays an important role for the appropriate function of the response regulator.
Table 2.
Differentially regulated genes in ARR2D80E compared to ARR2 overexpressors (selection)
| AGI no. | Putative function | Fold changea |
|---|---|---|
| Abscisic acid-related | ||
| AT4G26080 | Protein phosphatase ABI1 | 2.2 |
| AT5G13200 | ABA-responsive protein | 2.2 |
| AT5G57050 | Protein phosphatase ABI2 | 2.3 |
| AT5G59220 | ABA-induced protein phosphatase (PP2C) | 6.4 |
| Abiotic stress-related | ||
| AT1G08830 | Superoxidase dismutase | 2.4 |
| AT1G67970 | Heat shock transcription factor (HSF) | 2.7 |
| AT2G17820 | Histidine kinase (AtHK1) | 2.2 |
| AT2G46140 | Desiccation-related protein | 3.1 |
| AT3G53800 | Hsp70-binding protein HspBP1 | −2.7 |
| AT4G09350 | Heat shock protein (HSP) | −3.0 |
| AT4G15910 | Drought-induced protein | 2.1 |
| AT4G16660 | Heat shock protein (HSP) | 2.9 |
| AT4G18880 | Heat shock transcription factor (HSF) | 3.2 |
| AT4G24190 | Heat shock protein 90 (HSP90) | 2.8 |
| AT4G25100 | Superoxide dismutase | −2.7 |
| AT4G35970 | Ascorbate peroxidase | −2.0 |
| AT4G36680 | Salt-inducible protein | 2.3 |
| AT4G36990 | Heat shock transcription factor 4 (HSF4) | 6.2 |
| AT5G59610 | Heat shock protein 40 (HSP40) | −3.1 |
| AT5G05410 | DREB2A | 3.8 |
| AT5G09590 | Heat shock protein 70 (Hsc70-5) | 2.4 |
| AT5G49480 | NaCl-inducible Ca2+-binding protein | 2.4 |
| AT5G52300 | Low-temperature-induced 65 kDa protein | 2.3 |
| AT5G56010 | Heat shock protein 90 (HSP90) | 2.1 |
| AT5G60950 | Phytochelatin synthetase | 3.5 |
| Auxin-related | ||
| AT1G04250 | Auxin-induced protein | 2.7 |
| AT1G19840 | Auxin-induced protein | 3.6 |
| AT1G23080 | Auxin transport protein | −2.6 |
| AT1G29440 | Auxin-induced protein | −3.0 |
| AT1G75580 | Auxin-induced protein | 3.9 |
| AT1G77850 | Auxin response factor | 2.3 |
| AT1G80390 | Auxin-responsive protein IAA2 | −3.6 |
| AT2G21210 | Auxin-regulated protein | −2.6 |
| AT4G27260 | GH3-like protein | 2.8 |
| AT4G34750 | Auxin-regulated gene | 3.3 |
| AT4G34810 | Small auxin up RNA (SAUR-AC1) | 5.7 |
| AT4G37390 | Auxin-responsive GH3-like protein | 5.0 |
| AT4G38840 | Auxin-induced protein | −2.9 |
| AT4G38860 | Auxin-induced protein | −2.4 |
| AT5G13320 | Auxin-responsive protein | 6.2 |
| AT5G18010 | Auxin-induced protein | −3.2 |
| AT5G18050 | Auxin-induced protein | −4.5 |
| AT5G18080 | Auxin-induced protein | −3.2 |
| Cytokinin-related | ||
| AT1G19050 | Two-component response regulator (ARR7) | 2.0 |
| AT1G74890 | Two-component response regulator (ARR15) | 3.8 |
| AT2G07440 | Two-component response regulator (ARR24) | 2.6 |
| AT2G40670 | Two-component response regulator (ARR16) | 10.7 |
| AT2G41310 | Two-component response regulator (ARR8) | 2.4 |
| AT3G48100 | Two-component response regulator (ARR5) | 9.2 |
| AT3G56380 | Two-component response regulator (ARR4) | 6.0 |
| AT5G56970 | Cytokinin oxidase | 14.1 |
| Defense-related | ||
| AT1G05760 | Disease resistance protein RTM1 | −2.2 |
| AT1G05850 | Class I chitinase | −3.3 |
| AT1G53290 | Avr9 elicitor response protein-like | −2.2 |
| AT1G65390 | Disease resistance protein RPS4 | −4.5 |
| AT1G72230 | Blue copper protein | −2.2 |
| AT1G72920 | Virus resistance protein | 2.0 |
| AT1G72930 | Flax rust resistance protein | 3.3 |
| AT1G73260 | Trypsin inhibitor | 13.2 |
| AT1G75280 | NADPH oxidoreductase | −2.3 |
| AT2G21900 | WRKY-type DNA-binding protein | 3.1 |
| AT2G26010 | Antifungal protein | −2.6 |
| AT2G33050 | Leucine-rich repeat disease resistance protein | −2.2 |
| AT2G40000 | Nematode resistance protein | 2.0 |
| AT2G40740 | WRKY-type DNA-binding protein | 3.2 |
| AT2G43570 | Endochitinase | 13.4 |
| AT2G46400 | WRKY-type DNA-binding protein | 5.4 |
| AT3G01970 | WRKY-type DNA-binding protein | 9.0 |
| AT3G48720 | Hypersensitivity-related hsr201 protein | −2.9 |
| AT3G55470 | Elicitor-responsive protein (FIERG2) | 2.6 |
| AT3G56400 | WRKY-type DNA-binding protein 4 (WRKY4) | 4.0 |
| AT4G13920 | Disease resistance Cf-2-like protein | 2.2 |
| AT4G19530 | TMV resistance protein N-like | −2.4 |
| AT5G13080 | WRKY-type DNA-binding protein | 6.1 |
| AT5G14930 | Disease resistance protein EDS1 | 3.0 |
| AT5G26170 | WRKY-type DNA-binding protein 1 (WRKY1) | 3.3 |
| AT5G44420 | Antifungal protein (PDF1.2) | −2.6 |
| AT5G45110 | Regulatory protein NPR1-like | 2.1 |
| AT5G46350 | WRKY-type DNA-binding protein | 2.7 |
| AT5G62740 | Hypersensitive-induced response protein HIR3 | 3.2 |
| Degradation-related | ||
| AT1G20140 | SKP1 ASK1 (At4) | 3.1 |
| AT1G23410 | Ubiquitin extension protein | 2.9 |
| AT1G68050 | F-box protein (FKF1) | 2.3 |
| AT1G77000 | F-box protein (AtFBL5) | 3.3 |
| AT2G20160 | SKP1 ASK1 (At17) | 6.7 |
| AT3G17000 | E2 ubiquitin-conjugating enzyme | 2.1 |
| AT3G54650 | F-box protein (AtFBL17) | −2.3 |
| AT4G38930 | Ubiquitin fusion-degradation protein | 2.5 |
| AT5G24810 | Ubiquitin biosynthesis EIN AARF | 2.7 |
| AT5G27920 | F-box protein | 2.3 |
| AT5G55170 | Ubiquitin | 2.7 |
| AT5G57480 | AAA-type ATPase | 4.7 |
| AT5G57900 | SKP1 interacting partner 1 (SKIP1) | 2.0 |
| Development-related | ||
| AT1G01010 | NAC domain protein | 17.1 |
| AT1G02220 | NAM (no apical meristem) protein | 9.7 |
| AT1G02250 | NAM protein | 11.4 |
| AT1G52690 | Late embryogenesis-abundant protein | 8.9 |
| AT1G52890 | NAM protein | 7.2 |
| AT1G77450 | GRAB1-like protein | 2.1 |
| AT2G22850 | Embryo-abundant protein | −2.6 |
| AT4G00180 | YABBY3 axial regulator | −2.1 |
| AT5G06760 | Late embryogenesis-abundant protein | 4.7 |
| AT5G07190 | Embryo-specific protein 3 (ATS3) | −2.1 |
| AT5G20240 | PISTILLATA-like | 3.9 |
| AT5G39610 | NAM protein | 2.6 |
| Ethylene-related | ||
| AT3G12500 | Basic chitinase | 2.7 |
| AT3G16050 | Ethylene-inducible protein | 2.5 |
| AT3G23150 | Ethylene receptor (ETR2) | 2.4 |
| AT3G23240 | Ethylene response factor 1 (ERF1) | 3.9 |
| AT3G50260 | EREBP-3-like | 2.9 |
| AT4G23340 | ACC oxidase | −2.6 |
| AT4G26200 | ACC synthase | 2.4 |
| AT5G20400 | Ethylene-forming enzyme dioxygenase | 2.9 |
| AT5G43410 | EREBP-3-like | −3.1 |
| Folding-related | ||
| AT1G06330 | Copper chaperone | −13.9 |
| AT1G21750 | Protein disulfide isomerase | 3.8 |
| AT1G23100 | Chaperonin | 2.8 |
| AT2G47470 | Disulfide isomerase | 2.2 |
| AT3G18190 | Chaperonin | 2.0 |
| AT3G56070 | Peptidyl-prolyl isomerase | 3.0 |
| AT5G45680 | FKBP-type peptidyl-prolyl isomerase | −2.7 |
| AT5G55220 | Trigger factor | −2.4 |
| Gibberellin-related | ||
| AT1G22690 | GAST1-like protein | −4.5 |
| AT1G74670 | GAST1-like protein | −2.4 |
| Jasmonate-related | ||
| AT1G55020 | Lipoxygenase | 8.8 |
| AT3G16400 | Jasmonate-inducible protein | 6.0 |
| AT3G16420 | Jasmonate-inducible protein (lectin) | 5.5 |
| AT3G16430 | Jasmonate-inducible protein | 5.1 |
| AT3G16450 | Jasmonate-inducible protein (lectin) | 31.0 |
| AT3G16460 | Jasmonate-inducible protein (lectin) | 11.4 |
| AT5G48180) | Jasmonate-inducible protein | 3.0 |
| Light-related | ||
| AT1G06680 | OEC protein (23 kDa) | −2.1 |
| AT1G14280 | Phytochrome kinase substrate 1 | −3.4 |
| AT1G19150 | Chlorophyll a b-binding protein (LHCP) | −4.0 |
| AT1G51890 | Light-repressible receptor protein kinase | 2.6 |
| AT3G15570 | Nonphototropic hypocotyl protein | −2.1 |
| AT4G14110 | COP9 | 3.0 |
| AT5G11260 | HY5 | 2.3 |
| AT5G54190 | Protochlorophyllide oxidoreductase | −2.9 |
| AT5G58140 | Nonphototropic hypocotyl 1-like | −2.4 |
| aAverage channel intensity ratio of ARR2D80E overexpressors over ARR2 overexpressors. | ||
Phosphorylation modulates the transactivation capacity and stability of ARR2
The results presented in the previous sections and published data (Kakimoto, 2003) provide biochemical and functional evidence for a link of the histidine kinases ETR1 and CRE1 to ARR2 in mediating hormone signalling. Furthermore, Asp80 phosphorylation in the receiver domain of ARR2 is essential for its function. To determine how phosphorylation might regulate the activity of ARR2, we investigated whether the Asp80-to-Glu mutation affects its intracellular partitioning, DNA-binding transactivation capacity and stability. For the analysis of the intracellular distribution, we generated transgenic plants expressing ARR2-GFP and ARR2D80E-GFP fusion constructs. These plants displayed the same phenotype as described for ARR2- and ARR2D80E-expressing plants, respectively, indicating that the GFP fusion proteins are present and functional in vivo (Figure 5A). Epifluorescence images of hypocotyl cells from these plants revealed that ARR2-GFP as well as ARR2D80E-GFP is exclusively localized to the nucleus (Figure 5B).
Figure 5.
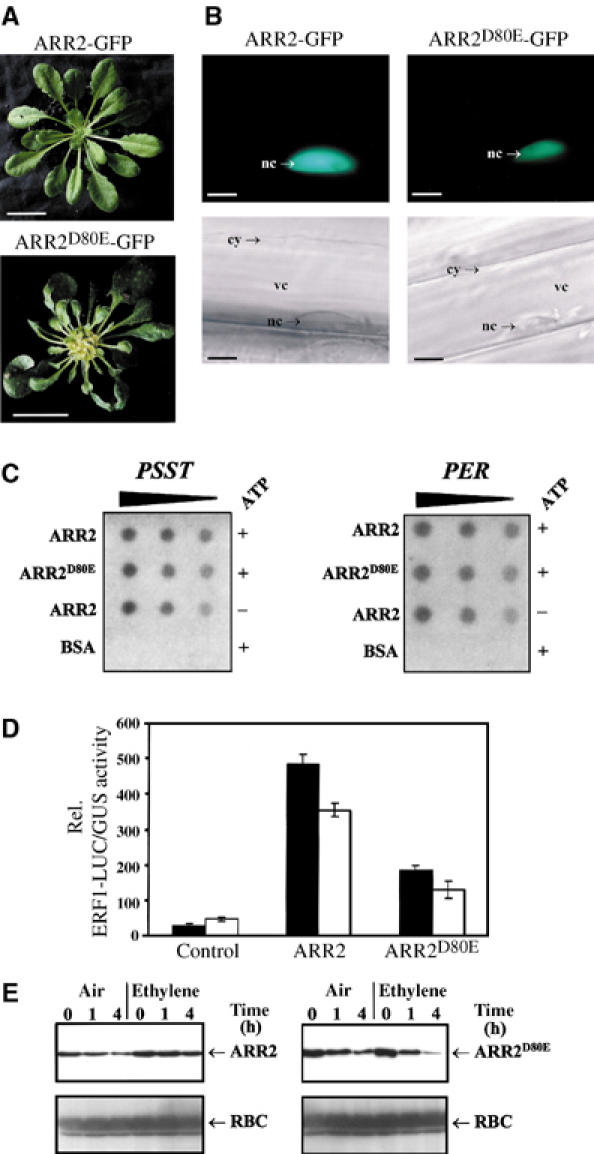
Asp80-to-Glu mutation regulates the transactivation capacity of ARR2. (A) Representative 40-day-old transgenic Arabidopsis plants (wild-type background) expressing either ARR2-GFP (left) or ARR2D80E-GFP (right). Bars, 2.5 cm. (B) ARR2-GFP and ARR2D80E-GFP are both localized to the nucleus. Presented are epifluorescence and bright-field images of hypocotyl cells from the transgenic plants described in (A). cy, cytoplasm; nc, nucleus; vc, vacuole. Bars, 10 μm. (C) In vitro phosphorylated and nonphosphorylated ARR2 and ARR2D80E display identical DNA-binding activities. Recombinant Strep-tagged ARR2 and ARR2D80E were treated in the cell-free phosphorelay system in the presence (+ATP) or absence of ATP (−ATP). After recovery by affinity purification, decreasing amounts of ARR2, ARR2D80E and, as a control, BSA were dotted onto PVDF membranes. The membranes were incubated with radioactive PSST and PER promoter fragments. Binding to DNA was analyzed by autoradiography. (D) Transactivation capacity of ARR2 is phosphorylation dependent. Protoplasts were transfected with UBQ10∷GUS (internal standard) and the ERF1∷LUC reporter construct and effector plasmids expressing either ARR2 or ARR2D80E (both HA tagged). Nonfunctional GFP was used as a control. The transfected protoplasts were incubated without hormone (black bars) or with ethylene (10 μl/l, upper panel, white bars). (E) Kinetics of ARR2 and ARR2D80E protein levels in Arabidopsis mesophyll protoplasts in the absence (air) or presence of ethylene (ethylene). The first sample (time 0 h) was harvested 30 min after protoplast transformation. ARR2 and ARR2D80E were detected using an HA-tag-specific antibody. RUBISCO (RBC) served as a control and was detected with a specific antiserum.
South-Western analyses were performed to investigate a possible phosphorylation dependence of the ARR2 DNA-binding activity. Strep-tagged ARR2 and ARR2D80E were incubated with ATP in the cell-free phosphorelay system derived from AtEP, affinity purified from the reaction mixture and dotted onto PVDF membranes. Nonphosphorylated but otherwise equally treated ARR2 and, as a control, ATP-treated BSA were dotted onto the membranes as well. The membranes were incubated with either a radioactively labelled PSST or PER promoter fragment and afterwards subjected to autoradiography. Independent of whether phosphorylated or nonphosphorylated ARR2 were present on the membrane, no differences in DNA binding were detected (Figure 5C). We also did not observe an altered affinity of dominant-active, nonphosphorylatable ARR2D80E to the PSST and PER promoter fragments, indicating that regulation of DNA-binding capacity by phosphorylation plays a minor role in ARR2 regulation.
Whether Asp80 phosphorylation interferes with the transactivation capacity of ARR2 was addressed in protoplasts using the ethylene-regulated ERF1∷LUC as reporter gene. As already presented in Figure 2C, expression of wild-type ARR2 in Arabidopsis mesophyll protoplasts resulted in basal expression of ERF1∷LUC. Application of ethylene inhibited the activity of ERF1∷LUC (Figures 2C and 5D). When ARR2D80E was present, we observed a strong repression of ERF1∷LUC gene in the absence of ethylene (Figure 5D). Treatment of protoplasts with ethylene further reduced ERF1∷LUC expression (Figure 5D).
To elucidate whether protein stability may play a role in the regulation of ERF1∷LUC activity, we determined ARR2 and ARR2D80E levels in the protoplasts. As shown in Figure 4E, the amounts of ARR2 and ARR2D80E decreased within the time period of 4 h with ARR2D80E appearing to be slightly less stable than ARR2 (Figure 5E). Ethylene treatment of protoplasts expressing ARR2 did not alter the destruction rate, whereas ARR2D80E seems to be less stable under these conditions.
As shown recently, ARR2 induces ARR6∷LUC reporter gene expression in protoplasts in a cytokinin-dependent way (Supplementary material; Hwang and Sheen, 2001). Remarkably, protoplasts expressing ARR2D80E displayed ARR6∷LUC activity, which was as high as in cytokinin-treated, ARR2-expressing cells. Exposure of ARR2D80E-expressing protoplasts to cytokinin further enhanced ARR6∷LUC activity (Supplementary material).
Taken into account that the DNA-binding activity and the intracellular distribution of ARR2 remained unaltered, these results indicate that phosphorylation of Asp80 regulates the transcriptional capacity and/or the stability of the response regulator.
Discussion
ARR2 functions as a transcriptional regulator in cytokinin signalling (Hwang and Sheen, 2001; Sakai et al, 2001) and binds to the GAT-box motif within the promoters of nCI genes (Lohrmann et al, 2001). Here we show that ARR2 is also able to bind to the PER promoter element of the ERF1 gene and mediates upregulation of ERF1∷LUC gene activity in Arabidopsis protoplasts. Application of exogenous ethylene results in repression of ERF1∷LUC in protoplasts. Because of ERF1 being an ethylene-triggered gene (Solano et al, 1998; Lorenzo et al, 2003), these data indicate that in Arabidopsis mesophyll protoplasts ARR2 negatively interferes with ethylene signal transduction. To further elucidate the contribution of ARR2 in ethylene signalling, we analyzed an Arabidopsis arr2 loss-of-function mutant in which the expression of the response regulator is abolished. In seedlings, the arr2 loss-of-function mutant renders plants less sensitive to cytokinin and ethylene. Furthermore, the overexpression of dominant-active ARR2D80E induces a triple response-like phenotype in the absence of ethylene and in the presence of the ethylene biosynthesis inhibitor AVG. Again, these results urge that ARR2 contributes to ethylene signalling in Arabidopsis. However, whereas ARR2 is a negative regulator of ethylene signalling in protoplasts, the hyposensitive hypocotyl growth phenotype of the recessive arr2 loss-of-function mutant implicates that ARR2 positively contributes to this response in seedlings. This effect becomes also visible when the activity of ARR2D80E is compared in protoplasts and transgenic plants. In protoplasts ARR2D80E represses ERF1∷LUC activity, while in ARR2D80E overexpressors ERF1 transcript accumulation is elevated. Furthermore, ARR2 and ARR2D80E positively regulate the activity of ARR6∷LUC in both protoplasts and transgenic plants. These data suggest that the way and direction (negative versus positive) of ARR2 action seem to depend on tissue-, cell- and even development-specific coacting factors that contribute to the activity and specificity of ARR2 function. In Arabidopsis protoplasts, which already produce ethylene and are responsive to the hormone (Yanagisawa et al, 2003), an additional exogenous supply in the presence of ARR2 may induce a negative feedback regulation. At the molecular level, this could result in inhibition of ARR2 transcription factor activity by an interacting repressor. Likewise, ARR2D80E appears to be less stable in protoplasts compared to wild-type ARR2. This instability may contribute to the reduction of the ERF1∷LUC activity and adds further complexity to the regulation of ARR2.
We established a plant cell-free phosphorelay system that implements a phosphotransfer from ATP to the conserved Asp80 in the receiver domain of ARR2. The microsomal activity for ARR2 phosphorylation derives from membrane-associated histidine kinases. The necessity for a soluble component may reflect the participation of HPt proteins in ARR2 modification. The cell-free phosphorelay system enabled us to investigate the functional interplay of ARR2 and ETR1. Compared to wild type, subcellular fractions from the etr1-7 mutant are significantly less efficient in phosphorylating ARR2. In extracts from etr1-7 plants expressing wild-type ETR1, phosphorylation of ARR2 was restored. These data indicate that the in vitro transfer of the phosphoryl group from ATP to the Asp80 of ARR2 depends to a certain extent on ETR1. These results not only suggest that ARR2 is involved in ethylene signalling, but also that the underlying mechanism is a two-component phosphorelay.
Our experiments suggest that the mutation of Asp80 to a Glu residue creates a constitutively active form of ARR2 (ARR2D80E). This finding is in agreement with observations in prokaryotic systems, where Asp-to-Glu mutations in the receiver domain of response regulators often generate constitutive active forms (Stock et al, 2000). Moreover, ARR2D80E expression also causes major phenotypic aberrations in transgenic plants. These results implicate that Asp phosphorylation of response regulators like ARR2 and, hence, phosphorelays play a crucial role in hormone signalling in plants.
Until now it has been assumed that ethylene signal transduction downstream of the receptors exclusively depends on CTR1 (Huang et al, 2003; Guo and Ecker, 2004). However, ctr1 null mutants still display residual ethylene responsiveness and an incomplete activation of ethylene-triggered responses (Guo and Ecker, 2003; Potuschak et al, 2003), suggesting the existence of an additional branch for ethylene signalling. Based on the evidence presented here, it is conceivable that a two-component signalling system represents such a branch. In this branch, ethylene binding may induce histidine kinase activity and autophosphorylation of ETR1. Phospho-ETR1 then could initiate a phosphorelay cascade that may include a shuttling HPt protein. The phosphotransfer could finally result in the transcriptional regulation of ARR2 inside the nucleus (Lohrmann and Harter, 2002). Thus, ETR1 may have a dual functional role in the initiation of ethylene signal transduction: whereas the CTR1-dependent pathway is negatively regulated (Guo and Ecker, 2004), the ARR2-dependent pathway is subject to positive regulation. The proposed phosphorelay is difficult to reconcile with observations that histidine kinase activity of ETR1 and ERS1 may not be required to mediate ethylene signalling (Guo and Ecker, 2004). However, it is of interest that the ethylene receptors with proven (ETR1) or proposed (ERS1) histidine kinase activity play a predominant role in the regulation of ethylene responses (Guo and Ecker, 2004). This relative importance of ETR1 and ERS1 could potentially be because of the presence of histidine kinase activity. Furthermore, it is impossible to observe an ARR2-dependent two-component mechanism in all of those ETR1 and ERS1 mutant plants, in which the receptors are present in a form that abolishes transphosphorylation capacity. Intramolecular transphosphorylation of the transmitter domains is a prerequisite for histidine kinases to initiate phosphorelay mechanisms (Stock et al, 2000; Ames et al, 2002). Thus, further biochemical and genetic experiments have to be carried out to clarify the relationship of the CTR1- and ARR2-dependent ethylene signal transduction pathways.
Genechip experiments were performed to investigate the expression network influenced by ARR2. We first compared the expression pattern of the arr2 null mutant with the corresponding wild type. Besides changes in the expression of genes related to ethylene signalling, we also observed alterations in the activity of genes associated with auxin, abscisic acid, jasmonic acid and gibberellin signal transduction. Although these genes may not be the primary targets of ARR2, their altered expression may explain the complex mutant phenotype of the arr2 loss-of-function plants that is different from those of plants solely impaired in ethylene (and cytokinin) perception. As a consequence of impaired hormone balance, genes related to defense and abiotic stress signalling and adaptation show altered expression as well (Zhong and Burns, 2003). Remarkably, significant changes in the expression of typical cytokinin-responsive genes like those encoding A-type response regulators (Rashotte et al, 2003) were not observed in the arr2 loss-of-function mutant. Obviously, in plants that have not been treated with exogenous cytokinin, absence of ARR2 does not cause significant alterations in the expression of early cytokinin-induced genes. Thus, endogenous variations in cytokinin levels may either induce slight, nondetectable changes in the accumulation of these cytokinin-induced genes or cytokinin signalling occurs by post-transcriptional mechanisms. Upregulation of type-A ARR genes in response to exogenous cytokinin may therefore reflect an adaptation process rather than a primary signalling event.
We also performed a comparative expression profiling with RNA from plants that express either wild-type ARR2 or ARR2D80E. The strongly altered expression of a variety of hormone-regulated genes indicates that the dominant-active ARR2D80E protein affects the entire two-component signalling network of Arabidopsis. As a consequence, the hormone-sustained homeostasis of gene expression pattern seems to be completely disturbed in ARR2D80E-expressing plants. This fundamental change in hormone homeostasis induces expression alterations of a multitude of genes related to many different signalling pathways. This also explains why the ARR2D80E-induced phenotype is distinct from the phenotype of the arr2 loss-of-function mutant. In conclusion, our expression studies suggest that the two-component network may directly or indirectly modulate many signal transduction pathways initiated by phytohormones and other endogenous and exogenous stimuli. An independent example for a modulating action of two-component systems is the stabilization of the active form of the phytochrome B photoreceptor by the type-A response regulator ARR4 (Sweere et al, 2001; To et al, 2004). In summary, these data suggest that two-component signalling systems may not always represent the primary mechanisms for signal transduction pathways but rather may establish a complex network that is predominantly responsible for maintaining fine-tuning and crosstalk of signalling pathways.
Materials and methods
Characterization of the arr2 insertion mutant
DNA and RNA manipulations and RT–PCR were performed as described previously (Lohrmann et al, 2001). The transposon insertion site within the ARR2 gene was verified by PCR using the DS-specific primers DS3′ and DS5′ (Parinov et al, 1999) and two ARR2-specific primers followed by sequencing of the PCR product.
Arabidopsis plants were grown for the indicated time periods on soil under short-day conditions (8 h light/16 h dark). Hormone treatments were performed as follows: seedlings were grown for 96 h on moisturized filter paper and treated with the indicated concentrations of hormones. For ethylene analyses, seedlings were germinated for 24 h on moisturized filter paper in open glass vessels. The vessels were closed and ethylene was injected. Seedlings were further grown for 72 h and ethylene concentration in the airspace was measured at the end of the 72 h growth period by gas chromatography. Ethylene gas was applied to protoplasts at a concentration of 10 μl/l for 9 h beginning 30 min after transfection. Air was applied as control. Physiological data are presented as the mean and standard deviation of at least 50 seedlings. Dose–response analyses were repeated at least three times and the means and the standard error of the mean are shown.
Plasmid construction and expression of recombinant proteins
For construction of binary constructs, wild-type ARR2 cDNA was cloned into the pPCV812 derivate pPCVB downstream of the (2 × 35S) promoter and upstream of the mGFP4 gene. For expression of ARR2 without C-terminal GFP tag, a stop codon was introduced in front of the mGFP4 gene. The ERF1∷LUC reporter construct was generated by fusing a 2.1 kbp 5′-flanking region of the ERF1 gene (At3g23240) to the firefly luciferase cDNA. The ARR6∷LUC construct has been described previously (Hwang and Sheen, 2001). Site-directed mutagenesis of Asp80 to Glu was performed using the QuickChange™ XL Mutagenesis System (Stratagene). PCR-generated cDNAs were verified by sequencing. Expression of Strep-tagged ARR2 and ARR2D80E in Escherichia coli and affinity purification on StrepTactin (IBA) were performed according to Lohrmann et al (2001).
Expression profiling
Total RNA from rosettes of four independently grown 30-day-old Arabidopsis plants was extracted using Qiagen RNeasy columns. Total RNA (5 μg) was used to synthesize cDNAs. Labelled cRNA, synthesized from the cDNA, was hybridized to the Arabidopsis whole genome exon GeneChip array according to the procedure described previously (Zhu et al, 2001). The custom Arabidopsis whole genome array used contains 42 Affymetrix control probe sets, three transgene control probe sets, four QC probe sets and 26 412 probe sets representing 26 367 Arabidopsis genes. On average, each gene contains 15 perfect match probes. These 25-mer oligonucleotide probes were selected from the 3′ end of 133 397 exons (Zhu, 2003). The hybridization signal of each probe set was quantified using the value of weighted average of all probes in a set, subtracting bottom 5% of average intensity of the entire array. The overall intensity of all probe sets of each array was further scaled to a target intensity of 100, so hybridization intensity of all arrays was equivalent. Genes corresponding to probe sets with a minimum detected expression level of 50 and a minimum change of 2.0-fold compared to wild type were selected and defined as genes with significant alteration of expression. The significant expression level threshold of 50 was determined based on the 100% above noise level from all negative controls on the mircoarray according to the determined scaling target 100. The reproducibility of the microarray was characterized by comparing data generated in parallel from 10 pairs of total RNA samples. The detection sensibility and specificity of the GeneChip microarray were characterized by spiking in a serial dilution of equimolar concentration of negative control transcripts from BioB, BioC and CreX to 10 μg Arabidopsis cRNAs and hybridizing to the 26K Arabidopsis whole genome array. The data are available at ArrayExpress with the accession numbers A-MEXP-79 for the array design and E-MEXP-125 for the experiment.
Generation of evacuolated protoplasts, cell fractionation and immunoblotting and transformation techniques
Protoplasts preparation, evacuolization of protoplasts, protein extraction and fractionation were carried out as described (Harter et al, 1994). The fractions were characterized by immunoblotting with antibodies against cytoplasmic GAPDH and the β subunit of plant G proteins (Obrdlik et al, 2000). Transformation of Arabidopsis plants was conducted according to Lohrmann et al (2001). Transfection of Arabidopsis mesophyll protoplasts has been described previously (Hwang and Sheen, 2001). Microscopic work was carried out as reported by Lohrmann et al (2001).
In vitro phosphotransfer assays
A 2 μg portion of recombinant Strep-tagged ARR2 was incubated with 5 μg of total extract, 1.5 μg of microsomal fraction or 4 μg of cytosolic fraction, and 20 μCi [γ-32P]ATP for 60 min at 4°C in a total volume of 75 μl standard phosphorylation buffer (50 mM Tris–HCl, 0.5 mM EDTA, 2 mM DTT, 50 mM KCl, 5 mM MgCl2, 5 mM MnCl2, 10% glycerol). The protein was recovered from the reaction mixture by affinity purification on StrepTactin beads (IBA). After washing three times with 0.5 ml of washing buffer (100 mM Tris–HCl pH 7.4, 150 mM NaCl), the ARR2 proteins were released from the beads by addition of 50 μl of washing buffer containing 2.5 mM desthiobiotine. The ARR2 proteins were subjected to SDS–PAGE, autoradiography and Western blot as described by Harter et al (1994).
EMSA and South-Western analysis
32P-labelling of the PSST (Lohrmann et al, 2001) and PER DNA probe (nucleotides −1213 to −1178 of the ERF1 promoter), EMSA and competition experiments were carried out as described previously (Lohrmann et al, 2001). For South-Western analyses, a dilution series (1:1, 1:2, 1:4) starting from 60 μl of affinity-purified, phosphorylated or control-treated ARR2 proteins was generated in 60 μl of washing buffer. The protein solutions were dotted on PVDF membranes. As a control, BSA (0.5, 1.0, 2.0 μg per dot) was dotted as well. The membrane was prehybridized for 2 h at 4°C in hybridization buffer (gel-shift binding buffer (Lohrmann et al, 2001) supplemented with 0.25% (w/v) BSA, 5 μg/ml salmon sperm DNA). Hybridization in 2 ml of hybridization buffer containing 0.4 ml of radioactively labelled PER or PSST promoter element was performed overnight at room temperature. Afterwards, the membrane was washed several times in hybridization buffer, dried and exposed to X-ray film (Kodak).
Supplementary Material
Supplementary File 1
Supplementary File 2
Supplementary File 3
Supplementary File 4
Supplementary File 5
Acknowledgments
We thank P Obrdlik and R Cerff for antibodies and EG Schaller for providing seeds. We are thankful to C Grefen, J Felix, H-S Chang, and L Cabrillac for excellent technical support, to D Wanke for help in image processing and to the Nottingham Stock Center for providing Arabidopsis lines. This work was supported by the DFG to ES (SFB388) and KH (SFB635, AFGN), the VW-Stiftung to JK and the Plant Signaling Natural Research Center, Korean Science and Engineering Foundation, to IH.
References
- Ames P, Studdert CA, Reiser RH, Parkinson JS (2002) Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc Natl Acad Sci USA 99: 7060–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancel JD, Larsen PB (2002) Loss-of-function mutations in the ethylene receptor ETR1 causes enhanced sensitivity and exaggerated response to ethylene in Arabidopsis. Plant Physiol 129: 1557–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae HS, Faure F, Kieber JJ (2003) The eto1, eto2, and eto3 mutations and cytokinin treatment increase ethylene biosynthesis in Arabidopsis by increasing the stability of ACS protein. Plant Cell 15: 545–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Chang H-S, Gupta R, Wang X, Zhu T, Luan S (2002) Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol 129: 661–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble RL, Qu X, Schaller GE (2002) Mutational analysis of the ethylene receptor ETR1. Role of the histidine kinase domain in dominant ethylene insensitivity. Plant Physiol 128: 1428–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2003) Plant responses to ethylene gas are mediated by SCFEBF1/EBF2-dependent proteolysis of EIN3 transcription factor. Cell 115: 667–677 [DOI] [PubMed] [Google Scholar]
- Guo H, Ecker JR (2004) The ethylene signalling pathway: new insights. Curr Opin Plant Biol 7: 40–49 [DOI] [PubMed] [Google Scholar]
- Harter K, Frohnmeyer H, Kircher S, Kunkel T, Mühlbauer S, Schäfer E (1994) Light induces rapid changes of the phosphorylation pattern in the cytosol of evacuolated parsley protoplasts. Proc Natl Acad Sci USA 91: 5038–5042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass C, Weinl S, Harter K, Kudla J (2004) Functional genomics of protein phosphorylation in Arabidopsis thaliana. In Plant Functional Genomics, Leister D (ed) Binghampton, NY: The Haworth Press (in press) [Google Scholar]
- Hua J, Meyerowitz EM (1998) Ethylene responses are negatively regulated by a receptor gene family in Arabidopsis thaliana. Cell 94: 261–271 [DOI] [PubMed] [Google Scholar]
- Huang Y, Li H, Hutchison CE, Laskey J, Kieber JJ (2003) Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J 33: 221–233 [DOI] [PubMed] [Google Scholar]
- Hwang I, Chen H-C, Sheen J (2002) Two-component signal transduction pathways in Arabidopsis. Plant Physiol 129: 500–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I, Sheen J (2001) Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature 413: 383–389 [DOI] [PubMed] [Google Scholar]
- Kakimoto T (2003) Perception and signal transduction of cytokinins. Annu Rev Plant Biol 54: 605–627 [DOI] [PubMed] [Google Scholar]
- Lohrmann J, Harter K (2002) Plant two-component signalling systems and the role of response regulators. Plant Physiol 128: 363–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohrmann J, Sweere U, Zabaleta E, Bäurle I, Keitel C, Kozma-Bognar L, Brennicke A, Schäfer E, Kudla J, Harter K (2001) The response regulator ARR2: a pollen-specific transcription factor involved in the expression of nuclear-encoded mitochondrial complex I genes. Mol Genet Genom 265: 2–13 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R (2003) Ethylene response factor 1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15: 165–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrdlik P, Neuhaus G, Merkle T (2000) Plant heterotrimeric G protein β subunit is associated with membranes via protein interaction involving coiled-coil formation. FEBS Lett 476: 208–212 [DOI] [PubMed] [Google Scholar]
- Ouaked F, Rozhon W, Lecourieux D, Hirt H (2003) A MAPK pathway mediates ethylene signalling in plants. EMBO J 22: 1282–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, De Y, Yang WC, Kumaran M, Sundaresan V (1999) Analysis of flanking sequences from dissociation insertion lines: a database for reverse genetics in Arabidopsis. Plant Cell 11: 2263–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potuschak T, Lechner E, Parmentier Y, Yanagisawa S, Grava S, Koncz C, Genschik P (2003) EIN3-dependent regulation ofn plant ethylene hormone signaling by two Arabidopsis F Box proteins: EBF1 and EBF2. Cell 115: 679–689 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Carson SDB, To JPC, Kieber JJ (2003) Expression profiling of cytokinin action in Arabidopsis. Plant Physiol 132: 1998–2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai H, Aoyama T, Oka A (2000) Arabidopsis ARR1 and ARR2 response regulators operate as transcriptional activators. Plant J 24: 703–711 [DOI] [PubMed] [Google Scholar]
- Sakai H, Honma T, Aoyama T, Sato S, Kato T, Tabata S, Oka A (2001) ARR1, a transcription factor for genes immediately responsive to cytokinins. Science 294: 1519–1521 [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signalling: a transcriptional cascade mediated by ethylene-insensitive3 and ethylene-response-factor1. Genes Dev 12: 3703–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, Goudreau PN (2000) Two-component signal transduction. Annu Rev Biochem 69: 183–215 [DOI] [PubMed] [Google Scholar]
- Sweere U, Eichenberg K, Lohrmann J, Mira-Rodado V, Bäurle I, Kudla J, Nagy F, Schäfer E, Harter K (2001) Interaction of the response regulator ARR4 with phytochrome B in modulating red light signalling. Science 294: 1108–1111 [DOI] [PubMed] [Google Scholar]
- Tajima Y, Imamura A, Kiba T, Amano Y, Yamashino T, Mizuno T (2004) Comparative studies on the type-B response regulators revealing their distinctive properties in the His-to-Asp phosphorelay signal transduction of Arabidopsis thaliana. Plant Cell Physiol 45: 28–39 [DOI] [PubMed] [Google Scholar]
- To JPC, Haberer G, Ferreira FJ, Deruere J, Mason MG, Schaller GE, Alonso JM, Ecker JR, Kieber JJ (2004) Type-A Arabidopsis response regulators are partially redundant negative regulators of cytokinin signalling in Arabidopsis. Plant Cell 16: 658–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hall AE, O'Malley R, Bleecker AB (2003) Canonical histidine kinase activity of the transmitter domain of the ETR1 ethylene receptor from Arabidopsis is not required for signal transmission. Proc Natl Acad Sci USA 100: 352–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa S, Yoo S-D, Sheen J (2003) Differential regulation of EIN3 stability by glucose and ethylene signalling in plants. Nature 425: 521–525 [DOI] [PubMed] [Google Scholar]
- Zhong GV, Burns JK (2003) Profiling ethylene-regulated gene expression in Arabidopsis thaliana by microarray analysis. Plant Mol Biol 53: 117–131 [DOI] [PubMed] [Google Scholar]
- Zhu T (2003) Global analysis of gene expression using GeneChip microarrays. Curr Opin Plant Biol 6: 418–425 [DOI] [PubMed] [Google Scholar]
- Zhu T, Budworth P, Han B, Brown D, Chang H-S, Zou G, Wang X (2001) Toward elucidating the global gene expression patterns of developing Arabidopsis: parallel analysis of 8300 genes by a high-density oligonucleotide probe array. Plant Physiol Biochem 39: 221–242 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary File 1
Supplementary File 2
Supplementary File 3
Supplementary File 4
Supplementary File 5


