Abstract
Most transport pathways between cell nucleus and cytoplasm are mediated by nuclear transport receptors of the importin β family. These receptors are in continuous circulation between the two compartments and transfer cargo molecules from one side of the nuclear envelope to the other. RanBP16 is a family member from higher eukaryotes of so far unknown function. We now show that it exports p50RhoGAP from the nucleus and thereby confines this activity to the cytoplasm. It also accounts for nuclear exclusion of 14-3-3σ, which in turn is known to anchor, for example, cyclin-dependent kinases in the cytoplasm. Our data further suggest that RanBP16 exports several additional cargoes. It thus appears to be a nuclear export mediator with broad substrate specificity and we will therefore refer to it as exportin 7 (Exp7). Finally, we demonstrate that Exp7-dependent nuclear export signals differ fundamentally from the leucine-rich, CRM1-dependent ones: First, they are not just short linear sequences, but instead include folded motifs. Second, basic residues are critical for Exp7 recruitment.
Keywords: 14-3-3, nuclear pore complex, p50RhoGAP, RanGTPase, Xpo7
Introduction
The nuclear envelope (NE) separates nucleus from cytoplasm and restricts nucleocytoplasmic exchange of material to nuclear pore complexes (NPCs). The selectivity of NPCs, in turn, allows eukaryotic cells to control this trafficking and hence the composition of the nucleus and the cytoplasmic compartment. NPCs can be traversed in a passive or a facilitated manner. The passive mode is efficient for small molecules but becomes increasingly restricted as the size of the object exceeds a limit of 20–40 kDa. By contrast, facilitated receptor-mediated translocation can accommodate objects of several MDa and a diameter of nearly 40 nm (for reviews see Mattaj and Englmeier, 1998; Görlich and Kutay, 1999; Conti and Izaurralde, 2001; Weis, 2003).
The superfamily of importin β-related factors represents the largest class of nuclear transport receptors (reviewed in Wozniak et al, 1998; Görlich and Kutay, 1999; Imamoto, 2000; Macara, 2001; Strom and Weis, 2001). Unlike the bulk of cellular proteins, they can cross the permeability barrier of nuclear pores rapidly and with little resistance (Ribbeck and Görlich, 2001). This property allows them to circulate continuously between nucleus and cytoplasm and to transfer cargo molecules between the two compartments.
These receptors occur in two flavours, mediators of import into the nucleus (importins) and export mediators (exportins). Both interact with RanGTP but respond to the nucleocytoplasmic RanGTP gradient (Görlich et al, 1996b; 2003; Izaurralde et al, 1997; Kalab et al, 2002; Smith et al, 2002) in diametrically opposed ways. Importins bind cargo at low RanGTP levels in the cytoplasm and release cargo upon RanGTP binding in the nucleus (Rexach and Blobel, 1995; Görlich et al, 1996b). In contrast, exportins recruit cargo at high RanGTP concentrations, as ternary cargo·exportin·RanGTP complexes, in the nuclear compartment and release cargo when the Ran-bound GTP molecule is hydrolysed in the cytoplasm (Bischoff and Görlich, 1997; Fornerod et al, 1997; Kutay et al, 1997a). This active control of cargo binding and release by the RanGTPase system constitutes the sole input of metabolic energy into these transport cycles and is sufficient to allow importins and exportins to accumulate cargoes actively against gradients of chemical activity (Weis et al, 1996; Kose et al, 1997; Schwoebel et al, 1998; Englmeier et al, 1999; Ribbeck et al, 1999; Görlich et al, 2003).
In terms of cellular function, it appears useful to distinguish four major categories of active nucleocytoplasmic transport. First, biosynthetic transport, which continuously supplies one compartment with macromolecules that had been produced in the other, examples being constitutive nuclear protein import or export of tRNA, mRNA and ribosomal subunits (reviewed in Mattaj and Englmeier, 1998; Görlich and Kutay, 1999). Second, recycling reactions that return cargo-free transport receptors or transport adaptors back to the compartment, in which the next cargo molecule should be loaded (Kutay et al, 1997a). Nuclear import of Ran also falls into this category (Ribbeck et al, 1998). Third, regulated transport, which is used to control key cellular processes (reviewed in Kaffman and O'Shea, 1999). For example, numerous transcription factors are held in the cytoplasm until adequate signals trigger their import and allow activation or repression of their respective target genes. Finally, the nuclear export machinery also counteracts the slow but steady leakage of cytoplasmic proteins into the nuclear compartment. For example, actin accumulates in the nucleus, unless expelled from there by exportin 6 (Exp6) (Stüven et al, 2003). Likewise, CRM1 mediates nuclear exclusion of the translation factors eIF2β, eIF2Bɛ, eIF5 and eRF1 (Bohnsack et al, 2002), while Exp5 keeps eEF1A cytoplasmic (Bohnsack et al, 2002; Calado et al, 2002).
While characterising a novel nuclear export pathway, we identified two additional examples for cytoplasmic proteins, whose nuclear exclusion is actively maintained: p50RhoGAP and 14-3-3σ. p50RhoGAP is a GTPase-activating protein for several Rho- and RacGTPases, but attenuation of signalling by activated (i.e. GTP-bound) Cdc42 appears to be the main function (Lancaster et al, 1994). Cdc42 in turn regulates several cellular processes, including actin polymerisation, cell shape changes, cell growth, endocytosis and vesicular transport through the Golgi (reviewed in Erickson and Cerione, 2001; Etienne-Manneville and Hall, 2002). 14-3-3σ is a member of a large protein family. 14-3-3 proteins occur as dimers and regulate diverse cellular processes, mainly through binding to phosphorylated ligands (reviewed in Fu et al, 2000; van Hemert et al, 2001; Tzivion and Avruch, 2002; Berg et al, 2003). 14-3-3σ becomes induced, for example, by DNA damage and then inhibits cell cycle progression apparently by sequestering CDK/cyclin complexes in an inactive form in the cytoplasm (Hermeking et al, 1997; Chan et al, 1999; Laronga et al, 2000).
Here we show that RanBP16/Exp7, encoded by the Xpo7 gene, functions as a nuclear export receptor that excludes p50RhoGAP, 14-3-3σ and apparently numerous other proteins from the nucleus. Exp7 therefore appears to define a general export pathway, comparable to the CRM1 pathway, and to play a broad role in maintaining the identities of the nuclear and the cytoplasmic compartments. We addressed the question as to how these different cargoes recruit Exp7 and found that the critical motif in eIF1 is a short acidic patch followed by a lysine-rich α-helix. Likewise, basic residues in p50RhoGAP appear essential for Exp7 binding; they might therefore constitute a general feature of Exp7-dependent export signals.
Results
Identification of Exp7-specific nuclear export cargoes
RanBP16/Exp7 was previously shown to interact specifically with RanGTP and NPCs, which qualified the protein as a bona fide member of the importin β family and suggested a function in nucleocytoplasmic transport (Kutay et al, 2000). However, Exp7-specific cargoes have not yet been identified and cellular functions of this receptor have so far remained elusive. To shed light on this issue, we used immobilised Exp7 to enrich potential cargoes from a cytoplasmic HeLa cell extract. As shown in Figure 1, several proteins bound strongly to Exp7 in the presence of RanGTP, but only weakly in its absence, and thus represented potential export substrates. Mass spectrometric analysis identified them as p50RhoGAP, eIF4AI, and the Vps35, Vps26 and Vps29 subunits of the mammalian retromer (Haft et al, 2000). All these factors fulfil cytoplasmic functions and their confinement to the cytoplasmic compartment through Exp7-mediated nuclear export therefore appeared an attractive possibility.
Figure 1.

Immobilised human Exp7 was used to recruit potential transport cargoes from a cytosolic HeLa extract. Binding was in the absence or presence of 5 μM GTP·RanQ69L (which mimics a nuclear environment). Bound proteins were separated by SDS–PAGE and detected by Coomassie staining. The following putative export substrates were identified by mass spectrometry: RhoGTPase-activating protein 1 (p50RhoGAP, NP-004299), eukaryotic translation initiation factor 4A-1 (eIF4AI, NP-001407) and three components of the retromer—human vacuolar sorting protein 35 (hVps35, AAF89953), hVps26 (AAF89954) and hVps29 (AAF89952).
p50RhoGAP is excluded from nuclei through a CRM1-independent mechanism
We then generated stably transduced cell lines expressing a GFP-p50RhoGAP fusion and observed that the protein is well excluded from nuclei, showing a strong cytoplasmic signal with significant enrichment at the Golgi (Figure 2). This localisation is consistent with the function of the p50RhoGAP substrate Cdc42 in Golgi transport. The inactivation of the CRM1 export pathway by leptomycin B (LMB) treatment (Nishi et al, 1994) did not affect this localisation, suggesting that CRM1 function is not required for nuclear exclusion of p50RhoGAP (see corresponding panels in Figure 2). A parallel control verified that the LMB treatment was effective in shifting the CRM1 cargo eIF2β from an exclusively cytoplasmic to a predominantly nuclear localisation (Figure 2; Bohnsack et al, 2002).
Figure 2.
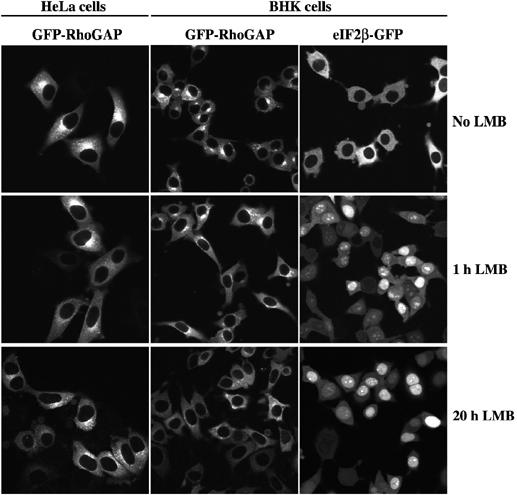
In either stably transduced HeLa or BHK cells, GFP-p50RhoGAP shows a cytoplasmic staining, with some enrichment at the Golgi. Inactivation of CRM1 by LMB (5 ng/ml) did not change this localisation, even after 20 h of treatment. In contrast, already 1 h of LMB treatment shifted the CRM1 cargo eIF2β-GFP from exclusively cytoplasmic to mainly nuclear. Images of the paraformaldehyde-fixed samples were taken by confocal laser scanning microscopy.
p50RhoGAP behaves like an Exp7 substrate
We so far demonstrated the potential of Exp7 to recognise p50RhoGAP (Figure 1). However, it was unclear to what extent other exportins would interact with this cargo. To clarify the issue, we assembled export complexes from a complete HeLa cell extract onto immobilised p50RhoGAP and analysed the composition of the bound fraction by immunoblotting. As expected, Exp7 was enriched strongly and in a RanGTP-stimulated manner (Figure 3A). In contrast, interactions with other export mediators, namely CRM1/Exp1, CAS/Exp2, Exp4, 5 or 6 (Figure 3A), exportin t or importin 13 (data not shown), were not detectable.
Figure 3.
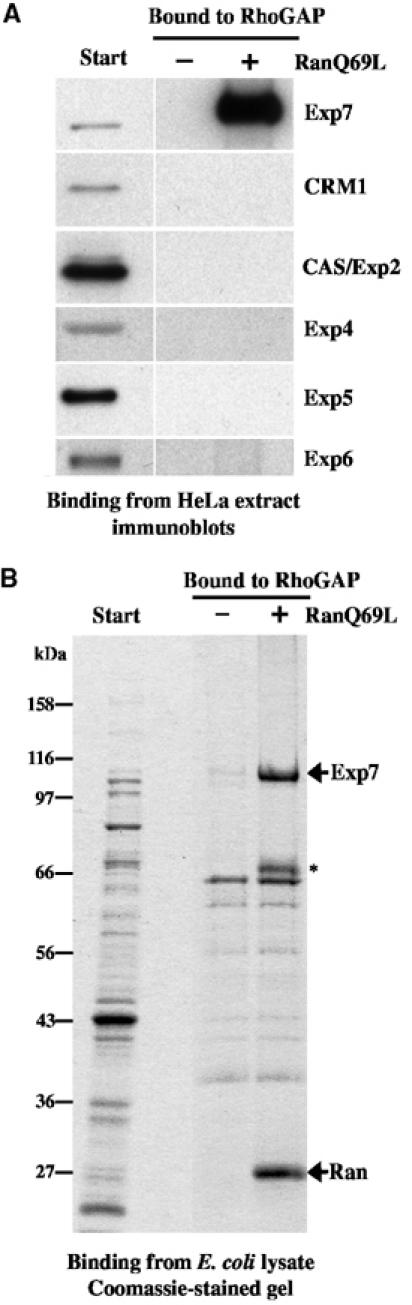
(A) Nuclear transport receptors were recruited from a cytosolic HeLa extract to immobilised p50RhoGAP and the behaviour of indicated exportins was analysed by immunoblotting. Exp7 bound to p50RhoGAP only in the presence of RanGTP. Binding of other exportins was not detectable. (B) Binding to immobilised p50RhoGAP using an E. coli lysate with recombinant human Exp7 as a source of transport receptors. Analysis was by SDS–PAGE followed by Coomassie staining. Also recombinant Exp7 bound p50RhoGAP specifically and in a RanGTP-dependent manner. ‘*' indicates a proteolytic Exp7 fragment.
The binding experiment just mentioned still leaves the possibility that the Exp7·p50RhoGAP interaction had been bridged by an additional factor present in the HeLa cell extract. We therefore repeated the experiment using a different source of Exp7, namely a lysate from an Escherichia coli strain that expressed the recombinant receptor. Also in this case, Exp7 bound selectively and in a RanGTP-dependent fashion (Figure 3B). As bacteria lack components of the nuclear transport machinery, which could possibly have mediated the cargo·exportin interaction, we can conclude that Exp7 recognises p50RhoGAP directly and thus behaves like a nuclear export receptor for this cargo.
Molecular cloning of Xenopus Exp7
We then decided to study nuclear export of p50RhoGAP by microinjections into the nuclei of Xenopus oocytes. Before this, however, we had to clarify whether Exp7 exists in Xenopus oocytes and, if so, how similar to the human protein it would be. We therefore searched the databases of the Xenopus laevis and Xenopus tropicalis EST projects (http://www.sanger.ac.uk), identified partial sequences that matched various parts of human Exp7 and used this collected sequence information to amplify the entire coding region from Xenopus oocyte mRNA by RT–PCR. Sequencing of the open reading frame revealed 93% amino-acid identity between human and X. laevis Exp7. Exp7 is thus less conserved between the two species than is CRM1 (97% identity), but considerably better conserved than CAS/Exp2 (89% identity), Exp6 (79% identity) or Exp5 (57% identity). As expected from the pseudo-tetraploidy of X. laevis, several allelic isoforms of Exp7 are evident.
Exp7 mediates nuclear export of p50RhoGAP
We next generated affinity-purified antibodies against Xenopus Exp7 and established by immunoblotting that Xenopus oocytes contain abundant amounts of the Exp7 protein (≈1–2 μM cellular concentration, data not shown). The antibody recognised a single band on blots of total oocyte extracts (not shown) and can thus be considered monospecific in immunoblot applications, that is, when the proteins are presented in a denatured form. We also established that the antibody recognises native Exp7 (not shown, but see below) and is thus suitable for selective inhibition experiments.
We then in vitro translated the potential Exp7 cargo p50RhoGAP in the presence of [35S]methionine and injected it along with labelled glutathione-S-transferase (GST, a control protein lacking import or export signals) and an NES-YFP fusion (a CRM1 export cargo) into the nuclei of Xenopus oocytes. After 2 h incubation, oocytes were dissected and the distribution of export cargoes between nuclear and cytoplasmic fractions was analysed. While the injection control GST remained nuclear, a nearly quantitative export of p50RhoGAP and NES-YFP to the cytoplasm was evident (Figure 4A).
Figure 4.
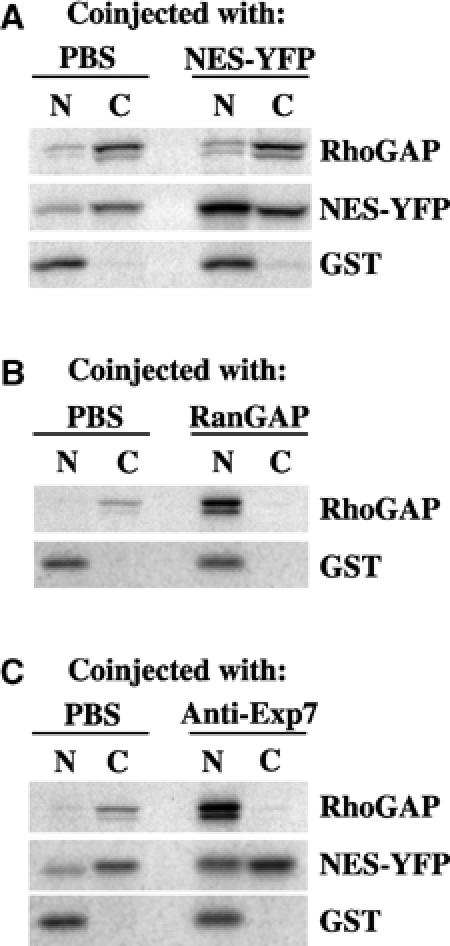
p50RhoGAP is actively exported from the nuclei of X. laevis oocytes. (A) 14 nl of a mixture of radiolabelled p50RhoGAP, NES-YFP and GST (nuclear injection control) was injected into the nuclei of stage IV–V oocytes. If indicated, 300 μM unlabelled NES-YFP competitor was coinjected. Oocytes were dissected 2 h postinjection into nuclear (N) and cytosolic (C) fractions and samples were analysed by 10% SDS–PAGE, followed by autoradiography. Each time point represents the average of five oocytes. (B) Oocytes injection was as in (A), but the injection mixture contained p50RhoGAP, GST and as indicated 10 μM (unlabelled) RanGAP for depletion of nuclear RanGTP. (C) Anti-Xenopus Exp7 antibodies (at 6 mg/ml in the injection mix) inhibited p50RhoGAP export 21-fold as compared to the control, where PBS had been coinjected.
Coinjection of 300 μM of unlabelled NES-YFP competed with export of the radiolabelled NES reporter, but not export of p50RhoGAP (Figure 4A). This confirms that p50RhoGAP is not exported via the CRM1 pathway and also explains why nuclear exclusion of GFP-p50RhoGAP is not sensitive to LMB (Figure 2). However, depletion of the nuclear RanGTP pool by nuclear RanGAP injection abolished the p50RhoGAP export completely (Figure 4B), indicating that the process is mediated by a RanGTP-dependent exportin.
We then coinjected the anti-Exp7 antibodies and observed strong inhibition of p50RhoGAP export (Figure 4C). The effect was highly specific as judged by two controls: first, anti-Exp7 antibodies did not interfere with export of NES-YFP (Figure 4C) and, second, anti-CRM1 antibodies inhibited NES-YFP export, but left export of p50RhoGAP unaffected (not shown). We can thus conclude that p50RhoGAP is actively confined to the cytoplasmic compartment and that this confinement is accomplished by Exp7-mediated nuclear export.
14-3-3σ is also exported by Exp7
In Figure 1, we used immobilised Exp7 to enrich potential nuclear export cargoes. The added RanGTP also appeared as a major band in the bound fraction and obscured comigrating export substrates. To avoid this problem, we employed a modified binding assay, where we immobilised RanGTP and assembled export complexes by adding Exp7 and a HeLa cell extract that had been prior depleted of endogenous transport receptors. This way, we identified 14-3-3σ as a factor that bound to the RanGTP column in a strictly Exp7-dependent manner (data not shown).
We next wanted to test if export complex formation of 14-3-3σ is specific for Exp7 (Figure 5A) and therefore performed a binding experiment using immobilised 14-3-3σ as a bait and a complete HeLa cell extract as a source of nuclear transport receptors. No interaction was detectable with the nuclear export mediators CRM1/Exp1, CAS/Exp2 or Exp4, 5 or 6. However, Exp7 was specifically recruited to the affinity matrix, provided RanGTP had been added to mimic a nuclear environment. This RanGTP-dependent binding could also be reproduced with recombinant Exp7, suggesting that the interaction is direct (Figure 5B).
Figure 5.
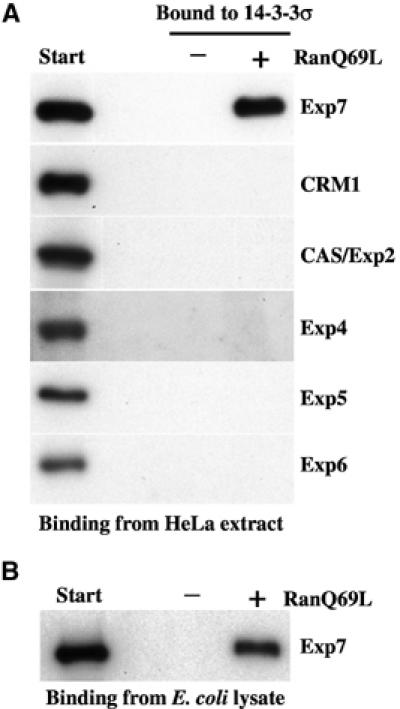
(A) Cognate nuclear export receptors were recruited from a cytosolic HeLa extract to an immobilised 14-3-3σ-zz fusion. Analysis was by immunoblotting. Exp7 bound to 14-3-3σ only in the presence of RanGTP (5 μM); binding of other exportins was undetectable. (B) Binding assay was performed as in (A), but an E. coli lysate with recombinant Exp7 was the source of transport receptors.
We could not find any evidence for an active nuclear import of 14-3-3σ and also the passive nuclear influx of the 14-3-3σ-GFP fusion occurs apparently only very slowly (not shown). Consistent with this, we observed a cytoplasmic localisation of the 14-3-3σ-GFP fusion and exclusion from nuclei (Figure 6; see also Hermeking et al, 1997; Chan et al, 1999; Yang et al, 2003; van Hemert et al, 2004). It should be noted that nuclear exclusion of 14-3-3σ is abolished by bulky N-terminal tags, such as GFP with too short a spacer (not shown). Such bulky N-terminal fusions interfere also with the Exp7·14-3-3σ interaction (not shown), which in turn suggests that this interaction is relevant for nuclear export.
Figure 6.
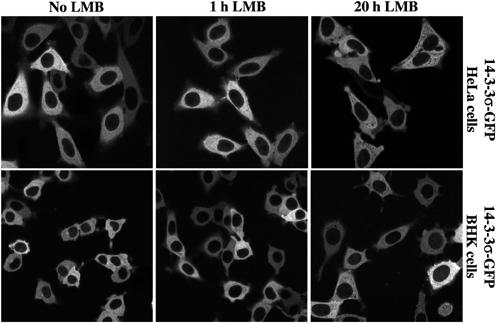
In either stably transduced BHK and HeLa cell lines, 14-3-3σ-GFP shows a cytoplasmic localisation. This did not change by incubating the cells for 1 or 20 h with LMB (5 ng/ml). For controls, see Figure 2.
LMB treatment did not increase the nuclear 14-3-3σ signal (Figure 6), pointing to a CRM1-independent nuclear exclusion mechanism and confirming that CRM1 fails to form export complexes with 14-3-3σ (Figure 5A).
However, microinjection of affinity-purified antibodies raised against human Exp7 abolished nuclear exclusion and 14-3-3σ then showed a clear nucleoplasmic signal with spared nucleoli (Figure 7). Two lines of control verified the specificity of this effect. First, injecting the same amount of antibodies recognising an unrelated antigen did not change the 14-3-3σ localisation (Figure 7). Second, the anti-human Exp7 antibodies did not increase the nuclear signal of unrelated reporters, such as eEF1A-GFP, eEF1A-GFP-NLS or eIF2β-GFP (not shown), which are excluded by Exp7-independent mechanisms (Bohnsack et al, 2002).
Figure 7.
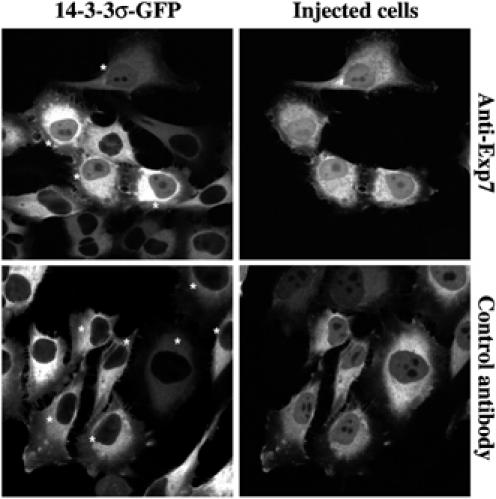
HeLa cells expressing 14-3-3σ-GFP were microinjected with affinity-purified anti-human Exp7 or control antibodies (anti-Drosophila Exp5), and fixed 18 h later. In anti-Exp7-injected cells, 14-3-3σ-GFP equilibrated between nucleus and cytoplasm and showed a clear nucleoplasmic signal with spared nucleoli. By contrast, the GFP fusion remained strictly excluded from the nuclei of noninjected cells or cells injected with the control antibody. Injected cells (indicated by ‘*') were identified by immunofluorescence detecting the injected antibody in the Red channel. Images were taken by sequential confocal scans detecting GFP and the Alexa 568-labelled secondary antibody.
We finally tested the behaviour of 14-3-3σ in the Xenopus oocyte system. When injected into nuclei, only slow export occurred (Figure 8) and this slow export was completely abolished by coinjection of anti-Xenopus Exp7 antibodies (not shown). Conversely, export was ∼10-fold stimulated by coinjection of recombinant Exp7, whereas coinjection of CRM1 at the same concentration had no effect (Figure 8). Exp7 is thus required and sufficient for 14-3-3σ export from both mammalian and Xenopus nuclei.
Figure 8.

35S-labelled 14-3-3σ and GST were generated by in vitro translation and injected into the nuclei of Xenopus oocytes. After 2 h, oocytes were dissected and the nucleocytoplasmic distribution of the two proteins was determined by SDS–PAGE followed by autoradiography. Coinjection of 15 μM human Exp7 stimulated 14-3-3σ export 10-fold but had no effect on GST.
The export signal in p50RhoGAP is multipartite
14-3-3σ and p50RhoGAP are very different in function and structure (Liu et al, 1995; Xiao et al, 1995; Barrett et al, 1997); they share no regions of significant homology. Yet, Exp7 recognises both of them as cargoes, and this poses the question for the corresponding nuclear export signals (NESs). To gain a first insight into this issue, we tested which parts of p50RhoGAP interact with the exportin. We found that at least three regions, namely the extreme N-terminus (residues 1–64), an internal fragment (residues 198–257) and the C-terminal RhoGAP domain (residues 257–439), play a role (see Supplementary data), although none of these is sufficient for a high-affinity interaction. In contrast to the CRM1-dependent leucine-rich NESs, the export signal in p50RhoGAP comprises therefore not just a single linear sequence. Instead, it is multipartite and includes at least one motif that is presented by the compactly folded RhoGAP domain, which, in turn, cannot be dissected further without disrupting folding.
Such complex NESs are very difficult to map and a complete description of the signal would probably require solving the structure of the p50RhoGAP·Exp7·RanGTP complex (as a side note it should be mentioned that not a single complex NES has been elucidated so far). In the meantime, however, we had identified numerous other proteins that bind to Exp7 in a RanGTP-dependent fashion and we chose the smallest of them, the translation initiation factor eIF1 (Supplementary Figure 1; see also Pestova and Kolupaeva, 2002; Majumdar et al, 2003, and references therein), for further analysis.
The Exp7-binding motif in eIF1 consists of aspartates and a lysine-rich α-helix
The solution structure of eIF1 has been solved by NMR (Fletcher et al, 1999). eIF1 consists of an extended N-terminal region, which we found to be dispensable for Exp7 binding (see Figure 9A), and a compactly folded C-terminal domain. This compactly folded domain contains 36 highly solvent-exposed residues, which were all candidates for making contacts to Exp7. We found that yeast eIF1 is also recognised by Exp7 (Figure 9A). Residues that are essential for Exp7 binding are therefore likely to be conserved between the two species. By this consideration, 19 charged residues remained as immediate candidates (Figure 9B).
Figure 9.
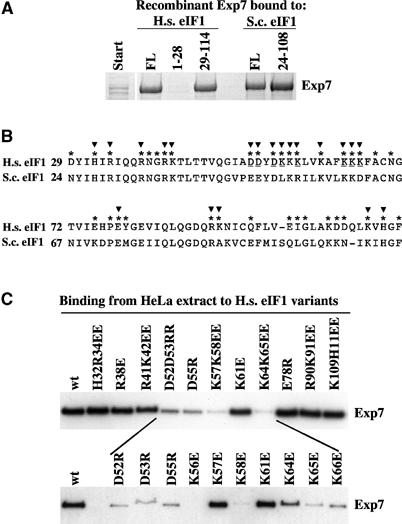
(A) An E. coli lysate containing recombinant human Exp7 was incubated with immobilised full-length human eIF1 (FL=residues 1–114), yeast eIF1 (FL=residues 1–108) or indicated eIF1 fragments. Binding was in the presence of 5 μM GTP·RanQ69L. (B) Alignment of the compactly folded domains from human (H.s.) and yeast (S.c.) eIF1. Highly solvent exposed residues (*) and residues mutagenised (▾) in panel C are indicated. Critical residues for Exp7 binding are underlined. (C) Wild-type (wt) or indicated mutant forms of human eIF1 were immobilised and used to bind Exp7 from complete cytosolic HeLa extract in the presence of 5 μM GTP·RanQ69L. Bound Exp7 was detected by immunoblot.
With a first round of mutagenesis on human eIF1, we established that H32, R34, R38, R41, E78, R90, K91, K109 and H111 do not detectably contribute to Exp7 binding (see binding assays in Figure 9C). In contrast, the D52D53 → RR, D55 → R, K57K65 → EE and K64K65 → EE mutations strongly reduced or even abolished the interaction. These crucial positions are in close proximity to each other. To map the signal at a single-amino-acid resolution, we finally mutagenised each of the exposed residues in this region and thereby identified a total of eight residues that are critical for Exp7 binding: D52, D53, D55, K56, K58, K64, K65 and K66 (Figure 9C, lower part). These critical residues are arranged in a very characteristic motif, namely a short, acidic extended region followed by an amphipathic helix, in which all hydrophilic, solvent-exposed residues are lysines and where the hydrophobic ones pack against the hydrophobic core of the domain (Fletcher et al, 1999).
The just described Exp7-binding motif follows, however, no rigid consensus. Instead, it appears that an unfavourable residue in one position can be compensated somewhere else in the molecule. This is exemplified by yeast eIF1, where the critical K56 is substituted by leucine and K66 by aspartic acid. Yet, yeast eIF1 binds Exp7, and its lower affinity becomes only apparent when eIF1 competes with other cargoes for Exp7 binding (not shown).
Positively charged residues appear to be a general feature of the Exp7 export signature
The binding data with eIF1 raised the possibility that positively charged residues mediate Exp7 binding also in other cargoes. To test this assumption, we mutated several basic patches in p50RhoGAP (Figure 10). None of the mutations interfered with folding as judged from the fact that the bacterially expressed mutant proteins were as soluble as the wild-type form. Three basic patches, namely R83K84, K152K153 and R282R283, turned out to be critical for the recruitment of Exp7 (see Figure 10). This loss of interaction was not just a matter of total charge, as mutating other basic residues (R149K150 or K156) had no effect. We do not yet know how the complete export signal in p50RhoGAP looks like; however, the data clearly indicate that positively charged patches are a genuine part of the signature by which Exp7 selects cargoes. Exp7-dependent export signals therefore differ fundamentally from CRM1-dependent ones.
Figure 10.

Immobilised p50RhoGAP wild type or the indicated variants were incubated with a complete cytosolic HeLa cell extract in the presence of 5 μM GTP·RanQ69L. Bound Exp7 was detected by immunoblot.
Discussion
A major goal of the nucleocytoplasmic transport field has been to obtain a comprehensive overview of the cellular nuclear transport pathways. In the yeast Saccharomyces cerevisiae, 13 importin β-type nuclear transport receptors are known and functions have been allocated to 12 of them (Fried and Kutay, 2003). The repertoire of higher eukaryotes is larger and comprises, for example, 20 family members in human cells. A total of 10 family members have been known to function in import, six in export and one in both. With this study, we have elucidated a novel human pathway and demonstrated that RanBP16/Exp7 represents a bona fide nuclear export receptor for at least two cytoplasmic proteins, p50RhoGAP and 14-3-3σ.
Nuclear export of 14-3-3σ was previously suggested to be CRM1 dependent (Brunet et al, 2002; van Hemert et al, 2004). Our data argue against such scenario: 14-3-3σ directly binds to Exp7 but not detectably to CRM1 (Figure 5); export is inhibited by blocking Exp7 (Figure 7) but not by inactivating CRM1 (Figure 6); and, finally, export is stimulated by exogenous Exp7 but not by CRM1 (Figure 8). How can these different conclusions be explained? One could assume that the problem originates, at least in part, from the use of LMB as a sole assessment. LMB selectively inactivates CRM1, but the consequences of this inactivation are ultimately lethal (Nishi et al, 1994) and thus pleiotropic. The exclusively cytoplasmic localisation of RanBP1 and RanGAP, for example, requires CRM1 (Richards et al, 1996 and our unpublished data). LMB will therefore impair the nucleocytoplasmic RanGTP gradient and thereby indirectly inhibit also those nuclear transport pathways that operate in parallel to CRM1. This could well explain why LMB could raise, under certain conditions, the nuclear concentrations of 14-3-3σ. Given the just described experience with 14-3-3σ, we would not be surprised if numerous other export events, which were previously attributed to CRM1, are in fact mediated by another transporter, such as Exp7.
Typical importins, such as importin β, transportin, importin 5 or importin 7, have a broad substrate specificity and are apparently able to transfer a large number of distinct cargoes into the nucleus. In contrast, most exportins appear specialised on a narrow group of cargoes or even on a single highly abundant export substrate, examples being CAS/Exp2, which retrieves importin α from the nucleus (Kutay et al, 1997a), exportin t, which mediates tRNA export (Arts et al, 1998; Kutay et al, 1998), Exp4, which exports eIF5A (Lipowsky et al, 2000), or Exp6, which exports actin from the nucleus (Stüven et al, 2003). CRM1 has so far been the only exception; it accounts for the export of several distinct RNPs and a large number of proteins or protein complexes (reviewed in Fornerod and Ohno, 2002; see also Tschochner and Hurt, 2003). It can thus be considered as a broad-spectrum exportin. Exp7 appears now to be the second example of an exportin with broad substrate specificity and we show in this study that it accounts for export of p50RhoGAP and nuclear exclusion of 14-3-3σ. The binding data from Figure 1 further suggest that it is also involved in nuclear export of eIF4AI and the retromer complex. In addition, we have preliminary evidence for a role of Exp7 in nuclear export of at least 12 further components (our unpublished observations).
The Exp7 cargoes described in this study are unrelated by sequence, structure and function and, yet, they are all recognised by this exportin. CRM1 also recognises many different cargoes, most of them through a small, transplantable peptide motif, the leucine-rich NES (Fischer et al, 1995; Wen et al, 1995; Bogerd et al, 1996). This raised the question as to whether Exp7 recognises an analogous motif. We have addressed this question first for eIF1, the so far smallest known protein that binds Exp7 in a RanGTP-dependent fashion. We found that in this case a highly charged motif is recognised: D52 DYDKKKLVKAFKKK66, whereby the underlined aspartic acid and lysine residues are required for a high-affinity interaction. This motif shows a characteristic fold. The acidic patch is extended, while the rest folds into an α-helix, such that the lysines are exposed to the solvent and the hydrophobic residues pack against the hydrophobic core of the domain (see Fletcher et al, 1999). Even though the binding motif is a comparably small peptide (16 residues), its fusion to a reporter protein is unlikely to be sufficient to confer Exp7 binding, simply because folding probably requires the context of the entire domain.
eIF1 (12.7 kDa) is so small that it traverses NPCs faster than typical transport receptor·cargo complexes, and we could not so far directly demonstrate that Exp7 confines eIF1 in the cytoplasm. This could just reflect technical complications. On the other hand, it is not certain that the eIF1·Exp7·RanGTP complex represents an export complex. It could as well form for a different reason, such as to directly quench eIF1 activity in the nuclear compartment and thus to suppress nuclear translation. In any case, it gave the hint that basic residues might be of general importance for cargo recognition by Exp7. However, the export signal in p50RhoGAP appears far more complex than the Exp7-binding site in eIF1. We have identified at least four distant regions of p50RhoGAP, which are required to recruit Exp7, namely three basic clusters (R38K84, K152K153 and R282R282; Figure 10) and the extreme N-terminus (residues 1–64; see Supplementary data). These data suggest that Exp7 recognises this cargo through multiple low-affinity interactions.
Out of the human 14-3-3 paralogues, the σ isoform is an Exp7 substrate (Figures 5, 6, 7 and 8), while 14-3-3β and ζ are not (our unpublished data). Thus, Exp7 does not just recognise the fold of 14-3-3 proteins but also regions of 14-3-3σ, which differ from the β and ζ isoforms. As expected from the eIF1 and p50RhoGAP signals, our preliminary data suggest that at least one basic patch in 14-3-3σ is involved in Exp7 binding.
p50RhoGAP, 14-3-3σ and the putative cargoes eIF4AI and the mammalian retromer fulfil, as far as we can tell, cytoplasmic functions only. eIF4AI mediates initiation of translation (reviewed in Pestova et al, 2001; Rogers et al, 2002). p50RhoGAP regulates Cdc42 activity (Lancaster et al, 1994). 14-3-3σ arrests the cell cycle, for example, after DNA damage by sequestering CDKs in the cytoplasm (Hermeking et al, 1997; Chan et al, 1999; Laronga et al, 2000), and the retromer mediates retrograde vesicular transport from the yeast prevacuolar compartment or the endosome of higher eukaryotes to the Golgi (see Arighi et al, 2004; Seaman, 2004, and references therein).
The nuclear exclusion of these components makes perfect biological sense; however, it poses the question as to why nuclear exclusion is not just achieved by a lack of active nuclear import. This could be because of the following three reasons: first, these factors might have cryptic import signals, such as a 14-3-3 ligand that contains a bona fide NLS. Second, the open mitosis in higher eukaryotes causes mixing of cytoplasmic and nuclear contents, which need to be re-separated after return to interphase conditions. Finally, not even in interphase, NPCs represent absolute barriers (Ribbeck and Görlich, 2001) and so leakage of any given cytoplasmic protein into the nuclear compartment is just a matter of time. Although protein synthesis takes place in the cytoplasm, it appears that an exclusive cytoplasmic localisation does not occur by default, but instead depends on cytoplasmic sorting signals, which in many cases confer exportin-mediated nuclear export. In fact, one should expect the number of proteins, which are actively excluded from the nuclear compartment, to be very large, possibly in the order of several or many thousands. How the nuclear export machinery should recognise such an extremely diverse set of proteins is yet unclear, but CRM1 and Exp7 are likely to play a key role.
Materials and methods
Expression constructs
The coding regions of the various proteins used in this work have been amplified by PCR using primers with appropriate restriction sites. Human and Xenopus Exp7 were amplified from HeLa and X. laevis cDNA, respectively, p50RhoGAP, 14-3-3σ and human eIF1 from clones obtained from the Resource Centre/Primary Database (RZPD), yeast eIF1 from S. cerevisiae complete genomic DNA and GST from pGEX-4T-1 (Amersham Biosciences).
The following vectors have been used in this work. For bacterial expression, four derivatives of pQE-80 (Qiagen) are used: pNhis1080 (provides an N-decahis tag), pChis1080 (provides a C-decahis tag), pNzztev80 (provides N-zz and C-hexahis tags) and pCzz80 (provides a C-zz-hexahis tag). For expression in mammalian cells, two derivatives of pRevTRE2 (BD Biosciences) are used: pNGFPrev and pCGFPrev (which provide N-GFP and C-GFP tags, respectively). For in vitro transcription–translation, pT7-60 (Görlich et al, 1996a) was used. This vector contains a T7 promoter and a β-globin 5′UTR for improved translation.
Human Exp7 was cloned in pNhis1080 and pNzztev80, Xenopus Exp7 in pNhis1080, p50RhoGAP in pNGFPrev, pNzztev80 and pT760, 14-3-3σ in pCzz80, pCGFPrev and pT7-60, yeast and human eIF1 in pNzztev80 and GST in pT7-60.
DNA coding for NES-YFP (a fusion between the NES of PKI (Wen et al, 1995) and EYFP) was cloned in pChis1080 and pT7-60.
All constructs were verified by DNA sequencing.
Protein expression and purification
Expression and purification of CRM1, RanQ69L and Rna1p (RanGAP) have been described previously (Izaurralde et al, 1997; Kutay et al, 1997b; Stüven et al, 2003). His-tagged proteins (human and Xenopus Exp7 and NES-YFP) were expressed in E. coli and purified by Ni-NTA agarose followed by gel filtration on Superdex 200. zz-tagged proteins (human Exp7, p50RhoGAP, yeast and human eIF1 and 14-3-3σ) were expressed in E. coli and immobilised directly from the 100 000 g cleared lysates to IgG-Sepharose.
In vitro transcription–translation
In vitro transcription–translation of p50RhoGAP, NES-YFP, 14-3-3σ and GST was performed from the corresponding T7 promoter constructs in the presence of [35S]methionine (Amersham) using the TNT reticulocyte lysate system (Promega). Reactions were performed at 30°C for 90 min in a 50 μl volume and stopped by adding 250 μl of ice-cold PBS. In order to remove free [35S]methionine and concentrate the proteins, the final volume of the samples was reduced to 15–20 μl by centrifugation through Nanosep Cenrifugal Devices (PALL Corporation) with a 10 kDa cutoff. Samples were then aliquoted and either frozen or directly used for oocyte microinjections.
Oocyte injection and analysis
Collagenase-treated X. laevis oocytes were prepared and injected as described before (Jarmolowski et al, 1994). After indicated time points, oocytes were dissected, proteins of the nuclear and cytoplasmic fractions precipitated and finally analysed by SDS–PAGE and autoradiography.
Binding assays
zz-tagged proteins were immobilised to IgG-Sepharose. A 25 μl portion of immobilised protein was incubated for 3 h, either in the absence or presence of 5 μM of purified GTP·RanQ69L (a GTPase-deficient mutant), with 0.8 ml of HeLa extract or a 100 000 g cleared lysate from an E. coli strain that expressed recombinant human Exp7 (both lysates adjusted to 50 mM Tris–HCl pH 7.5, 50 mM NaCl, 10 mM MgCl2). Bound proteins were eluted with 1.5 M MgCl2, 50 mM Tris–HCl pH 7.5, precipitated with 95% isopropanol, separated by SDS–PAGE and analysed by Coomassie staining or Western blotting.
Antibodies
All antibodies have been raised in rabbits and affinity purified on their respective immobilised antigens. Antibodies against human and Xenopus Exp7 were raised against C-terminal peptides. Antibodies against human Exp6 and mouse Exp4 were raised against the recombinant protein. Antibodies against CRM1, CAS/Exp2 and Exp5 have been described previously (Mingot et al, 2001; Bohnsack et al, 2002).
Microinjection of antibodies into mammalian cells and immunofluorescence
HeLa and BHK stable cell lines expressing 14-3-3σ-GFP or GFP-p50RhoGAP were generated by retroviral transduction, as described before (Bohnsack et al, 2002), using the appropriate pRevTRE2 constructs. The BHK stable cell line expressing eIF2β-GFP has been described before (Bohnsack et al, 2002).
Expression of the GFP-tagged proteins was induced with 100 ng/ml doxycycline for 18 h at 37°C and 5% CO2. Cells were then either fixed immediately or fixed after indicated incubations or microinjection with antibodies.
Microinjections of antibodies (6 mg/ml) were performed with the AIS 2 system (CellBiology Trading), which controls an Eppendorf Femtojet that is mounted onto a Zeiss Axiovert inverse microscope. After 18 h incubation at 37°C and 5% CO2, the cells were fixed and the injected antibodies were immunostained with an Alexa 568-labelled goat anti-rabbit antibody (Molecular Probes) following a protocol previously described (Bohnsack et al, 2002). Green and red fluorescence signals were then sequentially recorded with an SP2 confocal laser scanning microscope (Leica).
Fixation was performed in all cases at 37°C with 3% paraformaldehyde in PBS for 15 min.
Site-directed mutagenesis
Indicated mutations were introduced into the coding sequences of eIF1 (Figure 9) and p50RhoGAP (Figure 10) by the PCR-based QuickChange method (Stratagene). Plasmid DNA was purified from single clones and the presence of the mutations (as well as the absence of undesired mutations) was verified by DNA sequencing.
DDBJ/EMBL/GenBank accession no.
The nucleotide sequence of X. laevis Exp7 is listed under accession number AJ620730.
Supplementary Material
Supplementary Data
Acknowledgments
We gratefully thank Thomas Ruppert and Armin Bosserhoff for mass spectrometry, Petra Rübmann for excellent technical help, Kevin Czaplinski for introduction to oocyte injections, Katharina Ribbeck for initial experiments to block nuclear exclusion of 14-3-3σ, Rainer Saffrich for help with injection of cultured cells, Kathrin Regener for discovering that eFI1 binds Exp7 and the members of our laboratory for critical reading of the manuscript and stimulating discussions. This work received financial support from the Alfried Krupp-Foundation and the DFG (Graduiertenkolleg Molekulare Zellbiologie, SFBs 352 and 638).
References
- Arighi CN, Hartnell LM, Aguilar RC, Haft CR, Bonifacino JS (2004) Role of the mammalian retromer in sorting of the cation-independent mannose 6-phosphate receptor. J Cell Biol 165: 123–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arts GJ, Fornerod M, Mattaj IW (1998) Identification of a nuclear export receptor for tRNA. Curr Biol 8: 305–314 [DOI] [PubMed] [Google Scholar]
- Barrett T, Xiao B, Dodson EJ, Dodson G, Ludbrook SB, Nurmahomed K, Gamblin SJ, Musacchio A, Smerdon SJ, Eccleston JF (1997) The structure of the GTPase-activating domain from p50rhoGAP. Nature 385: 458–461 [DOI] [PubMed] [Google Scholar]
- Berg D, Holzmann C, Riess O (2003) 14-3-3 proteins in the nervous system. Nat Rev Neurosci 4: 752–762 [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Görlich D (1997) RanBP1 is crucial for the release of RanGTP from importin beta-related nuclear transport factors. FEBS Lett 419: 249–254 [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Benson RE, Hua J, Cullen BR (1996) Protein sequence requirements for function of the human T-cell leukemia virus type 1 Rex nuclear export signal delineated by a novel in vivo randomization-selection assay. Mol Cell Biol 16: 4207–4214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D (2002) Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J 21: 6205–6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Kanai F, Stehn J, Xu J, Sarbassova D, Frangioni JV, Dalal SN, DeCaprio JA, Greenberg ME, Yaffe MB (2002) 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J Cell Biol 156: 817–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Treichel N, Muller EC, Otto A, Kutay U (2002) Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J 21: 6216–6224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan TA, Hermeking H, Lengauer C, Kinzler KW, Vogelstein B (1999) 14-3-3Sigma is required to prevent mitotic catastrophe after DNA damage. Nature 401: 616–620 [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E (2001) Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol 13: 310–319 [DOI] [PubMed] [Google Scholar]
- Englmeier L, Olivo JC, Mattaj IW (1999) Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr Biol 9: 30–41 [DOI] [PubMed] [Google Scholar]
- Erickson JW, Cerione RA (2001) Multiple roles for Cdc42 in cell regulation. Curr Opin Cell Biol 13: 153–157 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420: 629–635 [DOI] [PubMed] [Google Scholar]
- Fischer U, Huber J, Boelens WC, Mattaj IW, Lührmann R (1995) The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 82: 475–483 [DOI] [PubMed] [Google Scholar]
- Fletcher CM, Pestova TV, Hellen CU, Wagner G (1999) Structure and interactions of the translation initiation factor eIF1. EMBO J 18: 2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, Ohno M (2002) Exportin-mediated nuclear export of proteins and ribonucleoproteins. Results Probl Cell Differ 35: 67–91 [DOI] [PubMed] [Google Scholar]
- Fornerod M, Ohno M, Yoshida M, Mattaj IW (1997) Crm1 is an export receptor for leucine rich nuclear export signals. Cell 90: 1051–1060 [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U (2003) Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci 60: 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H, Subramanian RR, Masters SC (2000) 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol 40: 617–647 [DOI] [PubMed] [Google Scholar]
- Görlich D, Henklein P, Laskey RA, Hartmann E (1996a) A 41 amino acid motif in importin alpha confers binding to importin beta and hence transit into the nucleus. EMBO J 15: 1810–1817 [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Kutay U (1999) Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Görlich D, Pante N, Kutay U, Aebi U, Bischoff FR (1996b) Identification of different roles for RanGDP and RanGTP in nuclear protein import. EMBO J 15: 5584–5594 [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Seewald MJ, Ribbeck K (2003) Characterization of Ran-driven cargo transport and the RanGTPase system by kinetic measurements and computer simulation. EMBO J 22: 1088–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft CR, de la Luz Sierra M, Bafford R, Lesniak MA, Barr VA, Taylor SI (2000) Human orthologs of yeast vacuolar protein sorting proteins Vps26, 29, and 35: assembly into multimeric complexes. Mol Biol Cell 11: 4105–4116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B (1997) 14-3-3 sigma is a p53-regulated inhibitor of G2/M progression. Mol Cell 1: 3–11 [DOI] [PubMed] [Google Scholar]
- Imamoto N (2000) Diversity in nucleocytoplasmic transport pathways. Cell Struct Funct 25: 207–216 [DOI] [PubMed] [Google Scholar]
- Izaurralde E, Kutay U, von Kobbe C, Mattaj IW, Görlich D (1997) The asymmetric distribution of the constituents of the Ran system is essential for transport into and out of the nucleus. EMBO J 16: 6535–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmolowski A, Boelens WC, Izaurralde E, Mattaj IW (1994) Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol 124: 627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A, O'Shea EK (1999) Regulation of nuclear localization: a key to a door. Annu Rev Cell Dev Biol 15: 291–339 [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R (2002) Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science 295: 2452–2456 [DOI] [PubMed] [Google Scholar]
- Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y (1997) Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J Cell Biol 139: 841–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Bischoff FR, Kostka S, Kraft R, Görlich D (1997a) Export of importin alpha from the nucleus is mediated by a specific nuclear transport factor. Cell 90: 1061–1071 [DOI] [PubMed] [Google Scholar]
- Kutay U, Hartmann E, Treichel N, Calado A, Carmo-Fonseca M, Prehn S, Kraft R, Görlich D, Bischoff FR (2000) Identification of two novel RanGTP-binding proteins belonging to the importin beta superfamily. J Biol Chem 275: 40163–40168 [DOI] [PubMed] [Google Scholar]
- Kutay U, Izaurralde E, Bischoff FR, Mattaj IW, Görlich D (1997b) Dominant-negative mutants of importin-beta block multiple pathways of import and export through the nuclear pore complex. EMBO J 16: 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Görlich D (1998) Identification of a tRNA-specific nuclear export receptor. Mol Cell 1: 359–369 [DOI] [PubMed] [Google Scholar]
- Lancaster CA, Taylor-Harris PM, Self AJ, Brill S, van Erp HE, Hall A (1994) Characterization of rhoGAP. A GTPase-activating protein for rho-related small GTPases. J Biol Chem 269: 1137–1142 [PubMed] [Google Scholar]
- Laronga C, Yang HY, Neal C, Lee MH (2000) Association of the cyclin-dependent kinases and 14-3-3 sigma negatively regulates cell cycle progression. J Biol Chem 275: 23106–23112 [DOI] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Schwarzmaier P, Kraft R, Kostka S, Hartmann E, Kutay U, Görlich D (2000) Exportin 4: a mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J 19: 4362–4371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Bienkowska J, Petosa C, Collier RJ, Fu H, Liddington R (1995) Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376: 191–194 [DOI] [PubMed] [Google Scholar]
- Macara IG (2001) Transport into and out of the nucleus. Microbiol Mol Biol Rev 65: 570–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R, Bandyopadhyay A, Maitra U (2003) Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a spreinitiation complex. J Biol Chem 278: 6580–6587 [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L (1998) Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem 67: 265–306 [DOI] [PubMed] [Google Scholar]
- Mingot JM, Kostka S, Kraft R, Hartmann E, Görlich D (2001) Importin 13: a novel mediator of nuclear import and export. EMBO J 20: 3685–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T (1994) Leptomycin B targets a regulatory cascade of crm1, a fission yeast nuclear protein, involved in control of higher order chromosome structure and gene expression. J Biol Chem 269: 6320–6324 [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG (2002) The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev 16: 2906–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 98: 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach M, Blobel G (1995) Protein import into nuclei: association and dissociation reactions involving transport substrate, transport factors, and nucleoporins. Cell 83: 683–692 [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Görlich D (2001) Kinetic analysis of translocation through nuclear pore complexes. EMBO J 20: 1320–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Kutay U, Paraskeva E, Görlich D (1999) The translocation of transportin-cargo complexes through nuclear pores is independent of both Ran and energy. Curr Biol 9: 47–50 [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Lipowsky G, Kent HM, Stewart M, Görlich D (1998) NTF2 mediates nuclear import of Ran. EMBO J 17: 6587–6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards SA, Lounsbury KM, Carey KL, Macara IG (1996) A nuclear export signal is essential for the cytosolic localization of the Ran binding protein, RanBP1. J Cell Biol 134: 1157–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW Jr, Komar AA, Merrick WC (2002) eIF4A: the godfather of the DEAD box helicases. Prog Nucleic Acid Res Mol Biol 72: 307–331 [DOI] [PubMed] [Google Scholar]
- Schwoebel ED, Talcott B, Cushman I, Moore MS (1998) Ran-dependent signal-mediated nuclear import does not require GTP hydrolysis by Ran. J Biol Chem 273: 35170–35175 [DOI] [PubMed] [Google Scholar]
- Seaman MN (2004) Cargo-selective endosomal sorting for retrieval to the Golgi requires retromer. J Cell Biol 165: 111–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AE, Slepchenko BM, Schaff JC, Loew LM, Macara IG (2002) Systems analysis of Ran transport. Science 295: 488–491 [DOI] [PubMed] [Google Scholar]
- Strom AC, Weis K (2001) Importin-beta-like nuclear transport receptors. Genome Biol 2, REVIEWS3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stüven T, Hartmann E, Görlich D (2003) Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J 22: 5928–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschochner H, Hurt E (2003) Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol 13: 255–263 [DOI] [PubMed] [Google Scholar]
- Tzivion G, Avruch J (2002) 14-3-3 proteins: active cofactors in cellular regulation by serine/threonine phosphorylation. J Biol Chem 277: 3061–3064 [DOI] [PubMed] [Google Scholar]
- van Hemert MJ, Niemantsverdriet M, Schmidt T, Backendorf C, Spaink HP (2004) Isoform-specific differences in rapid nucleocytoplasmic shuttling cause distinct subcellular distributions of 14-3-3 sigma and 14-3-3 zeta. J Cell Sci 117: 1411–1420 [DOI] [PubMed] [Google Scholar]
- van Hemert MJ, Steensma HY, van Heusden GP (2001) 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. BioEssays 23: 936–946 [DOI] [PubMed] [Google Scholar]
- Weis K (2003) Regulating access to the genome: nucleocytoplasmic transport throughout the cell cycle. Cell 112: 441–451 [DOI] [PubMed] [Google Scholar]
- Weis K, Dingwall C, Lamond AI (1996) Characterization of the nuclear protein import mechanism using Ran mutants with altered nucleotide binding specificities. EMBO J 15: 7120–7128 [PMC free article] [PubMed] [Google Scholar]
- Wen W, Meinkoth JL, Tsien RY, Taylor SS (1995) Identification of a signal for rapid export of proteins from the nucleus. Cell 82: 463–473 [DOI] [PubMed] [Google Scholar]
- Wozniak RW, Rout MP, Aitchison JD (1998) Karyopherins and kissing cousins. Trends Cell Biol 8: 184–188 [DOI] [PubMed] [Google Scholar]
- Xiao B, Smerdon SJ, Jones DH, Dodson GG, Soneji Y, Aitken A, Gamblin SJ (1995) Structure of a 14-3-3 protein and implications for coordination of multiple signalling pathways. Nature 376: 188–191 [DOI] [PubMed] [Google Scholar]
- Yang HY, Wen YY, Chen CH, Lozano G, Lee MH (2003) 14-3-3 sigma positively regulates p53 and suppresses tumor growth. Mol Cell Biol 23: 7096–7107 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data


