Abstract
The Escherichia coli origin of replication, oriC, comprises mostly binding sites of two proteins: DnaA, a positive regulator, and SeqA, a negative regulator. SeqA, although not essential, is required for timely initiation, and during rapid growth, synchronous initiation from multiple origins. Unlike DnaA, details of SeqA binding to oriC are limited. Here we have determined that SeqA binds to all its sites tested (9/11) and with variable efficiency. Titration of DnaA alters SeqA binding to two sites, both of which have overlapping DnaA sites. The altered SeqA binding, however, does not affect initiation synchrony. Synchrony is also unaffected when individual SeqA sites are mutated. An apparent exception was one mutant where the mutation also changed an overlapping DnaA site. In this mutant, the observed asynchrony could be from altered DnaA binding, as selectively mutating this SeqA site did not cause asynchrony. These results reveal robust initiation synchrony against alterations of individual SeqA binding sites. The redundancy apparently ensures SeqA function in controlling replication in E. coli.
Introduction
Chromosomal replication initiation in Escherichia coli is controlled by a combination of positive and negative regulators [1, 2]. The primary positive regulator is the initiator protein DnaA [3, 4]. A negative regulator, SeqA, was identified years later although an obligatory requirement for such a regulator was predicted earlier [1, 5, 6]. How the different regulators function in allowing replication once per cell cycle and at a particular time of the cell cycle is not fully understood [7].
Replication regulators in E. coli, including SeqA, modulate mainly the interaction of the initiator DnaA with the origin of replication, oriC [1, 8]. Binding sites of DnaA and SeqA are distributed throughout the chromosome, the density of both the sites being highest at oriC (11 sites of each within the 246 bp oriC) [9]. The affinity of DnaA for its binding sites in oriC varies, partly due to considerable variation of the site sequences from the 9-mer consensus sequence TTATC/ACACA [10]. The high affinity sites, whose sequences match the consensus sequence, are R1, R2 and R4 (Fig 1A). DnaA complexed with ATP or ADP binds equally well to these sites, and remains bound throughout the cell cycle [8]. In contrast, most of the low affinity sites such as τ1, R5, τ2, I1, I2, C3, C2, I3 and C1 show a preference for binding DnaA-ATP, the form that is required for initiation [9]. An exception is R5, which is a low affinity site but binds DnaA-ATP and DnaA-ADP equivalently, implying that low affinity and preference for DnaA-ATP are two different attributes [11]. Maximal binding to the weak sites occurs at the time of initiation when the level of DnaA-ATP peaks in the cell cycle [12]. Regulation of binding to these low affinity sites is the key regulatory strategy for controlling initiation in E. coli.
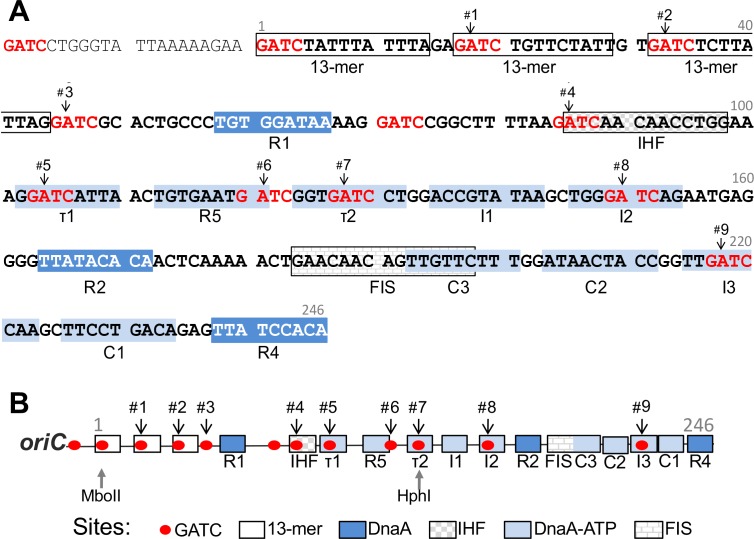
Fig 1. Sequence of oriC showing its major protein binding sites.
(A) Sequence of oriC that includes the minimal region (coordinates 1–246) required for origin function. The coordinate 1 correspondence to 3923744 of gb|U00096.3|. The region includes several GATC sites (shown in red) which are methylated by the Dam methylase enzyme. There are three 13-mer repeats of AT rich sequences where the origin initially unwinds. The remainder of the origin has mainly 9-mer DnaA binding sites as well as sites for binding IHF and FIS proteins. DnaA sites have either high (R1, R2 and R4) or low (τ1, R5, τ2, I1, I2, C3, C2, I3 and C1) affinity for DnaA. The numbers #1–9 mark the GATC sites studied here. A TaqI site (TCGA) overlapping each of the nine GATC sites was created by converting their two upstream bases to TC (NNGATC to TCGATC). (B) A linear map of oriC features described above. The map also shows location of sites for restriction enzymes MboII and HphI that naturally occurs in oriC.
DnaA-ATP is also required for binding to oriC in the region containing the three 13-mers [13] (Fig 1A). The strands of oriC initially open in this region allowing helicase loading. DnaA binds to one specific strand of the open region, which helps to stabilize the open state and facilitate helicase loading to the other stand [14–16].
The characteristics of SeqA binding to DNA are distinct from those of DnaA. SeqA binds to GATC sites, which are targets for adenine methylation by the deoxyadenosyl methyltransferase (Dam) enzyme. SeqA prefers to bind to hemimethylated (HM) GATC sites that are generated from fully methylated sites upon replication and SeqA binding prolongs the HM state [1]. The prolongation is particularly long (~1/3rd cell generation) at two loci, oriC and the promoter of the dnaA gene that also has a high density of GATC sites. Several of the low affinity DnaA binding sites of oriC have overlapping GATC sequences and DnaA binding to R5, I2 and I3 sites reduces in the presence of SeqA [8]. SeqA thus appears to delay initiation by interfering with DnaA binding to oriC.
During rapid growth, E. coli initiates DNA replication on a partially duplicated chromosome, causing overlapping rounds of replication and the presence of multiple origins in duplicating chromosomes. Nonetheless, replication initiates from all the origins within a narrow window of time and initiates from each origin only once per cycle (initiation synchrony) [17]. This requires the presence of SeqA protein, and in seqA mutants replication becomes highly asynchronous [1]. SeqA binding to newly generated HM origins that helps to prevent their additional firing in the same cell cycle is called sequestration. The sequestration of newly replicated origins also prevents them from competing with the yet-to-be replicated origins and thereby helps in their firing [18].
SeqA could inhibit initiation in additional ways: 1. By prolonging the HM state, which compromises oriC activity since the origin fires more efficiently when it is fully methylated [19]. 2. By sequestering the DnaA promoter, which reduces initiator synthesis following replication initiation and this helps to reduce premature reinitiation [20]. 3. By binding to fully methylated DNA, specifically to the left half of oriC in vitro [21, 22]. This is believed to inhibit initiation by interfering with origin opening 4. By absorbing negative supercoils, which are required for initiation [23]. This activity of SeqA does not require DNA to be methylated, a finding that is consistent with the fact that SeqA overproduction is inhibitory in dam mutant cells [1]. Thus, although primarily known as a HM DNA binding protein, SeqA can bind to fully methylated DNA and can also bind non-specifically to unmethylated DNA.
SeqA binding to oriC thus appears as complex as that of DnaA, and the requirements of SeqA binding in negative regulation of replication remain to be fully defined. Here we have focused on SeqA binding related to initiation synchrony. We show that 1) SeqA binds to all the GATC sites of oriC that we tested (9/11) and the extent of binding varies among the sites; 2) SeqA binding is altered to two of GATC sites upon titration of DnaA but this does not affect initiation synchrony; and 3) Initiation synchrony is also not affected by mutations in individual GATC sites. These results suggest that initiation synchrony is a robust process that does not depend on varied SeqA binding across oriC and integrity of any of the single GATC sites of oriC.
Materials and Methods
Bacterial strains, plasmids, primers and media
Bacterial strains, plasmids and primers used in this study are listed in Tables A-C in S1 File. E. coli was grown in either LB, or 1X M63 medium (KD Medical) supplemented with 0.005% thiamine, 0.2% glucose and additionally 0.1% or 0.5% casamino acids (CAA) depending on the experiment. Antibiotics were used at the following concentrations: ampicillin, 100 μg/ml; tetracycline, 12.5 μg/ml; chloramphenicol, 25 μg/ml and zeocin, 20 μg/ml.
Construction of E. coli mutants with mutated oriC
WT oriC (coordinates 3925715–3926016 of K12-MG1655 strain gb|U00096.3|) was amplified from E. coli genomic DNA from strain MG1655 (BR1703) by using primer pairs jj15+ jj16. The PCR product was digested with EcoRI and BamHI, and ligated to similarly digested pEM7-Zeo vector, resulting in plasmid pJJ04. Bases were substituted within oriC to create TaqI sites or alter GATC sites by site-directed mutagenesis (using Hifi-KAPA hot start ready mix PCR kit, KAPA-Biosystems), using pJJ04 as template and primers. The plasmids with new TaqI sites are pJJ06,-40,-41,-46,-47,-48,-49,-55 and -07, and with altered GATC sites are pJJ-385,-386,-387,-388,-389,-379,-391,-369,-370,-371,-372,-374,-377,-390,-415 and -416. Note that we created TaqI sites overlapping 9/11 GATC sites (Fig 1A). The GATC in the left 13-mer was probed earlier and here using MboII. Additionally, the GATC site next to R1 (called 3*) was probed in some of the experiments.
Transfer of oriC mutations from plasmid to chromosome by recombineering in E. coli
oriC (3925715–3926016) was amplified by PCR using pJJ04 or its mutant derivatives as templates and jj21+jj35 as primers. The 5’ and 3’ ends of these primers have homology with chromosomal flanks of oriC (zeo-mioC and gidA). The linear PCR products containing the WT or mutated oriC and the linked zeo cassette were transferred to the chromosome of the E. coli strain BR1703 (WT) or BR1704 (ΔseqA) by the lambda red recombineering method [24]. The strains were first made competent for recombineering by introducing the mini-λ tet plasmid carrying the Red recombination genes. The resulting strains CVC2073 and CVC2092 were used to introduce the linear PCR products by electroporation. After mutation transfer, the strains were cured of the mini-λ tet plasmid, and the sequence of the mutated origins was confirmed by PCR; the origin region (3925517–3926140) was amplified using primers jj40+jj42 and the amplified product was sequenced using the primer jj122 (3925620–3925642).
In order to delete the zeo cassette from the oriC-zeo#1–9 strains, they were transformed with pSIM5 plasmid. A PCR product containing FRT-Kan-FRT and flanks of ~40 bp with homology to the flanks of the zeo cassette was generated using pRFB105 plamid as template and jj184+jj185 as primers. This PCR product was electroporated into oriC-zeo#1–9 strains, and the transformants were screened for KanR and ZeoS. The Kan replacement was confirmed by PCR with Kan flanking primers jj40+jj42. The Kan marker was excised using pCP20 that left one FRT site (35 bp). The final oriC-FRT#1–9 strains with a single FRT scar were confirmed by PCR amplification using primers jj40+jj42 and sequencing the PCR products with jj122.
The oriC-FRT#1–9 strains were individually made dam minus by P1 transduction. The phage stock was derived from dam::Tn9 CmR strain, BR2786. The loss of dam was confirmed by flow cytometry.
Flow cytometry
Cultures of E. coli were grown in supplemented 1X M63 medium as described above. The cultures were grown to OD600 ~ 0.15 and processed for flow cytometry before and after replication run-out in the presence of rifampicin (100 μg/ml) and cephalexin (10 μg/ml) for four hours as described [25]. The peak fluorescence intensity of an overnight grown E. coli culture of BR1703 in M63 + 0.2% glycerol medium (without casamino acids) was taken to represent one full chromosome. From the flow cytometry profiles that represent multimodal distribution of cells, each peak of which represent cells with a fixed number of chromosomes, origins/cell was calculated from the product of the chromosome number and the fractional area of the total distribution representing that number of chromosomes, and summing the products from all the peaks of the distribution. The asynchrony index was calculated by measuring the area of the distribution containing three chromosomes compared to total area containing all the chrmosomes (two, three- and four chromosomes under the present experimental set up) [26]. Fraction of uninitiated cells is the present study is the fractional area of the distribution representing two-chromosome cells. This was used to measure initiation mass. 100,000 cells were analyzed in each panel using the Flow Cytometer BD LSR Forestessa SORP II (BD Biosciences). The triggering volt was set at 200 for the side scatter.
Southern blotting
Genomic DNA was isolated from LB grown log phase cultures (OD600 ~0.3) using the Genelute Bacterial Genomic DNA kit (Sigma). The DNA was digested for an hour with 3 units of MboII or HphI at 37°C, or TaqI at 65°C (New England Biolabs). Partial digestion of genomic DNA from dam minus strains was performed with 3 units of TaqI at 55°C for 10 min. The digested products were resolved in a 1.3% agarose gel. An oriC region (3925517–3925542) was PCR amplified using primers jj40+jj41 and the product was used as the probe. The probe for the HphI digested blot covered the region 3925275–3925633 and was amplified using primers jj168+ jj169. The probe for the oriC external-marker in lacZ covered the region 364871–365085 and was amplified using primers jj193+jj194. The probes were made radioactive using the Redi-PrimeII random primer labeling kit (GE Healthcare) and [α-32P] dCTP (Perkin Elmer). The band intensities were recorded and quantified as described earlier [27]. The blots were re-probed for an external marker (in lacZ) located ~365 kb away from oriC. As reported earlier, the level of HM DNA at lacZ was significantly lower compared to the levels at TaqI sites created in oriC (Fig 2B).
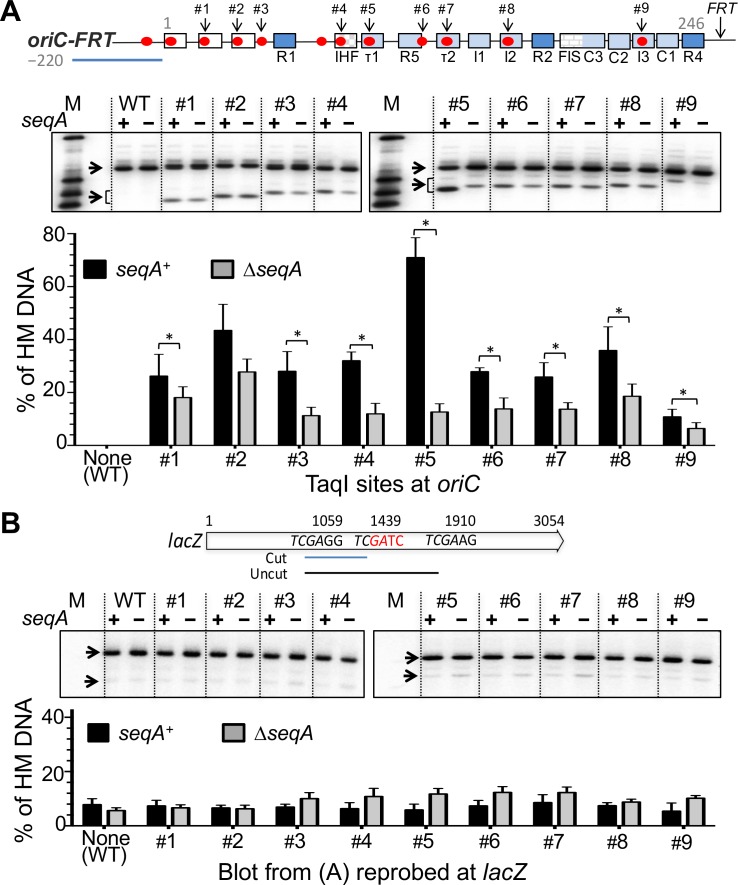
Fig 2. Quantification of hemimethylated DNA level at different GATC sites of oriC in seqA+ and ΔseqA strains.
(A) The top line shows a schematic map of oriC as in Fig 1B, which also includes the location of an FRT site that was present in all the strains used in this figure. The autorad is a representative Southern blot of genomic DNA from seqA+ and ΔseqA strains after digestion with TaqI. The blots were probed with a 220 bp PCR amplified fragment (uisng primers jj40+jj41) homologuous to the left flank of oriC (blue line). M represents oriC specific markers (1100bp, 460 bp, 330 bp and 240 bp). The arrows indicate the bands in which the test GATC sites were either fully methylated, thus resistant to TaqI digestion (upper bands), or HM and sensitive to TaqI digestion 50% of the time (lower bands), as explained in Figure A(A) in S1 File. The intensity of the lower bands was multiplied by a factor of two to calculate the relative level of HM DNA (lower panel). [Note that the separation of the two bands is gradually narrowing because of shifting position of TaqI site within oriC with respect to the two TaqI sites that flank oriC (see also Figure A(B) in S1 File). The upper fragment is generated when TaqI digests the two oriC flanking sites but not the TaqI site within oriC.] The black and gray bars represent mean HM DNA levels in seqA+ and ΔseqA strains, respectively, determined from three cultures innoculated with independent colonies (biological replicates). The error bar represents one standard deviation of the mean. Asterisks indicate pairwise comparisons that were statistically distinguishable (P < 0.05, students t-test). (B) Same as (A) except that the oriC probe was stripped off the blot, which was then reprobed with a PCR product (using primers jj193+jj194) from a region in lacZ (blue bar) [28]. The error bars represent variability in repeated measurements of the same blot (technical replicates).
Results
Introduction of new probing sites for SeqA binding across oriC
Semi-conservative replication of fully methylated oriC generates HM sister origins. The HM state is prolonged upon SeqA binding and this prolongation has been used as a proxy for SeqA binding in vivo [1, 28]. The detection of SeqA binding required the ingenious approach of utilising restriction enzymes whose sites overlap the GA of GATC sites and whose activities are inhibited when the adenine residue of GATC is methylated. However, for one of the two HM sister sites, the methylated adenine of the template strand falls outside of the restriction site, rendering the site sensitive to restriction digestion (Figure A(A) in S1 File). If HM state is prolonged at any site, a greater proportion of digested DNA is recovered from that site.
Restriction sites overlapping GATC and suitable for probing the HM state are available at only two of the 11 GATC sites in oriC: one overlaps with a MboII site and the other with a HphI site (Fig 1B). To probe the GATC sites across the entire oriC, we introduced GATC overlapping sites for an adenine methylation-sensitive enzyme, TaqI. TaqI was chosen because creating its site, TCGA, required making at most two changes to oriC. Note that changes outside of GATC are not known to affect SeqA binding [29–31]. We mutated one or two upstream bases of GATC to TC to create TCGATC sequences overlapping nine of the GATC sites of oriC (#1 - #9, Fig 1A and 1B; Figure A(B) in S1 File). The TaqI sites were initially created in a oriC plasmid, in which oriC was linked to a drug marker, zeo. The mutated origins with a TaqI site were designated oriC-zeo#1–9. We also deleted the zeo marker and replaced it with an FRT site (35bp). This set of mutants was designated oriC-FRT#1-9.
We tested the FRT set of mutants for initiation efficiency (origin/cell mass) and initiation synchrony by replication run-out using flow cytometry [17]. The origin/cell-mass of the mutants was 0.97 ± 0.05 compared to 1 for the WT, and the asynchrony index 7.8 ± 4.0% compared to 3.9% for the WT (Figure B in S1 File; Table 1). The latter result indicates that the mutants (mainly #1 and #3) may have become deficient by a few per cent only. Some initiation defect was evident in ΔseqA cells since the distribution shifted towards cells with fewer chromosomes (except may be for the mutant #4; Figure B in S1 File). The initiation defects, however, do not seem to have affected the growth rates significantly (Table 1).
Table 1. Cell cycle parameters of oriC-FRT mutants with TaqI sitesa.
| TaqI site next to GATC # | Strain No. | Gen. Timeb (min) | Origin/Cellc | Cell massd | Origin/Cell masse | Asyn. Indexc(%) | Frac. of uninitiatedCellsc | Initiation mass |
|---|---|---|---|---|---|---|---|---|
| None (WT) | 2239 | 33.4 | 3.49 | 1 | 1 | 3.9 | 0.20 | 0.39 |
| #1 | 2240 | 34.6 | 3.43 | 0.99 | 0.99 | 12.4 | 0.19 | 0.39 |
| #2 | 2241 | 33.6 | 3.48 | 0.96 | 1.04 | 7.2 | 0.20 | 0.37 |
| #3 | 2242 | 31.6 | 3.33 | 1.05 | 0.91 | 14.5 | 0.19 | 0.40 |
| #4 | 2243 | 33.7 | 3.49 | 0.97 | 1.03 | 4.1 | 0.19 | 0.37 |
| #5 | 2244 | 32.9 | 3.28 | 0.97 | 0.97 | 5.9 | 0.24 | 0.38 |
| #6 | 2245 | 34.1 | 3.34 | 1.01 | 0.95 | 9.6 | 0.22 | 0.39 |
| #7 | 2246 | 32.7 | 3.48 | 1.01 | 0.99 | 8.8 | 0.18 | 0.39 |
| #8 | 2247 | 33.6 | 3.31 | 0.99 | 0.95 | 2.4 | 0.28 | 0.40 |
| #9 | 2248 | 32.7 | 3.40 | 1.06 | 0.92 | 5.0 | 0.22 | 0.42 |
a Other than the generation time, cell cycle parameters were derived from Figure B in S1 File.
bMean generation time from four biological replicates for WT was 33.5 ± 2.2 min. The generation times of the mutants did not vary significantly from that of the WT in pairwise comparisons (p-values 0.18–0.89).
c Origin/cell, asyn. index and the fraction of uninitiated cells were determined from flow cytometry profiles after replication run-out. Origin/cell for WT among four biological replicates was 3.40 ± 0.09. The spread covers the range of values seen for the mutants. Further details are in Materials and Methods.
d Cell mass is the average light scatter from flow cytometric analysis normalized to 1 for WT cells. The values varied about 3% among repeated measurements of the same sample and about 8% among biological replicates.
e Values are normalized to 1 for WT cells.
The hemimethylated DNA varies at different GATC sites of oriC
The two GATC sites in oriC that have overlapping restriction sites suitable for probing HM DNA (MboII and HphI sites, Fig 1B) showed enriched HM DNA when compared to several other GATC sites outside of oriC [28]. In the present study, the enrichment was not significantly affected upon addition of the zeo or the FRT marker (Figure A(C) in S1 File). When our oriC mutants were assayed using the TaqI enzyme, the HM DNA amount was enriched at all nine GATC sites within oriC compared to a distal site in lacZ (Fig 2A and 2B). The level of enrichment varied among the sites: it was maximal in the case of mutant #5 and minimal in the case of mutant #9, where the HM DNA level approached to that of the site in lacZ. The results were essentially identical when the oriC mutants had the zeo marker instead of the FRT site (Fig 2 and Figure C(A) in S1 File). In ΔseqA cells, the amount of HM DNA was less compared to the isogenic seqA+ cells and the extent of reduction was variable (Fig 2A and Figure C(A) in S1 File). These results indicate that SeqA sequesters with variable efficiency all the GATC sites of oriC with the possible exception of site #9.
Although restriction enzymes are sequence specific, the efficiency with which they digest their target sites can be context dependent [32, 33]. To address the possibility that intrinsic variation in cutting efficiency, rather than SeqA binding, could account for the variable HM DNA levels, we tested whether TaqI digestion efficiency varies among the sites in genomic DNA from dam minus cells. GATC sites are not methylated in dam minus cells and are not expected to interfere with TaqI digestion. The digestion efficiency of TaqI did vary somewhat among the TaqI sites of oriC but the average pattern was not correlated with the pattern seen in our experimentalt (dam+) strains (Fig 2 vs. Figure A(D) in S1 File). For example, the #5 site, which showed maximal sensitivity in dam+ cells, was not any more sensitive than others in dam minus cells. It appears that the intrinsic cutting efficiency of the sites is unlikely to account for the variability seen in Fig 2A.
We next considered whether competition from other proteins that interact with oriC could explain the variability in HM DNA level. Among the proteins that interact with oriC, only DnaA has multiple binding sites scattered throughout oriC [9]. In fact, the GATC sites probed in mutants #1, 2 and 5–9 have overlapping DnaA binding sites. These sites are low-affinity DnaA binding sites, most requiring DnaA to be complexed with ATP [9]. The two sites showing extreme HM DNA values (#5 and #9; Fig 2A) are both low affinity DnaA binding sites, suggesting that the variability in the affinity of DnaA for binding to its sites in oriC is unlikely to be the sole cause of the variability in HM DNA level that we observed across oriC.
Finally, efficient SeqA binding in vitro was shown to require two GATC sites phased by two to three integral helical turns of 10.4 bp [29, 30, 34]. The GATC sites of oriC are not separated by integral helical turns (Figure A(E) in S1 File). Specifically, the #5 GATC site where the HM DNA level was maximal is separated from its two flanking GATC sites by 18 and 17 bp. Similarly, the #9 site where the HM DNA level was minimal is also not explained by phasing requirements because it has on its right flank a GATC site nearly optimally separated by three helical turns (33 bp). SeqA cooperatively binds to DNA in vitro, and can also aggregate on DNA [30]. It remains to be seen whether these higher-order interactions and/or binding of other proteins that interact with oriC caused the local variation in HM DNA level.
DnaA contributes to the HM DNA level at some of the GATC sites of oriC
SeqA can interfere with binding of DnaA to some of its low-affinity sites [8]. In contrast, DnaA appears to aid sequestration since titration of DnaA-ATP by an R1-datA plasmid reduces the amount of HM oriC DNA at the MboII site [35]. We asked whether the same R1-datA plasmid could affect the HM DNA level at other GATC sites of oriC. Introduction of the plasmid to our oriC-FRT#1–9 strains reduced origin/cell-mass in WT and in five out of nine mutants (#2, #5–7 and #9; Fig 3A; Tables D vs. E in S1 File). The origin/cell-mass for the WT reduced from 1 to 0.82 and for the five mutants from 1.05 ± 0.11 to 0.80 ± 0.04 in the presence of the R1-datA plasmid compared to the isogenic cells carrying the R1 vector. This indicates that the R1-datA plasmid was equally effective in reducing initiation efficiency of these mutants as it is for the WT origin. The reduction was expected from earlier studies, since DnaA-ATP level is crucial for oriC activity [36]. Why reduction was seen in only some of the mutants remains to be understood. The asynchrony indices reduced mainly in the mutants where the origin/cell-mass reduced. In other words, initiation synchrony was not disturbed any more than what cannot be explained by initiation defficiency. (Tables D vs. E in S1 File). In the ΔseqA background, all the mutants showed asynchrony and overinitiation, but the effect of DnaA titration on the mutants was not estimated because of large asynchrony (Figure D(A) in S1 File).
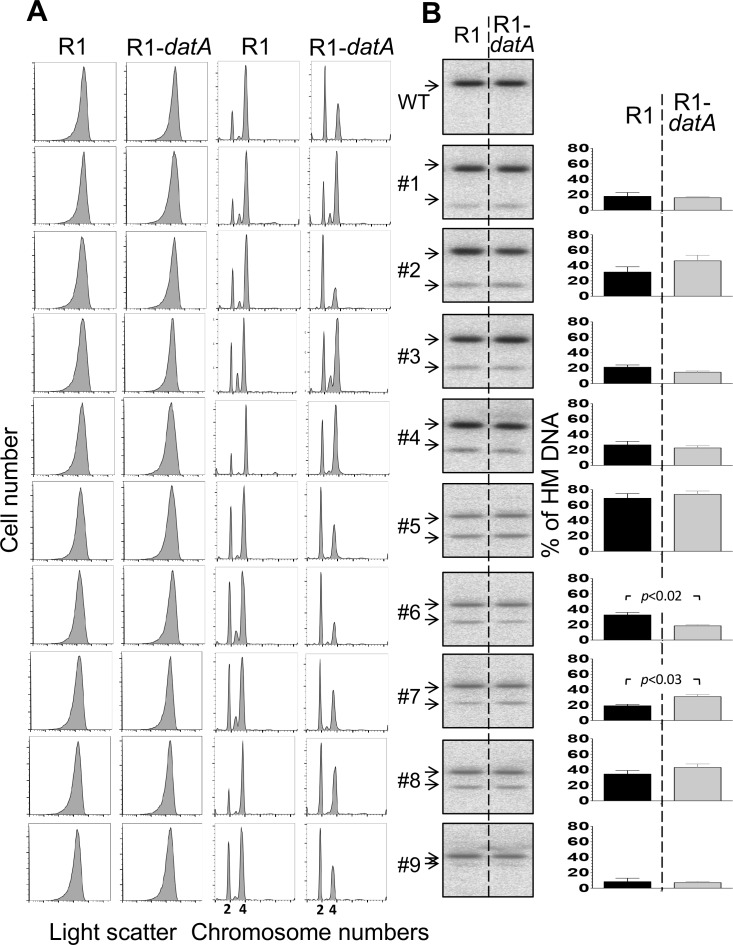
Fig 3. Effect of DnaA titration on cell size, initiation synchrony and HM DNA level at oriC.
(A) Cell size and chromosome content of FRT marked oriC cells without (WT) or with mutations creating TaqI sites #1–9. The cells had an R1 or R1-datA plasmid that was used as a vector control or for titrating DanA-ATP, respectively. Cells were grown at 37°C in supplemented 1X M63 medium up to an OD600 ~ 0.15, and then processed either before or after replication run-out for flow cytometry to measure cell size by light scattering or chromosome content by fluorescence emission, respectively. The multimodal distribution of fluorescence profiles represent cells with different integral number of chromosomes as stated in the abscissa. (B) Southern blots of genomic DNA from cultures in (A) before replication run-out. Black and gray bars represent HM DNA levels in seqA+ strain in the presence of R1 and R1-datA plasmid, respectively. The details are otherwise same as in Fig 2A. Note that band intensities were not quantified for the WT as no cut band was seen or expected because WT oriC does not have a TaqI site in the probed region. The difference in HM DNA levels in the presence of R1 and R1-datA plasmids was considered statistically significant in the case of mutants #6 and #7 only (P < 0.05).
To test the effect of the R1-datA plasmid on the HM DNA level, we assayed the amount of HM DNA from steady state cultures (Fig 3B). The effect of the plasmid was minimal in most cases, with the exception of mutant #6 in which HM DNA decreased, and mutant #7 in which the HM DNA increased. These results indicate that DnaA can help or hinder SeqA binding, depending upon the location of its binding site in oriC. While the increase in HM DNA in mutant #7 could result from reduced competition from DnaA, the decrease in mutant #6 may indicate that SeqA binding requires cooperation from DnaA. Alternate possibilities have also been considered in the Discussion. In the ΔseqA background, there was less origins/cell in the presence of the R1-datA plasmid compared to the presence of the R1vector for essentially all the mutants, indicating that titrating DnaA becomes more effective under over-replicating conditions (Figure D in S1 File).
Contribution of individual GATC sites to initiation synchrony and oriC sequestration
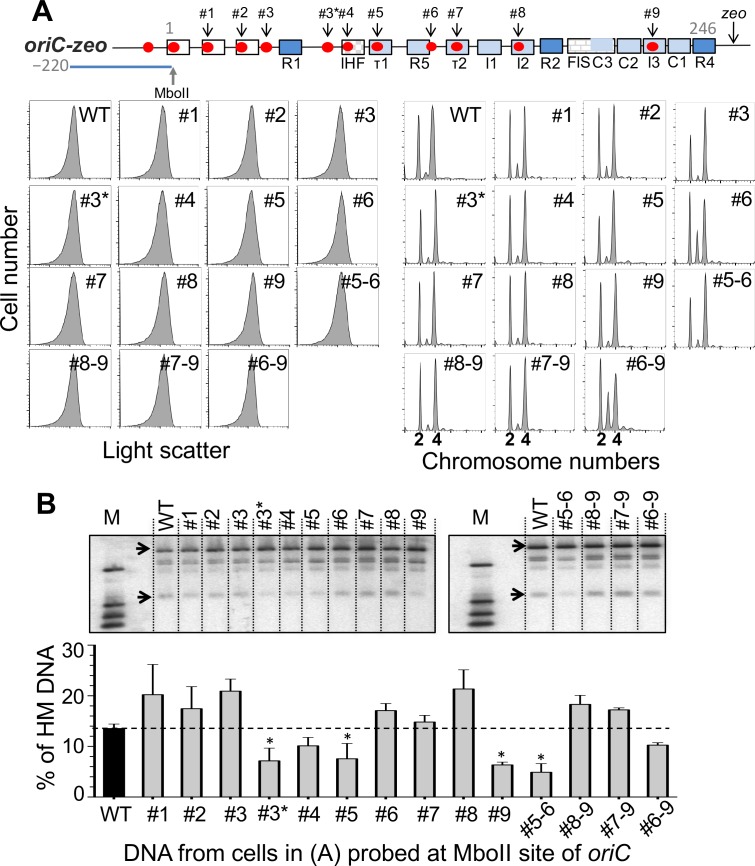
The results so far indicate that despite the variation in HM DNA across oriC, sequestration is nevertheless efficient enough to confer initiation synchrony (Fig 2 and Figure B in S1 File). This suggests that control by sequestration is rather robust and may not require efficient SeqA binding across all its sites in oriC. To test this inference further, we mutated individual GATC sites of oriC to GTTC and measured the effect on initiation synchrony and oriC sequestration (Fig 4A). Out of the ten mutated GATC sites that we tested, only the change within the R5 site (mutant #6) disturbed initiation synchrony. This particular change did not affect sequestration of oriC as measured at the natural MboII site (Fig 4B). GATC mutations at three other positions (#3*, #5 and #9), however, did reduce oriC sequestration without affecting initiation synchrony and growth rate (Table 2). Except for the #6 GATC mutant, these data support the view that sequestration is a robust process that does not require any one of the remaining GATC sites of oriC to be intact to support the extent of sequestration necessary for initiation synchrony.
Fig 4. Effect of GATC mutations in oriC on initiation synchrony and the level of HM DNA at the MboII site of oriC.
(A) The top line shows a schematic map of oriC as in Fig 1B except for an additional GATC site (#3*) included in this study. In these experiments oriC was marked with a zeo drugR cassette. GATC sites at positions #1–9 and #3* were individually mutated to GTTC and the mutant cells were analyzed by flow cytometry as in Fig 3A before and after replication run-out. Cells were also studied after combining some of the mutations (#5–6 etc.). (B) HM DNA levels as in Fig 2A except that the levels were measured at the MboII site for all. The black bar represents the HM DNA level in the WT strain (zeo marked but without any GATC mutation within oriC) and the grey bars the same strain with individual or multiple GATC mutations. The error bars were determined as in Fig 2B. Mutants whose HM DNA levels were significantly different from the WT (p-value <0.05) are marked by asterisks. The dashed line provides a visual aid for comparing HM DNA levels in different mutants relative to the WT.
Table 2. Cell cycle parameters of oriC-zeo mutants with mutated GATCa.
| oriC-zeo with mutated GATC | StrainNo. | Gen. Time(min) | Origin/Cell | Cell Mass | Origin/Cell Mass | Asyn. Index (%) | Frac. of Uninitiat.cells | Initiat. Mass |
|---|---|---|---|---|---|---|---|---|
| None (WT) | 2073 | 33.2 | 3.28 | 1.0 | 1.0 | 4.3 | 27.5 | 0.40 |
| #1GATC→GTTC | 2901 | 32.1 | 3.13 | 1.07 | 0.89 | 11.6 | 29.7 | 0.41 |
| #2 ,, → ,, | 2902 | 32.8 | 3.16 | 1.08 | 0.89 | 10.2 | 29.8 | 0.41 |
| #3 ,, → ,, | 2903 | 32.0 | 3.22 | 1.02 | 0.96 | 5.3 | 23.4 | 0.39 |
| #3* ,, → ,, | 2916 | 31.7 | 2.99 | 1.06 | 0.85 | 5.7 | 25.2 | 0.40 |
| #4 ,, → ,, | 2904 | 32.7 | 3.04 | 1.06 | 0.87 | 6.3 | 29.9 | 0.41 |
| #5 ,, → ,, | 2905 | 32.2 | 3.42 | 1.03 | 1.01 | 5.8 | 22.2 | 0.39 |
| #6 ,, → ,, | 2906 | 32.3 | 2.94 | 1.05 | 0.85 | 20.2 | 27.1 | 0.40 |
| #7 ,, → ,, | 2907 | 32.1 | 3.26 | 1.03 | 0.97 | 7.3 | 28.4 | 0.40 |
| #8 ,, → ,, | 2908 | 32.0 | 2.98 | 1.03 | 0.88 | 5.3 | 30.7 | 0.41 |
| #9 ,, → ,, | 2909 | 34.3 | 3.06 | 1.01 | 0.92 | 5.1 | 29.0 | 0.41 |
| #5–6 ,, → ,, | 2910 | 33.2 | 2.87 | 1.03 | 0.85 | 3.3 | 23.2 | 0.39 |
| #8–9 ,, → ,, | 2911 | 35.1 | 2.89 | 1.07 | 0.82 | 5.0 | 29.7 | 0.41 |
| #7–9 ,, → ,, | 2912 | 35.9 | 2.95 | 1.04 | 0.87 | 6.0 | 29.2 | 0.41 |
| #6–9 ,, → ,, | 2913 | 35.1 | 2.71 | 1.04 | 0.80 | 19.6 | 26.3 | 0.40 |
| #6 ,, →GATG | 2914 | 34.6 | 3.21 | 1.06 | 0.92 | 5.0 | 31.4 | 0.41 |
| #6 ,, →GAAC | 2915 | 34.7 | 3.44 | 1.08 | 0.97 | 6.7 | 19.8 | 0.38 |
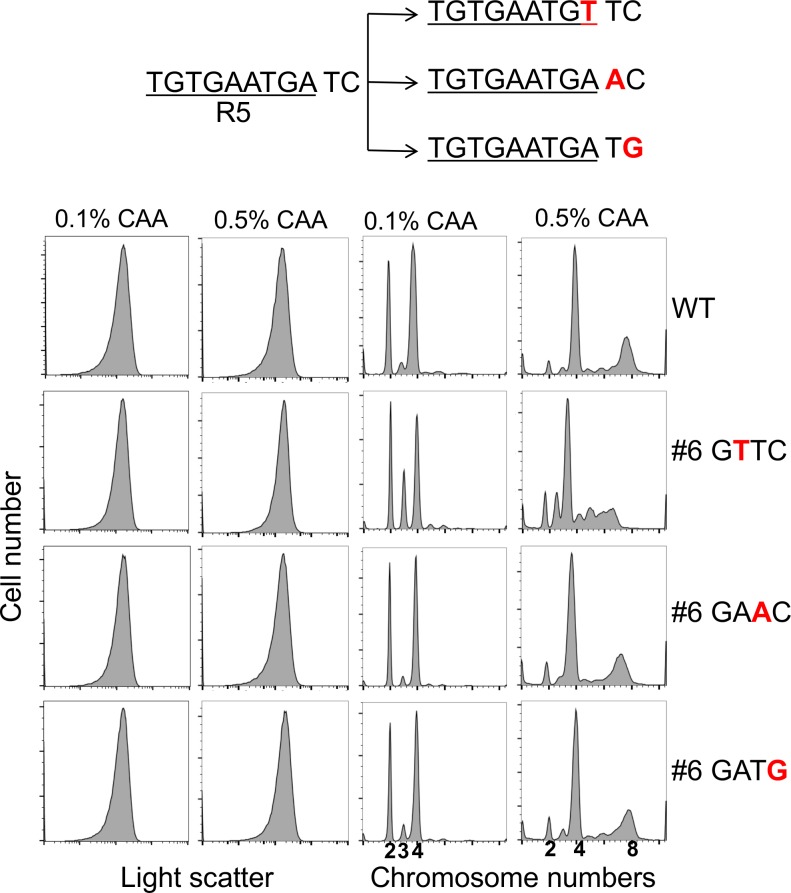
Since the adenine residue of the #6 GATC site is part of the R5 DnaA binding site, we noted that the asynchrony phenotype (20%; Table 2) of this particular mutant could result from a defect in DnaA binding rather than the lack of SeqA binding. To address this possibility, we mutated #6 GATC to GAAC or to GATG (Fig 5). Both of these base changes are outside of the canonical 9-bp DnaA binding site and therefore should abrogate methylation of the site without affecting the R5 DnaA binding site. These new GATC mutants no longer showed the asynchrony phenotype, suggesting that a defect in DnaA binding, rather than in SeqA binding, caused asynchrony when GATC was changed to GTTC in R5.
Fig 5. Importance of integrity of the DnaA binding site R5 and not its methylation in initiation synchrony.
The overlapping GATC site of R5 was mutated at three positions (shown in red), two of which reside outside of the canonical 9-mer DnaA binding site (underlined). Cell sizes and chromosome contents of the above GATC mutants were determined as in Fig 4A, using two different concentrations of casamino acids (CAA).
SeqA binding to the last three GATC sites (#7-#9) of oriC has the potential to control initiation
The importance of SeqA binding to the left half of oriC was previously examined by simultaneously mutating either three or seven GATC sites (up to the site in R5) [37]. Both sets of changes caused replication asynchrony and reduced HM DNA to nearly the level observed in ΔseqA cells, particularly evident in the mutant with seven changes. This suggested that the last three GATC sites (#7–9) in the right half of oriC do not significantly contribute to the sequestration required for initiation synchrony. When we combined the GTTC mutations in the last three GATC sites (#7–9), the triple mutant neither compromised synchrony nor reduced HM DNA at the MboII site (Fig 4A and 4B). However, when the triple mutant was combined with the #6 GATC mutation, the quadruple mutant with changes in #6–9 GATC sites showed reduced HM DNA and improved initiation efficiency at faster growth rates in the presence of 0.5% CAA, although the initiation remained asynchronous (Fig 4B and Figure E in S1 File). It appears that the last three GATC sites have the potential to negatively control initiation, but this becomes apparent only when the initiation is partially compromised. The initiation defect of the #6 mutant was also alleviated when mutations in the #5 and #6 GATC sites were combined (Fig 4 and Figure E in S1 File). Thus, although mutating individual GATC sites does not result in an initiation defect, eliminating SeqA binding even to a single site can help when the initiation efficiency is already suboptimal.
Discussion
During rapid growth when E. coli maintains multiple origins, sequestration of newly replicated origins is required for synchronous initiation from all the origins in a narrow window of time. Sequestration is mediated by binding of SeqA protein to GATC sites of the origin (oriC) when they are hemi-methylated (HM), as in newly replicated origins. To understand the process of sequestration better, we have individually probed most of the GATC sites of oriC for SeqA binding. SeqA is found to bind to all the GATC sites tested, indicating that sequestration normally involves binding to the full complement of GATC sites in oriC. However, synchronous initiation is still retained when the GATC sites were mutated individually, indicating the robustness of the sequestration process.
SeqA binding in vivo is inferred if it can prolong the HM state of a GATC site. By this criterion, SeqA binds to different GATC sites of oriC to different extents. Efficient SeqA binding in vitro requires cooperation from adjacent GATC sites that are on the same face of the DNA and phased by about three helical turns of B-DNA [21, 29, 30, 34]. One explanation for variable SeqA binding to different GATC sites is that the GATC sites in oriC are not phased by integral helical turns. The variation can also be rationalized by assuming that SeqA competes with many other proteins that bind to oriC. We tested the effect of titrating DnaA, the major protein that binds to oriC, on the HM DNA level. Significant but contrasting changes were seen at only two of the GATC sites. Both the observed increase at one site (mutant #6) and the decrease at the other (mutant #7) indicate that DnaA can cooperate as well as compete with SeqA binding, as was suggested earlier [8, 35]. The altered sequestration levels, however, did not change initiation efficiency (mutants #6 and #7, Fig 3A and 3B). Initiation thus seems buffered against small changes in sequestration.
The cooperation from DnaA for SeqA binding to #6 GATC site may not be the only explanation of our data. In the present study, DnaA was titrated using a R1-datA plasmid. The datA site reduces the availability of DnaA-ATP, thereby delay replication initiation [38]. The initiation delay will reduce the number of cells in the culture undergoing replication and, thereby, in the sequestration period. In this scenario the HM DNA level would also lower without a direct role of DnaA in controlling SeqA binding. The question, however, remains is why this scenario did not reduce HM DNA level at other sites. The presence of R1-datA plasmid also derepresses the dnaA promoter and increases the overall DnaA level [36]. The dnaA promoter is also sequestered like oriC because of the presence of six GATC sites in the region. However, when the GATC sites in the promoter region are mutated the initiation synchrony phenotype still persists, indicating that fluctuations in DnaA concentration are not critical for the phenotype [39].
The robustness of sequestration was most evident when we individually inactivated the GATC sites of oriC. The mutants with single mutated GATC sites showed normal level of initiation and initiation synchrony. Together with varying level of SeqA binding to different GATC sites (Fig 2A), these results suggest against cooperativity in SeqA binding. (We cannot rule out the possibility that because of cooperativity the mutated sites were still bound by SeqA.) There is also indication against cooperative binding since SeqA binding to one site does not seem to influence binding to neighboring sites [8]. The cooperativity, however, has been indicated in vitro [30]. Inactivation of the DnaA binding site I2 by mutation of GATC to GATT no longer allows SeqA to interfere with DnaA binding to I2, but SeqA still prevents DnaA binding to a distal site R5. Similarly, when a GATC was engineered into the R1 site, SeqA blocked DnaA binding to that site only.
Initiation synchrony was disturbed when the #6 GATC was changed to GTTC in the R5 DnaA binding site (mutant#6, Fig 4A). We believe that reduced DnaA binding, rather than lack of SeqA binding, is the basis of the observed asynchrony. SeqA binding is known to prevent DnaA binding to some of the lower affinity DnaA binding sites [40]. This is clearly the case both in vivo and in vitro in four of the low-affinity DnaA binding sites tested, including R5 (τ2, R5, I2 and I3) [8]. Reduced DnaA binding upon elimination of SeqA binding to R5 was therefore unexpected. This led us to speculate that the GTTC change reduced DnaA binding and thereby compromised initiation synchrony. Indeed, initiation synchrony was regained when the GATC was mutated to selectively affect SeqA binding (Fig 5). It is intriguing that among the many mutations that we have created in oriC, only the one in R5 showed a significant synchrony defect. R5 could be playing a more important role in initiation than other low-affinity DnaA binding sites, as we discuss below.
Chromosomal replication is disturbed mildly when individual protein binding sites of oriC are mutated along with inversion and scrambling of the R5 site sequence [41, 42]. However, compared to scrambling of R2 or R3, scrambling of the R5 sequence reduced replication more significantly and increased the asynchrony index about two-fold (from 10 to 20) [41]. R5 was also scrambled without disturbing the GATC site [43]. However, this was not sufficient for oriC activity in a plasmid-based replication assay, suggesting that binding of DnaA to R5 is required for initiation. It is also consistent that a viable minimal origin requires the R5 site, although the last 83 bp of oriC that includes the sites from R2 to R4 could be deleted [44]. More direct evidence for the importance of R5 was observed in an in vitro reconstituted system for origin opening [15]. In this assay, the opening could be obtained even when all oriC sequences beyond R5 were deleted, but not when the deletion was extended to include R5. This suggests that the nucleoprotein complex that opens DNA requires DnaA binding to R5. R5 was also required to nucleate DnaA-ATP binding to three weaker downstream sites τ2, I1 and I2 [9]. This cooperative binding requires the sites to be oriented in the same direction, explaining why previous attempts to invert R5 resulted in a non-functional oriC in a plasmid based assay [43].
How could R5 be important? Cooperative interactions that allow DnaA binding to low-affinity sites to the right of R5 appears important, but this cannot be its only role because those sites can be deleted for origin opening, whereas R5 is still required [8, 15]. At present, the role in nucleo-protein complex formation at oriC conducive to origin opening may distinguish R5 from other low-affinity DnaA binding sites of oriC.
We also find that the GATC sites to the right of R5 (sites #7–9) play an insignificant role in synchrony even when they are made defective simultaneously. The three sites alone also do not prevent overinitiation when the upstream GATC sites are mutated [37]. The sites are phylogenetically conserved indicating that their presence is important [45]. The functional significance of the sites #7–9 became evident when the GATC mutation in R5 was combined with GATC mutations of sites #7–9. The latter mutations more than compensated for the initiation defect of the R5 mutation (Figure E in S1 File). In other words, the sequestration of the last three GATC sites can contribute negatively to the regulation of oriC although this contribution appears to be redundant when R5 is not mutated. Similarly, although mutating the GATC site #5 (in τ1) did not significantly change the initiation phenotype on its own, it did rescue the initiation defect of the mutation in R5. In other words, reducing sequestration of the #5 site was helpful only when the initiation efficiency of oriC was compromised. These results lead us to believe that there is excess SeqA binding capacity at oriC to ensure its important role in controlling initiation.
Supporting Information
Table A. Bacterial strains. Table B. Plasmids. Table C. Primers. Table D. Cell cycle parameters of oriC-FRT mutants with TaqI sites carrying R1 plasmid. Table E. Cell cycle parameters of oriC-FRT mutants with TaqI sites carrying R1-datA. Figure A. Construction of strains with TaqI sites overlapping the GATC sites within oriC. (A) Restriction sites used for HM DNA analysis. Recognition sequences (in capital letters) of enzymes MboII and HphI naturally occur within oriC, and of TaqI were created in this study to overlap with the GATC sites. When fully methylated the sites resist enzyme digestion. Upon replication, one of the two sister sites become sensitive to digestion, the one in which the methylated adenine falls outside of the enzyme recognition sequence. (B) Schematic map of oriC as in Fig 1B marked with either an FRT site or a zeo cassette, inserted 27 bp away from the end of the R4 site. The primers (jj40+jj42) used to amplify the oriC region are shown by horizontal arrows at the two flanks of the origin, and the fragment used for probing Southern blots (blue line with dashed extension) at the left end of the origin is same as in Fig 2A. To verify the presence of TaqI sites, genomic DNA either FRT or Zeo marked seqA+ and ΔseqA cells was used to amplify the origin region by PCR, the amplified products were digested with TaqI and resolved on an 1.3% agarose gel. M represent mol. wt. markers (NEB) and WT represent cells without any TaqI site within minimal oriC sequence. The presence of a TaqI site is indicated if the upper band is split into two smaller bands. The change in the relative sizes of the digested bands confirm shifting positions of the TaqI site created within oriC in mutants #1–9. Note that PCR products are not methylated, thus are fully sensitive to TaqI digestion. (C) Comparison of HM DNA level in oriC, oriC-FRT and oriC-zeo strains that are either seqA+ or ΔseqA. The genomic DNA of the strains was digested with either MboII or HphI. Other details for probing and quatification of HM DNA were as described in Fig 2A. (D) Relative TaqI sensitivity of FRT marked genomic DNA from dam minus derivatives of oriC mutants #1–9. The genomic DNA was partially digested at 55°C for 10 min to monitor relative sensitivity of the TaqI sites created within oriC to TaqI digestion, otherwise the details are same as in Fig 2A. The band of interest (the lowest band of the gel) is generated by digestion of one TaqI site within oriC and the other in the left flank of oriC, and its intensity was quantified with respect to all other bands of the gel (the panel on the right). The mean intensity from three independent DNA preparations is shown with one standard deviation of the mean. (E) Distance (bp) separating different GATC sites of oriC. The distances are shown below the schematic map of oriC as in Fig 1B. Figure B. DNA histogram of FRT marked isogenic seqA+ and ΔseqA strains of oriC mutants #1–9. These experiments were done as in Fig 3A except these cells did not have the R1 plasmids. Figure C. HM DNA level at different GATC sites of oriC in seqA+ and ΔseqA strains. The experiments in (A) and (B) were done similarly to those in Fig 2A and 2B, except the strains were marked with zeo in place of the FRT site. Figure D. Effect of DnaA titration on initiation synchrony and HM DNA level at oriC. The experiments in (A) and (B) were done similarly to those in Fig 3A and 3B, except that the strains were ΔseqA derivatives of those used in Fig 3. Figure E. Overcoming of initiation defect due to a mutation in #6 GATC in R5 by GATC mutations in #5 or #7–9 GATC of oriC. Chromosome contents of the GATC mutants were determined as in Fig 4A, except that CAA concentration was 0.5% instead of 0.1%. Note that initiation becomes more efficient in the double mutant #5–6 and in the quadruple mutant #6–9 compared to the single mutant #6.
(PDF)
Acknowledgments
We thank Julia Grimwade, Anders Lϕbner-Olesen and an anonymous reviewer for many thoughtful comments, and to Jemima Barrowman for help with editing. The research was supported entirely by the Intramural Research Program of the Center for Cancer Research, NCI, NIH.
Data Availability
All relevant data are within the paper.
Funding Statement
This work was funded by the Intramural Research Program of the Center for Cancer Research, NCI, NIH.
References
- 1.Lu M, Campbell JL, Boye E, Kleckner N. SeqA: a negative modulator of replication initiation in E. coli. Cell. 1994;77(3):413–26. [DOI] [PubMed] [Google Scholar]
- 2.Boye E, Stokke T, Kleckner N, Skarstad K. Coordinating DNA replication initiation with cell growth: differential roles for DnaA and SeqA proteins. Proc Natl Acad Sci U S A. 1996;93(22):12206–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohiyama M, Cousin D, Ryter A, Jacob F. [Thermosensitive mutants of Escherichia coli K 12. I. Isolation and rapid characterization]. Ann Inst Pasteur (Paris). 1966;110(4):465–86. [PubMed] [Google Scholar]
- 4.Lϕbner-Olesen A, Skarstad K, Hansen FG, von Meyenburg K, Boye E. The DnaA protein determines the initiation mass of Escherichia coli K-12. Cell. 1989;57(5):881–9. [DOI] [PubMed] [Google Scholar]
- 5.Pritchard RH, Barth PT, Collins J, editors. Control of DNA synthesis in bacteria. XIX Symp Soc Gen Microbiol; 1969;263–97. [Google Scholar]
- 6.von Freiesleben U, Rasmussen KV, Schaechter M. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol Microbiol. 1994;14(4):763–72. [DOI] [PubMed] [Google Scholar]
- 7.Skarstad K, Katayama T. Regulating DNA replication in bacteria. Cold Spring Harb Perspect Biol. 2013;5(4):a012922 10.1101/cshperspect.a012922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nievera C, Torgue JJ, Grimwade JE, Leonard AC. SeqA blocking of DnaA-oriC interactions ensures staged assembly of the E. coli pre-RC. Mol Cell. 2006;24(4):581–92. 10.1016/j.molcel.2006.09.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rozgaja TA, Grimwade JE, Iqbal M, Czerwonka C, Vora M, Leonard AC. Two oppositely oriented arrays of low-affinity recognition sites in oriC guide progressive binding of DnaA during Escherichia coli pre-RC assembly. Mol Microbiol. 2011;82(2):475–88. 10.1111/j.1365-2958.2011.07827.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonard AC, Mechali M. DNA replication origins. Cold Spring Harb Perspect Biol. 2013;5(10):a010116 10.1101/cshperspect.a010116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGarry KC, Ryan VT, Grimwade JE, Leonard AC. Two discriminatory binding sites in the Escherichia coli replication origin are required for DNA strand opening by initiator DnaA-ATP. Proc Natl Acad Sci U S A. 2004;101(9):2811–6. 10.1073/pnas.0400340101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurokawa K, Nishida S, Emoto A, Sekimizu K, Katayama T. Replication cycle-coordinated change of the adenine nucleotide-bound forms of DnaA protein in Escherichia coli. Embo J. 1999;18(23):6642–52. 10.1093/emboj/18.23.6642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speck C, Messer W. Mechanism of origin unwinding: sequential binding of DnaA to double- and single-stranded DNA. Embo J. 2001;20(6):1469–76. 10.1093/emboj/20.6.1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duderstadt KE, Berger JM. A structural framework for replication origin opening by AAA+ initiation factors. Curr Opin Struct Biol. 2013;23(1):144–53. 10.1016/j.sbi.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozaki S, Katayama T. Highly organized DnaA-oriC complexes recruit the single-stranded DNA for replication initiation. Nucleic Acids Res. 2012;40(4):1648–65. 10.1093/nar/gkr832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jha JK, Ramachandran R, Chattoraj DK. Opening the Strands of Replication Origins-Still an Open Question. Front Mol Biosci. 2016;3:article 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Skarstad K, Boye E, Steen HB. Timing of initiation of chromosome replication in individual Escherichia coli cells [published erratum appears in EMBO J 1986 Nov;5(11):3074]. Embo J. 1986;5(7):1711–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skarstad K, Lϕbner-Olesen A. Stable co-existence of separate replicons in Escherichia coli is dependent on once-per-cell-cycle initiation. Embo J. 2003;22(1):140–50. 10.1093/emboj/cdg003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith DW, Garland AM, Herman G, Enns RE, Baker TA, Zyskind JW. Importance of state of methylation of oriC GATC sites in initiation of DNA replication in Escherichia coli. Embo J. 1985;4(5):1319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riber L, Lϕbner-Olesen A. Coordinated replication and sequestration of oriC and dnaA are required for maintaining controlled once-per-cell-cycle initiation in Escherichia coli. J Bacteriol. 2005;187(16):5605–13. 10.1128/JB.187.16.5605-5613.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Slater S, Wold S, Lu M, Boye E, Skarstad K, Kleckner N. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell. 1995;82(6):927–36. [DOI] [PubMed] [Google Scholar]
- 22.Skarstad K, Torheim N, Wold S, Lurz R, Messer W, Fossum S, et al. The Escherichia coli SeqA protein binds specifically to two sites in fully and hemimethylated oriC and has the capacity to inhibit DNA replication and affect chromosome topology. Biochimie. 2001;83(1):49–51. [DOI] [PubMed] [Google Scholar]
- 23.Torheim NK, Skarstad K. Escherichia coli SeqA protein affects DNA topology and inhibits open complex formation at oriC. EMBO J. 1999;18(17):4882–8. 10.1093/emboj/18.17.4882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Court DL, Swaminathan S, Yu D, Wilson H, Baker T, Bubunenko M, et al. Mini-lambda: a tractable system for chromosome and BAC engineering. Gene. 2003;315:63–9. [DOI] [PubMed] [Google Scholar]
- 25.Kim MS, Bae SH, Yun SH, Lee HJ, Ji SC, Lee JH, et al. Cnu, a novel oriC-binding protein of Escherichia coli. J Bacteriol. 2005;187(20):6998–7008. 10.1128/JB.187.20.6998-7008.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skarstad K, von Meyenburg K, Hansen FG, Boye E. Coordination of chromosome replication initiation in Escherichia coli: effects of different dnaA alleles. J Bacteriol. 1988;170(2):852–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Srivastava P, Fekete RA, Chattoraj DK. Segregation of the replication terminus of the two Vibrio cholerae chromosomes. J Bacteriol. 2006;188(3):1060–70. 10.1128/JB.188.3.1060-1070.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Campbell JL, Kleckner N. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell. 1990;62(5):967–79. [DOI] [PubMed] [Google Scholar]
- 29.Brendler T, Austin S. Binding of SeqA protein to DNA requires interaction between two or more complexes bound to separate hemimethylated GATC sequences. Embo J. 1999;18(8):2304–10. 10.1093/emboj/18.8.2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han JS, Kang S, Kim SH, Ko MJ, Hwang DS. Binding of SeqA protein to hemi-methylated GATC sequences enhances their interaction and aggregation properties. J Biol Chem. 2004;279(29):30236–43. 10.1074/jbc.M402612200 [DOI] [PubMed] [Google Scholar]
- 31.Bach T, Skarstad K. An oriC-like distribution of GATC sites mediates full sequestration of non-origin sequences in Escherichia coli. J Mol Biol. 2005;350(1):7–11. 10.1016/j.jmb.2005.04.055 [DOI] [PubMed] [Google Scholar]
- 32.Thomas M, Davis RW. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975;91(3):315–28. [DOI] [PubMed] [Google Scholar]
- 33.Chattoraj DK, Oberoi YK, Bertani G. Restriction of Bacteriophage P2 by the Escherichia coli R1 plasmid, and in vitro cleavage of its DNA by the EcoRI Endonuclease. Virology. 1977;81(2):460–70. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez-Romero MA, Busby SJ, Dyer NP, Ott S, Millard AD, Grainger DC. Dynamic distribution of SeqA protein across the chromosome of Escherichia coli K-12. MBio. 2010;1(1). e00012–10. 10.1128/mBio.00012-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bach T, Morigen, Skarstad K. The initiator protein DnaA contributes to keeping new origins inactivated by promoting the presence of hemimethylated DNA. J Mol Biol. 2008;384(5):1076–85. 10.1016/j.jmb.2008.09.042 [DOI] [PubMed] [Google Scholar]
- 36.Morigen, Lϕbner-Olesen A, Skarstad K. Titration of the Escherichia coli DnaA protein to excess datA sites causes destabilization of replication forks, delayed replication initiation and delayed cell division. Mol Microbiol. 2003;50(1):349–62. [DOI] [PubMed] [Google Scholar]
- 37.Bach T, Skarstad K. Re-replication from non-sequesterable origins generates three-nucleoid cells which divide asymmetrically. Mol Microbiol. 2004;51(6):1589–600. [DOI] [PubMed] [Google Scholar]
- 38.Kasho K, Katayama T. DnaA binding locus datA promotes DnaA-ATP hydrolysis to enable cell cycle-coordinated replication initiation. Proc Natl Acad Sci U S A. 2013;110(3):936–41. 10.1073/pnas.1212070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkinson TG 2nd, Kedar GC, Lee C, Guzman EC, Smith DW, Zyskind JW. The synchrony phenotype persists after elimination of multiple GATC sites from the dnaA promoter of Escherichia coli. J Bacteriol. 2006;188(12):4573–6. 10.1128/JB.00089-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wold S, Boye E, Slater S, Kleckner N, Skarstad K. Effects of purified SeqA protein on oriC-dependent DNA replication in vitro. Embo J. 1998;17(14):4158–65. 10.1093/emboj/17.14.4158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weigel C, Messer W, Preiss S, Welzeck M, Morigen, Boye E. The sequence requirements for a functional Escherichia coli replication origin are different for the chromosome and a minichromosome. Mol Microbiol. 2001;40(2):498–507. [DOI] [PubMed] [Google Scholar]
- 42.Riber L, Fujimitsu K, Katayama T, Lϕbner-Olesen A. Loss of Hda activity stimulates replication initiation from I-box, but not R4 mutant origins in Escherichia coli. Mol Microbiol. 2009;71(1):107–22. 10.1111/j.1365-2958.2008.06516.x [DOI] [PubMed] [Google Scholar]
- 43.Langer U, Richter S, Roth A, Weigel C, Messer W. A comprehensive set of DnaA-box mutations in the replication origin, oriC, of Escherichia coli. Mol Microbiol. 1996;21(2):301–11. [DOI] [PubMed] [Google Scholar]
- 44.Stepankiw N, Kaidow A, Boye E, Bates D. The right half of the Escherichia coli replication origin is not essential for viability, but facilitates multi-forked replication. Mol Microbiol. 2009;74(2):467–79. 10.1111/j.1365-2958.2009.06877.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zyskind JW, Smith DW. The bacterial origin of replication, oriC. Cell. 1986;46(4):489–90. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A. Bacterial strains. Table B. Plasmids. Table C. Primers. Table D. Cell cycle parameters of oriC-FRT mutants with TaqI sites carrying R1 plasmid. Table E. Cell cycle parameters of oriC-FRT mutants with TaqI sites carrying R1-datA. Figure A. Construction of strains with TaqI sites overlapping the GATC sites within oriC. (A) Restriction sites used for HM DNA analysis. Recognition sequences (in capital letters) of enzymes MboII and HphI naturally occur within oriC, and of TaqI were created in this study to overlap with the GATC sites. When fully methylated the sites resist enzyme digestion. Upon replication, one of the two sister sites become sensitive to digestion, the one in which the methylated adenine falls outside of the enzyme recognition sequence. (B) Schematic map of oriC as in Fig 1B marked with either an FRT site or a zeo cassette, inserted 27 bp away from the end of the R4 site. The primers (jj40+jj42) used to amplify the oriC region are shown by horizontal arrows at the two flanks of the origin, and the fragment used for probing Southern blots (blue line with dashed extension) at the left end of the origin is same as in Fig 2A. To verify the presence of TaqI sites, genomic DNA either FRT or Zeo marked seqA+ and ΔseqA cells was used to amplify the origin region by PCR, the amplified products were digested with TaqI and resolved on an 1.3% agarose gel. M represent mol. wt. markers (NEB) and WT represent cells without any TaqI site within minimal oriC sequence. The presence of a TaqI site is indicated if the upper band is split into two smaller bands. The change in the relative sizes of the digested bands confirm shifting positions of the TaqI site created within oriC in mutants #1–9. Note that PCR products are not methylated, thus are fully sensitive to TaqI digestion. (C) Comparison of HM DNA level in oriC, oriC-FRT and oriC-zeo strains that are either seqA+ or ΔseqA. The genomic DNA of the strains was digested with either MboII or HphI. Other details for probing and quatification of HM DNA were as described in Fig 2A. (D) Relative TaqI sensitivity of FRT marked genomic DNA from dam minus derivatives of oriC mutants #1–9. The genomic DNA was partially digested at 55°C for 10 min to monitor relative sensitivity of the TaqI sites created within oriC to TaqI digestion, otherwise the details are same as in Fig 2A. The band of interest (the lowest band of the gel) is generated by digestion of one TaqI site within oriC and the other in the left flank of oriC, and its intensity was quantified with respect to all other bands of the gel (the panel on the right). The mean intensity from three independent DNA preparations is shown with one standard deviation of the mean. (E) Distance (bp) separating different GATC sites of oriC. The distances are shown below the schematic map of oriC as in Fig 1B. Figure B. DNA histogram of FRT marked isogenic seqA+ and ΔseqA strains of oriC mutants #1–9. These experiments were done as in Fig 3A except these cells did not have the R1 plasmids. Figure C. HM DNA level at different GATC sites of oriC in seqA+ and ΔseqA strains. The experiments in (A) and (B) were done similarly to those in Fig 2A and 2B, except the strains were marked with zeo in place of the FRT site. Figure D. Effect of DnaA titration on initiation synchrony and HM DNA level at oriC. The experiments in (A) and (B) were done similarly to those in Fig 3A and 3B, except that the strains were ΔseqA derivatives of those used in Fig 3. Figure E. Overcoming of initiation defect due to a mutation in #6 GATC in R5 by GATC mutations in #5 or #7–9 GATC of oriC. Chromosome contents of the GATC mutants were determined as in Fig 4A, except that CAA concentration was 0.5% instead of 0.1%. Note that initiation becomes more efficient in the double mutant #5–6 and in the quadruple mutant #6–9 compared to the single mutant #6.
(PDF)
Data Availability Statement
All relevant data are within the paper.