Abstract
A number of proteins accumulate in the anaphase spindle midzone, but the interaction and precise role of these proteins in midzone organization remain obscure. Here, we found that the microtubule-bundling protein PRC1 bound separately to the three motor proteins, KIF4, MKLP1 and CENP-E, but not to the chromosomal passenger proteins. In KIF4-deficient cells, the central spindle was disorganized, and all midzone-associated proteins including PRC1 failed to concentrate at the midline, instead being dispersed along the loosened microtubule bundles of the central spindle. This suggests that KIF4 is essential for the organization of central spindles and for midzone formation. In PRC1-deficient cells, no midzone was formed, KIF4 and CENP-E did not localize to the disconnected half-spindle, and MKLP1 and chromosomal passenger proteins localized to discrete subdomains near microtubule plus ends in the half-spindle. Thus, PRC1 is required for interaction of the two half-spindles and for localization of KIF4 and CENP-E. These results suggest that KIF4 and its binding partner PRC1 play essential roles in the organization of central spindles and midzone formation.
Keywords: central spindle, KIF4, midzone, PRC1
Introduction
In late mitotic anaphase, an organized central spindle midzone forms between the two separating sets of chromatids. This consists of a dense network of overlapping antiparallel microtubules (Mastronarde et al, 1993). The spindle midzone plays an essential role in the initiation and completion of cell cleavage, and is the binding site for a number of proteins that play a part in cytokinesis (Cao and Wang, 1996; Wheatley and Wang, 1996; Glotzer, 1997; Robinson and Spudich, 2000). Proteins that have been reported to accumulate at the midzone during late mitosis include microtubule-bundling protein PRC1 (Jiang et al, 1998), chromosomal passenger proteins, inner centromere protein (INCENP) (Cooke et al, 1987), Survivin (Skoufias et al, 2000; Uren et al, 2000), Aurora B (Terada et al, 1998), TD-60 (Andreassen et al, 1991; Mollinari et al, 2003) and Borealin (Gassmann et al, 2004), protein kinase Plk (Carmena et al, 1998), Rho regulatory factors, CYK4/RhoGAP (Jantsch-Plunger et al, 2000; Mishima et al, 2002) and ECT2/RhoGEF (Prokopenko et al, 1999; Tatsumoto et al, 1999), and microtubule motor proteins MKLP1 (Sellitto and Kuriyama, 1988; Nislow et al, 1992), CENP-E (Yen et al, 1991) and Rab6-KIFL/MKLP2 (Hill et al, 2000; Neef et al, 2003). The mechanism of central spindle assembly and the functional roles of these midzone-associated proteins during late mitosis are still largely unknown.
Ase1 is a protein first identified in the budding yeast that binds and bundles microtubules and is required for anaphase spindle elongation. Overexpression of Ase1 induces spindle elongation in S phase (Pellman et al, 1995; Juang et al, 1997; Schuyler et al, 2003). Ase1 was proposed to form cross-bridges within the spindle midzone that are essential for maintenance of anaphase spindle integrity. PRC1, the mammalian homolog of Ase1 (Jiang et al, 1998), accumulates in the central spindle midzone during anaphase and midbody during telophase (Mollinari et al, 2002). Overexpressed PRC1 extensively bundles interphase microtubules, and a phosphorylation-null mutant causes extensive bundling of the prometaphase spindle. Suppression of PRC1 by siRNA causes a failure of microtubule interdigitation between half-spindles and the absence of a spindle midzone (Mollinari et al, 2002). The mechanism by which PRC1 is concentrated at the spindle midline during cytokinesis remains to be determined.
The kinesin superfamily proteins (KIFs) are a family of 45 (in human) microtubule-based motor proteins that transport membranous organelles, protein complexes and other cargoes to specific destinations in a microtubule- and ATP-dependent manner (Hirokawa, 1998; Miki et al, 2001, Schliwa and Woehlke, 2003). Chromokinesins are a subfamily of KIFs with amino-terminal motor domains that bind to chromosome arms. The chromokinesins identified to date can be divided into two main functional categories. Kid/Nod family proteins are required for chromosome alignment (Murphy and Karpen, 1995; Antonio et al, 2000; Funabiki and Murray, 2000), chromosome orientation and oscillation of chromosome arms (Levesque and Compton, 2001). A second chromokinesin subfamily is composed of Xenopus Xklp1 (Vernos et al, 1995), Drosophila Klp3A (Williams et al, 1995) and mammalian KIF4 (Sekine et al, 1994). Xenopus Xklp1 is required for chromosome positioning and bipolar spindle stabilization in egg extracts (Vernos et al, 1995), and chromatin–microtubule interactions in vitro (Walczak et al, 1998). Many functions of Drosophila Klp3A have been suggested. In spermatocytes these include central spindle formation, initiation of cytokinesis (Williams et al, 1995) and assembly of the actin-based contractile ring (Giansanti et al, 1998). Klp3A has been implicated in chromosome congression to the metaphase plate (Goshima and Vale, 2003) and spindle pole separation (Kwon et al, 2004) in S2 cells. KIF4 has multiple functions in membrane trafficking and cell division. In neurons, KIF4 is an anterograde motor that transports specific organelles involved in nerve cell morphogenesis (Sekine et al, 1994). Remarkably, KIF4 is also a chromokinesin that is associated with chromosome arms (Wang and Adler, 1995). The role of KIF4 during mitosis has not yet been determined.
In order to begin to dissect the mechanism of spindle midzone formation, we have identified proteins that bind to PRC1 during late mitosis. Immunoprecipitation with an anti-PRC1-specific antibody revealed a specific interaction between KIF4 and PRC1. We therefore studied the function of KIF4 and PRC1 during late mitosis in relation to other midzone-associated proteins including the two motor proteins MKLP1 and CENP-E and the chromosomal passenger proteins Aurora B, INCENP and Survivin. Our studies have revealed that both KIF4 and PRC1 have important roles in the formation of an organized spindle midzone and midbody.
Results
KIF4 is bound to PRC1 in vivo
To study the precise role of PRC1 during spindle midzone formation, we used immunoprecipitation to identify proteins that specifically associated with it in vivo. Affinity-purified anti-human PRC1-specific rabbit antibody was used to immunoprecipitate the endogenous protein from HeLa cell extracts at late mitosis. These immunoprecipitates contained a prominent band with an apparent molecular weight of 155 kDa in SDS–PAGE (Figure 1A, lane 2). Two lower molecular weight bands corresponded to PRC1 and the antibody used (Figure 1A, lane 2). No abundant polypeptides were seen in the immunoprecipitates with preimmune serum (Figure 1A, lane 1). Amino-acid sequence analysis of the potential PRC1 binding protein revealed it to be the chromokinesin KIF4. The other two bands of 175 and 110 kDa turned out not to be specific PRC1 binding proteins (data not shown).
Figure 1.
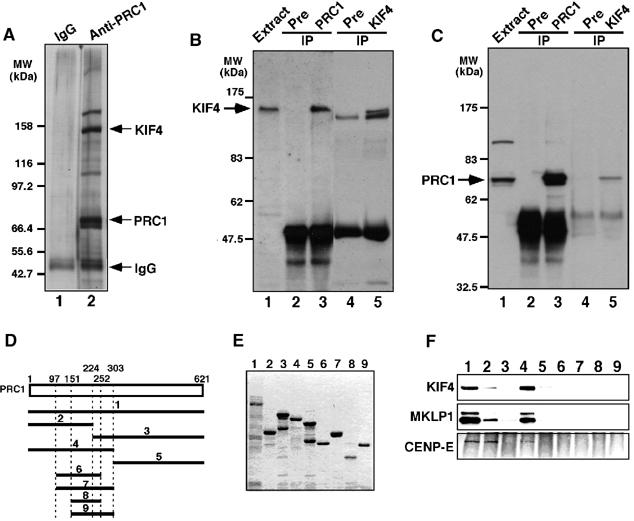
PRC1 specifically binds to KIF4 during late mitosis. (A) Immunoaffinity chromatography. HeLa cell extracts at late mitosis were fractionated by affinity chromatography using anti-PRC1-specific antibody, and bound proteins were separated by SDS–PAGE and silver-stained. (B) Immunoprecipitates with anti-PRC1 antibody contain KIF4. HeLa cell extracts at late mitosis were subjected to immunoprecipitation, with preimmune sera, anti-PRC1 or anti-KIF4, fractionated by SDS–PAGE and immunoblotted with anti-KIF4-specific antibody. The common band in the immunoprecipitates with preimmune serum and anti-KIF4 antibody is antibody-derived protein. (C) Immunoprecipitates with anti-KIF4 antibody include PRC1. (D–E) Pull-down experiments with a series of PRC1 fragments fused to GST. Schematic of PRC1 fragments used for pull-down experiments (D) and their SDS–PAGE pattern (E). The PRC1 fragments were incubated with HeLa cell extracts at late mitosis, and bound proteins separated by SDS–PAGE were probed with anti-KIF4, anti-MKLP1 and CENP-E antibodies (F).
This in vivo interaction between endogenous KIF4 and PRC1 was confirmed by immunoprecipitation using a rat serum specific for KIF4. Immunoblot analysis with this antiserum detected KIF4 as a single band of 155 kDa in HeLa cell extracts at late mitosis and in immunoprecipitates with anti-PRC1 antibody (Figure 1B). Furthermore, PRC1 was also readily detected in immunoprecipitates with the anti-KIF4 antiserum (Figure 1C). In controls, each protein was detected in immunoprecipitates with its cognate serum, but not in control immunoprecipitates with preimmune sera (Figure 1B and C).
We conclude from these experiments that PRC1 is specifically associated with KIF4 in vivo during late mitosis.
The amino-terminal half of PRC1 interacts with itself and the carboxyl-terminal half of KIF4
To further analyze the interaction between PRC1 and KIF4, we performed in vitro binding assays with KIF4 from HeLa late mitotic cell extracts and full-length or a series of truncated forms of human PRC1 (Figure 1D) fused to glutathione S-transferase (GST) and expressed in Escherichia coli. Figure 1E shows the SDS–PAGE patterns of the GST-fused PRC1 fragments used for binding assays. Both full-length PRC1 (Figure 1F, lane 1) and the amino-terminal half of the protein (amino acids 1–303) (Figure 1F, lane 4) bound to KIF4. The amino-terminal one-third of PRC1 weakly bound to KIF4 (Figure 1F, lane 2), but the other PRC1 fragments did not bind to KIF4. Yeast two-hybrid analyses confirmed that the amino-terminal half of PRC1 (amino acids 1–303) interacted both with PRC1 and with the carboxyl-terminal half of KIF4 (amino acids 663–1232) (Supplementary Figure 1).
Taken together, these results suggest that PRC1 associates with KIF4 through interaction between its amino-terminal half and the carboxyl-terminal half of KIF4.
Colocalization of PRC1 and KIF4 during late mitosis
Immunofluorescence analysis of HeLa cells with anti-KIF4-specific antibody showed that KIF4 was localized on the chromosomes throughout mitosis, and additionally to the spindle midzone during anaphase and the midbody during telophase and cytokinesis (Figure 2A). KIF4 was nuclear during interphase (data not shown).
Figure 2.
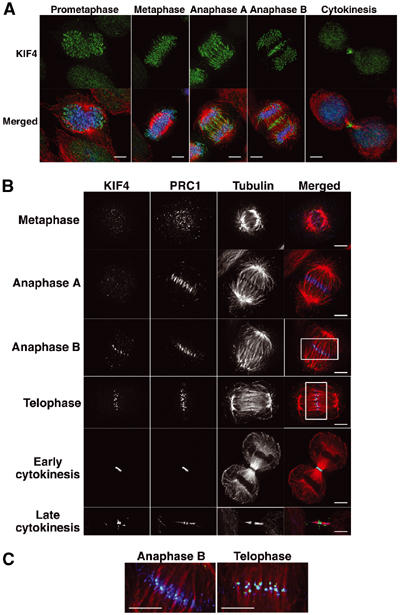
Colocalization of PRC1 and KIF4 during late mitosis. (A) Subcellular localization of KIF4 during mitosis. Immunofluorescence analyses of HeLa cells show KIF4 (green), α-tubulin (red) and DNA (blue). Scale bar, 5 μm. (B) PRC1 and KIF4 colocalize in the central spindle during late mitosis. HeLa cells at indicated mitotic stages were fixed with paraformaldehyde, and KIF4 (green), PRC1 (blue) and tubulin (red) were stained. Staining of KIF4 in chromosomes was weak due to the paraformaldehyde fixation. Scale bar, 5 μm. (C) The rectangular regions of (B) are enlarged. Scale bar, 5 μm.
As predicted by the biochemical analysis, PRC1 and KIF4 colocalize in late mitosis. This experiment was complicated by the fact that detection of PRC1 requires fixation with formaldehyde under conditions where the detection of KIF4 on chromosomes is relatively inefficient. PRC1 was distributed diffusely around the chromosomes and spindles until metaphase.
PRC1 colocalized with KIF4 from anaphase B to telophase (Figure 2B). PRC1 localized to the spindle midzone at anaphase A, but KIF4 localized to the midzone only later, during anaphase B (see below). When KIF4 reached the midzone, it accumulated in tight ball-like foci that appeared to link microtubule bundles from the two half-spindles. These KIF4 foci were surrounded by PRC1 in anaphase B, and completely coincided with PRC1 in telophase (Figure 2C). In late cytokinesis, KIF4 was localized in the center of the midbody (Flemming body), while some PRC1 was present on the flanking bundles of microtubules.
These data revealed that PRC1 shows significant colocalization with KIF4 during late mitosis.
KIF4 is localized at the spindle midzone at anaphase B but not at anaphase A
At anaphase in normal cells, PRC1 preceded KIF4 localization to the midzone (Figure 2B). To examine the timing of localization of KIF4 to the midzone in greater detail, cells at anaphase A and anaphase B were co-stained with KIF4, MKLP1 and Aurora B. At anaphase A in normal cells, KIF4 had not yet relocated to the central spindle, at a time when Aurora B and MKLP1 had already done so (Figure 3A—the blue signal represents KIF4 on chromatid arms). KIF4 subsequently localized to the midzone in anaphase B, at which point it colocalized with MKLP1 and was flanked by Aurora B on either side (Figure 3B). Therefore, KIF4 does not immediately relocate to the central spindle at the onset of anaphase. KIF4 and Aurora B also colocalized near the plus ends of microtubules at the periphery of normal cells at anaphase B. At these sites, MKLP1 localized to an extended region proximal to the plus ends (Figure 3C).
Figure 3.
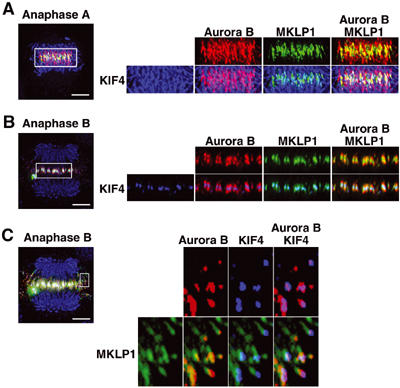
KIF4 is localized to the spindle midline at anaphase B but not at anaphase A. (A, B) Staining of KIF4 (blue), MKLP1 (green) and Aurora B (red) in normal cells at anaphase A (A) and at anaphase B (B). Scale bar, 5 μm. The rectangular regions are enlarged and shown in the right panels. (C) The peripheral region of the central spindle of a normal cell at anaphase B is enlarged in the right panels. Scale bar, 5 μm.
These data suggest that Aurora B and MKLP1 relocate directly to the spindle midzone, while KIF4 localizes first near the plus ends of the microtubules and then to the midzone as microtubules from opposite half-spindles are drawn together into bundles.
KIF4 depletion causes formation of aberrant midzone and midbody
We next analyzed KIF4 function during mitosis by using siRNA to deplete the protein in HeLa cells. Expression of KIF4 was effectively suppressed by 100 nM of KIF4 siRNA in transfection medium (Figure 4A), and KIF4 was either greatly reduced or absent from the chromosomes or midzone/midbody of the siRNA-transfected cells (Figures 4B and D (panels d and g) and 5A–G (panel c)). In all subsequent siRNA experiments, cells transfected with single-stranded sense RNA alone are shown as controls. The single-stranded KIF4 RNA had no effect on mitosis in control cells (Figures 4B and D (panel a) and 5A–G (panel a)).
Figure 4.
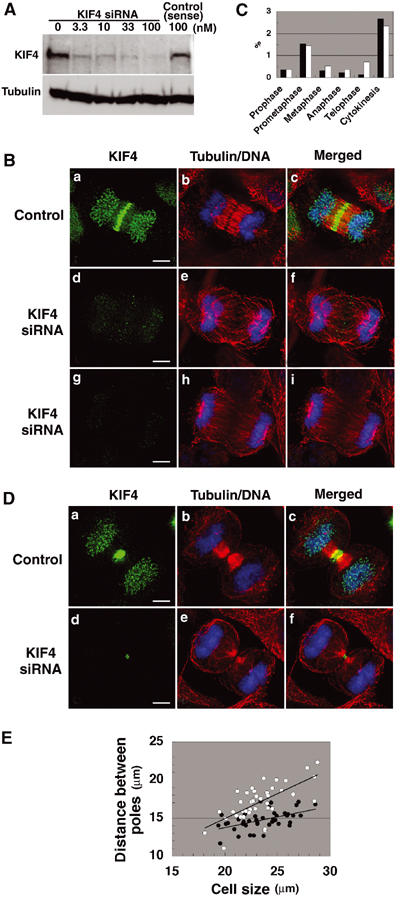
KIF4 deficiency results in aberrant midzone and midbody formation. (A) Immunoblot analysis of HeLa cells treated with various concentrations of KIF4 siRNA (0–100 nM) or with single-stranded sense RNA alone (100 nM). Cell extracts were probed with anti-KIF4 antibody and anti-tubulin antibody as an internal control. (B, D) Phenotypes of KIF4-deficient cells at anaphase (B) and telophase (D). HeLa cells were treated with single-stranded sense KIF4 RNA (panels a–c; control) or with KIF4 siRNA (panels d–i) for 48–55 h, and KIF4 (green), α-tubulin (red) and DNA (blue) were stained. Scale bar, 5 μm. (C) Percentage of each mitotic stage in KIF4-deficient cells (white) and control cells (black). (E) Plots of distance between the poles versus cell size in KIF4-deficient cells (open circles) and control cells (closed circles) at anaphase.
Figure 5.
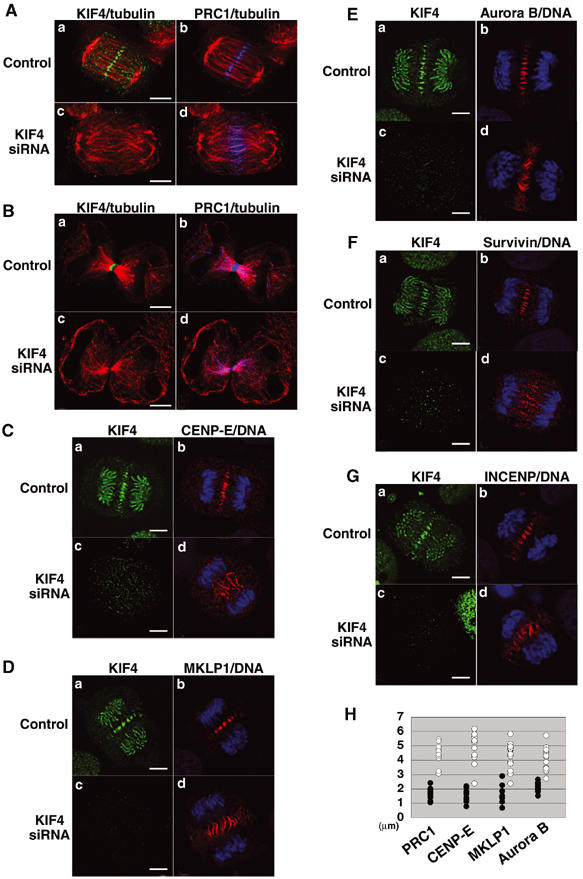
KIF4-deficient cells exhibit dispersed localization of PRC1, CENP-E, MKLP1, Aurora B, Survivin and INCENP. HeLa cells were treated with single-stranded sense KIF4 RNA (panels a and b; control) or with KIF4 siRNA (panels c and d), and cells at anaphase (A, C–G) and late telophase (B). (A, B) Cells were stained with anti-KIF4 (green), anti-PRC1 (blue) and tubulin (red) antibodies. (C–G) Cells were stained with anti-CENP-E (C, red), anti-MKLP1 (D, red), anti-Aurora B (E, red), anti-Survivin (F, red), anti-INCENP (G, red) antibodies, and with anti-KIF4 antibody (green) and DNA (blue). Scale bar, 5 μm. (H) Quantification of the lack of focus in the central spindle in KIF4-deficient cells. The graph shows the width of the region stained by PRC1, CENP-E, MKLP1 and Aurora B in KIF4-deficient cells (open circles), and normal cells (closed circles) are shown.
In KIF4-deficient cells, spindle formation, chromosome congression to a metaphase plate, sister chromatid segregation, interpolar microtubule elongation and cleavage furrow formation occurred normally, but the progression of these cells appeared to be delayed compared with normal cell division. The mitotic index of control cells was 5.2% and that of KIF4-deficient cells was 5.8%. Figure 4C shows the percentage of each mitotic stage in KIF4-deficient and control cells. The percentage of prophase, prometaphase and cytokinesis was about the same for both populations, but that of metaphase, anaphase and, especially, telophase was significantly increased. Although the phenotypes of KIF4-deficient cells at anaphase, telophase and cytokinesis were abnormal (see below), these cells clearly progressed through the late mitotic stages, albeit more slowly.
Immunofluorescence analysis with anti-α-tubulin antibody showed that the central spindle was disorganized at anaphase in KIF4-depleted cells (Figure 4B, panels e and h), in comparison with the organized central spindle in control cells (Figure 4B, panel b). We observed a tight correlation between the level of KIF4 and the degree of microtubule bundling. No organized spindle midzone was formed when KIF4 was strongly suppressed and could not be detected in equatorial regions (Figure 4B, panels g–i).
In KIF4-depleted late-anaphase cells, the separated sister chromatids were localized abnormally close to the plasma membrane (Figure 4B, panels f and i). The effect of KIF4 depletion on the length of the central spindle was quantified by measuring the distance between spindle poles in KIF4-deficient and control cells. Figure 4E shows the plots of the distance between the poles versus the cell size in KIF4-deficient cells (open circles) and control cells (closed circles) at anaphase. The central spindle was elongated by about 20% in KIF4-deficient cells compared with control cells.
At telophase in KIF4 siRNA-treated cells, in which KIF4 levels were significantly reduced, a much reduced midbody was formed (Figure 4D, panels d and f), and the midbody microtubules were less prominent and less organized (Figure 4D, panel e), compared with the robust midbodies in control cells (Figure 4D, panel b). We failed to observe cells in cytokinesis that were completely negative for KIF4. This could be either because cells lacking KIF4 fail to undergo cytokinesis or because even minute amounts of residual KIF4 become detectable when concentrated into the highly compact midbody structure. Regardless of the explanation, examination of cultures suggests that cytokinesis ultimately failed, as a significant increase in the number of binucleate cells (about 8%) was observed.
These observations suggest that KIF4 is required for the formation of an organized central spindle midzone and midbody and for successful cytokinesis.
KIF4 deficiency leads to mislocalization of PRC1, MKLP1, CENP-E and chromosomal passenger proteins
Since KIF4 binds to PRC1, we next determined whether PRC1 targeting late in mitosis is upstream or downstream of KIF4 function by examining the subcellular localization of PRC1 in KIF4-depleted cells. Instead of the compact midzone/midbody localization of PRC1 with KIF4 seen in control cells (Figure 5A and B, panel b), KIF4 deficiency caused PRC1 to be dispersed along microtubules throughout the central spindle during anaphase (Figure 5A, panel d). In KIF4-deficient cells during cytokinesis, PRC1 was not concentrated in the midbody but was dispersed all along the microtubule bundles (Figure 5B, panel d). These results suggest that PRC1 can bind to central spindle microtubules without KIF4, but that its concentration into compact foci at the midline of the central spindle requires the presence of KIF4.
Similarly, in KIF4-deficient cells, CENP-E, MKLP1 and the chromosomal passenger proteins, Aurora B, Survivin and INCENP, were all dispersed throughout the diffuse central spindle (Figure 5C–G), similar to PRC1 as shown in Figure 5A. The lack of focus in the central spindle in KIF4-deficient cells was quantified by measuring the width of the region stained by PRC1, CENP-E, MKLP1 and Aurora B antibodies. Figure 5H shows this width in KIF4-deficient cells (open circles) and control cells (closed circles). In KIF4-deficient cells, the value obtained was about 2.5 times larger than that of control cells at anaphase.
These results show that KIF4 is not required for this group of midzone-associated proteins to bind to midzone microtubules. However, it is required for those microtubules to form an organized central spindle, and for the midzone-associated proteins to concentrate into compact structures that will ultimately give rise to the midbody.
PRC1 is required for KIF4 and CENP-E to relocalize from chromosomes to microtubules during anaphase
We next examined the effects of PRC1 deficiency on the subcellular localization of this group of midzone-associated proteins. PRC1 could be effectively suppressed by 100 nM of siRNA (Figure 6A), and immunofluorescence analyses revealed that almost no PRC1 could be detected in PRC1-depleted cells (Figure 6B, panel d). No effect was detected in control cells transfected with single-stranded sense RNA (Figure 6B, panel b).
Figure 6.
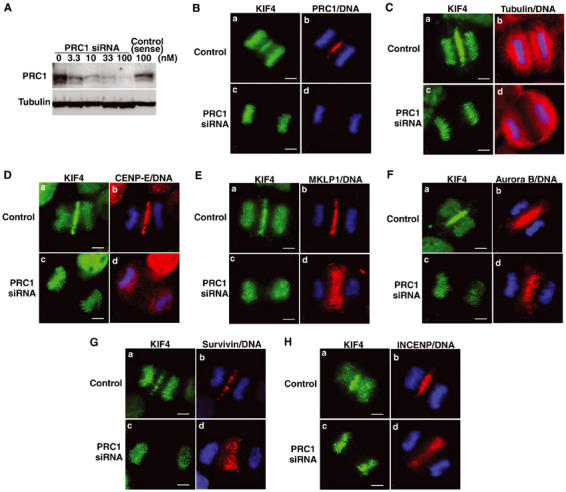
PRC1 deficiency causes loss of the spindle midzone and absence of KIF4 and CENP-E in the central spindle, but localization of MKLP1 and chromosomal passenger proteins in the half-spindle. (A) Immunoblot analysis of HeLa cells treated with various concentrations of PRC1 siRNA (0–100 nM) or single-stranded sense RNA alone (100 nM). Cell extracts were probed with anti-PRC1 antibody and anti-tubulin antibody as an internal control. (B–H) HeLa cells were treated with single-stranded sense PRC1 RNA (panels a and b; control) or with PRC1 siRNA (panels c and d), and PRC1 (B, shown in red), α-tubulin (C, red), CENP-E (D, red), MKLP1 (E, red), Aurora B (F, red), Survivin (G, red), INCENP (H, red), KIF4 (green) and DNA (blue) were stained in cells at anaphase. Scale bar, 5 μm.
In PRC1-depleted anaphase cells, KIF4 was absent from the central spindle, but appeared to be localized normally to chromosomes (Figure 6B, panel c). Failure of KIF4 relocation was not unlikely to be simply due to a loss of midzone structure since MKLP1 was localized to a diffuse midzone in PRC1-deficient cells (see below).
Because of the technical difficulty of double staining PRC1 together with other midzone-associated proteins, we subsequently exploited this change in the distribution of KIF4 as a monitor of the effectiveness of PRC1 depletion in particular cells (Figure 6B–H, panel c). This approach was validated by immunostaining of these cells with anti-α-tubulin antibody, which clearly demonstrated that an integrated spindle was not formed in anaphase cells judged to be PRC1-deficient as a result of their KIF4 distribution (Figure 6C, panel d). Instead, the half-spindles remained disconnected from one another, as reported previously (Mollinari et al, 2002).
CENP-E has been reported to redistribute during anaphase from kinetochores to the central spindle and later to the midbody at telophase (Yen et al, 1991). In PRC1-deficient cells, CENP-E remained on kinetochores or chromosomes as well as diffusely throughout the cells at anaphase (Figure 6D, panel d).
Together, these results suggest that KIF4 and CENP-E require PRC1 to exit from chromosomes and bind to central spindle microtubules during anaphase B.
MKLP1 and Aurora B localization to half-spindles does not require PRC1
MKLP1 and all three chromosomal passenger proteins were concentrated in equatorial regions of PRC1-depleted anaphase cells (Figure 6E–H). Double staining of Aurora B and tubulin clearly demonstrated that Aurora B was localized near the plus ends of the microtubules in the disconnected half-spindles (Figure 7A). MKLP1 was also localized near the plus ends of the same microtubules (Figure 7B), but did not exactly overlap with Aurora B (Figure 7C). Instead, it occupied an extended region proximal to the plus ends.
Figure 7.
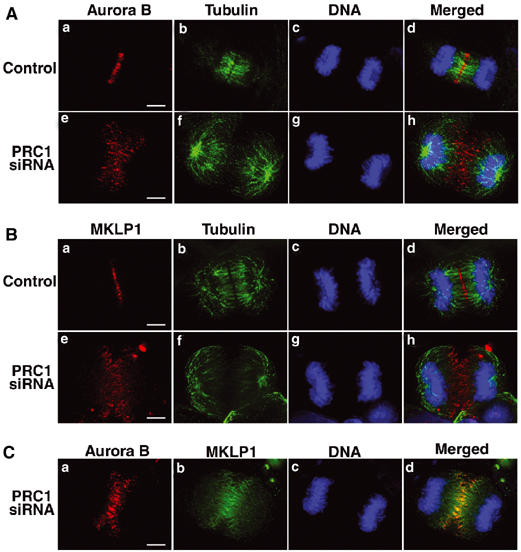
MKLP1 and Aurora B localize to the half-spindle of PRC1-deficient cells. (A) Staining of tubulin (green), Aurora B (red) and DNA (blue) in PRC1-deficient cells and control cells at anaphase. Scale bar, 5 μm. (B) Staining of tubulin (green), MKLP1 (red) and DNA (blue) in PRC1-deficient cells and control cells at anaphase. Scale bar, 5 μm. (C) Staining of MKLP1 (green), Aurora B (red) and DNA (blue) in PRC1-deficient cells at anaphase. Scale bar, 5 μm.
These distributions of MKLP1 and Aurora B in PRC1-depleted cells resemble what is seen for the peripheral microtubules in control cells early in anaphase B (Figure 3C). In those cells, MKLP1 and KIF4 concentrate in foci flanked by Aurora B in the compact central spindle; however at the periphery of the midzone, Aurora B can be seen near the plus ends of single microtubules surrounded by a more extensive domain of MKLP1 (Figure 3B and C). Apparently, MKLP1 only concentrates into foci as the microtubules are incorporated into the compact central spindle. These observations suggest that the phenotype of the PRC1 RNAi may correspond to an intermediate stage in the formation of a normal bipolar anaphase spindle, and that the targeting of Aurora B and MKLP1 on anaphase midzone microtubules does not require PRC1.
When anaphase cells were subjected to a brief treatment with colcemid, central spindles persisted but astral microtubules disappeared (Supplementary Figure 2). In these cells, KIF4 and CENP-E remained associated with chromosomes and/or kinetochores, respectively. In contrast, PRC1, MKLP1 and Aurora B were localized in the central spindle remnants. This experiment shows that KIF4 and CENP-E exhibit different requirements for targeting and/or binding to central spindles than do PRC1, MKLP1 and Aurora B. Binding of KIF4 and CENP-E to central spindles may require dynamic microtubules.
When a YFP-fused phosphorylation-null PRC1 mutant was overexpressed in HeLa cells, a number of bundled microtubules were detected in cells at mitosis (Supplementary Figure 3A) and interphase, as previously reported (Mollinari et al, 2002). This microtubule bundling was unaffected by depletion of KIF4 with siRNA (Supplementary Figure 3A and B). These data appear to suggest that KIF4 is not required for the aberrant microtubule bundling induced by dominant-negative PRC1, and therefore that this mutation can over-ride the normal KIF4-dependent regulation of PRC1 function.
PRC1 binds separately to the three motor proteins KIF4, MKLP1 and CENP-E in cell extracts
Because PRC1 was required for the relocalization of CENP-E from chromosomes to the central spindle during anaphase, we also checked for physical interactions between these two proteins, as well as for interactions between PRC1 and MKLP1 or a chromosomal passenger protein, Survivin. We analyzed interactions between the endogenous proteins by immunoprecipitation with anti-PRC1 followed by immunoblotting for the other proteins. This analysis revealed that PRC1 associates in extracts not only with KIF4 but also with MKLP1 and CENP-E, and not with Survivin (Figure 8). In complementary experiments, PRC1 was also detected in immunoprecipitates with anti-KIF4, anti-MKLP1 and anti-CENP-E, but not anti-Survivin (Figure 8, fifth panel). However, no interactions between the three motor proteins were detected by co-immunoprecipitation (Figure 8). These interactions are likely to be direct, as in vitro binding assays also confirmed that the amino-terminal half of PRC1 binds not only to KIF4 but also MKLP1 and CENP-E (Figure 1F). These data indicate that PRC1 binds to KIF4, MKLP1 and CENP-E during late mitosis; however, it apparently does not interact simultaneously with more than one of these motor proteins.
Figure 8.
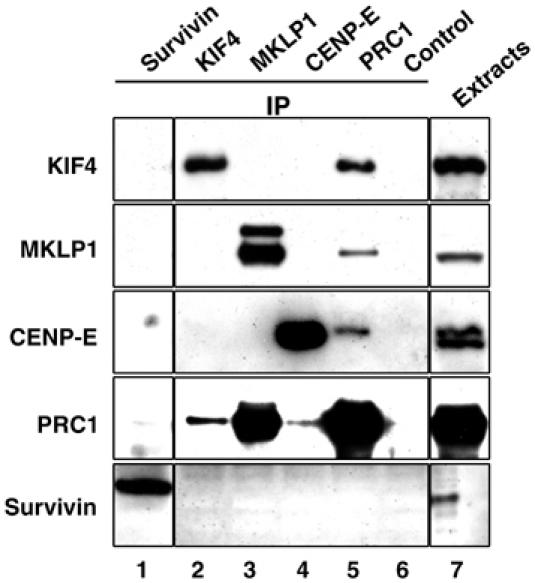
PRC1 binds separately to the three motor proteins KIF4, MKLP1 and CENP-E but not to Survivin. Immunoprecipitates of late mitotic HeLa cell extracts with anti-PRC1, anti-KIF4, anti-MKLP1, anti-CENP-E and anti-Survivin antibodies were probed with the same set of antibodies after SDS–PAGE. Preimmune serum (lane 6) was also used as control.
Discussion
In recent years, it has emerged that the central spindle has a key role in the localization and assembly of the cleavage furrow and in the completion of mitosis. There has thus been considerable interest in the identification of central spindle components, characterization of the interactions between them and determination of their respective roles in central spindle assembly. This has proven to be challenging, in part because the central spindle forms only during a very brief temporal window during anaphase, and also because many of the components of this structure are highly insoluble.
Here we have used biochemical fractionation to detect interactions between PRC1, a microtubule-binding protein of the central spindle, and three very different kinesin-related proteins, all of which are also concentrated in the central spindle during anaphase. We have found that endogenous PRC1 binds to chromokinesin KIF4, MKLP1/CHO1 and CENP-E in mitotic cell extracts. That these interactions are likely to be direct was confirmed by in vitro binding experiments using GST-PRC1 constructs. The interactions between PRC1 and the KIFs all involve the amino-terminal half of PRC1 and, in the case of KIF4, the carboxyl-terminal half of the protein. We were unable to detect tertiary or quaternary complexes of these proteins in lysates, but this could be either because such complexes do not form or because they are insoluble. Indeed, such complexes could constitute the stem body matrix, which is known to be an integral component of the central spindle (Buck and Tisdale, 1962).
Functional analysis by RNAi revealed that KIF4 and its binding partner PRC1 play important roles in the formation of the central spindle. Without PRC1, there is no central spindle, as the two half-spindles do not fuse to form an integrated structure (see also Mollinari et al, 2002). We found that PRC1 is required for two of its binding partners, KIF4 and CENP-E, to leave the chromosomes at anaphase, but not for the chromosomal passengers to do so. Importantly, Aurora B and MKLP1 concentrate near the spindle midline in the absence of PRC1 even though the half-spindles do not fuse. Thus, spindle polarity is still maintained and the midline determined in the absence of an integrated bipolar structure—otherwise Aurora B and MKLP1 would have been expected to adopt a more radial distribution around the aster. Our results together with those of Mollinari et al (2002) suggest that either PRC1 itself or a downstream activity has a crucial role in linking half-spindles together.
The distribution of Aurora B and MKLP1 near microtubule plus ends following PRC1 knockdown is indistinguishable from that seen on microtubules at the periphery of the cell during normal early anaphase prior to their incorporation into the organized central spindle. This suggests that the spindle morphology in PRC1-depleted cells may correspond to a transient state in the normal assembly of the anaphase spindle. As diagrammed in Figure 9, during the early stages of anaphase spindle reorganization, Aurora B becomes concentrated near the plus ends of the microtubules within a more extended zone of MKLP1 binding. In contrast, in the mature anaphase midzone and in the midbody, MKLP1 is concentrated at the midline, where it is flanked by Aurora B on either side. This suggests that as microtubules make lateral contact and begin to form bundles, MKLP1 becomes concentrated in a zone near the microtubule plus ends, where it becomes fixed as part of the stem body matrix. Either microtubule sliding or assembly could then produce the mature array, with Aurora B remaining near the plus ends and flanking the midline.
Figure 9.
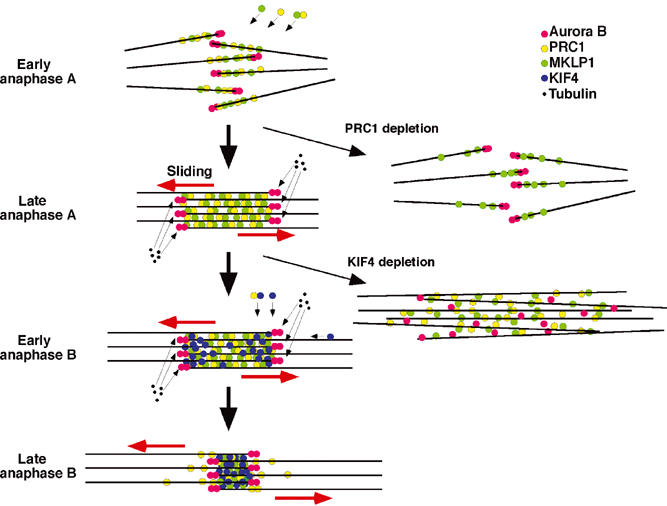
Model of midzone formation by KIF4 and PRC1. For details, see the text. The red arrows indicate the movement of the microtubules sliding toward the poles. Phenotypes observed following depletion of PRC1 or KIF4 are indicated.
Although this is the first description of Aurora B binding near microtubule plus ends in mammalian cells, it was reported previously that yeast Ipl1 localizes to microtubule plus ends (Buvelot et al, 2003). The mechanism of chromosomal passenger targeting to microtubule plus ends remains unknown. The differences in Aurora B and MKLP1 localization observed in normal and PRC1-depleted cells together with our inability to demonstrate any physical interactions between the chromosomal passengers and KIF4, MKLP1, CENP-E or PRC1 in cell lysates suggest that the latter proteins are not involved in this targeting. INCENP can bind directly to microtubules (Wheatley et al, 2001), although it has not been demonstrated previously to show any preference for plus ends. It will be important in the future to look for interactions between the chromosomal passengers and the +TIP proteins identified to date (Schuyler and Pellman, 2001).
In contrast to its binding partner PRC1, KIF4 is not required for integration of the two half-spindles. In cells depleted of KIF4, we find that the two half-spindles become linked to form a bipolar structure, but that microtubule bundling is subsequently defective. As a result, other midzone-associated proteins, although they target to the spindle midzone, remain dispersed along the unbundled microtubules and do not concentrate into a focused stem body matrix. Thus, although PRC1 is required for transfer of KIF4 and CENP-E to microtubules, KIF4 is then subsequently required for concentration of PRC1, CENP-E, MKLP1 and the chromosomal passengers into a stem body matrix. It is possible that KIF4 promotes stem body matrix formation through modification of the microtubule lattice, as has been observed in one recent study of the Xenopus homolog of KIF4 (Bringmann et al, 2004). In our studies we saw no evidence of spindle collapse or failure of chromosomes to interact with microtubules, as was seen in a previous study in which antibodies were used to inhibit the Xenopus homolog of KIF4 (Walczak et al, 1998). The eventual consequence of KIF4 depletion is that mitosis appears to be delayed, and cytokinesis frequently fails.
MKLP1 by itself can bundle microtubules, and depletion of CHO1, a splice variant of MKLP1, by siRNA yields phenotypes very similar to those observed here for KIF4 depletion (Matuliene and Kuriyama, 2002). Thus it has been proposed that one of the major functions of MKLP1/CHO1 is to organize the central spindle. However, our data reveal that even though it still binds to microtubules in PRC1- or KIF4-deficient cells, MKLP1 is not sufficient to interconnect and bundle the two half-spindles. It is possible that KIF4 and MKLP1 might act cooperatively in bundling the central spindle and organizing the midzone and midbody in the presence of PRC1, even though interactions between KIF4 and MKLP1 through PRC1 were not detected in our study.
Depletion of KIF4 by siRNA caused not only a disorganization of the central spindles but also an aberrant localization of the separated daughter nuclei, which migrated abnormally close to the plasma membrane at the poles of the telophase cell. The failure of stable interactions between polar microtubules and the cortex could cause this phenotype. KIF4 was localized near the microtubule plus ends in peripheral regions of mitotic cells, thereby raising the possibility that it might play a role in the regulation of microtubule plus-end dynamics, as has been shown for the Xenopus homolog of KIF4 in in vitro studies (Bringmann et al, 2004). A perturbation of microtubule dynamics in the spindle midzone during late anaphase could also explain the abnormal elongation of central spindles observed in KIF4-deficient cells.
Because KIF4 is a chromokinesin (Miki et al, 2001) that binds to the chromosome arms and Xenopus KIF4/Xklp1 had been shown to be required for chromatin association with the spindle (Walczak et al, 1998), we expected that the protein might have an essential role in alignment of the chromosomes on the metaphase plate during prometaphase. However, we did not observe any significant defects in chromosome biorientation and congression in KIF4-deficient cells. It was recently reported that another chromosome-associated kinesin, Drosophila Nod, an ortholog of mammalian Kid, plays a functionally redundant role with Drosophila Klp3A, an ortholog of mammalian KIF4, in prometaphase chromosome movement (Goshima and Vale, 2003). A similar functional redundancy in human cells could explain our results. Drosophila Klp3A mutants exhibited cytokinesis defects in testes (Williams et al, 1995), although inactivation of Klp3A by RNAi did not cause cytokinesis defects in the Drosophila S2 cell line (Goshima and Vale, 2003). Therefore, KIF4 and Kid may have redundant roles as chromokinesins, but the role of KIF4 as a central spindle kinesin at late mitosis could be unique.
In summary, the experiments shown here have revealed physical and functional interactions between PRC1 and KIF4 during formation of the central spindle in late mitotic cells. PRC1 is required for formation of an integrated bipolar spindle during anaphase. KIF4 is required for microtubule bundling and formation of an organized central spindle within this bipolar structure. PRC1 is required for KIF4 recruitment to the central spindle, but then KIF4 is required for PRC1 concentration at the midline. Although much remains to be learned, this study has provided insight into the complex events involved in the assembly of the stem body matrix and organization of the central spindle.
Materials and methods
Antibody preparation
The GST-fused carboxyl-terminal region of human KIF4 (amino acids 858–1232) cDNA in pGEX-4T (Amersham Pharmacia Biotech) was expressed in ER2566 with 0.1 mM isopropyl β-D-thiogalactoside. The amino-acid sequences of human KIF4 in this region have little homology to other KIF members. The GST-KIF4 was purified by eluting from glutathione-Sepharose (Amersham Pharmacia Biotech) with 20 mM glutathione, and used for immunization of rats. The full-length human PRC1 cDNA in pET-23d was expressed as a T7- and His-tagged protein, purified by His-Bind resin, and injected into a rabbit. The anti-PRC1-specific antibody was affinity-purified with recombinant PRC1 coupled to HiTrap NHS-activated Sepharose (Amersham Pharmacia Biotech). Rabbit anti-INCENP serum (1186) was prepared as described (Eckley et al, 1997).
Synchronization, immunoprecipitation and immunoblot analysis
HeLa cells were cultured with DMEM supplemented with 10% fetal calf serum and 2 mM glutamine. For synchronization to obtain the mitotic cells, cells were treated with 1 μg/ml aphidicolin (Wako) for 15 h, washed with PBS twice, incubated with fresh medium for 4 h, and then blocked with medium containing 12 ng/ml nocodazole (Calbiochem) for 7 h. To obtain late mitotic cells, mitotic cells were mechanically shaken off from the culture flasks and collected, and washed twice with PBS prewarmed at 37°C, and incubated in prewarmed fresh medium for 45 min, and then harvested. The cell extracts at late mitosis were subjected to immunoprecipitation and/or immunoblot analysis as described previously (Kurasawa and Todokoro, 1999).
Binding assay
Full-length or a series of truncated forms of human PRC1 cDNA inserted in pGEX-4T1 was expressed in ER2566 and the recombinant proteins were purified. The purified GST-fused proteins (5 μg) bound to glutathione-Sepharose beads were incubated with 0.6 mg precleared HeLa cell mitotic extracts for 2 h at 4°C. Beads were washed four times with lysis buffer and the bound proteins were subjected to immunoblot analysis.
Yeast two-hybrid analysis
For yeast two-hybrid screening, human PRC1 cDNA corresponding to the amino-terminal half region (nucleotides 1–909) was inserted into the vector pGBKT7, and was used as a bait to screen the HeLa cDNA library constructed in the vector pGADT7-Rec (Clontech) in AH109 reporter strain. Positive 52 clones were obtained from the screening of 4 × 105 clones, and the sequences of these clones were determined.
siRNA and transfection
Suppression of KIF4 or PRC1 by siRNA was performed by the modified method of Harborth et al (2001). For annealing, oligomers were heated at 90°C for 1 min, followed by gradual cooling to 65°C (0.5°C/s) and 37°C (0.05°C/s), incubation at 37°C for 60 min and cooling to 4°C. The targeted sequence for PRC1 was 5′-AAATATGGGAGCTAATTGGGA-3′ and that for KIF4 was 5′-AAGCAGATTGAAAGCCTAGAG-3′. The double-stranded oligomers or single-stranded sense oligomers (60 pmol) were transfected into HeLa cells on coverslips in a 24-well plate using Oligofectamine (Gibco-BRL), and fixed after 40–55 h culture. For immunoblotting, transfected cells were trypsinized, washed twice in ice-cold PBS and solubilized in SDS sample buffer. Protein from the transfected cells of half-well was analyzed by immunoblotting.
Immunofluorescence microscopy
HeLa cells on coverslips were washed with PHEM (60 mM PIPES, 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, pH 6.9 with KOH), fixed with 2% paraformaldehyde in PHEM at room temperature for 15 min, followed by 0.4% Triton X-100 in PBS at room temperature for 10 min, or fixed with methanol at –20°C for 4 min and 30 s. The KIF4 staining was performed by methanol fixation, but the cells had to be fixed with paraformaldehyde to see KIF4 and other proteins simultaneously, and under these conditions KIF4 staining was not optimal. Cells were then stained with primary antibodies for 1 h at 37°C. The dilutions of primary antibodies were as follows: rat anti-KIF4 antiserum, 1:100; purified rabbit anti-PRC1 antibody, 1:70; monoclonal mouse anti-α-tubulin antibody (Sigma) 1:150; rat anti-tubulin antibody (Harlan Sera-Lab) (YL1/2) 1:150; mouse anti-Aurora B (AIM-1) antibody (BD Biosciences), 1:150; rabbit anti-Survivin antibody (Santa Cruz Biotechnology), 1:100; rabbit anti-INCENP antiserum (1186), 1:800; rabbit anti-MKLP1 antibody (N-19) (Santa Cruz Biotechnology), 1:100; and goat anti-CENP-E antibody (C-19) (Santa Cruz Biotechnology), 1:150. After rinsing with PBS, cells were incubated with secondary antibodies for 1 h at 37°C. The secondary antibodies used were as follows: Cy3-conjugated anti-rabbit, mouse or goat antibody (Jackson Immuno Research Laboratories), Cy5-conjugated anti-rat antibody (Jackson Immuno Research Laboratories), Alexa Fluor 488-conjugated goat anti-rat or rabbit IgG (Molecular Probes) and Alexa Fluor 568-conjugated goat anti-mouse IgG (Molecular Probes). The slides were washed with PBS and mounted in 90% glycerol/PBS containing 0.1% p-phenylenediamine and 200 ng/ml 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Sigma). Cells were observed on a Delta Vision system based around an Olympus IX-70 inverted microscope, and operated by the SoftWorx system. A total of 14 image planes were captured for each cell, deconvolved on a dual processor SGI Octane computer, and projected onto a single plane using the maximum intensity algorithm.
Supplementary Material
Supplementary Figure 1.
Supplementary Figure 2.
Supplementary Figure 3.
Acknowledgments
We thank Mr Tomokazu Maruno and Dr Toshinari Onogi for technical assistance. This work was supported by a grant from the Ministry of Education, Science, Sports and Culture of Japan and by a grant of Uehara Memorial Foundation. Work in the WCE laboratory is supported by the Wellcome Trust, of which he is a Principal Research Fellow.
References
- Andreassen PR, Palmer DK, Wener MH, Margolis RL (1991) Telophase disk: a new mammalian mitotic organelle that bisects telophase cells with a possible function in cytokinesis. J Cell Sci 99: 523–534 [DOI] [PubMed] [Google Scholar]
- Antonio C, Ferby I, Wilhelm H, Jones M, Karsenti E, Nebreda AR, Vernos I (2000) Xkid, a chromokinesin required for chromosome alignment on the metaphase plate. Cell 102: 425–435 [DOI] [PubMed] [Google Scholar]
- Bringmann H, Skiniotis G, Spilker A, Kandels-Lewis S, Vernos I, Surrey T (2004) A kinesin-like motor inhibits microtubule dynamic instability. Science 303: 1519–1522 [DOI] [PubMed] [Google Scholar]
- Buck R, Tisdale JM (1962) An electron microscopic study of the development of the cleavage furrow in mammalian cells. J Cell Biol 13: 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvelot S, Tatsutani SY, Vermaak D, Biggins S (2003) The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J Cell Biol 160: 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao LG, Wang YL (1996) Signals from the spindle midzone are required for the stimulation of cytokinesis in cultured epithelial cells. Mol Biol Cell 7: 225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmena M, Riparbelli MG, Minestrini G, Tavares AM, Adams R, Callaini G, Glover DM (1998) Drosophila polo kinase is required for cytokinesis. J Cell Biol 143: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke CA, Heck MM, Earnshaw WC (1987) The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J Cell Biol 105: 2053–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckley DM, Ainsztein AM, Mackay AM, Goldberg IG, Earnshaw WC (1997) Chromosomal proteins and cytokinesis: patterns of cleavage furrow formation and inner centromere protein positioning in mitotic heterokaryons and mid-anaphase cells. J Cell Biol 136: 1169–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funabiki H, Murray AW (2000) The Xenopus chromokinesin Xkid is essential for metaphase chromosome alignment and must be degraded to allow anaphase chromosome movement. Cell 18: 411–424 [DOI] [PubMed] [Google Scholar]
- Gassmann R, Carvalho A, Henzing AJ, Ruchaud S, Hudson DF, Honda R, Nigg EA, Gerloff DL, Earnshaw WC (2004) Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J Cell Biol (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giansanti MG, Bonaccorsi S, Williams B, Williams EV, Santolamazza C, Goldberg ML, Gatti M (1998) Cooperative interactions between the central spindle and the contractile ring during Drosophila cytokinesis. Genes Dev 12: 396–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M (1997) The mechanism and control of cytokinesis. Curr Opin Cell Biol 9: 815–823 [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD (2003) The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol 162: 1003–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K (2001) Identification of essential genes in cultured mammalian cells using small interfering RNAs. J Cell Sci 114: 4557–4565 [DOI] [PubMed] [Google Scholar]
- Hill E, Clarke M, Barr FA (2000) The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J 19: 5711–5719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N (1998) Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science 279: 519–526 [DOI] [PubMed] [Google Scholar]
- Jantsch-Plunger V, Gonczy P, Romano A, Schnabel H, Hamill D, Schnabel R, Hyman AA, Glotzer M (2000) CYK-4: a Rho family GTPase activating protein (GAP) required for central spindle formation and cytokinesis. J Cell Biol 149: 1391–1404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Jimenez G, Wells NJ, Hope TJ, Wahl GM, Hunter T, Fukunaga R (1998) PRC1: a human mitotic spindle-associated CDK substrate protein required for cytokinesis. Mol Cell 2: 877–885 [DOI] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D (1997) APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science 275: 1311–1314 [DOI] [PubMed] [Google Scholar]
- Kurasawa Y, Todokoro K (1999) Identification of human APC10/Doc1 as a subunit of anaphase promoting complex. Oncogene 18: 5131–5137 [DOI] [PubMed] [Google Scholar]
- Kwon M, Morales-Mulia S, Brust-Mascher I, Rogers GC, Sharp DJ, Scholey JM (2004) The chromokinesin, KLP3A, drives mitotic spindle pole separation during prometaphase and anaphase, and facilitates chromatid motility. Mol Biol Cell 15: 219–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levesque AA, Compton DA (2001) The chromokinesin Kid is necessary for chromosome arm orientation and oscillation, but not congression, on mitotic spindles. J Cell Biol 154: 1135–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronarde DN, McDonald KL, Ding R, McIntosh JR (1993) Interpolar spindle microtubules in PTK cells. J Cell Biol 123: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuliene J, Kuriyama R (2002) Kinesin-like protein CHO1 is required for the formation of midbody matrix and the completion of cytokinesis in mammalian cells. Mol Biol Cell 13: 1832–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki H, Setou M, Kaneshiro K, Hirokawa N (2001) All kinesin superfamily protein, KIF, genes in mouse and human. Proc Natl Acad Sci USA 98: 7004–7011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishima M, Kaitna S, Glotzer M (2002) Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell 2: 41–54 [DOI] [PubMed] [Google Scholar]
- Mollinari C, Kleman JP, Jiang W, Schoehn G, Hunter T, Margolis RL (2002) PRC1 is a microtubule binding and bundling protein essential to maintain the mitotic spindle midzone. J Cell Biol 157: 1175–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinari C, Reynaud C, Martineau-Thuillier S, Monier S, Kieffer S, Garin J, Andreassen PR, Boulet A, Goud B, Kleman JP, Margolis RL (2003) The mammalian passenger protein TD-60 is an RCC1 family member with an essential role in prometaphase to metaphase progression. Dev Cell 5: 295–307 [DOI] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH (1995) Interactions between the nod+ kinesin-likegene and extracentromeric sequences are required for transmission of a Drosophila minichromosome. Cell 81: 139–148 [DOI] [PubMed] [Google Scholar]
- Neef R, Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, Barr FA (2003) Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol 162: 863–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR (1992) A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature 359: 543–547 [DOI] [PubMed] [Google Scholar]
- Pellman D, Bagget M, Tu YH, Fink GR, Tu H (1995) Two microtubule-associated proteins required for anaphase spindle movement in Saccharomyces cerevisiae. J Cell Biol 130: 1373–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokopenko SN, Brumby A, O'Keefe L, Prior L, He Y, Saint R, Bellen HJ (1999) A putative exchange factor for Rho1 GTPase is required for initiation of cytokinesis in Drosophila. Genes Dev 13: 2301–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson DN, Spudich JA (2000) Towards a molecular understanding of cytokinesis. Trends Cell Biol 10: 228–237 [DOI] [PubMed] [Google Scholar]
- Schliwa M, Woehlke G (2003) Molecular motors. Nature 422: 759–765 [DOI] [PubMed] [Google Scholar]
- Schuyler SC, Liu JY, Pellman D (2003) The molecular function of Ase1p: evidence for a MAP-dependent midzone-specific spindle matrix. Microtubule-associated proteins. J Cell Biol 160: 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuyler SC, Pellman D (2001) Microtubule ‘plus-end-tracking proteins': the end is just the beginning. Cell 105: 421–424 [DOI] [PubMed] [Google Scholar]
- Sekine Y, Okada Y, Noda Y, Kondo S, Aizawa H, Takemura R, Hirokawa N (1994) A novel microtubule-based motor protein (KIF4) for organelle transports, whose expression is regulated developmentally. J Cell Biol 127: 187–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellitto C, Kuriyama R (1988) Distribution of a matrix component of the midbody during the cell cycle in Chinese hamster ovary cells. J Cell Biol 106: 431–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias DA, Mollinari C, Lacroix FB, Margolis RL (2000) Human survivin is a kinetochore-associated passenger protein. J Cell Biol 151: 1575–1582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T (1999) Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol 147: 921–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terada Y, Tatsuka M, Suzuki F, Yasuda Y, Fujita S, Otsu M (1998) AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J 17: 667–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren AG, Wong L, Pakusch M, Fowler KJ, Burrows FJ, Vaux DL, Choo KH (2000) Survivin and the inner centromere protein INCENP show similar cell-cycle localization and gene knockout phenotype. Curr Biol 10: 1319–1328 [DOI] [PubMed] [Google Scholar]
- Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C (1995) Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell 81: 117–127 [DOI] [PubMed] [Google Scholar]
- Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R (1998) A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol 8: 903–913 [DOI] [PubMed] [Google Scholar]
- Wang SZ, Adler R (1995) Chromokinesin: a DNA-binding, kinesin-like nuclear protein. J Cell Biol 128: 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley SP, Kandels-Lewis SE, Adams RR, Ainsztein AM, Earnshaw WC (2001) INCENP binds directly to tubulin and requires dynamic microtubules to target to the cleavage furrow. Exp Cell Res 262: 122–127 [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Wang Y (1996) Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol 135: 981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Riedy MF, Williams EV, Gatti M, Goldberg ML (1995) The Drosophila kinesin-like protein KLP3A is a midbody component required for central spindle assembly and initiation of cytokinesis. J Cell Biol 129: 709–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW (1991) CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO J 10: 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1.
Supplementary Figure 2.
Supplementary Figure 3.


