Abstract
SRY, a Y chromosome-encoded DNA-binding protein, is required for testis organogenesis in mammals. Expression of the SRY gene in the genital ridge is followed by diverse early cell events leading to Sertoli cell determination/differentiation and subsequent sex cord formation. Little is known about SRY regulation and its mode of action during testis development, and direct gene targets for SRY are still lacking. In this study, we demonstrate that interaction of the human SRY with histone acetyltransferase p300 induces the acetylation of SRY both in vitro and in vivo at a single conserved lysine residue. We show that acetylation participates in the nuclear localisation of SRY by increasing SRY interaction with importin β, while specific deacetylation by HDAC3 induces a cytoplasmic delocalisation of SRY. Finally, by analysing p300 and HDAC3 expression profiles during both human or mouse gonadal development, we suggest that acetylation and deacetylation of SRY may be important mechanisms for regulating SRY activity during mammalian sex determination.
Keywords: acetylation, mammals, nuclear localisation, sex determination, SRY
Introduction
In mammals, testis determination results from a cascade of genetic events that are controlled by the master regulatory Y-chromosomal gene SRY (Berta et al, 1990). Sry expression in the somatic cell precursors of the undifferentiated bipotential genital ridge directs Sertoli cell differentiation leading to testicular cord formation and subsequent male development (Koopman et al, 1990; Albrecht and Eicher, 2001). SRY (sex-determining region of the Y chromosome) gene codes for a protein containing a centrally located 79-amino-acid ‘high-mobility group' (HMG) domain (HMG box). By this HMG box, SRY binds to DNA minor groove via A-T-rich sequences (Harley et al, 1992), inducing sharp angle to DNA (Ferrari et al, 1992). Mutations within the HMG box of the SRY gene from XY sex reserval patients affect either its DNA-binding (Hawkins et al, 1992), DNA bending (Pontiggia et al, 1994) or nuclear localisation activity (Li et al, 2001). In mouse SRY, the glutamine-rich motif present in the C-terminal part of the protein is crucial for its activity (Bowles et al, 1999), whereas no function has been assigned to the regions of the human SRY protein outside the HMG box, except an interaction with a PDZ domain protein SIP-1 (Poulat et al, 1997). Currently, little is known about the mechanism by which SRY mediates testis determination. The earliest identified effects of Sry expression are an increase in cell proliferation in the male coelomic epithelium (Schmahl et al, 2000) along with a cell migration from the mesonephros to the differentiating gonad (Tilmann and Capel, 1999).
These observations suggest that SRY, like other HMG box proteins, may regulate gene expression through architectural effects that convert promoter regions to a higher order of structure allowing the assembly of regulatory complexes similar to that reported for LEF-1 (Eastman and Grosschedl, 1999). Without any intrinsic ability to modulate transcription, SRY (as for other SOX proteins; Bowles et al, 2000) can be viewed as a chromosomal docking site for auxiliary proteins. Target gene specificity would then be the result of SRY–protein interactions in addition to protein–DNA interactions. Furthermore, the activity of the resulting combinatorial transcription complex could be modulated by post-translational modifications of different partners in each situation. Phosphorylation by the cyclic AMP-dependent protein kinase has been previously described to modulate the DNA-binding ability of human SRY protein (Desclozeaux et al, 1998). However, acetylation of that factor still remains to be investigated.
The reversible acetylation of histones and also of numerous transcription factors is widely recognised as a common way to regulate gene activity, and numerous nuclear histone acetyltransferases (HATs) have so far been identified (Narlikar et al, 2002). Among them, adenovirus E1A-associated p300 and the closely related CREB-binding protein (CBP) can form larger protein complexes including other acetylases and serve as coactivators for transcription factors, linking them to the basal transcription machinery (Torchia et al, 1997; Chen et al, 1999). Numerous studies have shown that CBP/p300 regulates transcription factor activities via direct interaction and acetylation process (Ogryzko et al, 1996; Bannister et al, 2000; Goodman and Smolik, 2000; Sterner and Berger, 2000; Chen et al, 2001). On the other hand, histone deacetylase (HDAC) activities that reverse the acetylation process are known to repress transcription (Wolffe, 1996). An increasing number of HDACs are now characterised in higher eukaryotes and grouped into class I (HDACs 1–3) and class II (HDACs 4–8). The repression of the activity of several transcription factors such as Sp1 (Doetzlhofer et al, 1999), nuclear receptors NcoR and SMRT (Nagy et al, 1997), and GATA-2 (Ozawa et al, 2001) has been ascribed to direct interaction with HDACs. LEF1, another HMG protein, acts as a repressor in the absence of Wnt signalling by recruiting HDAC1 activity to DNA (Billin et al, 2000). These data, together with others (Gwack et al, 2001; Yao et al, 2001), suggest that HAT and HDAC activities regulate the equilibrium between repression and activation of several transcription factors.
In this report, we have demonstrated that human sex-determining factor SRY interacts with the p300 acetylase both in vitro and in vivo. This interaction leads to SRY acetylation on one lysine residue (K136) that modifies SRY nuclear sublocalisation. We also identified that HDAC3 associated with and then deacetylated SRY. Expression of p300 and HDAC3 in somatic cells of human and mouse genital ridge at the time of SRY expression may involve these proteins in the regulation of SRY activity during mammalian sex determination.
Results
SRY and p300 can associate in cells and in vitro
To determine whether SRY was complexed with p300 in cells, p300 was immunoprecipitated from NT2/D1 cell extracts, a cell line known to express SRY protein (Poulat et al, 1995). Immune complexes were subsequently resolved by SDS–PAGE analysis followed by Western blotting. Figure 1A shows that the p300-precipitated complex contained detectable levels of SRY, thus demonstrating the in vivo interaction between both proteins. In a reverse experiment, co-precipitation of p300 using the anti-SRY polyclonal rat antibody was also observed (Figure 1A).
Figure 1.

SRY interacts with p300. (A) NT2/D1 cell extracts were used for IP with anti-SRY (SRY) or anti-p300 (p300) antibodies and preimmune antisera (preI). Immunoprecipitated proteins were resolved by SDS–PAGE and analysed by Western blotting with α-SRY and α-p300 antibodies as shown. (B) Schematic representation of SRY fragments, which were amplified by PCR and cloned into pGEX vectors. The 10 lysines comprised in the HMG box are indicated; numbers refer to the position of the lysines within the SRY primary sequence. (C) p300 interacts directly with the SRY HMG domain in GST pull-down assays using purified GST-SRY fusion proteins and in vitro-translated 35S-labelled p300 protein. Similar GST-SRY protein amounts were used, and interaction between SRY and p300 was analysed by SDS–PAGE of the pull-down reactions and autoradiography. The input contains 10% of the p300 protein. (D) To confirm the direct interaction of SRY and p300, the same GST pull-down experiments were carried out using 50 and 150 ng of purified p300. Interaction was visualised by Western blot with anti-p300 antibody (0.5 μg/ml).
To assess that this interaction was direct and to define the region of SRY that interacts with p300, different fragments of SRY (Figure 1B) were expressed as recombinant glutathione S-transferase (GST) fusion proteins and incubated with in vitro-translated 35S-labelled p300. As wild-type (wt) SRY, SRY HMG box was shown to bind p300 protein, whereas no interaction was seen with the Nter and Cter parts of SRY protein (Figure 1C). Incubation with the region spanning amino acids 107–139 of the HMG box (box107/139 construct) was shown to be sufficient to allow interaction with p300 (Figure 1C). In contrast, no interaction was observed with box100/133 fragment, delineating the SRY/p300 interaction region on the last six amino acids of the SRY HMG box. Direct interaction between GST-SRY and p300 was confirmed using purified p300 in GST pull-down experiments (Figure 1D), while no interaction was detected with GST protein (data not shown).
Acetylation of SRY by p300 in vitro
Since SRY could form physical complexes with p300, we determined whether p300 could acetylate SRY in vitro. To address this question, purified human SRY protein was incubated with purified p300 or HAT-inactive p300 (p300 ΔHAT) proteins in the presence of [14C]acetyl CoA. Figure 2A demonstrates that p300 but not p300 ΔHAT was able to acetylate SRY in vitro. Autoacetylated p300 indicates that HAT protein was enzymatically active. Using the GST fusion proteins box, Nter and Cter as substrates (Figure 1B), we mapped SRY acetylation site on SRY HMG box that contains most of the target lysine residues (Figure 2A). Using derived truncated box fragments (Figure 1B), we narrowed down the acetylated target to the five putative lysines (K115, K123, K128, K134 and K136) on SRY box107/139 fragment (data not shown). A 35-amino-acid-derived peptide (P35) including these multiple sites and located at the C-terminus of the SRY HMG box was synthesised and subjected before and after p300 acetylation to mass spectrometry. Figure 2B shows that the mock-acetylated P35 peptide revealed a molecular weight of 4712 shifting to 4754 after acetylation with p300, a shift corresponding to one additional acetyl group. Additional peaks of mass 4696 and 4738 correspond to oxidative forms of mock-acetylated and acetylated peptides, respectively. Our results strongly suggest that one lysine residue located between residues 107 and 139 within the SRY HMG box undergoes acetylation by p300.
Figure 2.
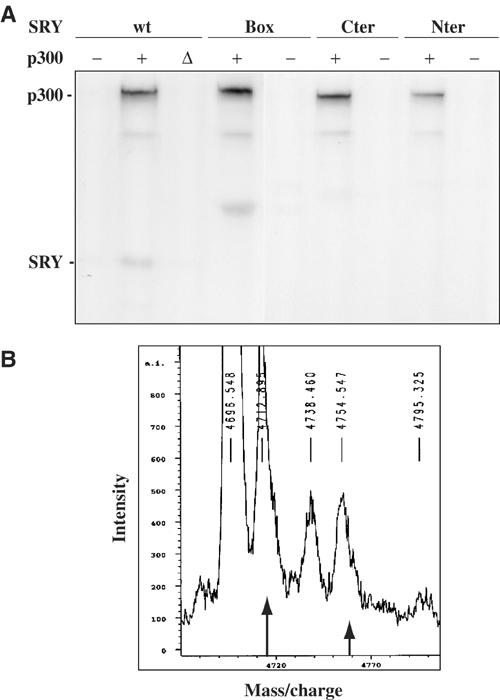
p300 acetyltransferase acetylates SRY in vitro. (A) Purified human SRY (wt) and GST-SRY fusion proteins (box, Cter, Nter) were in vitro acetylated after incubation of SRY proteins (500 μg) with 14C-acetylCoenzymeA in the absence of p300 (−) and presence of 50 ng of recombinant p300 (+) or HAT-inactive p300 (Δ). Reaction products were separated by SDS–PAGE and visualised by autoradiography. Autoacetylated p300 and acetylated SRY are indicated. (B) The synthetic peptide P35 corresponding to SRY (amino acids 107–139) was in vitro acetylated using p300 and submitted to mass spectra. The unmodified peptide (long arrow) has an Mr of 4712, and the peak of Mr 4754 corresponds to a monoacetylated peptide (short arrow). Other peaks correspond to oxidative forms of acetylated and unacetylated peptides.
Acetylation of SRY by p300 in vivo
We next examined whether SRY might be covalently modified in vivo by acetylation. HeLa cells that express HA-tagged SRY proteins were labelled for 1 h with [3H]sodium acetate or [35S]methionine. The cells were then lysed and SRY was detected using anti-HA immunoprecipitation (IP). The specific in vivo acetylation signal observed (Figure 3A, panel 3H, wt) was then used to delineate which lysine from the human SRY primary sequence forms the target of this post-translational modification. Systematic point mutations of each lysine codon into arginine (K115, K123, K128, K134 and K136) were produced. The corresponding SRY proteins were expressed at similar levels in HeLa cells, as judged by using [35S]Met labelling (Figure 3A, panel 35S). All of them, except SRY K136R (Figure 3A, panel 3H), were still labelled by [3H]sodium acetate, suggesting that the lysine residue at position 136 in the human SRY sequence was the substrate for acetylation. This motif located close to the second nuclear localisation signal (NLS) of the HMG domain is conserved in mouse Sry protein (K81). To confirm that p300 itself was able to in vivo acetylate human (h) and mouse (m) SRY proteins on their respective K136 and K81 sites, HeLa cells were transfected with p300 cDNA together with wt and mutant SRY cDNAs. Figure 3B shows that hSRY and mSry acetylation was increased in cotransfection experiments with p300 cDNA compared to the acetylation level observed with endogenous p300 protein (Figure 3B, panel 3H), whereas no signal was detected with K136 and K81 mutants. These results indicated that indeed p300 is able to in vivo acetylate hSRY and mSry, respectively, on their K136 and K81 sites. The lack of K136 mutant acetylation was not due to modified interaction with p300 prior to the enzymatic reaction. K136R SRY mutant still interacts with p300 with the same efficiency as wt SRY in GST pull-down experiments (Figure 3C).
Figure 3.
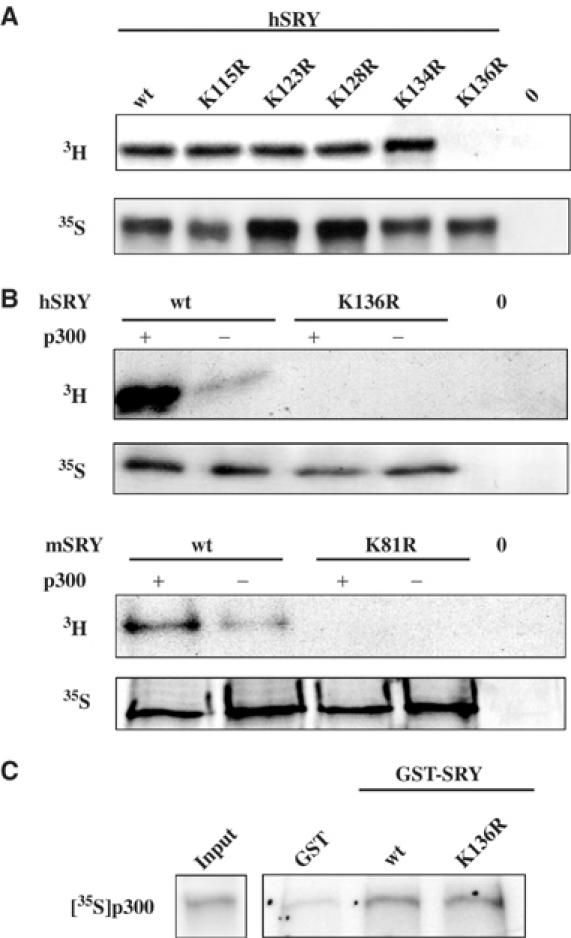
SRY is acetylated in vivo by p300 on the lysine residue K136. (A) HeLa cells were transfected with empty vector (0), HA SRY (wt) and HA SRY mutant (K115R, K123R, K128R, K134R and K136R) expression vectors. SRY proteins were in vivo labelled by [3H]acetate (panel 3H) or [35S]Met (panel 35S) and cell extracts were submitted to IP with anti-HA monoclonal antibody. Immunoprecipitated proteins were analysed by SDS–PAGE and autoradiography. (B) p300 was identified as the acetyltransferase that acetylates SRY in vivo in HeLa cells. After cotransfection of human HA-SRY (h), mouse HA-Sry (m) and respective mutant K136R and K81R expression vectors with (+) or without (−) p300 expression vector, cells were in vivo labelled by [3H]acetate (panel 3H) or by [35S]Met (panel 35S). Panel 3H indicates that hSRY and mSry were in vivo acetylated by p300 and panel 35S verifies expression of SRY proteins. (C) hSRY acetylation mutant K136R still interacted with p300. GST pull-down experiments were performed using GST fusion proteins SRY (wt) and SRY (K136R) or GST as a control, with in vitro-translated 35S-labelled p300 protein. Proteins were analysed by SDS–PAGE and the gel was subjected to autoradiography. Input represents 10% of the p300 protein used in the experiments.
Interaction with p300 increases SRY DNA-binding activity
To assess the functional consequences of interaction with p300 and acetylation on SRY DNA-binding activity, we next compared the DNA-binding capacity of wt and K136R SRY proteins using band-shift experiments. Recombinant GST-SRY fusion proteins were in vitro incubated in the presence or absence of p300 and/or acetyl CoA, and submitted to binding reaction with labelled TCF probe (Harley et al, 1992). Binding of K136 SRY protein was similar to that of wt SRY in the absence of p300, indicating that K136R mutation kept the SRY DNA-binding activity intact (Figure 4). However, p300 in the presence or absence of acetyl CoA increased DNA-binding activity of both proteins by approximately two-fold (Figure 4). Reactions without acetyl CoA indicated that the sole interaction of p300 with wt and K136R SRY was able to increase their binding affinities (Figure 4). Both complexes observed were supershifted with SRY antibody (data not shown) indicating that lower complex corresponds to degraded GST-SRY protein during the purification process. These results suggested that independent of acetylation the interaction with p300 modulates the affinity of SRY for its DNA target site.
Figure 4.
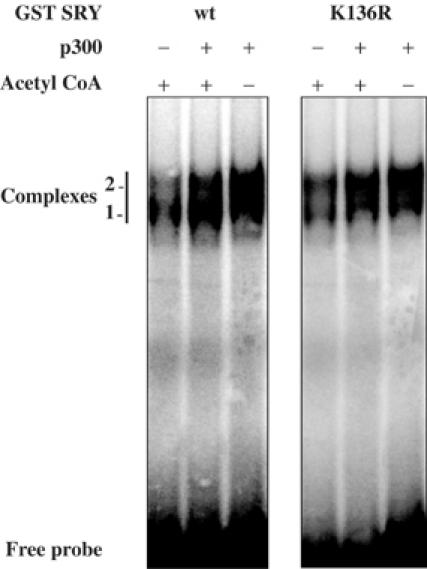
Interaction with p300 modifies SRY DNA-binding activity. The GST/SRY (wt) and (K136R) fusion proteins were bacterially expressed and purified. Equivalent amounts of SRY proteins were incubated in in vitro acetylation reaction in the presence (+) or absence (−) of p300 and acetyl CoA and submitted to binding reaction in EMSA experiments with 32P-labelled TCF probe. Reaction products were resolved by electrophoresis and visualised by autoradiography. Specific SRY complexes are indicated.
Acetylation of SRY contributes to its nuclear localisation
Nuclear localisation of SRY has been well described in different cell lines (Poulat et al, 1995; Sudbeck and Scherer, 1997). Two NLSs at the N- and C-termini of the HMG domain have been identified within the human SRY protein (Poulat et al, 1995; Sudbeck and Scherer, 1997). However, in the 8-week human male embryonic gonad, SRY displayed a cytoplasmic localisation in the somatic cell compartment (Figure 5A). The implication of acetylation on SRY subcellular localisation was thus analysed in vivo. NT2/D1 cells endogenously expressing SRY were treated with trichostatin A (TSA), a potent inhibitor of different HDAC activities (Yoshida et al, 1995), and immunostained with an SRY antibody. Whereas NT2/D1 cells showed an SRY labelling in both nuclear (punctuated) and cytoplasmic (perinuclear patch) cell compartments (Figure 5B), TSA treatment induced an exclusive SRY nuclear staining and the perinuclear SRY patch was abolished (Figure 5B). These data indicated that acetylation of SRY might directly or indirectly contribute to nuclear localisation of SRY. As a control, implication of SRY acetylation on its subcellular localisation was analysed in transfected HeLa cells. In this case, wt SRY protein was exclusively nuclear, whereas K136R SRY mutant showed a partial cytoplasmic localisation (Figure 6A). To confirm that physiological levels of p300 contributes to SRY nuclear localisation, small interfering RNA (siRNA) experiments were performed using p300 siRNA already described (Bres et al, 2002). Since no efficient p300 ‘knock-down' has been measured in NT2/D1 cells using these siRNAs, experiments were performed in nuclear SRY-expressing HeLa cell lines that contained an integrated hSRY gene (HeLa B3 clone) (Poulat et al, 1995). As shown in Figure 6B, transfection of p300 siRNA led to a dramatic decrease in p300 expression. In these cells, SRY subcellular localisation was modified and a perinuclear SRY patch was seen, similar to that observed in NT2/D1 cells endogenously expressing SRY (Figure 5B). Interestingly, no perinuclear patch was observed in HeLa cells transiently cotransfected with HDAC3 and SRY, suggesting that this particular structure could only occur in ‘long-term' SRY-expressing cells. Altogether, these data indicated that acetylation via p300 greatly contributed to nuclear localisation of SRY.
Figure 5.
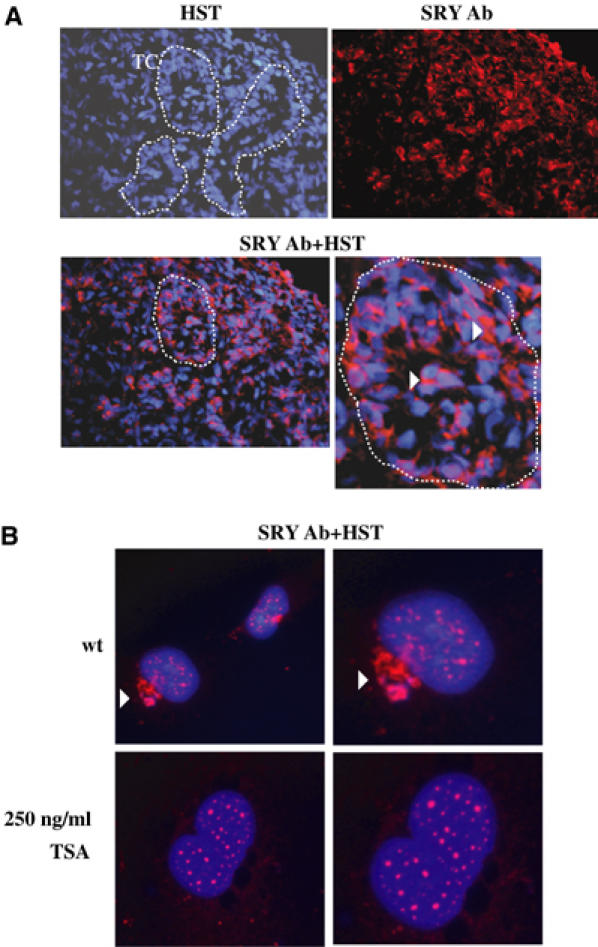
Acetylation of SRY contributes to its nuclear localisation. (A) Sections of male human embryonic gonads at 8 weeks stage were submitted to immunostaining with anti-SRY antibody (SRY Ab) (Poulat et al, 1995) and nuclear staining was accomplished with Hoescht dye (HST) (magnification × 40). Testicular cords (TCs) are delimited by broken lines. One of these is zoomed and cytoplasmic SRY is indicated by an arrowhead. (B) NT2/D1 cell line endogenously expressing SRY was cultured on glass coverslips and treated for 18 h with 250 ng/ml TSA. Cells were fixed and SRY protein was detected by immunostaining with anti-SRY Ab. Nuclei were visualised by Hoescht dye staining (HST). The right panels represent zoomed nuclei from the left panels. An arrowhead indicates the SRY perinuclear patch that is inhibited after TSA treatment. Cells were visualised at magnification × 60.
Figure 6.

(A) K136 mutation induces a partial cytoplasmic localisation. HeLa cells were transfected with HA SRY (wt SRY) or HA SRY K136 (K136R SRY) expression vectors and fixed. SRY localisation was analysed by anti-HA antibody (HA Ab) immunostaining. Nuclei were visualised by HST. (B) Endogenous p300 is required for SRY nuclear localisation in HeLa B3 cells. Stably SRY-transfected HeLa cells (clone B3) were transfected with p300 siRNA and were colabelled with SRY and p300 antibodies. Cells transfected by p300 siRNA (indicated by an arrow) showed a modified SRY subcellular localisation. Cells were visualised at magnification × 60. (C) Purified SRY (100 ng) was incubated in in vitro acetylation reaction in the presence (+) or absence (−) of p300 and submitted to GST pull-down experiments with GST (T) and GST-importin β (Impβ). Interaction between SRY and importin β was analysed by SDS–PAGE of the pull-down reactions and Western blot with anti-SRY antibody. (D) The same GST pull-down experiments were performed using GST (T) and GST- importin β (impβ) fusion proteins and wt and K136R SRY-transfected HeLa cell extracts. Interaction between importin β and SRY proteins was visualised by Western blot with anti-SRY antibody.
Lysine K136 was located close to the second NLS, which has been implicated in SRY nuclear import via direct interaction with importin β (Forwood et al, 2001). Here, we demonstrated that in vitro acetylation of SRY prior to the interaction reaction with importin β increased SRY–importin β interaction three-fold (Figure 6C). Moreover, GST pull-down performed with recombinant GST-importin β- and wt or K136R SRY-transfected HeLa cell extracts showed that lysine K136 was essential for SRY interaction with importin β (Figure 6D). These results indicated that SRY acetylation on lysine K136 next to the second SRY NLS was involved in SRY nuclear import via importin β.
HDAC3 associates and deacetylates SRY
Sensitivity of SRY subcellular localisation to the deacetylase inhibitor TSA in NT2/D1 cells led us to hypothesise that SRY acetylation state was the result of an equilibrium between p300 acetylation and deacetylation by HDACs. To establish whether a known deacetylase can deacetylate SRY, in vivo acetylation experiments were performed in HeLa cells. HA-tagged SRY expression vector was cotransfected with Flag-tagged HDAC (1–6) expression vectors, cells were in vivo labelled with [3H]sodium acetate and anti-HA IP evaluated the acetylated SRY signal. This signal was specifically inhibited by cotransfection with HDAC3 expression vector (Figure 7A, panel 3H), whereas SRY was still acetylated in the presence of HDAC1, 2 (Figure 7A, panel 3H), 4, 5 and 6 (data not shown) expression, indicating that SRY can be specifically deacetylated by HDAC3.
Figure 7.
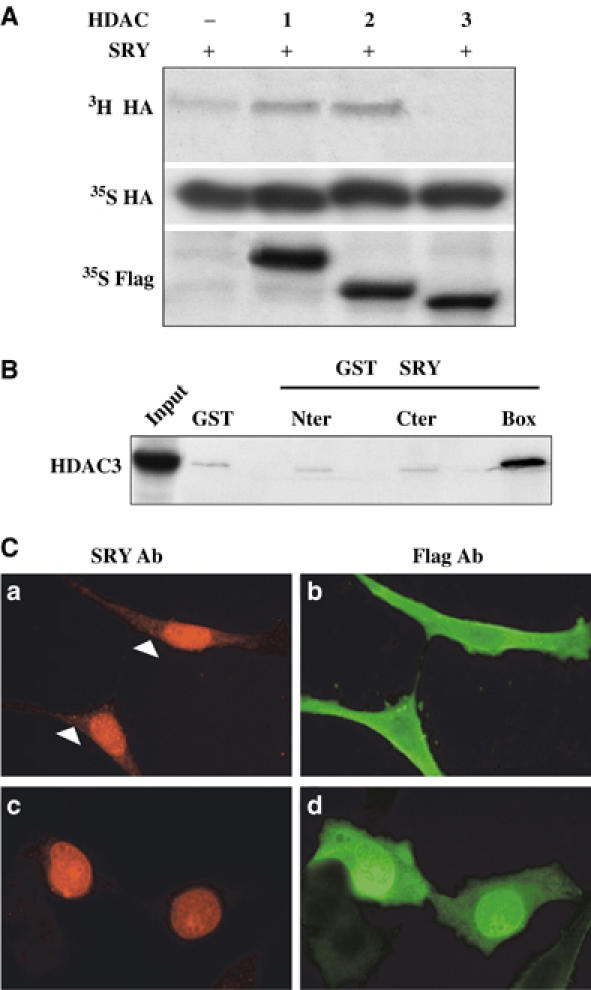
HDAC3 interacts with SRY and modulates its subcellular localisation. (A) HDAC3 deacetylated SRY in vivo in HeLa cells. Cells were transfected with HA SRY expression vector and Flag-tagged HDAC1–3 expression vectors and in vivo labelled by [3H]acetate or [35S]Met. 3H cell extracts were submitted to anti-HA IP to evaluate SRY acetylation state in the absence (−) or presence of HDAC1, 2 or 3 (panel 3H HA). Presence of SRY expression or HDAC1–3 expression in cell lysates was assessed by 35S labelling and anti-HA or anti-Flag IP, respectively (panels 35S HA and 35S Flag). (B) SRY and HDAC3 interacted in vitro in GST pull-down experiments using GST-SRY fusion proteins (Nter, Cter and Box) or GST as a control, and in vitro-translated 35S-labelled HDAC3. Input (20%) HDAC3 protein is represented in the first lane. (C) HDAC3 induced a partial cytoplasmic localisation in SRY. HeLa cells were cultured on glass coverslips and cotransfected with HASRYpJ3Ω and Flag-HDAC3pcDNA or Flag-HDAC1pcDNA expression vectors. The cells were fixed, and SRY and HDAC proteins were localised by immunostaining with anti-SRY antibody (SRY Ab) (a, c) and anti-Flag antibody (Flag Ab) (HDAC3: b; HDAC1: d). Magnification × 60.
Interaction between SRY and HDAC3 protein was analysed by GST pull-down experiments using GST-SRY fusion proteins and in vitro-translated 35S-labelled HDAC3. Figure 7B shows interaction between HDAC3 protein and SRY HMG box, indicating that deacetylation of SRY by HDAC3 might implicate the previous interaction between both proteins. As treatment by the deacetylase inhibitor TSA induced an exclusive nuclear localisation of endogenous SRY in NT2/D1 cells, we asked the question whether deacetylase HDAC3 could affect SRY subcellular localisation. SRY localisation was followed in HeLa cells transfected with SRY expression vector together with HDAC3 or HDAC1 (as a control) expression vectors. SRY protein found to be exclusively nuclear in these cells showed a partial cytoplasmic staining after cotransfection with HDAC3 expression vector (Figure 7Ca), similar to that observed with K136R SRY mutant (Figure 6A), whereas cotransfection with HDAC1 did not modify SRY nuclear localisation (Figure 7Cc). These results confirm that acetylation of SRY contributed to its nuclear localisation.
Expression of p300 and HDAC3 in the embryonic gonad
To attest the presence of p300 in the gonadal tissue during the sex determination steps, we first performed Western blot analysis on male human embryonic gonads at different stages. Figure 8A demonstrates the presence of p300 in the developing gonad from 7 to 12 weeks at the time of testis cord formation. Immunofluorescence staining of 8-week human embryonic gonadal sections showed a strong nuclear p300 expression in cells outside the seminiferous tubules, presumably in the future Leydig, which strongly expressed SF-1 (Figure 8B). A weaker signal was also present in the differentiating Sertoli cells inside the cords, in which SF-1 staining decreases at this developmental stage (Figure 8B).
Figure 8.
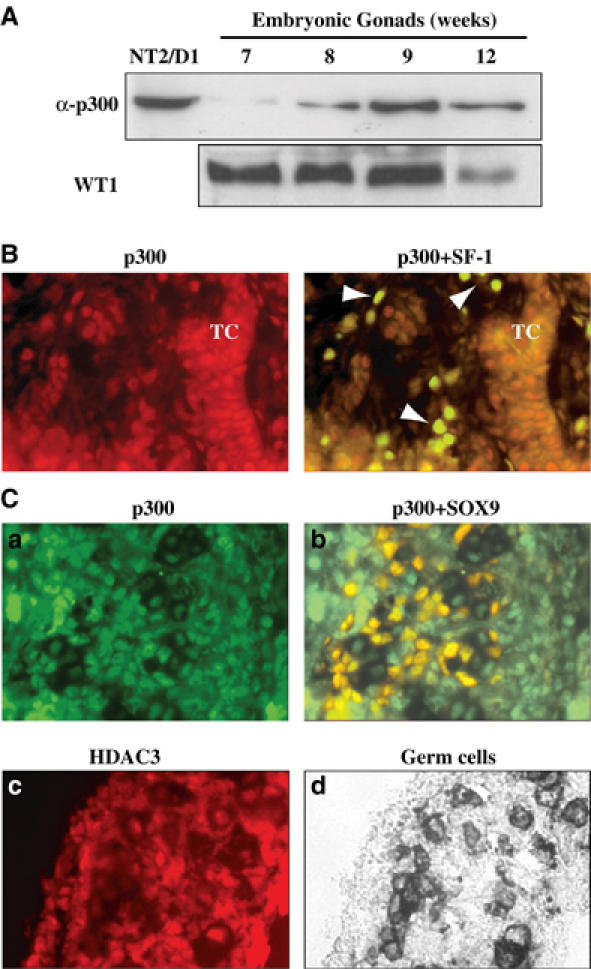
p300 and HDAC3 are expressed in the male embryonic gonad at the time of sexual differentiation. (A) Male human embryonic gonads at 7, 8, 9 and 12 weeks development stage and NT2/D1 as a control were lysed. Proteins (20 μg) were resolved by SDS–PAGE and blotted onto a nitrocellulose membrane. Immunoblots were performed using anti-p300 polyclonal (1 μg/ml) and anti-WT-1 (1/500) antibodies and were revealed using the ECL kit (Amersham). (B) An 8-week human male gonad section was co-immunostained with SF-1 antibody (de Santa Barbara et al, 1998) (in green) and p300 antibody (1/100, in red). At this stage, cells colabelled with both antibodies were identified as Leydig cells (indicated by an arrowhead), whereas Sertoli cells within the testicular cord (TC) were stained by p300 antibody only (magnification × 100). (C) p300 and HDAC3 proteins are expressed in 12.5 dpc male mouse gonads. p300 immunostaining (in green) was detected in most of the gonadal cells and colocalised with SOX9 (in red) in Sertoli cells. HDAC3 immunostaining (in red) was detected in the different gonadal cell types. Alkaline phosphatase staining of primordial germ cells (in black) localised gonadal tissue (magnification × 40).
On 12.5 dpc mouse embryo sections, p300 appears to be uniformly expressed in the nucleus of different cell compartments of the male embryonic gonad (Figure 8Ca), particularly in SOX9-expressing Sertoli cells (Figure 8Cb). HDAC3 immuno-staining shows the same widespread expression pattern in the nucleus of male embryonic gonadal cells (Figure 8Cc) located in germ cells region (Figure 8Cd). These data demonstrated that p300 and HDAC3 are expressed in embryonic Sertoli cells during the sex determination process. This observation reinforces the hypothesis that SRY/p300/HDAC3 is involved in the same regulatory loop.
Discussion
Histone acetylation has been suggested to regulate gene expression (Wolffe and Pruss, 1996; Grunstein, 1997). In addition to histones, many transcription factors have been shown to be acetylated (Bannister et al, 2000; Kouzarides, 2000; Sterner and Berger, 2000). In most cases, the functional relevance of this modification in gene activation has been demonstrated, and the underlying mechanism usually involved acetylation-dependent increase in sequence-specific DNA binding. HAT activities are borne by several coactivators such as p300/CBP or p300/CBP-associated factors (P/CAFs) (Bannister and Kouzarides, 1996; Ogryzko et al, 1996).
Here we have shown that human SRY protein interacted with and was acetylated by p300 in in vitro acetylation experiments. This acetylation status was confirmed in vivo and occurred on the K136 lysine that was preceded by an alanine as one of the acetylated sites in histone H4. An alignment of known CBP/p300 targets indicated that the only detectable preference is a glycine or a serine residue directly preceding the acetylated lysine (Bannister et al, 2000) even though no consensus acetylation sites have been assigned. The SRY acetylated motif (K136) within the second NLS was well conserved among primates as well as mouse, bovine, sheep or goat SRY proteins (Table I) (Nagai, 2001) and was conserved within groups B, C and G of the SOX proteins (Bowles et al, 2000). Indeed, we have shown that mouse Sry protein was also an in vivo p300 acetylated protein.
Table 1.
Conservation of acetylated lysine among SRY proteins
| NLS2 | ||
|---|---|---|
| Human | K136 | KY RPRRKAKMLP |
| Gorilla | K136 | KY RPRRKAKMLP |
| Orangutan | K136 | KY RPRRKAKMLQ |
| Bison | K131 | KY RPRRRAKRPQ |
| Bovine | K131 | KY RPRRRAKRPQ |
| Goat | K131 | KY RPRRKAKRPQ |
| Sheep | K141 | KY RPRRKAKRPQ |
| Marsupial | K110 | KY QPRRKTKSFL |
| Horse | K136 | KY RPRRKAKMPQ |
| Mouse | K81 | KY QPHRRAKVSQ |
| Sequences of SRY proteins were compared using BCM Search Launcher, Multiple Sequence Alignments Program. Conservation of acetylated human and mouse lysine is indicated in bold. Accession numbers for SRY proteins are A47533 (human), P48046 (gorilla), Q28783 (orangutan), Q27949 (bison), Q03256 (bovine), Q03256 (goat), Q03257 (sheep), P36395 (marsupial), Q05738 (mouse) and P36389 (horse). Position of the second NLS of the SRY HMG box is represented. | ||
Due to the proximity of acetylated lysine to the C-terminal NLS, we investigated the role of acetylation in SRY nuclear import. SRY protein contains two functional NLSs (Poulat et al, 1995; Sudbeck and Scherer, 1997) within its DNA-binding HMG domain that can function independently to target a carrier protein to the nucleus. Forwood et al (2001) showed that nuclear import of SRY was dependent on the recognition of the C-terminal NLS by nuclear import factor importin β. We demonstrated that SRY interaction with importin β needed the integrity of K136 residue, which might belong to the SRY interaction domain with p300. This mutation may also perturb NLS2 function by conformational changes even though lysine K136 is not directly involved in the basic NLS. Furthermore, SRY acetylation was shown to enhance its binding to importin β. These results suggested that contribution of acetylation to SRY nuclear localisation might occur directly via SRY and possibly indirectly via acetylation of different nuclear import factors. Acetylation of importin α by CBP led us to hypothesise that nuclear import machinery may be subject to regulation by acetylation (Bannister et al, 2000).
Acetylation enhances nuclear retention of HNF-4, E1A adenovirus-transforming protein and POP-1, the Caenorhabditis elegans LEF/TCF homologue, by increasing nuclear import and blocking nuclear export of these transcription factors (Soutoglou et al, 2001; Madison et al, 2002; Gay et al, 2003). Besides nuclear localisation, interaction with p300 independent of acetylation affected SRY DNA-binding affinity as demonstrated by electrophoretic mobility shift assay (EMSA) experiments. We were not able to test the functional consequence of SRY acetylation and the increase in its DNA binding as no target genes for SRY were identified.
HDACs are involved in transcriptional regulation by deacetylation of histones and transcription factors. HDAC enzyme activities are inhibited by TSA, a potent antitumor agent (Finnin et al, 1999; Furumai et al, 2001) involved in induction of cell differentiation and apoptosis (Marks et al, 2000). TSA increases the acetylation level and transcriptional activity of p53 (Luo et al, 2000), TAT (Kiernan et al, 1999) or nuclear receptor SF-1 (Jacob et al, 2001). Even though nuclear localisation of SRY is well documented (Poulat et al, 1995; Sudbeck and Scherer, 1997), we found cytoplasmic SRY in NT2/D1 cell line. In these cells, TSA treatment led to an exclusive nuclear localisation and to an increased punctuate clustering of SRY in the nucleus. Sensitivity of SRY to TSA led us to hypothesise that SRY acetylation state is relevant to an equilibrium between acetylation by p300 and deacetylation by an HDAC. We identified HDAC3 that can interact with SRY protein and can deacetylate SRY in vivo. We showed that cotransfection of HDAC3 and SRY cDNAs mimicked the subcellular localisation of K136R SRY mutant and induced a partial cytoplasmic localisation confirming a role of acetylation in SRY nuclear localisation. Our results are in accordance with the recent study showing that TSA treatment and consequently enhancement of acetylation status have no negative effect on the earliest steps of male sex determination (Mizukami et al, 2004).
Acetylation and deacetylation of SRY observed in vivo in transfected cells and expression of p300 and HDAC3 proteins in primordial gonads suggested that these differential modifications might be involved in the regulation of SRY activity during the sex-determining process that takes place in the undifferentiated gonad. In human embryos, SRY showed a cytoplasmic localisation in the somatic cell compartment. At later stages of male sex determination process (8–9 weeks), while the testis cords were already organised and anti-Müllerian hormone (AMH) was produced by Sertoli cells (Morais da Silva et al, 1996), this cytoplasmic localisation of SRY might be part of a mechanism that blocks SRY outside of the nucleus to inhibit any further action. This observation could explain the persistence of SRY expression in most of the mammals, while in rodents this expression is extremely transient. p300/CBP has been previously implicated in diverse developmental processes (Yao et al, 1998). Homozygous p300- and CBP-deficient mice died at around E9.5–11.5 and E10.5–12.5, respectively. p300-deficient embryos displayed specific transcriptional defects and a major cell proliferation deficiency (Yao et al, 1998). Thus, p300 might contribute to the earliest function of SRY, that is, the cell proliferation in the coelomic epithelium observed within the undifferentiated gonad (Schmahl et al, 2000). Even though the implication of p300 in gene transactivation dependent on SOX2 and SOX9, two SRY-related SOX transcription factors, has been described (Nowling et al, 2000; Tsuda et al, 2003), these data reported the first regulation of a SOX protein activity by acetylation/deacetylation. Acetylation of architectural HMG proteins has been well documented (Sterner et al, 1979; Munshi et al, 2001). Factor UBF, an RNA polymerase I activator, associates to CBP and Rb-HDAC with two out of its three HMG boxes (Pelletier et al, 2000). These authors demonstrated that CBP activation and HDAC1 suppression of ribosomal transcription by the recruitment of UBF are mutually exclusive, regulating in vivo PolI transcription through an acetylation–deacetylation ‘flip-flop'. As SRY expression is tightly regulated in a tissue-specific and developmental manner, one can expect that acetylation by HAT and deacetylation by HDAC might activate or repress SRY function in combination with other specific regulatory factors. As we demonstrated for the first time, a conserved post-translational modification between human and mouse SRY proteins and since, until now, no cellular systems are available to test SRY protein activity, in the future, only transgenic approaches should confirm the impact of SRY acetylation on gonadal organogenesis.
Materials and methods
Cell culture and transfections
NT2/D1 were grown in DMEM/F12 medium (Invitrogen), and HeLa cells and HeLa B3 clone (Poulat et al, 1995) were grown in DMEM medium (Invitrogen) supplemented with 10% fetal bovine serum, 2 mM glutamine and penicillin–streptomycin at 37°C in a 5% CO2 atmosphere. TSA (250 ng/ml) (Sigma Aldrich) was added for 18 h to the culture medium when indicated. HeLa cells were transfected using LipofectAMINE PLUS ™ reagent (Invitrogen) and luciferase activities were measured as previously described (de Santa Barbara et al, 2001). p300 siRNA experiments in HeLa B3 cells were performed as previously described, using oligofectamine (Invitrogen) (Bres et al, 2002).
Plasmids and constructs
HA-tagged human (h) SRY pJ3omega (HAhSRY-pJ3Ω) expression vector and GST-SRY fusion constructs (pGEX4T3-SRYwt and pGEX4T3-SRYbox) have been described by Desclozeaux et al (1998). HA-tagged mouse (m) SRY pJ3omega (HAmSry-pJ3Ω) expression vector was constructed. GST-SRYNter and GST-SRYCter were constructed by PCR amplification of 1–175 and 409–617 bp SRY ORF fragments, respectively, and cloning in pGEX4T1 vector. SRY box constructs (amino acids 59/88, 81/114, 100/133 and 107/139) were generated by PCR and cloned in the pGEX4T3 vector to produce GST-SRY fusion proteins. N-flagged p300 expression vector (pcDNA) has been previously described (Kiernan et al, 1999). The HAhSRY-pJ3Ω (K115R, K123R, K128R, K134R and K136R) and the HAmSry-pJ3Ω (K81R) mutated plasmids were constructed by site-directed mutagenesis (Quickchange kit, Stratagene) and confirmed by sequencing the SRY HMG box. The construct pGEX21T-importin β to produce a GST fusion protein (Imamoto et al, 1995) was a gift from Dr Yoneda (Osaka University Medical School, Japan).
Antibodies
Polyclonal human SRY-specific rat sera were raised against the GST-SRYNter and GST-SRYCter fusion proteins containing amino acids 1–55 and 137–204, respectively, from the human SRY protein. The fusion proteins were expressed in Escherichia coli strain BL21 and purified as previously described (de Santa Barbara et al, 1998). Rats were injected with purified protein mixed with complete Freund's adjuvant (Eurogentec Laboratories). SRY antibody specificity was checked by immunostaining of NT2/D1 nuclear extracts and SRY fusion protein transferred to nitrocellulose. Rabbit polyclonal antibodies, p300 (N-15) raised against N-terminal amino acids, HDAC3 (H99) and WT-1 (C19), were purchased from Santa Cruz Biotechnology. Monoclonal mouse anti-HA tag and anti-Flag tag antibodies were from Roche Diagnostics and Sigma Aldrich, respectively.
In vivo co-immunoprecipitation, immunoblotting and in vivo labelling of SRY
Interaction between p300 and SRY was studied in vivo in the NT2/D1 cells as per the procedure described by de Santa Barbara et al (1998). The rat SRYNter (3 μl/assay) (and the corresponding preimmune rat serum) or the rabbit p300 (1 μg) (and a preimmune rabbit serum) antibodies conjugated to protein G–Sepharose were used to immunoprecipitate endogenous SRY and p300, respectively (de Santa Barbara et al, 1998). After several washes, the final immunoprecipitated proteins were subjected to SDS–PAGE and the proteins were transferred to nitrocellulose (SRY detection) or PVDF membranes (p300 detection) with a Trans-Blot apparatus (Bio-Rad). Western blot analyses were performed using p300 antibody (0.5 μg/ml) after SRY IP and SRY antibody (1/2000) after p300 IP, and revealed by the ECL method (Amersham).
In vivo acetylation of SRY was performed in HA-tagged SRY-transfected HeLa cells. In vivo SRY labelling was accomplished by incubating the cells in DMEM containing 1 mM sodium butyrate, 1 mCi/ml [3H]acetic acid (Amersham Pharmacia) or 100 μCi/ml [35S]Met (in vitro 35S-cell labelling mix, Amersham Pharmacia) for 1 h at 37°C (Kiernan et al, 1999). Cell lysates were prepared (de Santa Barbara et al, 1998) and SRY was immunoprecipitated using anti-HA antibody. Immunoprecipitated proteins were resolved by SDS–PAGE and gels were fixed, dried and exposed to an X-ray film.
Recombinant proteins, in vitro HAT assay, in vitro binding assays and EMSA experiments
GST protein and GST-SRY fusion proteins were prepared as pre-viously described (de Santa Barbara et al, 1998), and purified SRY protein was eluted by thrombin digestion and dialysed (Desclozeaux et al, 1998). GST-importin β fusion protein was produced and purified according to Imamoto et al (1995). Proteins thus produced were checked by SDS–PAGE analysis and immunoblotting. Peptide P35 (EAEKWPFFQEAQKLQAMHREKYPNYKYRPRRKAKM) mapping the C-terminal part of the SRY box (35 amino acids) was newly synthesised.
For in vitro acetylation assays, purified SRY protein and GST-SRY fusion proteins were extensively dialysed against HAT X1 buffer (Kiernan et al, 1999) at 4°C for 12 h and concentrated using Centricon columns. The quality and quantity of each protein were assessed by running an SDS–PAGE electrophoresis gel and staining by Coomassie blue. Full-length p300 and HAT-inactive p300 were expressed as Flag-tagged fusion proteins using baculovirus transfer vectors and were affinity-purified as described previously (Ogryzko et al, 1996; Kiernan et al, 1999). In vitro assays for protein acetylation were conducted in the presence of 1 μCi/reaction 14C-acetylCoenzymeA (Amersham) as described (Kiernan et al, 1999).
[35S]Met-labelled p300 protein was generated using the coupled reticulocyte lysate system (TNT, Promega). For in vitro binding studies, equal amounts of GST or GST fusion proteins (SRYs or importin β) were incubated with in vitro-translated [35S]Met-labelled or purified (50 and 150 ng) p300 protein or with transfected HeLa cell extracts (100 μg proteins) in TBST buffer (de Santa Barbara et al, 1998) containing Complete ™ Protease Inhibitor Cocktail (Roche Diagnostics) for 1 h, followed by four washes in the same buffer. Bound proteins were analysed by SDS–PAGE and autoradiography.
EMSAs were performed as previously described (de Santa Barbara et al, 2001) using the TCF oligonucleotide (5′-AACAAT) as a probe. Labelled oligonucleotide was incubated with 20 ng of recombinant SRY protein, previously submitted to in vitro acetylation reaction.
Immunofluorescence staining of cultured cells and human embryo sections
SRY staining of NT2/D1 cells or transfected HeLa cells cultured on glass coverslips was performed as previously described (de Santa Barbara et al, 1998) using anti-SRY (1/100) and anti-HA (1/1000) antibodies. Immunohistochemistry using rat SF-1 antibody (1/100 dilution) (de Santa Barbara et al, 1998), p300 antibody (1 μg/ml) or HDAC3 antibody (1 μg/ml) was accomplished on human or mouse embryo sections at different developmental stages (8 weeks and 12.5 dpc, respectively) according to de Santa Barbara et al (2000).
Acknowledgments
We thank Patrick Atger for high-quality photography processing and René Seyer (LGF, CNRS UPR 2580, Montpellier) for HPLC assistance. We also thank Dr Rosemary Kiernan for critical reading of the manuscript. This work was supported by the European Economic Community through the fifth framework Program no. GLG2-CT-1999-00741. LT was the recipient of the Ligue Départementale de l'Hérault contre le Cancer. MB's laboratory was supported by the ACI and Human Frontiers.
References
- Albrecht KH, Eicher EM (2001) Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev Biol 240: 92–107 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Kouzarides T (1996) The CBP co-activator is a histone acetyltransferase. Nature 384: 641–643 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Miska EA, Gorlich D, Kouzarides T (2000) Acetylation of importin-alpha nuclear import factors by CBP/p300. Curr Biol 10: 467–470 [DOI] [PubMed] [Google Scholar]
- Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN, Fellous M (1990) Genetic evidence equating SRY and the testis-determining factor. Nature 348: 448–450 [DOI] [PubMed] [Google Scholar]
- Billin AN, Thirlwell H, Ayer DE (2000) Beta-catenin–histone deacetylase interactions regulate the transition of LEF1 from a transcriptional repressor to an activator. Mol Cell Biol 20: 6882–6890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowles J, Cooper L, Berkman J, Koopman P (1999) Sry requires a CAG repeat domain for male sex determination in Mus musculus. Nat Genet 22: 405–408 [DOI] [PubMed] [Google Scholar]
- Bowles J, Schepers G, Koopman P (2000) Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev Biol 227: 239–255 [DOI] [PubMed] [Google Scholar]
- Bres V, Tagami H, Peloponese JM, Loret E, Jeang KT, Nakatani Y, Emiliani S, Benkirane M, Kiernan RE (2002) Differential acetylation of Tat coordinates its interaction with the co-activators cyclin T1 and PCAF. EMBO J 21: 6811–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Xie W, Wilpitz D, Evans RM (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98: 675–686 [DOI] [PubMed] [Google Scholar]
- Chen L, Fischle W, Verdin E, Greene WC (2001) Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science 293: 1653–1657 [DOI] [PubMed] [Google Scholar]
- de Santa Barbara P, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Sudbeck P, Scherer G, Poulat F, Berta P (1998) Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Mol Cell Biol 18: 6653–6665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Santa Barbara P, Mejean C, Moniot B, Malcles MH, Berta P, Boizet-Bonhoure B (2001) Steroidogenic factor-1 contributes to the cyclic-adenosine monophosphate down-regulation of human SRY gene expression. Biol Reprod 64: 775–783 [DOI] [PubMed] [Google Scholar]
- de Santa Barbara P, Moniot B, Poulat F, Berta P (2000) Expression and subcellular localization of SF-1, SOX9, WT1, and AMH proteins during early human testicular development. Dev Dyn 217: 293–298 [DOI] [PubMed] [Google Scholar]
- Desclozeaux M, Poulat F, de Santa Barbara P, Capony JP, Turowski P, Jay P, Mejean C, Moniot B, Boizet B, Berta P (1998) Phosphorylation of an N-terminal motif enhances DNA-binding activity of the human SRY protein. J Biol Chem 273: 7988–7995 [DOI] [PubMed] [Google Scholar]
- Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, Wintersberger E, Seiser C (1999) Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol 19: 5504–5511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman Q, Grosschedl R (1999) Regulation of LEF-1/TCF transcription factors by Wnt and other signals. Curr Opin Cell Biol 11: 233–240 [DOI] [PubMed] [Google Scholar]
- Ferrari S, Harley VR, Pontiggia A, Goodfellow PN, Lovell-Badge R, Bianchi ME (1992) SRY, like HMG1, recognizes sharp angles in DNA. EMBO J 11: 4497–4506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, Breslow R, Pavletich NP (1999) Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401: 188–193 [DOI] [PubMed] [Google Scholar]
- Forwood JK, Harley V, Jans DA (2001) The C-terminal nuclear localization signal of the sex-determining region Y (SRY) high mobility group domain mediates nuclear import through importin beta 1. J Biol Chem 276: 46575–46582 [DOI] [PubMed] [Google Scholar]
- Furumai R, Komatsu Y, Nishino N, Khochbin S, Yoshida M, Horinouchi S (2001) Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc Natl Acad Sci USA 98: 87–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay F, Calvo D, Lo MC, Ceron J, Maduro M, Lin R, Shi Y (2003) Acetylation regulates subcellular localization of the Wnt signaling nuclear effector POP-1. Genes Dev 17: 717–722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman RH, Smolik S (2000) CBP/p300 in cell growth, transformation, and development. Genes Dev 14: 1553–1577 [PubMed] [Google Scholar]
- Grunstein M (1997) Histone acetylation in chromatin structure and transcription. Nature 389: 349–352 [DOI] [PubMed] [Google Scholar]
- Gwack Y, Byun H, Hwang S, Lim C, Choe J (2001) CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J Virol 75: 1909–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley VR, Jackson DI, Hextall PJ, Hawkins JR, Berkovitz GD, Sockanathan S, Lovell-Badge R, Goodfellow PN (1992) DNA binding activity of recombinant SRY from normal males and XY females. Science 255: 453–456 [DOI] [PubMed] [Google Scholar]
- Hawkins JR, Taylor A, Goodfellow PN, Migeon CJ, Smith KD, Berkovitz GD (1992) Evidence for increased prevalence of SRY mutations in XY females with complete rather than partial gonadal dysgenesis. Am J Hum Genet 51: 979–984 [PMC free article] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y (1995) In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J 14: 3617–3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob AL, Lund J, Martinez P, Hedin L (2001) Acetylation of steroidogenic factor 1 protein regulates its transcriptional activity and recruits the coactivator GCN5. J Biol Chem 276: 37659–37664 [DOI] [PubMed] [Google Scholar]
- Kiernan RE, Vanhulle C, Schiltz L, Adam E, Xiao H, Maudoux F, Calomme C, Burny A, Nakatani Y, Jeang KT, Benkirane M, Van Lint C (1999) HIV-1 tat transcriptional activity is regulated by acetylation. EMBO J 18: 6106–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R (1990) Expression of a candidate sex-determining gene during mouse testis differentiation. Nature 348: 450–452 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2000) Acetylation: a regulatory modification to rival phosphorylation? EMBO J 19: 1176–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Zhang W, Chan G, Jancso-Radek A, Liu S, Weiss MA (2001) Human sex reversal due to impaired nuclear localization of SRY. A clinical correlation. J Biol Chem 276: 46480–46484 [DOI] [PubMed] [Google Scholar]
- Luo J, Su F, Chen D, Shiloh A, Gu W (2000) Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature 408: 377–381 [DOI] [PubMed] [Google Scholar]
- Madison DL, Yaciuk P, Kwok RP, Lundblad JR (2002) Acetylation of the adenovirus-transforming protein E1A determines nuclear localization by disrupting association with importin-alpha. J Biol Chem 277: 38755–38763 [DOI] [PubMed] [Google Scholar]
- Marks PA, Richon VM, Rifkind RA (2000) Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst 92: 1210–1216 [DOI] [PubMed] [Google Scholar]
- Mizukami T, Fujisawa M, Kanai Y, Kurohmaru M, Hayashi Y (2004) Effects of trichostatin a, a histone deacetylase inhibitor, on mouse gonadal development in vitro. J Reprod Dev 50: 227–235 [DOI] [PubMed] [Google Scholar]
- Morais da Silva S, Hacker A, Harley V, Goodfellow P, Swain A, Lovell-Badge R (1996) Sox9 expression during gonadal development implies a conserved role for the gene in testis differentiation in mammals and birds. Nat Genet 14: 62–68 [DOI] [PubMed] [Google Scholar]
- Munshi N, Agalioti T, Lomvardas S, Merika M, Chen G, Thanos D (2001) Coordination of a transcriptional switch by HMGI(Y) acetylation. Science 293: 1133–1136 [DOI] [PubMed] [Google Scholar]
- Nagai K (2001) Molecular evolution of Sry and Sox gene. Gene 270: 161–169 [DOI] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM (1997) Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89: 373–380 [DOI] [PubMed] [Google Scholar]
- Narlikar GJ, Fan HY, Kingston RE (2002) Cooperation between complexes that regulate chromatin structure and transcription. Cell 108: 475–487 [DOI] [PubMed] [Google Scholar]
- Nowling TK, Johnson LR, Wiebe MS, Rizzino A (2000) Identification of the transactivation domain of the transcription factor Sox-2 and an associated co-activator. J Biol Chem 275: 3810–3818 [DOI] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V, Howard BH, Nakatani Y (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell 87: 953–959 [DOI] [PubMed] [Google Scholar]
- Ozawa Y, Towatari M, Tsuzuki S, Hayakawa F, Maeda T, Miyata Y, Tanimoto M, Saito H (2001) Histone deacetylase 3 associates with and represses the transcription factor GATA-2. Blood 98: 2116–2123 [DOI] [PubMed] [Google Scholar]
- Pelletier G, Stefanovsky VY, Faubladier M, Hirschler-Laszkiewicz I, Savard J, Rothblum LI, Cote J, Moss T (2000) Competitive recruitment of CBP and Rb-HDAC regulates UBF acetylation and ribosomal transcription. Mol Cell 6: 1059–1066 [DOI] [PubMed] [Google Scholar]
- Pontiggia A, Rimini R, Harley VR, Goodfellow PN, Lovell-Badge R, Bianchi ME (1994) Sex-reversing mutations affect the architecture of SRY–DNA complexes. EMBO J 13: 6115–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulat F, Barbara PS, Desclozeaux M, Soullier S, Moniot B, Bonneaud N, Boizet B, Berta P (1997) The human testis determining factor SRY binds a nuclear factor containing PDZ protein interaction domains. J Biol Chem 272: 7167–7172 [DOI] [PubMed] [Google Scholar]
- Poulat F, Girard F, Chevron MP, Goze C, Rebillard X, Calas B, Lamb N, Berta P (1995) Nuclear localization of the testis determining gene product SRY. J Cell Biol 128: 737–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahl J, Eicher EM, Washburn LL, Capel B (2000) Sry induces cell proliferation in the mouse gonad. Development 127: 65–73 [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Viollet B, Vaxillaire M, Yaniv M, Pontoglio M, Talianidis I (2001) Transcription factor-dependent regulation of CBP and P/CAF histone acetyltransferase activity. EMBO J 20: 1984–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner DE, Berger SL (2000) Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev 64: 435–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner R, Vidali G, Allfrey VG (1979) Studies of acetylation and deacetylation in high mobility group proteins. Identification of the sites of acetylation in HMG-1. J Biol Chem 254: 11577–11583 [PubMed] [Google Scholar]
- Sudbeck P, Scherer G (1997) Two independent nuclear localization signals are present in the DNA-binding high-mobility group domains of SRY and SOX9. J Biol Chem 272: 27848–27852 [DOI] [PubMed] [Google Scholar]
- Tilmann C, Capel B (1999) Mesonephric cell migration induces testis cord formation and Sertoli cell differentiation in the mammalian gonad. Development 126: 2883–2890 [DOI] [PubMed] [Google Scholar]
- Torchia J, Rose DW, Inostroza J, Kamei Y, Westin S, Glass CK, Rosenfeld MG (1997) The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387: 677–684 [DOI] [PubMed] [Google Scholar]
- Tsuda M, Takahashi S, Takahashi Y, Asahara H (2003) Transcriptional co-activators CREB-binding protein and p300 regulate chondrocyte-specific gene expression via association with Sox9. J Biol Chem 278: 27224–27229 [DOI] [PubMed] [Google Scholar]
- Wolffe AP (1996) Histone deacetylase: a regulator of transcription. Science 272: 371–372 [DOI] [PubMed] [Google Scholar]
- Wolffe AP, Pruss D (1996) Targeting chromatin disruption: transcription regulators that acetylate histones. Cell 84: 817–819 [DOI] [PubMed] [Google Scholar]
- Yao TP, Oh SP, Fuchs M, Zhou ND, Ch'ng LE, Newsome D, Bronson RT, Li E, Livingston DM, Eckner R (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell 93: 361–372 [DOI] [PubMed] [Google Scholar]
- Yao YL, Yang WM, Seto E (2001) Regulation of transcription factor YY1 by acetylation and deacetylation. Mol Cell Biol 21: 5979–5991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Horinouchi S, Beppu T (1995) Trichostatin A and trapoxin: novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays 17: 423–430 [DOI] [PubMed] [Google Scholar]


