Abstract
Identification and characterization of cancer subtypes are important areas of research that are based on the integrated analysis of multiple heterogeneous genomics datasets. Since there are no tools supporting this process, much of this work is done using ad-hoc scripts and static plots, which is inefficient and limits visual exploration of the data. To address this, we have developed StratomeX, an integrative visualization tool that allows investigators to explore the relationships of candidate subtypes across multiple genomic data types such as gene expression, DNA methylation, or copy number data. StratomeX represents datasets as columns and subtypes as bricks in these columns. Ribbons between the columns connect bricks to show subtype relationships across datasets. Drill-down features enable detailed exploration. StratomeX provides insights into the functional and clinical implications of candidate subtypes by employing small multiples, which allow investigators to assess the effect of subtypes on molecular pathways or outcomes such as patient survival. As the configuration of viewing parameters in such a multi-dataset, multi-view scenario is complex, we propose a meta visualization and configuration interface for dataset dependencies and data-view relationships. StratomeX is developed in close collaboration with domain experts. We describe case studies that illustrate how investigators used the tool to explore subtypes in large datasets and demonstrate how they efficiently replicated findings from the literature and gained new insights into the data.
1. Introduction
The discovery, refinement, and characterization of cancer subtypes are the basis for targeted treatment and have implications for patient outcomes and patient well-being. Lately, much of the research on cancer subtypes is being performed with data from large-scale projects such as The Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov), which are generating comprehensive genomic and clinical datasets for thousands of patients. Recent studies [VHP*10, NWD*10] have shown that an integrated analysis of different molecular data types generated by the TCGA project can indeed be used to discover subtypes and suggest molecular alterations relevant for therapeutic approaches.
Interactive visualization tools are crucial to fully exploit the potential of these large and heterogeneous datasets for cancer subtype characterization. Such tools can greatly increase the efficiency of investigators, who currently are relying mainly on ad-hoc scripts and static plots, making the process of exploring the data and checking hypothesis a tedious task. From a visualization research perspective, the conceptual and technical hurdles to provide seamless data visualization across the boundaries of individual heterogeneous datasets are not yet overcome, although they have been discussed for over a decade [UAB*98]. It stands to reason that there will be no all-encompassing heterogeneous data visualization concept available anytime soon, but investigators urgently need solutions for integrated visual analysis to make progress in their specific domains.
In this paper, we present an integrated solution for the visual exploration needs arising during the classification of cancer subtypes in large-scale, heterogeneous genomics data. Besides a task analysis elicited in semi-structured interviews with investigators, we contribute two novel visual encodings supporting these tasks. The first is StratomeX, which employs a column-based layout to represent datasets, with bricks in those columns encoding potential subtypes or stratifications (partitionings into homogeneous subsets) of the data. Bricks can embed different visualizations and StratomeX enables investigators to interactively refine these bricks. Contextual information from other data sources, such as biological pathways and clinical variables, are seamlessly integrated as dependent columns and provide information critical for interpretation. Another challenge that arises when working with large numbers of complex datasets is the coordination of the datasets and stratifications, as well as their assignment to views. This is addressed by another contribution, the Data-View Integrator, a meta visualization that shows relationships between datasets and allows investigator to interactively assign stratifications and datasets to views.
Our approach is validated in case studies with investigators who are domain experts. We report on findings, in which data from TCGA for glioblastoma multiforme (GBM) [The08] was used to characterize subtypes. Investigators were able to quickly reproduce known results from the literature and to gain further insights into the data.
2. Biological Background and Data
Cancer is a family of complex diseases that are caused by the accumulation of molecular alterations that are either genomic and affect the DNA sequence or epigenomic and affect other inheritable characteristics, such as methylation patterns of the DNA. These alterations can lead to abnormal cell growth, which results in tumor formation, invasion of nearby tissue, and often in growth of metastases in distant parts of the body.
Traditionally, cancers have been classified and named after the tissue or cell type where they originate, such as “breast ductal carcinoma” or “lung squamous cell carcinoma”. However, cancers that originate from the same tissue or cell type are often not homogeneous with respect to their histology or the underlying genomic and epigenomic alterations, which gives rise to the notion of cancer subtypes. Cancer subtypes are highly relevant for patient treatment and prognosis, since the efficacy of cancer drugs can vary greatly between cancer subtypes, and patients with different subtypes often have very different survival chances. In recent years, the identification and characterization of subtypes has focused increasingly on genome-wide molecular data, which is now becoming available also for large numbers of patients through the work of consortia such as The Cancer Genome Atlas project. Our collaborators are analyzing data from TCGA, which is a large-scale study designed to identify and catalog the molecular changes that are recurrent in large cohorts of cancer patients and therefore implied to drive tumor formation. TCGA aims to collect samples from at least 500 patients for each of over 20 different cancer types for a total of more than 10,000 patients. Several dozens of clinical parameters are collected for each patient, and all samples are subjected to extensive molecular profiling. The data generated for each sample includes genome-wide gene mutation status, copy number alterations, mRNA gene expression levels, DNA methylation levels, and microRNA expression levels.
Gene mutations are mutations in the genomic DNA of a cell in the region of the gene that determines the sequence of the protein encoded by the gene. Such mutations can lead to changes in the structure or function of the protein, which can have serious effects, for instance, if they affect tumor suppressor genes. Copy number alterations are another category of genomic mutations that can occur, for instance, if the genomic DNA of a cell is copied incorrectly during cell division. Whereas gene mutations only affect single or a very small number of consecutive positions in the genome, these alterations may affect hundreds to tens of thousands of positions and even whole chromosomes. Regions of the genome may be either amplified, resulting in an increased number of copies of the genes in that region, or lost, resulting in a decreased number of copies of genes. Since normal cells carry only two copies of each gene, they can either lose one copy – a “heterozygous deletion” – or both copies, resulting in a “homozygous deletion”. On the other hand, there is no theoretical limit to the number of times a gene can be amplified.
Gene expression is the process in which a gene is transcribed from the genomic DNA into an mRNA molecule, which can subsequently be translated into a protein. By measuring the abundance of such mRNA molecules – the “gene expression level” – the activity of a gene can be determined. For genome-wide studies the gene expression level is typically used as an indicator for the amount of protein that is being produced for the corresponding gene. Gene expression levels are controlled by various regulatory mechanisms. For instance, DNA methylation, an epigenomic modification, is known to suppress transcription if present in the regulatory region of a gene. In cancer, gene expression levels are also often affected by copy number alterations. An increased number of copies of a gene, for instance, often leads to increased gene expression levels and vice versa. Another part of the gene regulatory machinery are microRNAs, which are short RNA molecules that unlike mRNA are not translated into proteins, but regulate the translation of mRNAs into proteins by binding to mRNA molecules.
TCGA data generation centers are using either microarray or next-generation sequencing technologies to generate aforementioned data types. The consortium maintains Firehose (http://gdac.broadinstitute.org), a data analysis pipeline that is used to automatically preprocess the data and to perform a range of bioinformatics analyses. The analyses are performed jointly for all samples from patients with a particular cancer type and include clustering algorithms for mRNA, microRNA, and methylation data, as well as identification of mutated genes and copy number changes.
Investigators who are working on cancer subtype identification and characterization use three types of results from the analysis pipeline: (1) Quantitative data matrices, such as gene expression matrices with measurements for all genes in all patient samples. (2) Clusterings on these matrices that stratify patients into mutually exclusive subsets. (3) Categorical data matrices, containing information such as the copy number status (ordinal) – homozygously or heterozygously deleted, normal, lowly, or highly amplified – or mutation status (nominal) – mutated or not mutated – for each gene in each patient. Entries for individual genes in these matrices can be used to stratify the patients.
In addition to the output from the data analysis pipeline, investigators include quantitative clinical parameters, such as “time until death”, in their analyses. They may also include patient stratifications in their analyses that were computed outside the main data analysis pipeline. Furthermore, pathways, models of biological processes, are used to investigate the role gene products play in molecular interactions.
3. Tasks
To understand the requirements of our collaborators for subtype analysis, we conducted a series of semi-structured interviews and evaluated recent publications that report findings of subtype analyses on TCGA data, for example, [VHP*10] and [NWD*10], to complement the requirements elicited from the interviews. We also presented an abstract on StratomeX at a TCGA-internal workshop [LPG11] to gather feedback.
Our working definition of a (candidate) subtype is a subset of patients obtained from one or more stratifications and we use the terms subset and subtype interchangeably.
The exploratory analysis can be roughly divided into two phases. In Phase 1, the investigators try to find stratifications of patients that are derived from multiple data types, for example, an mRNA gene expression clustering that correlates with the mutation status of a particular gene. In Phase 2, they evaluate these subsets with respect to their functional and clinical implications. Tasks from Phase 1 and Phase 2 are addressed in an iterative fashion. More specifically, in Phase 1, investigators need to:
Select combinations of stratifications and data matrices from different data types for visualization.
Evaluate how well two or more stratifications support each other.
View and explore mRNA and microRNA expression or DNA methylation matrices as stratified by candidate subtypes. If different patient subsets exhibit distinct patterns, this is an indicator that there might be supporting evidence for these subtypes.
Refine stratifications by combining information from two data types, for instance by splitting a gene expression cluster based on the mutation status of a gene.
In Phase 2, investigators focus on the following tasks:
Review the effect of a stratification on clinical outcomes, such as patient survival or tumor recurrence. If there are notable differences between subtypes, there might be clinical relevance.
Determine if the subtypes have a functional impact by viewing stratified molecular profiling data in the context of biological pathways. As an example, investigators are interested in pathways that are generally activated but deactivated in some subtypes.
In addition, investigators will also perform quality control tasks, for example, by comparing different clusterings (same algorithm but different parameters; different algorithms) for a particular data type to evaluate how stable the clusters are.
4. Related Work on Comparative Subset Visualization
The common element of the tasks listed above is their comparative nature. This is a direct implication of our application, as by comparing different stratifications, it is not only possible to pinpoint the most sensible subset across different datasets as a “candidate subtype”, but also to investigate the functional effects of these possible subtypes within pathway visualizations, as well as their effect on clinical parameters. Since subset membership can be treated like an additional categorical variable, visualization methods for comparing categorical data would be suitable representations for this case. The literature describes two principle approaches to categorical data visualization: conversion of the categorical visualization problem to a quantitative problem in data space, as well as categorical representation approaches in view space. A recent study suggests that both variants have their place in visual data analysis, as each of them is suited best for specific visual analysis tasks [JFJ11]. The following will briefly describe the related work for both approaches.
Categorical data can be either ordinal or nominal. In the ordinal case, the categories are inherently ordered, while in the nominal case, an order of the categories can be determined, e.g., by Correspondence Analysis [Gre07, RRB*04] or clustering-based approaches [MH99, BPM01]. The reasoning behind most of these approaches follows Friendly's mantra of Effect Ordering: “Sort the data by the effects to be observed” [FK03]. A second step then computes a spacing between the categories to convey the degree of similarity between the categories. An established method to achieve this is the Optimal Scaling approach [RRB*04], which is able to use the output of a correspondence analysis for deriving a spacing. After this transformation, a visualization using commonly available techniques for quantitative data can be used. Parallel coordinates, for example, have been proposed to compare different clusters [HSPW06].
If categorical data is not transformed to quantitative data, there exist two general ways of visualizing it. The first is by utilizing relative positions. For example, slice and dice subdivisions of the drawing area are used to create Mosaic Plots [Hof00], where each region occupies an area with a size relative to the number of data records that fall into the category it represents. Other techniques use different positional arrangements, such as the Mosaic Plot Matrix [Fri99], which, instead of using a single multivariate plot, puts multiple bivariate Mosaic Plots in an arrangement similar to scatter plot matrices. This makes for a simpler reading of the plots and also prevents unfavorable aspect ratios of mosaic tiles from affecting the reading on lower levels. While Mosaic Plots are not often used for cluster comparison, there exist examples of their use for comparing a clustering of records across different categories [Hof08].
The second visualization option is explicitly drawn, ribbon-like links between the categories in the sense that links split up in width proportional to the amount of records shared with other categories. This basic idea has been adapted, among others, to parallel coordinates, yielding Parallel Sets [KBH06, Kos10]. Given a clean edge routing, ribbons make the distribution of records across different categories easy to follow and analyze. These techniques have been used extensively for subset comparison – for example, in CComViz [ZKG09], Matchmaker [LSP*10], VisBricks [LSS*11], and others [TPRH11].
Only few studies have been reported on visualizations of categories from multiple, heterogeneous data sources. One of them is the D-Dupe software [KGS*08], which clusters multiple datasets and then matches up the results in a visualization to identify duplicates between both datasets. In the biological domain, interactive visualization of heterogeneous data centers mainly on pathways as the frame of reference in which the different sources of data are integrated. A recent example is Pathline [MWS*10], which integrates multiple quantitative data sources along a linearized pathway for comparative analysis. Visualization resembling the row/column structure of Mosaic Plots can also be found in the biological field, as the aforementioned publication on cancer subtype classification shows [VHP*10, Fig.3]. Yet, the figure in this paper is only a static, specifically produced representation to illustrate the findings.
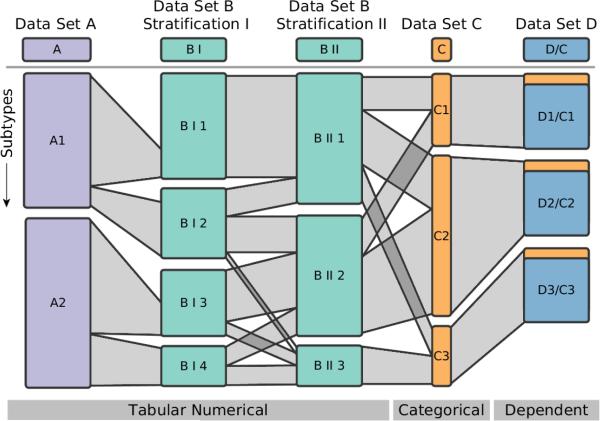
Figure 3.
Schematic comparison of five columns. The first three columns show groupings of tabular data, where the second and third show the same data only with different stratifications. The fourth, orange column represents a categorization. The rightmost column illustrates the concept of dependent subsets, where the groups are based on the stratification of another column. The ribbons between the subsets indicate how many patients are shared between them. For instance, all patients of BI1 are contained in BII1. BII1, however, also contains patients from BI2.
We found that the state-of-the-art does not provide a technique for interactive visual subset comparison across dataset boundaries in the biological domain. Since we intend to combine visualization of underlying data with the encoding of categories, data space techniques are not suitable for our tasks. We have decided against employing relative positions, since they do not easily integrate with embedded visualization due to unfavorable aspect ratios for smaller categories. Hence, we decided for a ribbon-based technique, which scales better in this regard.
5. Data-View Integrator
Dealing with many different datasets, each with several stratifications that can be displayed in several views is challenging. Consequently, it is common to show the relationships between the datasets. Examples are relational database schemas. North et al. extend this idea to views, as they envision DataFaces, interactive connections of visualization and data schemas, as future work [NCS02]. This approach was recently realized in Stack'n'flip [SSL*11] as well as in HIVE [RKS11]. We take up this idea and extend it to accommodate multiple stratifications.
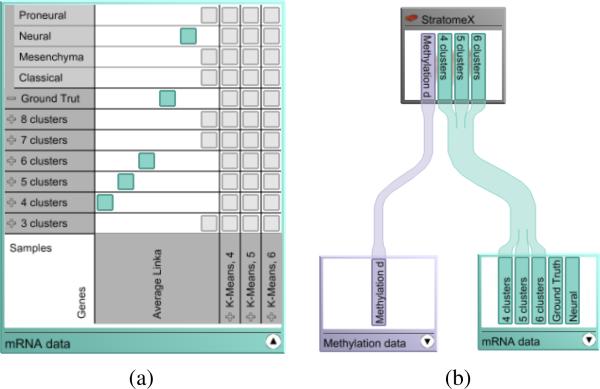
The Data-View Integrator is a meta visualization that serves two purposes. First, it orients the user by providing an overview of the datasets and the relationships among them. Second, it allows the user to dynamically configure combinations of stratifications and assign them to the views in which they can be analyzed. By default, the Data-View Integrator shows a representation of the data model as a graph where the nodes correspond to the individual datasets and the edges represent the shared identifiers between the datasets. A unique patient ID serves as the primary key for referencing patients across all datasets. In addition, datasets such as mRNA and methylation data both contain patient IDs as rows and genes as columns and are therefore linked twice in the model. The nodes representing the datasets can be visualized in two modes. The compact overview mode shows only a caption for the dataset. The detail mode, shown in Figure 1 (a), also shows the associated stratifications. In this example, multiple clustering results are loaded for both patient samples and genes, in addition to an external patient stratification. As stratifications themselves are one-dimensional, views can only show combinations of patient stratifications with record lists, for example, gene clusterings. Possible combinations are shown in a matrix in the detail mode. By selecting a matrix cell, the user can indicate that he is interested in this combination, which is then highlighted and shown in an additional column.
Figure 1.
The two modes of the dataset nodes in the Data-View Integrator. (a) In the detail mode, the patient stratifications and gene clusterings are displayed as a matrix of possible combinations. By selecting one of the gray matrix cells, the user can interactively create a combination (cyan). (b) A view node connected to two dataset nodes that are in compact mode, listing only the existing combinations.
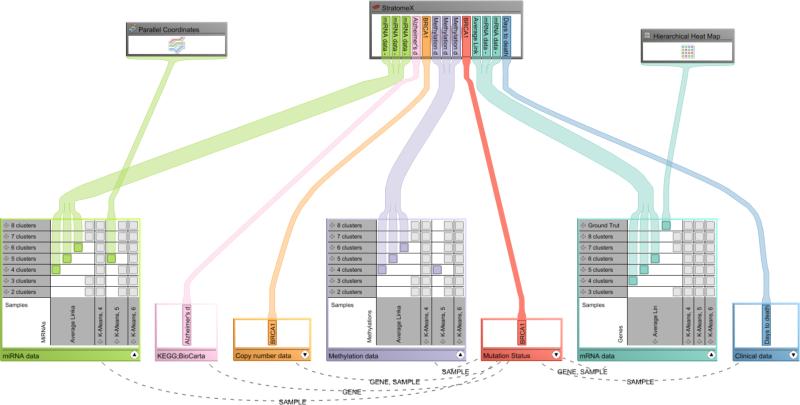
In addition to datasets, views are represented in the graph. The user can directly assign which stratification combination he wants to explore in a view by using drag-and-drop. Figure 1 (b) shows an example where the dataset node is in compact mode. Figure 2 shows a more complex scenario with multiple datasets and stratifications, as they would be used for cancer subtype analysis.
Figure 2.
The Data-View Integrator showing the relationships between datasets as well as their association to views for the application scenario. Datasets and stratifications are shown at the bottom with the views placed above. Relationships between a selected dataset and all others are shown. Note that some views can show only one stratification, while others, like StratomeX, can show multiple.
6. StratomeX - Subtype Visualization
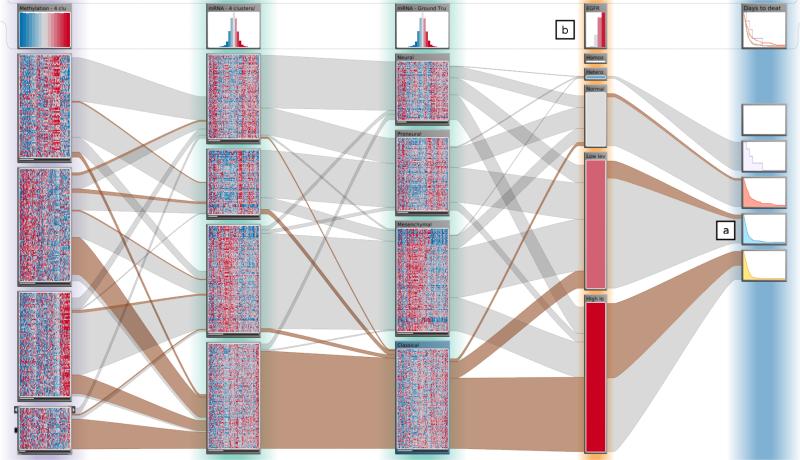
The visual encoding used in StratomeX employs the basic strategies used in Parallel Sets [KBH06], Matchmaker [LSP*10], and VisBricks [LSS*11]. As shown in Figures 3 and 4, stratifications of datasets are arranged as columns side-by-side. The columns are split up into disjoint bricks representing either candidate subtypes, clusters, or categories – depending on the data type and stratification loaded. The data inside a brick can be encoded using various visualization techniques such as heatmaps, parallel coordinates plots, or histograms, which can be switched on demand. When showing heatmaps, the height of a brick encodes the number of patients it contains. Ribbons connect bricks of neighboring columns. Their width encodes how many patients they share. This is illustrated in Figure 3.
Figure 4.
StratomeX configured as illustrated in Figure 3. The heatmaps in the bricks allow the investigator to judge the homogeneity of the subsets. The header bricks at the top show the name of the column and an overview of the data in the column. In the fourth column, a stratification based on the categories for copy number variation of EGFR can be seen. The rightmost column shows Kaplan-Meier plots for “days to death” as dependent bricks for the copy number-based stratification. Note that patients with amplifications of EGFR have far worse outcomes compared to patients with two copies.
As different data sets can contain disjoint sets of patients, the height of the bricks can not be used to compare absolute values. We have chosen relative heights, since investigators are primarily interested in the relative relationships. Additionally, relative heights optimally utilize the available space. This is valid if the data set constitutes a representative subset of the population. As long as two neighboring columns contain the same patients, the outer edges of the ribbons connecting them will be parallel. For disjoint sets of patients, however, the height at the beginning of a ribbon may not be the same as at its end, as shown between the first and second column in Figure 3. In this example, Data Type B contains more patients than Data Type A, leaving parts of the sides of the bricks unconnected. StratomeX also supports static sizes for bricks to accommodate embedded visualizations, which only work for a specific aspect ratio.
6.1. Column Classes
One aspect that distinguishes StratomeX from previous work is that it can deal with multiple heterogeneous datasets. We have identified three classes of columns that are needed for the cancer subtype analysis tasks in StratomeX:
Table Columns
In this class we use the stratifications to group tabular, quantitative datasets. The bricks in the columns contain a visualization of the underlying data. For the subtype identification task, the heatmap representation is best suited and therefore chosen as the default. The stratification into subsets is in most cases not fixed, often alternative stratifications exist. This can make manual refinement of the stratifications necessary. The plausibility of a particular stratification is judged by investigators using the embedded views and the relationships to other stratifications in StratomeX. Figure 3 shows a stratification for one dataset in the first column, and two stratifications for another dataset in the second and third column.
Categorical Columns
Categorical columns represent an unambiguous stratification of patients based on a single attribute. An example is the mutation status of one particular gene of interest – mutated or not mutated. Categorical columns contain no visualization of the underlying data other than a constant color, but have permanently visible labels showing the name of the category.
Dependent Columns
In many cases, it is of great interest to explore the effect of a stratification of one dataset on another one. StratomeX allows the user to do this by introducing dependent columns. The dependent columns use the same stratification of patients as their source column, but show the data of the dependent dataset. As a consequence the ribbons connecting the source column always connect exactly two bricks. An example for a dependent column is shown on the far right in Figure 3. Dependent columns are crucial for two tasks in this application context: to explore clinical data and to investigate pathways. By using multiple Kaplan-Meier curves [RNP*10] next to candidate subtypes, investigators can explore whether the stratification has effects on the clinical status of patients. The small multiples of the Kaplan-Meier curves could, for example, show that the disease-free survival in one subtype is significantly lower than that of another. In Figure 4 at (a) for example, we can see that patients with a normal copy number status of the EGFR gene appear to have a better chance of living longer than those who have EGFR amplifications. Dependent pathway columns can be used to judge whether there is different behavior between subtypes in the biological processes that the pathways represent. By placing multiple small thumbnails of pathways, one for each subset, next to an mRNA expression dataset, and overlaying the average expression of the group onto the gene nodes of the pathways [GOB*10], investigators can easily compare the effects of the subtype on the pathway. To visually amplify and make them stand out even in the thumbnail-sized small multiples, we enlarge the expression overlays. An example of pathway small multiples is shown on the right of Figure 7.
Figure 7.
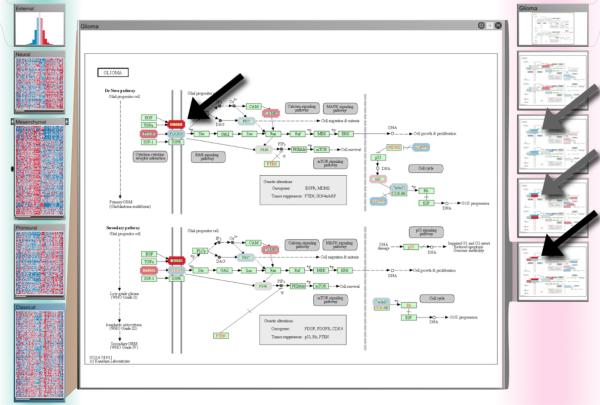
Subtypes in the context of pathways. Column 1 shows mRNA gene expression subtypes identified by Verhaak et al. (from top: neural, mesenchymal, proneural, classical). The dependent Column 2 shows small multiples of the “Glioma” pathway from KEGG overlaid with the average gene expression levels for each subtype. The detail view in the center shows the same pathway enlarged with the gene expression levels for the classical subtype. The arrows indicate a part of the pathway where we observed notable differences in gene expression levels between the subtypes. Note that not all genes in the pathway have been mapped since the gene expression data matrix only contained a subset of the most variable 1,500 genes in the dataset.
6.2. Visual Encoding Details
Beyond the high-level visual encoding strategy described above, StratomeX contains a series of additional encodings that support the analysis tasks. StratomeX is designed to follow the visual information seeking mantra – Overview First, Zoom and Filter, Details on Demand [Shn96].
Overview
To facilitate the association between the columns in StratomeX and the dataset nodes in the Data-View Integrator, we use a combination of color coding and labels. The columns have a halo in a color that corresponds to the color of the dataset node in the Data-View Integrator. The header brick, shown, for example, at (b) in Figure 4, contains a summary view representing the whole dataset. The type of view shown depends on the dataset and user preference. For tabular and categorical data the header brick shows a histogram by default. Pathway columns show the pathway with the average expression encoding of the whole dataset overlaid. Clinical survival data uses a summary Kaplan-Meier plot that overlays the survival curves of each subtype.
Zoom and Filter, Interaction
As subtypes are rarely based on only one factor (and therefore one data type), it is crucial to be able to refine candidate subtypes by splitting and merging bricks. StratomeX supports interactive splitting of bricks based on the ribbons connecting them to other columns, as well as merging of multiple bricks of the same column. The user can add labels for candidate subtypes, which are then shown at the top of the subtype brick. StratomeX allows users to arbitrarily arrange both columns, and bricks within the columns. The former facilitates the comparison of multiple columns, the latter can be used to minimize crossings of the ribbons.
Details on Demand
While the process of characterizing subtypes is conducted mainly by investigating global trends in the overview, it is often also necessary to explore some part of the data in detail. If, for example, the small multiples of the pathways show differences in the mapping on the genes between the subsets, a details on demand strategy is necessary to identify the genes. StratomeX facilitates this by enabling investigators to create focus-duplicates of arbitrary bricks as illustrated in Figure 7.
7. Implementation
StratomeX and the Data-View Integrator are implemented in Caleydo (http://caleydo.org), a visual analytics framework for molecular biology. In addition to the ability to load datasets individually, a scripting interface allows creating predefined data-setups, e.g., in the process of running analyses in a bioinformatics pipeline such as Firehose. The software is written in Java and uses OpenGL for rendering. For the case studies described below, seven datasets were loaded. With the exception of the pathways, each contained between 300 and 550 samples, with 1,500 genes each for the expression datasets, and between 5,000 and 6,000 each for copy-number and mutation status data, for a total of roughly 6 million data points, making it a very effective visualization tool for the visual analysis of large-scale data. The embedded views switch automatically from texture-based, static views to a fully interactive visualization if enough space is available. The main limitation in terms of scalability is the number of subsets, where about 20 are feasible.
8. Case Studies
StratomeX was designed in collaboration with a group of domain experts. To evaluate our approach, we asked two investigators, who were not involved in the design process, to use StratomeX to explore data from one of the TCGA cancer types that they are currently analyzing. We prepared datasets for glioblastoma multiforme (GBM) and breast invasive carcinoma (BRCA) based on the output of the Firehose pipeline and additional, “external” stratifications provided by the investigators. The case studies were conducted in two hourlong sessions with each investigator and were recorded for later analysis. Here we only report case studies from the GBM dataset with 529 patients, as the findings for the BRCA dataset are unpublished. The following observations and findings were made during the evaluation sessions with our collaborators.
Comparing Clusterings
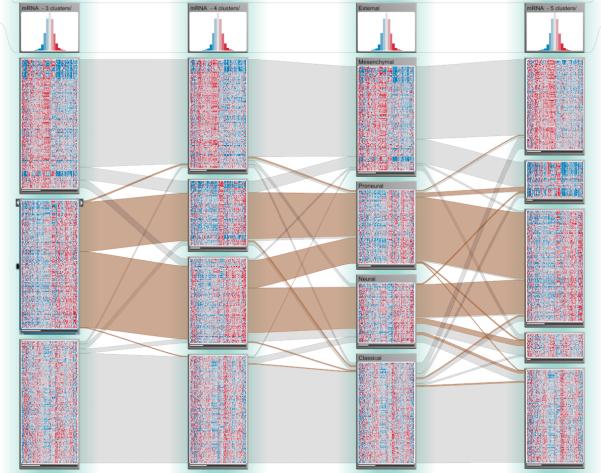
Even though the TCGA analysis pipeline reports a single “best” clustering for each mRNA, microRNA, and DNA methylation data matrix, clusterings with different numbers of clusters are also available. Since Verhaak et al. [VHP*10] identified four mRNA gene expression subtypes, but the analysis pipeline reported three clusters as the best result for mRNA expression data based on one of the implemented clustering algorithms, we were interested in how the clustering for three, four, and five clusters compared to an updated classification based on the one by Verhaak et al. (see Figure 5). The first observation that we made based on the salient ribbon patterns was that one of the subsets from the three-cluster solution was split into two clusters in the four-cluster solution, but that almost all patients from these two clusters make up a single cluster in the five-cluster solution. This might be a biologically meaningful result because of a second observation that we made: said two clusters in the four-cluster solution are a mix of the neural and proneural subtypes identified by Verhaak et al., whereas the other two clusters almost exactly correspond to the classical and mesenchymal subtypes. This indicates that the clustering computed by the analysis pipeline is a reasonable and meaningful solution. Verhaak et al. also reported that the tumors in the neural and proneural subtypes exhibit similar gene expression patterns, which are not found in the other two subtypes. This is one possible explanation for why neural and proneural subtypes are hard to separate by clustering mRNA data.
Figure 5.
Clustering comparisons. Columns 1, 2, and 4 show clusterings from the analysis pipeline with three, four, and five clusters respectively. Column 3 shows a stratification of the patients based on subtypes identified by Verhaak et al. (from top: mesenchymal, proneural, neural, classical).
Combining Gene Mutation Status and Methylation Data
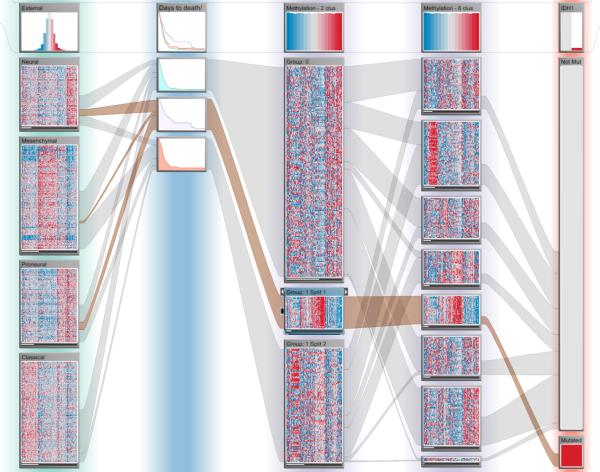
Noushmehr et al. [NWD*10] used clustering of DNA methylation profiles to identify three GBM subtypes, one of which is based on hypermethylation of certain regions of the genome, implicating that gene expression in those regions is repressed. They also found that this subtype is associated with mutations of IDH1 and mostly falls within the proneural subtype. When in one of our evaluation sessions our collaborator was interested in studying this methylation subtype, he realized quickly that it had not been detected in the clustering from the analysis pipeline that created three clusters. None of the clusters was strongly associated with either IDH1 mutations or the proneural subtype. Using the Data-View Integrator, we easily added the other clusterings to the view. Our collaborator pointed out that one of the clusters from the eight clusters case had a distinct methylation pattern and only contained patients with an IDH1 mutation. This was also the only cluster with such mutations (see Figure 6). We then used this cluster to split the methylation clustering with two clusters into three clusters. We hypothesized that the newly created patient subset contained many patients with the Noushmehr et al. subtype, both due to its strong association with the IDH1 mutation and the large overlap with the proneural subtype. Our collaborator suggested to confirm this using the survival data, which showed that the newly created patient subset indeed seemed to have better survival outcomes than patients in the two other subsets (see Figure 6), as reported by Noushmehr et al. This example emphasizes the importance of interactive refinements of stratifications that is supported by StratomeX.
Figure 6.
Methylation subtypes. Column 1 shows mRNA gene expression subtypes identified by Verhaak et al. Column 2 shows patient survival outcomes (days to death) and was created as a dependent column of Column 3, which shows a stratification of methylation data. The stratification of Column 3 was created by splitting off a part of the original clusters based on the mutation status of IDH1, shown in Column 5, which reveals a characteristic expression pattern overlooked by the algorithm. Only in the eight-cluster case, shown in Column 4, the clustering algorithm was able to detect this pattern.
Evaluating the functional impact of subtypes
In one of our evaluation sessions we looked into the effect of the Verhaak et al. gene expression subtypes on molecular processes that are known to play a role in gliomas, which is the family of brain cancers that GBM is part of. We opened the “glioma” pathway from KEGG (Kyoto Encyclopedia of Genes and Genomes) [KAG*08] as a dependent column to see if there are any differences in the expression levels of these pathways when stratified according to the subtypes. The small multiples showed very clearly that the glioma pathway indeed has different activation patterns across the four subtypes (see Figure 7). In particular, we noted that there was a striking difference between the proneural and the classical subtype in the left part of the pathway. With the help of a detailed view of the pathway that StratomeX provides as a focus duplicate of the small multiples, we were able to identify the genes that are showing the most notable differences between the classical and proneural subtypes in the glioma pathway: EGFR and PDGF are upregulated whereas PDGFRA is downregulated in classical GBM, and vice versa in the proneural subtype. This observation is probably due to a finding that Verhaak et al. reported, namely that increased EGFR copy numbers are a hallmark of the classical subtype, whereas copy number amplifications of PDGFRA are a characteristic of the proneural subtype. These increases in copy number are likely responsible for the increased gene expression levels that we observed here.
In general, our collaborators noted that the brick and ribbon metaphor to visualize patient subsets and their relationships across different stratifications feels natural and intuitive. They also told us that the combination of small multiples with details on demand is very useful, in particular for the pathway maps. A very positive outcome of the evaluation sessions with our collaborators was that in all cases they asked us to load further data that they wanted to explore with StratomeX. They also made suggestions on how to improve the tool by integrating further analyses, for example, to cluster data matrices on the fly or to compute statistical significance values for observed differences in patient outcomes.
9. Conclusion and Future Work
In this paper we presented StratomeX, which was developed to address the visual analysis needs of investigators who are performing cancer subtype characterization based on large-scale genomics data. In a series of case studies we documented the validity of our approach and its potential to generate new insights.
From a visualization research perspective, we contribute a task analysis for subtype characterization, as well as a method for comparative subset visualization of heterogeneous datasets that share at least one common identifier. We also described the Data-View Integrator, a method that supports the configuration of multiple datasets and stratifications as part of the exploration process.
In the future we aim for a tighter integration of StratomeX into the overall cancer genome analysis workflow by incorporating additional analyses and further data sources suggested by our collaborators. We are also planning to integrate information to guide the exploration, for example, correlation scores that could suggest stratifications that support each other. We also aim to deepen our understanding of the complex analysis process by making the software available to a larger group of investigators in TCGA. We intend to conduct longitudinal observations on how StratomeX is used in these scenarios. Ultimately, we will distribute a public release of StratomeX.
Supplementary Material
Acknowledgements
We would like to thank I. Watson and S. Quayle from the Dana-Farber Cancer Institute as well as A. McKenna, A. Cherniak, and M. Noble from the Broad Institute for their input and feedback on this project. We also acknowledge the Broad Institute's TCGA Genome Data Analysis Center led by L. Chin and G. Getz for providing access to the data and analysis results. The StratomeX project is supported by the United States National Cancer Institute (U24 CA143867), the Caleydoplex project (P22902) funded by the Austrian Science Fund and the inGeneious project (385567) funded by the Austrian Research Promotion Agency.
References
- Beygelzimer A, Perng C, Ma S. Proceedings ofthe ACM SIGKDD Conference on Knowledge Discovery and Data Mining (KDD '01) ACM; 2001. Fast ordering of large categorical datasets for better visualization. pp. 239–244. [BPM01], 3. [Google Scholar]
- Friendly M, Kwan E. Effect ordering for data displays. Computational Statistics & Data Analysis. 2003;43(4):509–539. [FK03], 3. [Google Scholar]
- Friendly M. Extending mosaic displays: Marginal, conditional, and partial views of categorical data. Journal of Computational and Graphical Statistics. 1999;8(3):373–395. [Fri99], 4. [Google Scholar]
- Gehlenborg N, O'Donoghue SI, Baliga NS, Goesmann A, Hibbs MA, Kitano H, Kohlbacher O, Neuweger H, Schneider R, Tenen-baum D, Gavin A. Visualization of omics data for systems biology. Nature Methods. 2010;7(3):56–68. doi: 10.1038/nmeth.1436. [GOB*10], 6. [DOI] [PubMed] [Google Scholar]
- Greenacre M. Correspondence Analysis in Practice. 2nd ed. Chapman & Hall/CRC; 2007. [Gre07], 3. [Google Scholar]
- Hofmann H. Exploring categorical data: interactive mosaic plots. Metrika. 2000;51(1):11–26. [Hof00], 4. [Google Scholar]
- Hofmann H. Mosaic plots and their variants. In: Chen C.-h., Haerdle W, Unwin A., editors. Handbook of Data Visualization. Springer; 2008. pp. 617–642. [Hof08], doi:10.1007/978-3-540-33037-0_24. 4. [Google Scholar]
- Havre SL, Shah A, Posse C, Webb-Robertson B. Proceedings of the SPIE Conference on Visualization and Data Analysis (VDA '06) Vol. 6060. SPIE; 2006. Diverse information integration and visualization. pp. 60600M–60600M-11. [HSPW06], 4. [Google Scholar]
- Johansson Fernstad S, Johansson J. Proceedings ofthe Conference on Information Visualisation (IV '11) IEEE; 2011. A task based performance evaluation of visualization approaches for categorical data analysis. pp. 80–89. [JFJ11], 3. [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. Nucleic Acids Research. Vol. 36. Database-Issue; 2008. KEGG for linking genomes to life and the environment. pp. 480–484. [KAG*08], 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosara R, Bendix F, Hauser H. Parallel sets: interactive exploration and visual analysis of categorical data. IEEE Transactions on Visualization and Computer Graphics. 2006;12(4):558–568. doi: 10.1109/TVCG.2006.76. [KBH06], 4, 5. [DOI] [PubMed] [Google Scholar]
- Kang H, Getoor L, Shneiderman B, Bilgic M, Licamele L. Interactive entity resolution in relational data: A visual analytic tool and its evaluation. IEEE Transactions on Visualization and Computer Graphics. 2008;14(5):9991014. doi: 10.1109/TVCG.2008.55. [KGS*08], 4. [DOI] [PubMed] [Google Scholar]
- Kosara R. Turning a table into a tree: Growing parallel sets into a purposeful project. In: Steele J, Iliinsky N, editors. Beautiful Visualization: Looking at Data through the Eyes of Experts. O'Reilly; 2010. pp. 193–204. [Kos10], 4. [Google Scholar]
- Lex A, Park PJ, Gehlenborg N. Proceedings of The Cancer Genome Atlas' 1st Annual Scientific Symposium: Enabling Cancer Research Through TCGA. Washington, D.C., USA: Nov, 2011. Supporting subtype characterization through integrative visualization of cancer genomics data sets. [LPG11], 3. [Google Scholar]
- Lex A, Streit M, Partl C, Kashofer K, Schmalstieg D. Comparative analysis of multidimensional, quantitative data. IEEE Transactions on Visualization and Computer Graphics (InfoVis '10) 2010;16(6):1027–1035. doi: 10.1109/TVCG.2010.138. [LSP*10], 4, 5. [DOI] [PubMed] [Google Scholar]
- Lex A, Schulz H, Streit M, Partl C, Schmalstieg D. VisBricks: multiform visualization of large, inhomogeneous data. IEEE Transactions on Visualization and Computer Graphics (InfoVis '11) 2011;17(12):2291–2300. doi: 10.1109/TVCG.2011.250. [LSS* 11], 4, 5. [DOI] [PubMed] [Google Scholar]
- Ma S, Hellerstein J. Ordering categorical data to improve visualization. Proceedings ofthe IEEE Information Visualization Symposium (InfoVis '99) Late Breaking Hot Topics. 1999:15–18. [MH99], 3. [Google Scholar]
- Meyer M, Wong B, Styczynski M, Munzner T, Pfister H. Pathline: A tool for comparative functional genomics. Computer Graphics Forum (EuroVis '10) 2010;29(3):1043–1052. [MWS*10], 4. [Google Scholar]
- North C, Conklin N, Saini V. Proceedings of the IEEE Symposium on Information Visualization (InfoVis 02) iEEE; 2002. Visualization schemas for flexible information visualization. pp. 15–22. [NCS02], 4. [Google Scholar]
- Noushmehr H, Weisenberger DJ, Diefes K, Phillips HS, Pujara K, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [NWD*10], 1,3,8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohn H, Klukas C, Schreiber F. Creating views on integrated multidomain data. Bioinformatics. 2011;27(13):1839–1845. doi: 10.1093/bioinformatics/btr282. [RKS11], 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich JT, Neely JG, Paniello RC, Voelker CCJ, Nussenbaum B, Wang EW. A practical guide to understanding Kaplan-Meier curves. Otolaryngology—Head and Neck Surgery. 2010;143(3):331–336. doi: 10.1016/j.otohns.2010.05.007. [RNP*10], 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario GE, Rundensteiner EA, Brown DC, Ward MO, Huang S : Mapping nominal values to numbers for effective visualization. Information Visualization. 2004;3(2):80–95. [RRB*04], 3. [Google Scholar]
- Shneiderman B. Proceedings ofthe IEEE Symposium on Visual Languages (VL 96) iEEE; 1996. The eyes have it: A task by data type taxonomy for information visualizations. pp. 336–343. [Shn96], 6. [Google Scholar]
- Streit M, Schulz H, Lex A, Schmalstieg D, Schumann H. Model-Driven design for the visual analysis of heterogeneous data. IEEE Transactions on Visualization and Computer Graphics PP. 2011;99:1–1. doi: 10.1109/TVCG.2011.108. [SSL* 11], 4. [DOI] [PubMed] [Google Scholar]
- The Cancer Genome Atlas Research Network: Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008 Oct;455(7216):1061–1068. doi: 10.1038/nature07385. [The08], 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkay C, Parulek J, Reuter N, Hauser H. integrating cluster formation and cluster evaluation in interactive visual analysis. Proceedings ofthe Spring Conference on Computer Graphics (SCCG 11) 2011 [TPRH11], 4. [Google Scholar]
- Uselton S, Ahrens J, Bethel W, Treinish L, State A. Proceedings ofthe IEEE Conference on Visualization (Vis 98) iEEE; 1998. Multi-Source data analysis challenges. pp. 501–504. [UAB*98], 1. [Google Scholar]
- Verhaak RG, Hoadley KA, Purdom E, Wang V, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010 Jan;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [VHP* 10], 1, 3, 4, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Konecni S, Grinstein G. Proceedings ofthe SPIE Conference on Visualization andData Analysis (VDA 09) SPiE; 2009. Visually comparing multiple partitions of data with applications to clustering. pp. 72430J–72430J-12. [ZKG09], 4. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.