Abstract
Amyloid plaques and neurofibrillary tangles are the pathological hallmarks of Alzheimer's disease. However, there has been a long-standing discussion on the dynamic relations between Aβ and tau proteins, partially due to the lack of a tool to track protein dynamics in individual live neurons at the early stage of Aβ generation and tau phosphorylation. Here, we developed nanoplasmonic fiber tip probe (nFTP) technology to simultaneously monitor Aβ42 generation and tau phosphorylation (at serine 262) in living, single neuroblastoma cells over 12 h. We observed that Aβ42 generation, under clinically relevant anesthetic treatment, preceded tau phosphorylation, which then facilitated Aβ42 generation. This observation is also supported by measuring proteins in cell lysates using the ultrasensitive label-free photonic crystal nanosensors. nFTP therefore provides an advanced method to investigate protein expression and post-translational modification in live cells and determine outcomes of intervention of Alzheimer's disease and other neurodegenerative disorders.
Keywords: nanoplasmonic fiber tip probe (nFTP), protein dynamics, Aβ, tau, Alzheimer's disease, photonic crystal nanosensor
Graphical Abstract
Aβ hypothesis posits that Aβ generation triggers a cascade of neurotoxic events, including tau phosphorylation, which leads to neurofibrillary tangles.1 However, tau hypothesis suggests that tau phosphorylation leads to Aβ accumulation and eventually the formation of amyloid plaques.2,3 Debates still remain on the precedence and causal relations between Aβ generation and tau phosphorylation,4 partially due to the lack of a technique to simultaneously measure the dynamics of Aβ and phosphorylated tau proteins in the same individual neuron over time at their native conditions. Current techniques that measure intracellular proteins either are destructive to cells (e.g., enzyme-linked immunoassay, Western blot) or require transfected cell lines or transgenic animals (e.g., fluorescent imaging).5,6 Recent development in nanosensors demonstrated measurement of action potentials using nanowire field-effect transistors,7 optical endoscopy with nanofibers,8–10 carbon nanotubes,11,12 nanowires,13 and nanobeam cavities.14 Gold nanoparticles have been used to visualize localization of proteins and RNAs in live cells.15,16 However, these approaches do not analytically quantify the sensitive changes of protein levels. These limitations impose a major hurdle in studying protein dynamics in living cells and clinically obtained primary samples. Here we demonstrate direct observation of dynamic change of Aβ42 and phosphorylated tau (at serine 262) proteins in living single neuroblastoma cells using the nanoplasmonic fiber tip probe (nFTP) technology. Our measurements suggest a hierarchical and circular pattern between Aβ generation and tau phosphorylation: Aβ generation precedes and leads to tau phosphorylation, and phosphorylated tau also promotes Aβ generation. This conclusion is also supported by measuring protein concentrations from lysed cells using photonic crystal label-free nanosensors, thus establishing nFTP as a reliable method for live cell research.
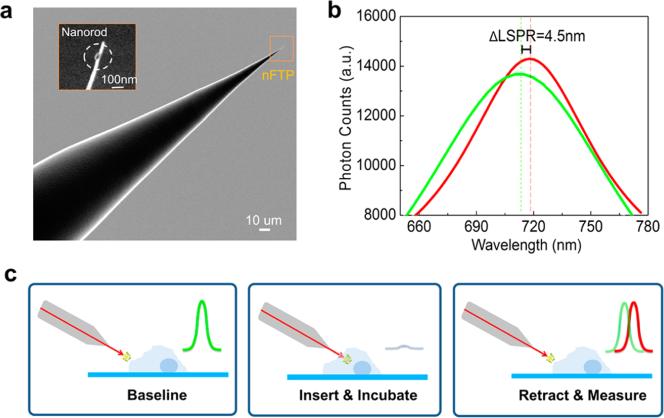
The nFTP was comprised of a nanoscale optical fiber with a single gold nanorod biosensor on its tip (Figure 1a). A broadband white light source (tungsten-halogen lamp) excited the localized surface plasmon resonance (LSPR) of the gold nanorod through the optical fiber (Supporting Information Figure S1). The LSPR spectrum displayed a wavelength shift when proteins bound to the gold nanorod (Figure 1b), which was detected from the inverted microscope (Olympus IX73) and analyzed with a spectrometer (Princeton Instrument). The method and fabrication of nFTP was specified in our previous work.17 In the current research, our primary focus is to establish nFTP as a reliable method to study the Aβ-tau dynamic changes. First, blocking nonspecific binding is proved to be an essential step for intracellular measurements (Supporting Information Figure S2). Prior to the single cell measurements, each functionalized nFTP was tested in the cell culture medium containing premixed solution of proteins. A positive spectrum shift, if observed, would verify the successful functionalization of the antibodies on the sensor surface (Supporting Information Figure S3). To dynamically measure protein expressions in the same cell, we used glycine solution (pH 2.5 tuned with hydrogen chloride) to unbind proteins from the antibodies while preserving the antibodies on the sensor (Supporting Information Figure S4). Figure 1c illustrates one measurement cycle: nFTP was sequentially incubated in the regeneration and blocking solution. It was then brought into proximity to the cell, controlled by the motorized stages. The baseline spectra were first taken outside of the cell. The nFTP was then inserted into the cell, incubated for 2 min and retracted out of the cell. The signal spectra were taken at the same focal position as the baseline spectra. The LSPR shift between the baseline and the signal spectra, induced by the captured proteins (e.g., tau-PS262), quantified their intracellular concentrations. This regenerate–block–baseline–insert–incubate–retract–signal completed one measurement cycle, and could be repeatedly performed in the same single live neuron or cell.
Figure 1.
Nanoplasmonic fiber tip probe (nFTP). (a) Scanning electron micrograph (SEM) image of nFTP. Inset shows a single gold nanorod attached to the nFTP. (b) Localized surface plasmon resonance (LSPR) spectra measured before (green) and after (red) inserting the nFTP into the cell. LSPR spectrum was collected using an electron multiplying charge coupled device (EMCCD) spectrometer (Princeton Instrument) on an inverted microscope (Olympus IX73). (c) During each measurement cycle, nFTP was first regenerated in pH 2.5 glycine solution. 1% bovine serum albumin (BSA) mixed into the cell culture medium (without fetal bovine serum) was then used as the blocking solution. The baseline LSPR spectra were taken in close proximity to the cell. The nFTP was then inserted into the cytoplasm, incubated for 2 min, and retracted from the cell. The signal LSPR spectra were immediately taken at the same focus spot as the baseline spectra. The LSPR shift from the baseline spectra to the signal spectra [ΔLSPR = 4.5 nm in (b)] quantified the intracellular tau-PS262 level.
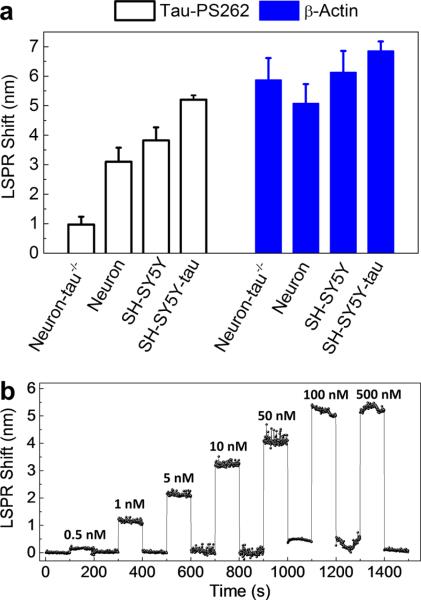
We measured both tau-PS262 and β-Actin levels in the wild-type neuroblastoma SH-SY5Y cell, tau overexpressed SH-SY5Y cell (SH-SY5Y-tau cell), mouse cortex neuron and tau knockout mouse cortex neuron (Neuron-tau−/−). All these cells were first treated with anesthetic isoflurane to trigger tau-PS262 generation. Results summarized in Figure 2a showed clear difference in tau-PS262 expressions between different cell types, whereas similar levels of β-Actin levels were observed as control. LSPR shifts can be converted to protein concentrations by calibrating nFTP using known concentrations of tau-PS262 solutions. As shown in Figure 2b, the sensor has a detection range from 0.5 nM to 0.5 μM, equivalent to approximately 300 to 300 000 copies of proteins in the volume of a typical cell (~10 μm size). This dynamic range of proteins falls in the biologically relevant concentration of intracellular proteins.18
Figure 2.
Specificity and sensitivity. (a) nFTP sensors were functionalized with anti-tau-PS262 (sc-32828, Santa Cruz) and anti-β-Actin (ab13822, Abcam), and were employed to measure tau-PS262 and β-Actin proteins in different cell types after the cells were treated with isoflurane. The error bar comes from three measurements on the same cell. (b) LSPR shift at different concentrations of tau-PS262 solutions. 0.5, 1, 5, 10, 50, 100, 500 nM tau-PS262 (Santa Cruz, sc-32828p) in the cell culture medium (without FBS) were prepared, and sequentially tested. At each concentration, 1,000 spectra were measured. The nFTP was regenerated between two different concentrations.
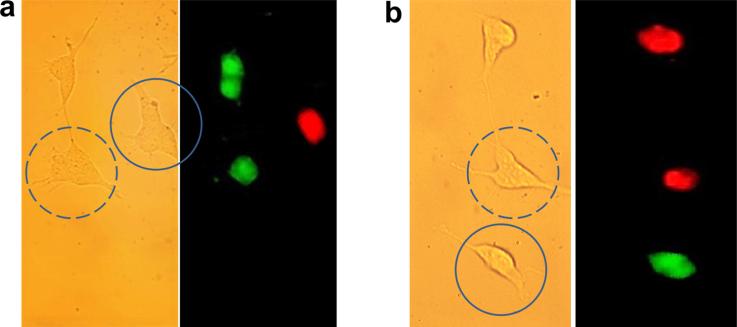
We assessed the cell membrane integrity, enzymatic and metabolic activity after the cell was punctured by nFTP once every hour, consecutively for 12 h. First, we used the cell viability/cytotoxicity assay (L3224, Invitrogen): 2 μM calcein-AM and 4 μM ethidium homodimer-1 (ethD-1) were applied to the cell. After incubating for 40 min, cell medium was replaced by 1 mL of Dulbecco's phosphate-buffered saline (DPBS) and cells were imaged under fluorescent microscope. The calcein-AM is a green fluorescent dye that indicates intracellular esterase activity. EthD-1 is a red fluorescent dye that stains nucleic acid, indicating a compromised cell membrane. As shown in Figure 3a, the dash-circled cell was punctured by the nFTP and showed green fluorescence and no red fluorescence. The solid-circled cell, punctured by a microscale fiber showed red fluorescent color and no green fluorescence. These results indicated that puncturing by the nFTP (but not microscale probe) preserved esterase activity and membrane integrity.
Figure 3.
Cell viability and vitality assay on SH-SY5Y-tau cells. The dash-circled cell was punctured by nFTP once every hour for 12 h. The solid-circled cell was punctured by a microscale fiber. (a) Cell viability/cytotoxicity (L3224, Invitrogen) assay. Green-fluorescence indicated active esterase condition. Red-fluorescence indicated loss of cell membrane integrity. (b) Cell vitality (L34951, Invitrogen) assay. Green-fluorescence indicated loss of cell membrane integrity. Red-fluorescence indicated active metabolic condition.
Next, we performed cell vitality assay (L34951, Invitrogen): 500 nM C12-resazurin and 10 nM SYTOX DNA binding dye were applied to the cell and incubated for 15 min. Reduction of C12-resazurin to red-fluorescent C12-resorufin indicates active metabolic condition. SYTOX is a cell-impermeant, green-fluorescent nucleic acid stain that indicates compromised cell membrane. In Figure 3b, the dash-circled cell was punctured by the nFTP and the solid-circled cell was punctured by a microscale probe. The presence of red fluorescent color and absence of green color in the dash-circled cell indicated that the nFTP (but not microscale probe) puncturing preserves cell metabolic activity and membrane integrity.
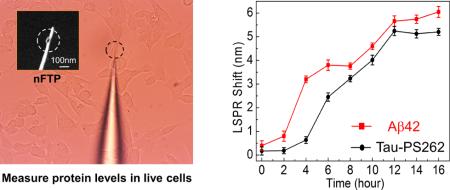
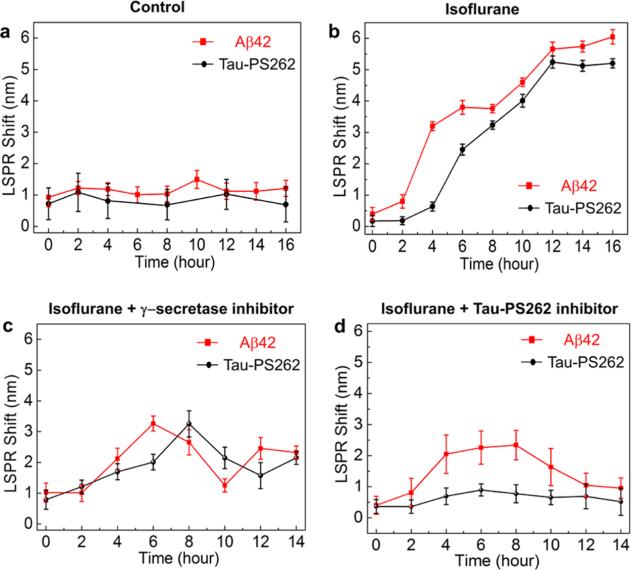
Now to study Aβ42 and tau-PS262 dynamics simultaneously in the same cell, we built the mechanically revolving dual-nFTP (Supporting Information Figure S6) and functionalized the gold nanorod surfaces with Aβ42 antibodies and tau-PS262 antibodies, respectively. Inhalation anesthetic isoflurane has been shown to induce Aβ accumulation19 and tau phosphorylation (increase in tau-PS262 levels without significant changes in total tau levels),20 which provides a clinically relevant tool to simultaneously induce both Aβ generation and tau phosphorylation. We first performed a control experiment in the SH-SY5Y-tau cells without the treatment of isoflurane. As shown in Figure 4a, neither Aβ42 nor tau-PS262 levels increased under this control condition. Each data point in all of the plots represents the averaged value of three consecutive measurements in the same cell. Next, we treated SH-SY5Y-tau cells with 2% isoflurane for 2 h, and then measured Aβ42 and tau-PS262 levels simultaneously every 2 h up to 12 h. In contrast to the control condition, isoflurane significantly increased the Aβ42 and tau-PS262 levels (Figure 4b). Moreover, the time to reach a 50% increase in Aβ42 level (approximately 4 h) was less than that needed for a 50% increase in tau-PS262 level (approximately 7 h). These data suggest that Aβ42 generation precedes tau phosphorylation (at serine 262).
Figure 4.
Aβ42 and tau-PS262 dynamics. (a) Aβ42 and tau-PS262 levels were monitored in the same SH-SY5Y-tau cell when no isoflurane was applied (control condition). (b) Aβ42 and tau-PS262 levels were monitored after the treatment of isoflurane in the SH-SY5Y-tau cell up to 16 h. (c, d) Dynamic Aβ42 and tau-PS262 levels in SH-SY5Y-tau cells, which were preconditioned with 0.5 μM γ-secretase inhibitor (c) or 3 μM phosphorylation kinase inhibitor K252a (d) prior to the isoflurane treatment.
Next, we set out to study whether Aβ generation and tau phosphorylation could interfere with each other. We treated the SH-SY5Y-tau cells with 0.5 μM γ-secretase inhibitor (L-685,458, Sigma L1790) 1 h before the isoflurane treatment, which has been demonstrated to suppress Aβ accumulation.19 In the same cell, we observed simultaneous decrease in the tau-PS262 level (Figure 4c). We also used 3 μM tau phosphorylation inhibitor (K252a, Abcam ab120419) to treat SH-SY5Y-tau cells for 1 h before the isoflurane treatment, which has been shown to suppress tau-PS262 generation.21,22 Western blot was also carried out to verify K252a efficacy (Supporting Information Figure S7). In the same cell, we observed Aβ42 level first increased but finally reduced after 10 h (Figure 4d). These data indicate that inhibition of tau phosphorylation led to a partial reduction in Aβ42 levels. Taken together, these results suggest a hierarchical and circular pattern between Aβ generation and tau phosphorylation: Aβ generation precedes and leads to tau phosphorylation, and phosphorylated tau also promotes Aβ generation.
Our studies are the first direct measurements of intracellular Aβ generation and tau phosphorylation in living cells. To further verify our major conclusion, that is, Aβ precedes tau phosphorylation, we prepared multiple cell cultures, treated them with isoflurane, lysed them, and harvested their intracellular proteins at different times after the isoflurane treatment. As proteins were significantly diluted during this process, we used the ultrasensitive photonic crystal label-free nanosensors to measure the Aβ and tau-PS262 levels in each sample.
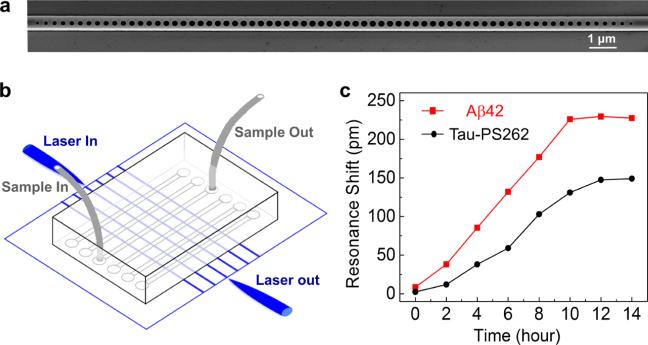
The photonic crystal nanosensor is a silicon waveguide with periodic structures deterministically designed and patterned along the waveguide (Figure 5a).23,24 These structures provide constructive interference and create a high-Q optical resonance, which is sensitive to any proteins that bind on the sensor surface.25,26 Therefore, protein concentrations in the cell lysates can be quantified by measuring the optical resonance shift. Precise fabrication of the designed structures was achieved using electron beam lithography and reactive ion etching processes (see Supporting Information). A polydimethylsiloxane (PDMS) microfluidic channel was bonded on the chip for fluid delivery.27 We functionalized the surface of the photonic crystal nanosensor with Aβ42 antibody (ab10148, Abcam) and phospho-specific tau antibody (anti-tau-PS262, sc-32828, Santa Cruz), respectively (see Supporting Information). Cell lysates taken at different times after the isoflurane treatment were flowed into the microfluidic channel (Figure 5b). The resonance shifts observed from the photonic crystal nanosensors are highly consistent with the live-cell measurement by nFTP: as shown in Figure 5c, the time to reach a 50% increase in Aβ42 level (approximately 5 h) was less than that needed for a 50% increase in tau-PS262 level (approximately 7 h). The nFTP approach, therefore, is proved to be a reliable method that has the unique capability of revealing protein dynamics in live, single cells. The limitation of nFTP includes low throughput to interrogate large amounts of cells, which would be improved by multiplexing single probes into probe arrays. For the same reason, we did not perform statistical analysis for the data obtained; however, the findings were confirmed in repeated experiments.
Figure 5.
Aβ42 and tau-PS262 detection in the SH-SY5Y-tau cell lysates using photonic crystal nanosensors. (a) Scanning electron microscope (SEM) image of the photonic crystal nanobeam cavity. (b) Schematics of the sensor chip, where light paths are denoted in blue and fluid paths are denoted in gray. The sensor chip is fabricated in silicon-on-insulator (SOI) platform. Telecom lasers were coupled to the edge of the chip and collected from the opposite side. Microfluidic channels were bonded on the chip for sample delivery. (c) Resonance shifts of the photonic crystal nanosensors measured in different cell lysates collected at different times after the isoflurane was applied.
In conclusion, nFTP technique brings traditional immunoassay to the nanoscale dimension and nanomolar sensitivity by integrating a nanoplasmonic sensor on a nanoscale optical fiber tip. This technology, at the same time, enables the detection of post-translational modifications of proteins in live cells, which traditionally have been measured by first separating the proteins via electrophoresis, chromatography, or immunoprecipitation and detecting them with mass spectrometry or fluorescent-labeling methods.28–31 The nFTP approach is particularly powerful to study protein dynamics and post-translational modifications in live cells, as well as to determine the outcomes of interventions of neurodegenerative diseases.32–36 Its high-sensitivity, free-of-labels, and single cell capability make it a unique tool to diagnose clinically obtained, limited, primary cells.
MATERIALS AND METHODS
Nanoplasmonic Fiber Tip Probe (nFTP) Fabrication
The nFTP was fabricated by wet-etching an optical fiber (SM28, Thorlabs Inc.) with hydrogen fluoride chemistry by precisely monitoring and controlling the etch time.17 Gold nanorods were dispersed on the coverslip and then picked up onto the nanosize tip of the nFTP by using a micromanipulator under the dark field of an inverted microscope. A UV-curing optical adhesive (Norland NOA 128) was used to adhere a nanorod to the tip. The nFTP had a tip size of approximately 50 nm; the gold nanorod was 86 nm long, and had a 25 nm diameter cross-section (Nanopartz Inc.).
Surface Functionalization
One milliliter of cetrimonium bromide (CTAB)-capped gold nanorods (Nanopartz Inc.) was mixed with 100 μL of a 20 mM solution of 11-mercaptoundecanoic acid (11-MUA, Sigma-Aldrich) prepared in ethanol. The mixture was sonicated for 90 min at 55 °C and kept at room temperature overnight. These nanorods were centrifuged at 5500 rcf for 10 min and redispersed in water to remove the excess 11-MUA. A single nanorod was immobilized on the tip of the nFTP. The assembled nFTP was incubated in 100 mM 1-ethyl-3-(3-(dimethylamino)propyl)carbodiimide (EDC, Sigma-Aldrich) with 100 nM of antibodies to the target proteins. In this work, we used anti-Aβ42 (ab10148, Abcam), anti-tau-PS262 (sc-32828, Santa Cruz) and anti-β-Actin (ab13822, Abcam). To block the nonspecific binding, the nFTP was incubated in the cell culture medium in which fetal bovine serum (FBS) was replaced by 1% bovine serum albumin (BSA).
Cell Culture
The wild-type human neuroblastoma SH-SY5Y cells [CRL-2266, American Type Culture Collection (ATCC)] and tau overexpressed SH-SY5Y-tau cells (generous gift of Dr. Luc Buee from Inserm UMR1172, France) were cultured at 37 °C in a humidified incubator with 5% CO2 in a DMEM F-12 medium (ATCC, 30-2006) supplemented with 10% fetal bovine serum (ATCC, 30-2020), 100 U/ml penicillin, 100 μg/mL streptomycin (Sigma, P4333). For SH-SY5Y-tau cells, we added additional 1% (v/v) minimum essential medium (MEM) non-essential amino acid solution (Sigma, M7145).
Anesthetic Isoflurane Treatment
A mixture of 21% oxygen, 5% carbon dioxide, 2% isoflurane, and balanced nitrogen were delivered from an anesthesia vaporizer to a sealed plastic box in a 37 °C incubator containing plates seeded with cells for 2 h. A mixture of 21% oxygen, 5% carbon dioxide, and balanced nitrogen without isoflurane were used in the control condition.19
Supplementary Material
ACKNOWLEDGMENTS
The authors are thankful for helpful discussions with Diane Schaak at Rowland Institute at Harvard University. The authors thank Fang Fang and Lining Huang from Massachusetts General Hospital and Harvard Medical School for their help in harvesting cortex neurons of mice. The cost of isoflurane was generously provided by the Department of Anesthesia, Critical Care and Pain Medicine, Massachusetts General Hospital and Harvard Medical School. The research was supported in part by the Rowland Junior Fellowship Award at Rowland Institute at Harvard University, Cambridge, Massachusetts (to Q.Q.), and in part by R01 GM088801, R01 AG041274, and R01 HD086977 from the National Institutes of Health, Bethesda, Maryland (to Z.X.).
Footnotes
Author Contributions
Q.Q., Z.X., Y.Z., and F.L. designed the research. F.L. performed the experiments with contributions from Y.Z., W.H., and Y.D. prepared the mice primary neurons. Q.Q., Z.X., F.L., and Y.Z. analyzed the experimental results and wrote the manuscript.
The authors declare no competing financial interest.
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.6b00320.
Details of the surface functionalization, regeneration, calibration and mechanical design of the revolving dual-nFTP and Western blot validation of tau phosphorylation inhibitor K252a. It also includes the fabrication and surface functionalization of the photonic crystal nanosensors. (PDF)
REFERENCES
- 1.Hardy J, Selkoe DJ. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 2.Duff K. Biochem. Soc. Symp. 2001;67(67):195–202. doi: 10.1042/bss0670195. [DOI] [PubMed] [Google Scholar]
- 3.Chin J. Methods Mol. Biol. 2010;670:169–89. doi: 10.1007/978-1-60761-744-0_13. [DOI] [PubMed] [Google Scholar]
- 4.Small SA, Duff K. Neuron. 2008;60(4):534–42. doi: 10.1016/j.neuron.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lichtman JW, Conchello JA. Nat. Methods. 2005;2(12):910–9. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 6.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. Science. 2006;312(5771):217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 7.Tian B, Cohen-Karni T, Qing Q, Duan X, Xie P, Lieber CM. Science. 2010;329(5993):830–834. doi: 10.1126/science.1192033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan W, Shi ZY, Smith S, Birnbaum D, Kopelman R. Science. 1992;258:778–81. doi: 10.1126/science.1439785. [DOI] [PubMed] [Google Scholar]
- 9.Vo-Dinh T, Alarie JP, Cullum BM, Griffin GD. Nat. Biotechnol. 2000;18(7):764–767. doi: 10.1038/77337. [DOI] [PubMed] [Google Scholar]
- 10.Vo-Dinh T, Kasili P. Anal. Bioanal. Chem. 2005;382(4):918–925. doi: 10.1007/s00216-005-3256-7. [DOI] [PubMed] [Google Scholar]
- 11.Singhal R, Orynbayeva Z, Kalyana Sundaram RV, Niu JJ, Bhattacharyya S, Vitol EA, Schrlau MG, Papazoglou ES, Friedman G, Gogotsi Y. Nat. Nanotechnol. 2011;6(1):57–64. doi: 10.1038/nnano.2010.241. [DOI] [PubMed] [Google Scholar]
- 12.Spiller DG, Wood CD, Rand DA, White MR. Nature. 2010;465:736–45. doi: 10.1038/nature09232. [DOI] [PubMed] [Google Scholar]
- 13.Yan R, Park JH, Choi Y, Heo CJ, Yang SM, Lee LP, Yang P. Nat. Nanotechnol. 2011;7(3):191–196. doi: 10.1038/nnano.2011.226. [DOI] [PubMed] [Google Scholar]
- 14.Shambat G, Kothapalli SR, Provine J, Sarmiento T, Harris J, Gambhir SS, Vuckovic J. Nano Lett. 2013;13:4999–5005. doi: 10.1021/nl304602d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dreaden EC, Alkilany AM, Huang X, Murphy CJ, El-Sayed MA. Chem. Soc. Rev. 2012;41(7):2740–79. doi: 10.1039/c1cs15237h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee K, Cui Y, Lee LP, Irudayaraj J. Nat. Nanotechnol. 2014;9(6):474–80. doi: 10.1038/nnano.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong W, Liang F, Schaak D, Loncar M, Quan Q. Sci. Rep. 2014;4:6179. doi: 10.1038/srep06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xie XS, Choi PJ, Li GW, Lee NK, Lia G. Annu. Rev. Biophys. 2008;37:417–44. doi: 10.1146/annurev.biophys.37.092607.174640. [DOI] [PubMed] [Google Scholar]
- 19.Xie Z, Dong Y, Maeda U, Moir RD, Xia W, Culley DJ, Crosby G, Tanzi RE. J. Neurosci. 2007;27(6):1247–54. doi: 10.1523/JNEUROSCI.5320-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong Y, Wu X, Xu Z, Zhang Y, Xie Z. PLoS One. 2012;7(6):e39386. doi: 10.1371/journal.pone.0039386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le Corre S, Klafki HW, Plesnila N, Hubinger G, Obermeier A, Sahagun H, Monse B, Seneci P, Lewis J, Eriksen J, Zehr C, Yue M, McGowan E, Dickson DW, Hutton M, Roder HM. Proc. Natl. Acad. Sci. U. S. A. 2006;103(25):9673–8. doi: 10.1073/pnas.0602913103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hubinger G, Geis S, LeCorre S, Muhlbacher S, Gordon S, Fracasso RP, Hoffman F, Ferrand S, Klafki HW, Roder HM. J. Alzheimers Dis. 2008;13(3):281–94. doi: 10.3233/jad-2008-13306. [DOI] [PubMed] [Google Scholar]
- 23.Quan QM, Deotare PB, Loncar M. Appl. Phys. Lett. 2010;96(20):203102. [Google Scholar]
- 24.Quan QM, Loncar M. Opt. Express. 2011;19(19):18529–18542. doi: 10.1364/OE.19.018529. [DOI] [PubMed] [Google Scholar]
- 25.Quan QM, Floyd DL, Burgess IB, Deotare PB, Frank IW, Tang SKY, Ilic R, Loncar M. Opt. Express. 2013;21(26):32225–32233. doi: 10.1364/OE.21.032225. [DOI] [PubMed] [Google Scholar]
- 26.Liang F, Quan QM. ACS Photonics. 2015;2(12):1692–1697. [Google Scholar]
- 27.Liang F, Clarke N, Patel P, Loncar M, Quan Q. Opt. Express. 2013;21(26):32306–12. doi: 10.1364/OE.21.032306. [DOI] [PubMed] [Google Scholar]
- 28.Thingholm TE, Jensen ON, Larsen MR. Proteomics. 2009;9(6):1451–1468. doi: 10.1002/pmic.200800454. [DOI] [PubMed] [Google Scholar]
- 29.Macek B, Mann M, Olsen JV. Annu. Rev. Pharmacol. Toxicol. 2009;49:199–221. doi: 10.1146/annurev.pharmtox.011008.145606. [DOI] [PubMed] [Google Scholar]
- 30.Dephoure N, Gould KL, Gygi SP, Kellogg DR. Mol. Biol. Cell. 2013;24(5):535–542. doi: 10.1091/mbc.E12-09-0677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steen H, Mann M. Nat. Rev. Mol. Cell Biol. 2004;5(9):699–711. doi: 10.1038/nrm1468. [DOI] [PubMed] [Google Scholar]
- 32.Tanzi RE, Bertram L. Cell. 2005;120(4):545–555. doi: 10.1016/j.cell.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 33.Querfurth HW, LaFerla FM. N. Engl. J. Med. 2010;362(4):329–44. doi: 10.1056/NEJMra0909142. [DOI] [PubMed] [Google Scholar]
- 34.Huang Y, Mucke L. Cell. 2012;148(6):1204–22. doi: 10.1016/j.cell.2012.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Israel MA, Yuan SH, Bardy C, Reyna SM, Mu YL, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, Carson CT, Laurent LC, Marsala M, Gage FH, Remes AM, Koo EH, Goldstein LS. B. Nature. 2012;482(7384):216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D'Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY. Nature. 2014;515(7526):274–8. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.