Abstract
We have established a uniform procedure for the expression and purification of the cyclin-dependent kinases CDK7/CycH/MAT1, CDK8/CycC and CDK9/CycT1. We attach a His6-tag to one of the subunits of each complex and then co-express it together with the other subunits in Spodoptera frugiperda insect cells. The CDK complexes are subsequently purified by Ni2+-NTA and Mono S chromatography. This approach generates large amounts of active recombinant kinases that are devoid of contaminating kinase activities. Importantly, the properties of these recombinant kinases are similar to their natural counterparts (Pinhero et al. 2004, Eur J Biochem 271:1004-14). Our protocol provides a novel systematic approach for the purification of these three (and possibly other) recombinant CDKs.
Keywords: Cyclin-Dependent Kinases, Baculoviridae, Isolation and Purification
Introduction
Cyclin-dependent kinases (CDKs) play important roles in the regulation of eukaryotic cell cycle and gene expression. They represent a family of Ser/Thr protein kinases that are critically dependent on their association with a cyclin partner (1). In addition, these kinases are regulated by a variety of mechanisms including phosphorylation events, association with inhibitory proteins and assembly factors, and protein degradation (1).
Three CDKs (CDK7, CDK8 and CDK9) have been implicated in the phosphorylation of the carboxyl-terminal domain (CTD) of the largest subunit of RNA polymerase II (RNA pol II) (2, 3). The phosphorylation of this domain is essential for several post-initiation events during the synthesis of mRNA in all eukaryotes (2, 3). It is important that CDK7, CDK8 and CDK9 are biochemically and functionally distinct kinases (4, 5) that execute non-redundant functions during the transcription reaction (2). CDK7 exists in several forms including a core tri-partite CDK7/CycH/MAT1 complex known as CAK (cyclin-dependent kinase activating kinase), as a component in the general transcription factor TFIIH and as a component of larger complexes containing pol II and other transcription factors (2). CDK8/CycC exists as a bipartite complex, but has also been found in a variety of complexes that contain pol II and general pol II transcription factors (2). CDK9 was initially identified as P-TEFb (Positive Transcription Elongation Factor-b) (6). Independently, CDK9/CycT1 has been isolated as the HIV-tat associated kinase TAK (7).
CDK7/CycH/MAT1 acts as a cyclin-dependent kinase activating kinase that phosphorylates and activates different CDKs in Xenopus and mammalian extracts (8). As a component of the general transcription factor TFIIH, this complex also phosphorylates the carboxyl-terminal domain of pol II. The retinoic acid receptors (RAR)-α-1 and (RAR)-γ (9, 10), the estrogen receptor-α (11), the transactivator Oct-1 (12) and the tumor suppressor p53 (13) have also been identified as substrates of CDK7. The substrate preferences of CDK8/CycC and CDK9/CycT1 have not been extensively characterized.
All of the previous studies on CDK7, CDK8 and CDK9 were carried out with kinases that were immunoprecipitated from cell extracts or by epitope-tagged recombinant kinases that were purified by analytical scale affinity schemes (for references see (14)). In many cases, the homogeneity of the preparations and the presence of contaminating kinase activities were not systematically addressed. In order to circumvent these problems and the variance that could result from the differences in the purification strategies, we designed a uniform procedure for the purification of recombinant CDK7, CDK8 and CDK9 complexes. Using these recombinant kinases, we identified additional differences in the preference of these kinases towards different parts of the pol II CTD (14). In this manuscript, we provide extensive details on the expression/purification protocols for the three recombinant kinases that will be of use in future attempts to prepare these as well as other cyclin-dependent kinases.
Materials and Methods
Expression vectors
a. Baculovirus for the expression of His6-CDK8
The GibcoBRL Bac-to-Bac baculovirus expression system was used to generate recombinant baculovirus for the expression of His6-CDK8. The human CDK8 coding sequence was PCR amplified from pBSCDK8 (a gift from E. Lees) using primers for the T7 promoter and the C-terminus of CDK8 (5’-GCC GAA TTC GAC TAT GAC TTT AAA GTG AAG CTG AGC-3’) and sub-cloned into the donor plasmid pFastBacHTa (Invitrogen). To generate the desired recombinant baculovirus, the donor plasmid (pFastBacHTa/CDK8) was transformed into DH10BAC cells, which contain the bacmid (Bmon14272, Invitrogen). Recombinant bacmids were isolated from the blue colonies and the presence of the coding sequence of CDK8 was confirmed by PCR. A bacmid containing the His6-CDK8 sequence was transfected into Spodoptera frugiperda (Sf9) cells using cellfectin. The baculoviruses were amplified and used for protein expression.
b. Baculovirus expressing His6-CDK9/CycT1
Recombinant baculovirus for the expression of His6-CDK9/CycT1 was produced using the Novagen Baculovirus expression system. This system utilizes a Bacvector-3000 (Novagen) triple cut virus DNA, which is a modified form of the AcNPV genome. A transfer plasmid cassettes for the expression of His6-CDK9 and CycT1 (pBAC-CDK9/CycT1, a gift from D. Price (15)) and Bacvector-3000 DNA were co-transfected into Sf9 cells. The recombinant baculovirus was amplified and used for protein expression.
c. Baculovirus expressing His6-CycH
The coding sequence of human CycH (from pBS/CycH, a gift from D. Morgan) was subcloned into pBlueBac2 (Invitrogen). The plasmid and the transfection module B825-03 (digested AcNPV DNA, Invitrogen) were co-transfected into Sf9 cells. Purification of blue plaques (containing recombinant virus) was conducted as in reference (16). The presence of the CycH encoding sequence was confirmed by Northern blot.
The baculoviruses for the expression of CDK7 and MAT1 were gifts from D. Morgan (17). The baculovirus for the expression of human CycC was provided by Dr. E. Lees (5).
Growth and maintenance of Sf9 cells
Sf9 cells were cultured at 28°C in tissue culture flasks (175 cm2, SARSTEDT) in Grace’s Medium/1X yeastolate solution/1X lactalbumin hydrolysate solution/1X antibiotic/antimycotic solution (GibcoBRL), and 10% fetal bovine serum (Cansera).
Amplification of recombinant viruses
High titer viral stocks for the production of recombinant proteins were prepared from low passage (1 or 2) viral stocks by a two-step amplification procedure. Sf9 cells at density of 0.1-0.3x106 cells/ml were infected with individual viruses at a multiplicity of infection (MOI) of 0.1-1. The infected cells were incubated for 5-6 days. The efficiency of infection was monitored by the loss of adherence and the larger size of the cells. The supernatant from these cultures was used to infect a larger Sf9 culture of density 0.5-1x106 cells/ml at a MOI of 1. The culture was incubated for 4-5 days, cells were spun and the supernatant was immediately used or stored at -80°C for up to six months. This procedure typically produced viral stocks of ~1x108 pfu/ml.
Expression of recombinant kinases
Expression of recombinant CDK complexes was conducted by co-infecting about 1.5-2x109 Sf9 cells (1.5-2x106 cells/ml) with the appropriate combination of baculoviruses at MOI 4 for each individual virus. The cells were harvested after 48 hours by spinning at 275 g for 5 minutes at 4°C, washed with PBS and frozen (-80°C) in 15 ml of lysis buffer (10 mM Tris.HCl pH 7.5, 10 mM NaCl, 2 mM b-mercaptoethanol, 0.5 mM EDTA, 10 mM 2-glycerophosphate, 0.5 mM Na-vanadate, 2 mM NaF, 2 μg/ml leupeptin, 2 μg/ml aprotonin, 2 μg/ml pepstatin, 0.2 % (v/v) NP-40, 50 μg/ml PMSF). Alternatively, the cells were immediately lysed in lysis buffer by 10 strokes with a Dounce homogenizer. After cell lysis, the proteins were extracted by adding 0.5M NaCl and 5 mM imidazole, and rocking for 30 min at 4°C. The extract was clarified by spinning in a SW50.1 rotor (Beckman) at 75000 g for 30 min and immediately processed by metal (Ni2+) affinity chromatography.
Ni2+-NTA pull-down assay
Pull down assays were performed with 250 μl aliquots of Sf9 cell extracts and 50 μl of 50% (v/v) Ni2+-NTA agarose beads equilibrated with 10 mM Tris.HCl, pH 7.6, 0.5 M NaCl, 5 mM imidazole, 50 μg/ml PMSF and 10% (v/v) glycerol (buffer A). The suspension was rocked on a nutator for 1 h and the beads were pelleted by spinning for 1 minute at 3000 rpm. The beads were then washed five times with 1ml of buffer A + 0.1M NaCl, boiled for 5 minutes in SDS-sample buffer and further analyzed by Western blot or silver staining.
Purification of CDK complexes by Ni2+-NTA chromatography
The cell extract from about 1 liter of infected cells was mixed with 1ml of Ni2+-NTA agarose beads (Qiagen) that were equilibrated with 10 mM Tris.HCl pH 7.6, 0.5 M NaCl, 5 mM imidazole, 50 μg/ml PMSF and 10% (v/v) glycerol, and rocked on a nutator for 1 h. The beads were washed once in the equilibration buffer and transferred to a disposable 10 ml column (Amersham). Bound proteins were step-wise eluted with 15, 25, 100 and 400 mM imidazole in 10 mM Tris-HCl pH 7.6, 0.1 M NaCl, 50 μg/ml PMSF and 10% (v/v) glycerol. The fractions containing the recombinant protein kinases were identified by SDS-PAGE/Coomassie Brilliant Blue R-250 staining, pooled and stored at -80°C.
Purification of CDK complexes by Mono S chromatography
The pooled protein fractions from Ni2+-NTA chromatography were buffer exchanged in PD10 columns (Amersham) to 25 mM HEPES pH 7.6, 0.1 mM EDTA, 1 mM DTT, 5% (v/v) glycerol, 50 μg/ml PMSF and 80 mM NaCl. The proteins were loaded on a tandem of two 5 ml Econo-Pac Mono S cartridges (BioRad) and eluted with a linear 0.08-0.5M NaCl gradient in 25mM HEPES pH 7.6, 0.1 mM EDTA, 1 mM DTT, 50 μg/ml PMSF, and 5% (v/v) glycerol. The fractions containing recombinant protein kinases were identified by SDS-PAGE/silver staining and stored at -80°C.
Kinase substrates
Glutathione-S-transferase carboxyl terminal domain (GST-CTD) was expressed and purified as described (18). Highly purified myelin basic protein (MBP) from bovine brain was a gift from Dr. G. Harauz.
Kinase assays
The kinase assays were performed in a volume of 20 μl containing 20 mM Tris.HCl, pH 8, 50 mM KCl, 7 mM MgCl2, 5 mM 2-glycerophosphate, 100 μg/ml BSA (2μg), 10 μM ATP, 2 μCi (7.4 x 104 Bq) α-32P-ATP (ICN), 40μg/ml (800 ng) GST-CTD or MBP and about 100-400 ng/ml (2-8 ng) of purified kinase. These amounts correspond to 100-500 fold molar excess of substrate versus kinase. It is important to note that the GST-CTD molecule has at least 52 sites of phosphorylation (52 repeats with a consensus YSPTSPS) on a single molecule, thus additionally increasing the kinase/substrate ratio. MBP also contains multiple sites of phosphorylation. Under the described conditions the kinase reactions are linear for at least three hours (data not shown). The kinase reactions were incubated for 30 minutes at 30°C and terminated by the addition of SDS-PAGE loading buffer and then boiled for 5 minutes. Aliquots were analyzed by SDS-PAGE gels and autoradiography. The incorporation of ATP in GST-CTD (1-52) and MBP (pmol of ATP/min/mg of protein) was determined according to the procedure in (19). Kinase assays were also performed in the presence of kinase inhibitors, DRB (5,6-dichlorobenzimidazole riboside), and roscovitine (2-(1-ethyl-2-hydroxyethylamino)-6-benzylamino-9-isopropylpurine). DRB was dissolved at 50 mM in 95% ethanol and stored at -20°C. Working dilutions of 800, 200 and 40 μM in water were prepared at the time of assay and immediately added to the kinase reaction. Roscovitine was dissolved in DMSO at 50 mM and stored at -20°C. Dilutions in water were made prior to the reactions and immediately added to the kinase reaction.
Results and Discussion
Expression of recombinant CDK kinases
We produced recombinant CDK7/His6-CycH/MAT1, His6-CDK8/CycC and His6-CDK9/CycT1 complexes by co-infecting Sf9 cells with the appropriate combination of baculoviruses. Each virus was applied at MOI of 4. Preliminary experiments have shown that this MOI generates the highest level of expression for all the CDKs and that further increase in MOI did not yield more protein. The expression of the different subunits of the CDK complexes was verified by Western blot with antibodies against CDK7, CycH, MAT1, CDK8, CycC and CDK9. These westerns were not quantitative and do not necessarily mean that all peptides were expressed at similar levels.
Monolayer Sf9 cells in large T-flasks were infected at high density (1.5-2x106 cells/ml). We found that this high cell density did not lead to poorer expression or protein degradation as compared to infection at lower cell density (0.5-1 x106 cells/ml). Considering the high cost of growth medium, this approach is more economical. We have not tested if the same trend applies to infecting Sf9 cells that are maintained in suspension.
An important consideration derived from the Western blot analyses was the choice of individual expression of the CDKs and cyclins and their subsequent reconstitution in vitro versus co-expression and assembly in vivo. Both approaches were applied in the past (4, 5, 18, 20-22). Although the Westerns blot analyses were not quantitative, we noticed that a higher level of expression of individual peptides was achieved by co-infection as compared to infection with individual viruses. These observations suggest that complex formation in vivo stabilizes the individual peptides and increases the yields of recombinant proteins. Therefore, we recommend that the production of these kinase complexes is carried out by co-infection rather than individual expression and mixing of the extracts in vitro. It is also important to mention that the mixing of the extracts in vitro poses the risk of inadequate modifications of the individual peptides in vivo that might lead to altered properties of the reconstituted kinases (18, 20).
Confirmation of complex formation by Ni2+-NTA pull-down assay
We confirmed the formation of complexes between the recombinant proteins by Ni2+-NTA pull-down assay. Cell extracts were incubated with Ni 2+-NTA beads and the retained proteins were analyzed by Western blot (Fig. 1A). The results showed that His6-CycH pulls down CDK7 and MAT1, His6-CDK8 pulls down CycC and His6-CDK9 pulls down CycT1 (Fig. 1A).
Fig. 1. Expression and assembly of CDK complexes in Sf9 cells.

A: Pull down assays were performed with 250 μl Sf9 cell lysates and 25 μl Ni2+-NTA agarose beads. The proteins were separated on 10% SDS gels, transferred to PVDF membrane and detected by Western blot with antibodies against the polypeptides shown on the left. B: Proteins from the pull down assay was separated on a 10% gel. Silver stained gel is shown. A prominent band of about 87 kDa most likely corresponds to CycT1, whereas the 43 kDa corresponds to His6-CDK9.
Antibodies against CycT1 were not available. However, a prominent band of about 87 kDa most likely corresponding to CycT1 was detected in Ni2+-NTA pull-down fractions from His6-CDK9/CycT1 infected cells by silver staining (Fig. 1B). Thus, the recombinant proteins form complexes in vivo. Another important conclusion from the image in Fig. 1B is that the pull-down fractions are unclean and should be used only for confirmation of expression and complex formation, but not for fine analyses.
Purification of CDK complexes by Ni2+-NTA chromatography
All the CDK complexes were uniformly purified by a two-step process involving Ni2+-NTA agarose chromatography followed by Mono S chromatography (Fig. 2).
Fig. 2. A strategy for the purification of CDK complexes.
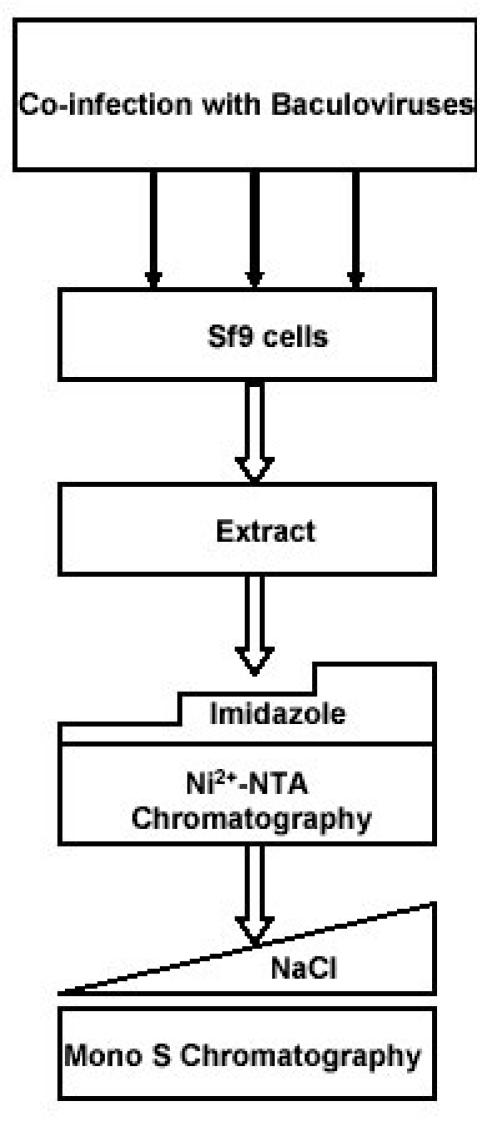
Sf9 cells were co-infected with the baculoviruses expressing cyclin dependent kinases and their corresponding cyclin partners. Cell extract was loaded on Ni2+-NTA beads and eluted with stepwise imidazole gradients. Fractions containing the recombinant kinases were pooled, buffer exchanged on PD10 column and loaded on Mono S beads and purified by a linear 0.08-0.5 M NaCl gradient.
During Ni2+-NTA chromatography, CDK7/His6-CycH/MAT1 complexes were eluted at a low imidazole concentration of 25 mM (Fig. 3A), whereas His6-CDK8/CycC and His6-CDK9/CycT1 were eluted at higher imidazole concentrations of 100 and 400 mM, respectively (Figs. 3B and C). At this stage, the fractions containing different recombinant kinases were contaminated with different sets of peptides that in certain cases contributed to estimated 40% of the total protein. We did not investigate the nature of these proteins. Also, we detected a very significant CTD kinase activity in crude extracts and in the Ni2+-NTA fractions from the uninfected cell extracts, which precludes the quick and reliable detection of the expression of the kinases by kinase assays. This observation also indicates that the Ni2+-NTA fractioned kinases are inappropriate for analysis of their kinase activity or other fine assays. This necessitated further purification by ion exchange chromatography to remove the contaminating proteins.
Fig. 3. Purification of CDK complexes by Ni2+-NTA agarose chromatography.
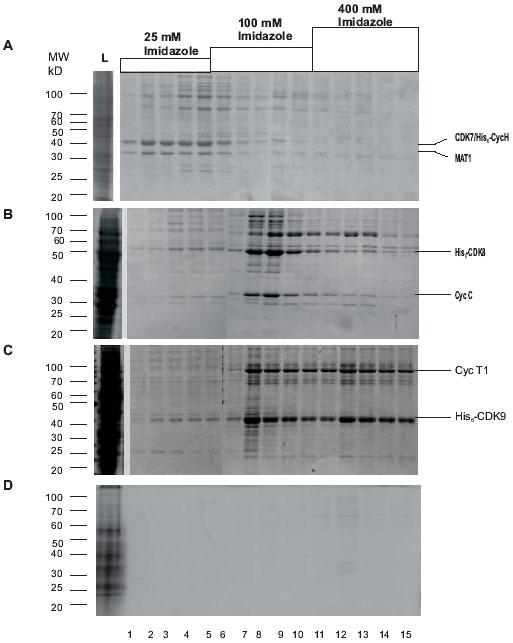
A: CDK7/His6-CycH/MAT1; B: His6-CDK8/CycC; C: His6-CDK9/CycT1; D: Proteins from uninfected Sf9 cells. Coomassie stained gels with different fractions from the Ni2+-NTA agarose chromatography of cell extracts are shown. Molecular weight markers are shown on the left. The bands corresponding to the kinases and the cyclin partners are shown on the right. L, load. The concentration of imidazole in the corresponding fractions is shown on the top of the figure.
Purification of CDK complexes by Mono S chromatography
All the fractions from the Ni2+-NTA agarose chromatography that contained the CDK complexes of interest were pooled and purified by Mono S chromatography. This additional step produced significantly cleaner CDK complexes (Figs. 4A, B and C).
Fig. 4. Purification of CDK complexes from pooled Ni2+-NTA fractions by Mono S chromatography.
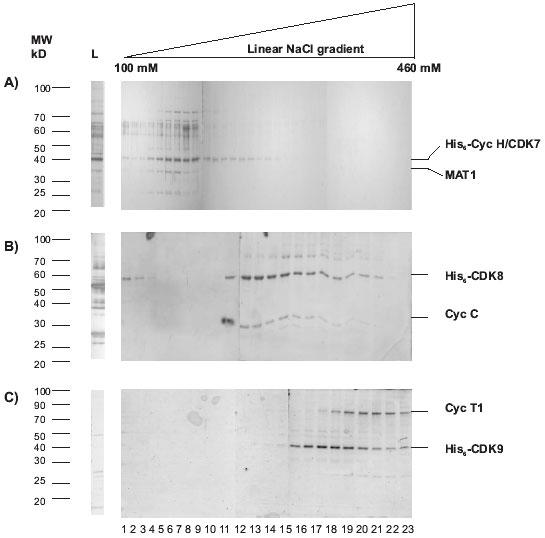
A linear gradient of 80-500 NaCl was applied. The 100-460 mM range of the gradient is shown. A: CDK7/His6-CycH/MAT1; B: His6-CDK8/CycC; C: His6-CDK9/CycT1. Silver stained gels with different fractions are shown. Molecular weight markers are shown on the left. The bands corresponding to the kinases and the cyclin partners are shown on the right. L, load.
In the case of CDK7/His6-CycH/MAT1, a peak consisting of two bands with molecular weights of approximately 40 and 36 kDa was eluted at a low salt concentration of around 100 mM NaCl (Fig. 4A, lanes 1-9). Western blot analyses (not shown) indicated that the 40 kDa band is a doublet of CDK7 and His6-CycH, while the 36 kDa corresponds to MAT1. It should be noted that the peak was followed by a trail of the 40 kDa band that appeared to possess the CDK7/His6-CycH doublet but lack the MAT1 band (Fig. 4A, lanes 10-15). Our results suggest that besides the tripartite CDK7/His6-CycH/MAT1 complex, a bipartite CDK7/His6-CycH complex and possibly monomeric His6-CycH were purified during the Ni2+-NTA agarose step. Earlier reports have established that the tripartite and bipartite complexes differ in their substrate specificity (18, 20). Thus, the Mono S purification step is necessary not only to exclude the contaminating peptides from the Ni2+-NTA agarose step, but also to resolve these two complexes.
His6-CDK8/CycC (Fig. 4B, lanes 12-21) and His6-CDK9/CycT1 (Fig. 4C, lanes 15-23) were eluted at higher salt concentrations from the Mono S column. His6-CDK8, CycC and His6-CDK9 were estimated to be approximately 53, 36 and 43 kDa, respectively by Western blot analyses (data not shown). The peak of His6-CDK8/CycC (Fig. 4B, lanes 12-17) was followed by a trail of fractions containing His6-CDK8 only (Fig. 4B, lanes 19-22). In the case of His6-CDK9/CycT1, the peak containing the complex (Fig. 4C, lanes 19-23) was preceded by fractions that contained predominantly His6-CDK9 (Fig. 3C, lanes 15-18). Again, the profile of peptides in the Mono S chromatography fractions clearly indicates that the affinity chromatography step purifies a mixture of assembled complexes and monomeric tagged polypeptides. This observation should be considered if the properties of the monomeric subunits versus the assembled complex are to be analyzed.
Recovery of recombinant CDKs
We estimate that, on an average, from 1 liter culture of infected Sf9 cells, we purified about 115 mg, 880 mg and 1.2 mg of pure CDK7/His6-CycH/MAT1, His6-CDK8/CycC and His6-CDK9/CycT1 respectively. We suspect that the lower yield of CDK7/ His6-CycH/MAT1 is stemming from its poorer affinity towards Ni2+-NTA agarose as exemplified by its elution at lower imidazole concentration (Fig. 3A).
Activity of the kinases
The functional activities of the purified kinases were studied using GST-CTD and MBP as substrates ((14) and Fig. 5)). All the kinases phosphorylated GST-CTD and MBP (Fig. 5). CDK7/His6-CycH/MAT1 transferred 1.6 and 0.94 nmols of ATP/min/mg protein with GST-CTD and MBP as substrates, respectively. The His6-CDK8/CycC preparations showed somewhat lower specific activities of 0.1 and 0.15 nmols of ATP/min/mg protein with GST-CTD and MBP, whereas the specific activities for His6-CDK9/CycT1 were 0.38 and 3.74 nmols of ATP/min/mg protein. No kinase activity was detected in the corresponding fractions from non-infected Sf9 cell. Importantly, the purified kinases displayed distinct patterns of CTD phosphorylation, which were reported elsewhere (14).
Fig. 5. Phosphorylation of GST-CTD and MBP by CDK7/His6-CycH/MAT1, His6-CDK8/CycC and His6-CDK9/CycT1.
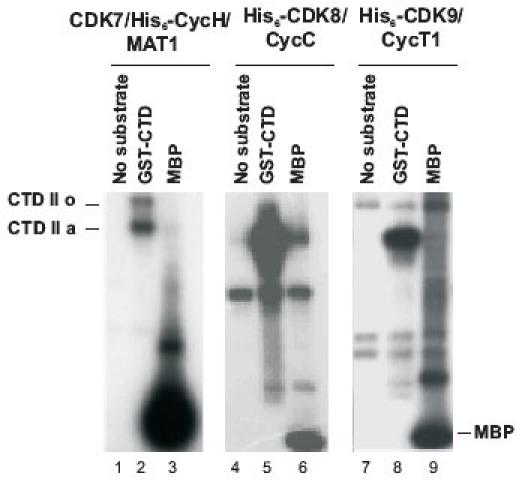
Lanes 1, 4, and 7 are kinases without any substrate. CDK8 and CDK9 have background bands that resulted from their autophosphorylation. CDK7 produced two bands with GST-CTD, the low mobility band corresponds to the hyperphosphorylated form (IIo), and the higher mobility band, the hypophosphorylated IIa. CDK8 and CDK9 produced only the IIa form.
As reported by others, roscovitine is a potent inhibitor of CDK7 and CDK9, but not CDK8 (23-25), whereas DRB inhibits CDK9 at lower concentrations as compared to CDK7 (26). We used this information and performed several experiments to confirm the catalytical purity of the kinases. In Fig. 6, we show that, in agreement with the literature, roscovitine strongly inhibits CDK7/His6-CycH/MAT1 (Fig. 6, upper panel, lanes 1-5), but not His6-CDK8/CycC (Fig. 6, upper panel, lanes 6-10). In addition, we also show that His6-CDK9/CycT1 (Fig. 6, upper panel, lanes 11-15) is equally sensitive to roscovitine as compared to CDK7/His6-CycH/MAT1 (Fig. 6, upper panel, lanes 1-5, see also the graph in the lower panel of the figure). DRB is a potent inhibitor of His6-CDK9/CycT1 (Fig. 6, lower panel, lanes 11-15) and a moderate inhibitor of CDK7/His6-CycH/MAT1 (Fig. 6, lower panel, lanes 1-5). His6-CDK8/CycC showed intermediate sensitivity to DRB (Fig. 6, lower panel, lanes 6-10). Together, this data shows the expected inhibition patterns of these kinases with two independent kinase inhibitors thus supporting the notion of their catalytic purity.
Fig. 6. Effect of roscovitine and DRB on the phosphorylation of GST-CTD by CDK7/His6-CycH/MAT1, His6-CDK8/CycC and His6-CDK9/CycT1.

Roscovitine and DRB were added to the kinase reaction mixtures at concentrations indicated and kinase reactions were carried out as described in Materials and Methods. Lanes 1, 6 and 11 are kinases without substrate. The graph shows the percentage of the kinase activity as compared to their corresponding activity without inhibitors.
Significance and novelty of the proposed purification procedure
The significance of this uniform procedure is rationalized as follows. When CDK complexes are obtained by different expression/purification methods, there is a real risk for the introduction of contaminating kinases and other peptides. Consequently, the comparison of such complexes can be impaired by the nature of the impurities. This concern is especially valid when working with immunoprecipitated complexes, which may contain other kinases that are difficult to track. If the immunoprecipitates are derived from crude extracts, it is also possible that the experiment will be conducted with multiple forms of these complexes that share the same CDK component (see (2) for reference).
Many of the studies that have characterized CDK7, CDK8 and CDK9 have used either immunoprecipitated enzyme complexes (4, 27-29) or different methods for purification of different kinases (4, 5, 18, 20, 30). In order to achieve a high precision in the comparison of the three CDK complexes, we expressed and purified them by a uniform procedure. This procedure generates large amounts of kinases that are practically devoid of other kinase activities. It is important that this procedure enabled us to identify distinct preferences of CDK7, CDK8 and CDK9 towards different parts of pol II CTD (14). This analysis is demanding. The CTD consists of 52 repeats of a Ser/Thr-rich consensus heptapeptide (2) which might attract promiscuous kinase activities. We believe that our success in these analyses demonstrates the high value of the proposed procedure.
We did not detect measurable CTD kinase activities in the corresponding fractions from uninfected cells, meaning that our preparations are not contaminated with independent co-purifying CTD kinase activities. At the same time, we observed different patterns of contaminating peptides in the preparations of CDK7, CDK8, and CDK9. Our interpretation is that these peptides co-purified probably because of interactions with the complexes. We are not sure how these peptides contributed to the activity and specificity of each kinase. The inhibitory studies with roscovitine and DRB (see Fig. 6) and juglone (14) suggest that most if not all of the CTD kinase activities should be attributed to the recombinant kinases. However, we can not exclude the possibility that minor amounts of very active contaminating kinases could alter the results with other substrates. We recommend that a set of inhibitory studies is preformed with each substrate to minimize these risks.
A similar, but not identical procedure for the purification of His-CDK9/CycT1 was used in an earlier study (15). The procedures for the purification of His6-CDK8/CycC and CDK7/His6-CycH/MAT1 are novel.
We believe that our systematic approach can be applied effectively to characterize other CDKs. In addition, we believe that the presented details on the purification of the three CDKs can be of use when other finer analyses of CDK7, CDK8 and CDK9 are conducted.
Acknowledgments
We thank Drs. D. Morgan, E. Lees and D. Price for providing baculoviruses and vectors for the expression of the recombinant kinases. MBP was a gift from Dr. G. Harauz. This study was supported by grants to K. Y. from the Natural Sciences and Engineering Research Council of Canada (NSERC #217548) and the Ontario Genomics Institute (OGI #043567).
Abbreviations
- CDK
cyclin-dependent kinase
- CTD
carboxyl-terminal domain of RNA polymerase II
- Cyc
cyclin
- MBP
myelin basic protein
Appendix
Protocols
Amplification of baculovirus stocks
Infect 10 ml Sf9 insect culture in a 25 cm2 T-flask (SARSTEDT) at a density of 0.1-0.3x106 cells/ml with 0.1 ml of virus stock passage 1 or passage 2 (MOI 0.1-1.).
Incubate the cells for 5-6 days at 28°C. We consider that 100% infection is achieved when all the cells are detached from the substrate. Normally, 100% of the infection is observed by day 4.
Pellet the cells by spinning at 275xg for 5 minutes and collect the supernatant.
Use 1 ml of this supernatant to infect a large culture of 75 ml at a density of 0.5-1x106 cells/ml in a 175 cm2 flask (SARSTEDT). Incubate the cells for 4-5 days and collect the supernatant.
Use the supernatant from step 4 for infecting the Sf9 culture for large scale protein expression. The stocks can be stored at 4°C for a week or -80°C for up to six months. These stocks (Passage 4) typically contain about 1x108 pfu/ml.
Infecting and harvesting insect cells
Grow Sf9 cells to a density of 1.5-2x106 cells/ml in several (5-8) 175 cm2 T-flasks.
Infect each flask with 2 ml of passage 4 of each of the appropriate viruses and incubate for 48 hours at 28°C. Check the cells for infection under microscope. 100% infection (large floating cells) should be observed 30 hours post infection.
Harvest the cells after 48 hours by spinning at 275x g for 5 minutes at 4°C.
Pour off the supernatant, loosen the pellet by gently shaking it and wash the pellets with 15 ml of phosphate buffered saline.
Add approximately 3 pellet volumes of Lysis buffer (10 mM Tris, pH 7.5, 10 mM NaCl, 2 mM β-merkaptoethanol, 0.5 mM EDTA, 10 mM 2-glycerophosphate, 0.5 mM Na vanadate, 2 mM NaF, 2 μg/ml leupeptin, 2 μg/ml aprotonin, 2 μg/ml pepstatin, 0.2% NP-40, 50 μg/ml PMSF) and freeze at -80°C.
Preparation of cell lysate
Thaw the re-suspended cells in an ice bath by gently swirling the tube. Add fresh PMSF.
Leave the lysate on ice for 10 minutes.
Pass the lysate through a dounce homogenizer 10 times. Check for cell lysis under a microscope.
Add imidazole and NaCl to a final concentration of 5 mM and 0.5 M, respectively.
Rock the lysate for 30 minutes in the cold room.
Spin at 75000 x g at 4°C in SW50.1 rotor (Beckman) for 30 minutes.
Transfer the supernatant to a fresh pre-chilled tube; add glycerol and MgCl2 to a final concentration of 10% and 3 mM, respectively.
Note: You can freeze the sample at this point. If planning to load to the N2+-NTA agarose column on the same day, do not add glycerol.
Ni2+-NTA chromatography
Equilibrate 1 ml Ni2+-NTA agarose beads (Qiagen) with 10 ml of buffer A (10 mM Tris-HCl pH 7.6, 0.5 M NaCl, 5 mM imidazole, 50 μg/ml PMSF, 10% glycerol) in a 50 ml falcon tube.
Spin the beads at 3000 rpm for 2 minutes in a table-top centrifuge and aspire the supernatant.
Add the cell extract from 1 liter of infected cells. Rock the tube for 1 h in the cold room.
Pellet the beads at 3000 rpm for 2 minutes in a table-top centrifuge, collect the supernatant (flow through) by aspiration, and store at -80°C. If there is any problem with binding, the supernatant can be used again.
Add 10 ml buffer A to the beads and transfer to a 15 ml disposable BioRad column.
Wash the column with 10 ml buffer A + 0.1 M NaCl in the cold room.
Elute the proteins as 5 x 1ml fractions in steps using
10 mM Tris-HCl pH 7.6, 0.1 M NaCl, 50 μg/ml PMSF, 10% glycerol and 15 mM imidazole.
10 mM Tris-HCl pH 7.6, 0.1 M NaCl, 50 μg/ml PMSF, 10% glycerol and 25 mM imidazole.
10 mM Tris-HCl pH 7.6, 0.1 M NaCl, 50 μg/ml PMSF, 10% glycerol and 100 mM imidazole.
10 mM Tris-HCl pH 7.6, 0.1 M NaCl, 50 μg/ml PMSF, 10% glycerol and 400 mM imidazole.
Run samples of the fractions on SDS-PAGE and stain with Coomassie Brilliant Blue. Pool the fractions containing the proteins of interest and store at -80°C.
Mono S chromatography
Equilibrate a PD10 column (Amersham Pharmacia Biotech) with 25 ml of 25 mM HEPES, pH 7.6, 5% glycerol, 0.1 mM EDTA, 0.08M NaCl, 1 mM DTT, 50 μg/ml PMSF.
Buffer exchange the pooled fractions from the Ni2+-NTA chromatography step as per manufacturer’s instruction.
Attach two 5 ml Econo-Pac Mono S cartridges (BioRad) in tandem and equilibrate as follows:
5 ml of buffer A (25 mM HEPES, pH 7.6, 5% glycerol, 0.1 mM EDTA, 1 mM DTT, 50 μg/ml PMSF) at 0.5 ml/min.
5 ml of buffer A at 1ml/min.
10 ml of 1M NaCl in buffer A.
5 ml of gradient 1M - 80 mM NaCl in buffer A at 1 ml/min.
15 ml of 80 mM NaCl in buffer A.
Load the column at 1ml/min and collect the flow through and save.
Wash the column with 5 ml of 80 mM NaCl in buffer A at 1 ml/min.
Elute with a linear gradient of 20 ml of 80-500 mM NaCl in buffer A at 1 ml/min.
Collect 1 ml fractions in 1.5 ml tubes containing 100 μl of 80% glycerol.
Analyze the samples of the fractions by SDS-PAGE and silver staining. Pool the fractions of interest and store at -80°C.
Equipment
Tissue culture hood
Basic radiation safety equipment and Geiger counter
Ultracentrifuge, centrifuge (Beckman)
Mini-PROTEAN II electrophoresis cell (BioRad)
Microcentrifuge, incubator, standard Molecular Biology equipments, gel dryer
References
- Morgan DO. Cyclin-dependent kinases: engines, clocks, and microprocessors. The dynamics of cyclin dependent kinase structure. Annu Rev Cell Dev Biol. 1997;13(6):261–291. doi: 10.1146/annurev.cellbio.13.1.261. [DOI] [PubMed] [Google Scholar]
- Kobor MS, Greenblatt J. Regulation of transcription elongation by phosphorylation. Biochim Biophys Acta. 2002;1577(2):261–275. doi: 10.1016/s0167-4781(02)00457-8. [DOI] [PubMed] [Google Scholar]
- Palancade B, Bensaude O. Investigating RNA polymerase II carboxyl-terminal domain (CTD) phosphorylation. Eur J Biochem. 2003;270(19):3859–3870. doi: 10.1046/j.1432-1033.2003.03794.x. [DOI] [PubMed] [Google Scholar]
- Ramanathan YS, Rajpara M, Reza SM, Lees E, Shuman S, Mathews MB, Pe'ery T. Three RNA polymerase II carboxyl-terminal domain kinases display distinct substrate preferences. J Biol Chem. 2001;276(14):10913–10920. doi: 10.1074/jbc.M010975200. [DOI] [PubMed] [Google Scholar]
- Rickert P, Corden JL, Lees E. Cyclin C/CDK8 and cyclin H/CDK7/p36 are biochemically distinct CTD kinases. Oncogene. 1999;18(4):1093–1102. doi: 10.1038/sj.onc.1202399. [DOI] [PubMed] [Google Scholar]
- Price DH. P-TEFb, a cyclin-dependent kinase controlling elongation by RNA polymerase II. Mol Cell Biol. 2000;20(8):2629–2634. doi: 10.1128/MCB.20.8.2629-2634.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping YH, Rana TM. Tat-associated kinase (P-TEFb): a component of transcription preinitiation and elongation complexes. J Biol Chem. 1999;274(11):7399–7404. doi: 10.1074/jbc.274.11.7399. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Cyclin-dependent kinase-7 - at the cross-roads of transcription, DNA-repair and cell-cycle control. Curr Opin Cell Bio. 1996;8(3):312–317. doi: 10.1016/s0955-0674(96)80003-2. [DOI] [PubMed] [Google Scholar]
- Rochette-Egly C, Adam S, Rossignol M, Egly JM, Chambon P. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 1997;90(1):97–107. doi: 10.1016/S0092-8674(00)80317-7. [DOI] [PubMed] [Google Scholar]
- Bastien J, Adam-Stitah S, Riedl T, Egly JM, Chambon P, Rochette-Egly C. TFIIH interacts with the retinoic acid receptor gamma and phosphorylates its AF-1-activating domain through cdk7. J Biol Chem. 2000;275(29):21896–21904. doi: 10.1074/jbc.M001985200. [DOI] [PubMed] [Google Scholar]
- Chen D, Riedl T, Washbrook E, Pace PE, Coombes RC, Egly JM, Ali S. Activation of estrogen receptor alpha by S118 phosphorylation involves a ligand-dependent interaction with TFIIH and participation of CDK7. Mol Cell. 2000;6(1):127–137. doi: 10.1016/S1097-2765(00)00014-9. [DOI] [PubMed] [Google Scholar]
- Inamoto S, Segil N, Pan ZQ, Kimura M, Roeder RG. The cyclin-dependent kinase-activating kinase (CAK) assembly factor, MAT1, targets and enhances CAK activity on the POU domains of octamer transcription factors. J Biol Chem. 1997;272(47):29852–29858. doi: 10.1074/jbc.272.47.29852. [DOI] [PubMed] [Google Scholar]
- Ko LJ, Shieh SY, Chen X, Jayaraman L, Tamai K, Taya Y, Prives C, Pan ZQ. p53 is phosphorylated by CDK7-cyclin H in a p36MAT1-dependent manner. Mol Cell Biol. 1997;17(21):7220–7229. doi: 10.1128/mcb.17.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhero R, Liaw P, Bertens K, Yankulov K. Three cyclin-dependent kinases preferentially phosphorylate different parts of the C-terminal domain of the large subunit of RNA polymerase II. Eur J Biochem. 2004;271(5):1004–1014. doi: 10.1111/j.1432-1033.2004.04002.x. [DOI] [PubMed] [Google Scholar]
- Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12(5):755–762. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Banville M, Lalumiere M, Vialard J, Meighen EA. Bacterial luciferase produced with rapid-screening baculovirus vectors is a sensitive reporter for infection of insect cells and larvae. Intervirology. 1992;34(4):213–227. doi: 10.1159/000150285. [DOI] [PubMed] [Google Scholar]
- Fisher R, Jin P, Chamberlin H, Morgan D. Alternative mechanisms of CAK assembly require an assembly factor or an activating kinase. Cell. 1995;83(1):47–57. doi: 10.1016/0092-8674(95)90233-3. [DOI] [PubMed] [Google Scholar]
- Yankulov KY, Bentley DL. Regulation of CDK7 substrate specificity by MAT1 and TFIIH. Embo J. 1997;16(7):1638–1646. doi: 10.1093/emboj/16.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa U, Minakuchi R, Takai Y, Nishizuka Y. Calcium-activated, phospholipid-dependent protein kinase (protein kinase C) from rat brain. Methods Enzymol. 1983;99:288–298. doi: 10.1016/0076-6879(83)99064-x. [DOI] [PubMed] [Google Scholar]
- Rossignol M, Egly JM. Substrate specificity of the cdk-activating kinase (CAK) is altered upon association with TFIIH. Embo J. 1997;16(7):1628–1637. doi: 10.1093/emboj/16.7.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RP, Morgan DO. A novel cyclin associates with MO15/CDK7 to form the CDK-activating kinase. Cell. 1994;78(4):713–724. doi: 10.1016/0092-8674(94)90535-5. [DOI] [PubMed] [Google Scholar]
- Larochelle S, Chen J, Knights R, Pandur J, Morcillo P, Erdjument-Bromage H, Tempst P, Suter B, Fisher RP. T-loop phosphorylation stabilizes the CDK7-cyclin H-MAT1 complex in vivo and regulates its CTD kinase activity. Embo J. 2001;20(14):3749–3759. doi: 10.1093/emboj/20.14.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungman M, Paulsen MT. The cyclin-dependent kinase inhibitor roscovitine inhibits RNA synthesis and triggers nuclear accumulation of p53 that is unmodified at Ser15 and Lys382. Mol Pharmacol. 2001;60(4):785–789. [PubMed] [Google Scholar]
- Schang LM. Cyclin-dependent kinases as cellular targets for antiviral drugs. J Antimicrob Chemother. 2002;50(6):779–792. doi: 10.1093/jac/dkf227. [DOI] [PubMed] [Google Scholar]
- Taylor SL, Kinchington PR, Brooks A, Moffat JF. Roscovitine, a cyclin-dependent kinase inhibitor, prevents replication of varicella-zoster virus. J Virol. 2004;78(6):2853–2862. doi: 10.1128/JVI.78.6.2853-2862.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano M, Abdellatif M, Oh H, Xie M, Bagella L, Giordano A, Michael LH, DeMayo FJ, Schneider MD. Activation and function of cyclin T-Cdk9 (positive transcription elongation factor-b) in cardiac muscle-cell hypertrophy. Nat Med. 2002;8(11):1310–1317. doi: 10.1038/nm778. [DOI] [PubMed] [Google Scholar]
- Zhou M, Halanski MA, Radonovich MF, Kashanchi F, Peng J, Price DH, Brady JN. Tat modifies the activity of CDK9 to phosphorylate serine 5 of the RNA polymerase II carboxyl-terminal domain during human immunodeficiency virus type 1 transcription. Mol Cell Biol. 2000;20(14):5077–5086. doi: 10.1128/MCB.20.14.5077-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois MF, Vincent M, Vigneron M, Adamczewski J, Egly JM, Bensaude O. Heat-shock inactivation of the tfiih-associated kinase and change in the phosphorylation sites on the c-terminal domain of RNA-polymerase-II. Nucleic Acids Research. 1997;25:694–700. doi: 10.1093/nar/25.4.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee A, Wu L, Liu L, Kobayashi R, Xiong Y, Hall FL. Biochemical characterization of the human cyclin-dependent protein kinase activating kinase. J Biol Chem. 1996;271(1):471–477. doi: 10.1074/jbc.271.44.27201. [DOI] [PubMed] [Google Scholar]
- Adamczewski JP, Rossignol M, Tassan JP, Nigg EA, Moncollin V, Egly JM. Mat1, cdk7 and cyclin-H form a kinase complex which is uv light-sensitive upon association with TFIIH. Embo J. 1996;15(8):1877–1884. [PMC free article] [PubMed] [Google Scholar]


