Abstract
This study aimed to identify novel loci influencing the antihypertensive response to hydrochlorothiazide monotherapy. A genome-wide meta-analysis of blood pressure response to hydrochlorothiazide was performed in 1739 white hypertensives from six clinical trials within the International Consortium for Antihypertensive Pharmacogenomics Studies (ICAPS), making it the largest study to date of its kind. No signals reached genome-wide significance (P<5×10−8) and the suggestive regions (P<10−5) were cross-validated in two black cohorts treated with hydrochlorothiazide. Additionally, a gene-based analysis was performed on candidate genes with previous evidence of involvement in diuretic response, in blood pressure regulation or in hypertension susceptibility. Using the genome-wide meta-analysis approach, with validation in blacks, we identified two suggestive regulatory regions linked to GJA1 and FOXA1, relevant for cardiovascular and kidney function. With the gene-based approach, we identified HSD3B1 as significantly associated with blood pressure response (P<2.28×10−4). HSD3B1 encodes the 3beta-HSD enzyme and plays a crucial role in the biosynthesis of aldosterone and endogenous ouabain. By amassing all of the available pharmacogenomic studies of blood pressure response to hydrochlorothiazide, and using two different analytical approaches, we identified three novel loci influencing blood pressure response to hydrochlorothiazide. The gene-based analysis, never before applied to pharmacogenomics of antihypertensive drugs to our knowledge, provided a powerful strategy to identify a locus of interest, and which was not identified in the single-SNP analysis due to high allelic heterogeneity. These data pave the way for future investigations on new pathways and drug targets to enhance the current understanding of personalized antihypertensive treatment.
Keywords: Hydrochlorothiazide, blood pressure response, pharmacogenomics, genome-wide meta-analysis, gene-based approach, thiazide diuretics, ICAPS
INTRODUCTION
Hypertension is a major global risk factor for stroke, coronary heart disease, renal failure, and heart failure, and is the most common chronic disease for which medications are prescribed.1 The primary goal of hypertension treatment is the reduction of blood pressure (BP), which is strongly associated with the prevention of adverse cardiovascular outcomes. However, only approximately 50% of the treated hypertensive patients achieve BP control, despite the availability of many classes of antihypertensive drugs.2,3
Thiazide diuretics (TD), inhibitors of the Na+-Cl− symporter, are first line agents to treat uncomplicated hypertension as they are effective, relatively safe and well tolerated.4 However, as with other anti-hypertensive drugs, there is substantial inter-individual variation in BP response to TD which can be at least partially attributed to genetic differences among individuals.5 Pharmacogenomics, the study of the influence of genomic variations on drug response, could be a useful tool to select the most effective antihypertensive therapy for an individual, based on genetic profile, once replicated drug-gene pairs have been discovered.6 Many groups have conducted pharmacogenetic studies on BP response to TD using candidate gene(s)7–11 or genome wide association studies (GWAS).12–15 These studies, recently reviewed6,16–18, have advanced the knowledge surrounding hypertension pharmacogenomics and suggest several genetic variants that may be important determinants of response to TD. Nevertheless, only a small percentage of the variability in BP response has been explained to date.
The International Consortium for Antihypertensive Pharmacogenomics Studies (ICAPS) (icaps-htn.org) was created to promote collaboration between independent research groups, share knowledge and increase the likelihood for genetic discoveries in the field of anti-hypertensive pharmacogenomics.
Here, we present the largest genome-wide meta-analysis of BP response to hydrochlorothiazide (HCTZ, a TD) monotherapy to date in whites with uncomplicated hypertension, from six clinical trials included in ICAPS. Because all the currently available clinical trials for HCTZ monotherapy in whites with genome-wide genetic data were included in our meta-analysis, we used cohorts of black individuals for validation. This validation approach among multiple race groups, increases the confidence of a functional finding. Moreover, as a useful complement to the GWAS, we conducted a gene-based analysis focused on 219 candidate genes with prior evidence of involvement in diuretic response or blood pressure regulation, selected after a comprehensive literature search.
METHODS
Study Participants and Inclusion Criteria
Six study cohorts contributed data to the meta-analysis: the Genetics of Drug Responsiveness in Essential Hypertension Study (GENRES)15,19; the Genetic Epidemiology of Responses to Antihypertensives study (GERA-1)20; The Milan Hydrochlorothiazide study (HCTZ-Milan)13; the Nordic Diltiazem study (NORDIL)9,21; the Pharmacogenomic Evaluation of Antihypertensive Responses study (PEAR-1)22 and the Pharmacogenomics of Hydrochlorothiazide Sardinian Study (PHSS).13 Detailed information regarding each of six cohorts is included in the Supplement. Across all studies, participants with uncomplicated hypertension were included if they had a baseline untreated BP level (i.e., pre-HCTZ treatment) in the hypertensive range (systolic BP (SBP) >140 mmHg or diastolic BP (DBP) >90 mmHg). Participants previously taking antihypertensive medications underwent a wash-out period during which all antihypertensive medications were withdrawn. All participants were treated with HCTZ as monotherapy for at least 4 weeks. All participants voluntarily signed ethics committee approved informed consent forms and all clinical trials were conducted in accordance with regulations set forth by the Declaration of Helsinki and local regulatory agencies.
Blood Pressure Response Phenotype
We used the most precise measure of BP response available for each study. In the GENRES and NORDIL studies, BP response was determined as the difference between the averaged BP measurements prior to and at the end of HCTZ treatment. In the other studies, in which BP was measured at least once between the baseline and the end of the treatment period, or by multiple methodologies (e.g., office, home, ambulatory BP), we used a single model to take into account all available BP measurements, including the intermediate time points and multiple methods of measurement. PEAR-1 generated a weighted average of the office, home, ambulatory daytime and nighttime BP responses calculated on the sums of the inverse of the inter-method covariance matrices.23 In the GERA-1, HCTZ-Milan and PHSS studies, office BP was measured at one intermediate time point between the baseline and the final measurement. These data were fit to a general linear model that included baseline BP, sex, age, the first ten principal components and time point. The residuals from this model represent adjusted measurements of treatment response. There were typically two such residuals per individual, calculated for the two time points, which were combined using a weighted average.
Genotyping and imputation
Genome-wide genotyping was done on commercially available platforms from Illumina or Affymetrix. All genotype data were imputed to HapMap CEU II (build 36, version 22), and standard quality control procedures were applied. Details of the genotyping and quality control procedures are included in the Supplement.
Statistical Analysis
Continuous variables are presented as mean and standard deviation, categorical variables as numbers and percentages. Between-group comparison of continuous variables was performed using one-way analysis of variance (ANOVA) and Tukey’s “Honestly Significant Difference" post-hoc test. Categorical data were compared between groups using the chi-squared test.
A total of 1739 white individuals from the six study cohorts are included in the GWAS for BP response. Using mach2qtl24 or SNPtest25 software we performed linear regressions of the BP response phenotype on SNP dosages adjusting for sex, age, pre-diuretic treatment BP and principal components. All GWAS results underwent quality control using the EasyQC package26. Genome-wide SNP meta-analysis was performed using the software METAL27. Details of these analysis procedures are included in the Supplement.
SNPs with P<5×10−8 were considered genome-wide significant and those with P<10−5 were considered suggestive.
We then performed a trans-ethnic validation in black hypertensives treated with HCTZ from the GERA-1 and PEAR-1 studies (Table S1) for the suggestive SNPs with P<10−5. We tested the genetic regions harboring the suggestive signals as well, since we did not necessarily expect to observe the same SNPs, due to the differences in linkage disequilibrium (LD) across the genome between white and black populations. Neighboring SNPs were not required to have effects in the same direction as, due to differences in LD and allele frequency, neighboring SNPs could tag the same unknown causal variant.28
We conducted a candidate gene-based association analysis in each cohort separately using the VEGAS program29 and then performed a meta-analysis of the results applying the Fisher’s method30 using the sumlog R function31. We selected the candidate genes beginning with the catalogue recently reviewed by Padmanabhan et al32 with the inclusion of additional genes found in PubMed by using the key search terms “diuretic response,” “hypertension” and “blood pressure regulation”. Following this comprehensive literature search, 219 autosomal genes (Table S2) were identified based on evidence from candidate studies or GWAS of involvement in diuretic response, in BP regulation or in hypertension susceptibility, and included in the gene-based analysis.
RESULTS
Demographic and baseline characteristics of the six cohorts included in the GWAS meta-analysis are summarized in Table 1. All participants were white and from the USA or Europe. With the exception of GENRES, all cohorts included a majority or exclusively males. Overall, mean age and BMI were significantly different among the cohorts. GENRES, HCTZ-Milan and PEAR-1 had similar pre-treatment SBP (P=0.09) whereas pre-treatment DBP levels were similar between HCTZ-Milan and PEAR-1 (P=0.19) and between GENRES and PHSS (P=0.80).
Table 1.
Baseline characteristics of study participants *
| Characteristic† | GENRES ‡ (n=192) |
GERA-1 (n=282) |
HCTZ-Milan (n=207) |
NORDIL (n=381) |
PEAR1 (n=228) |
PHSS (n=449) |
|---|---|---|---|---|---|---|
| Men/women | 192/0 | 161/121 | 177/30 | 148/233 | 137/91 | 293/156 |
| Age, y | 50.8±6.2 | 46.3±8.1 | 45.7±7.98 | 61.5±6.7 | 50.0±9.5 | 50.9±10.1 |
| BMI, kg/m2 | 26.8±2.6 | 30.9±5.5 | 26.14±3.05 | 28.4±4.7 | 30.3±4.9 | 27.6±4.0 |
| Pretreatment SBP, mmHg |
152.3±12.2 | 142.4±12.5 | 149.76±12.5 | 172.5±15.6 | 151.8±12.4 | 158.1±13.0 |
| Pretreatment DBP, mmHg |
100.2±6.1 | 95.4±5.4 | 98.96±7.76 | 103.0±4.5 | 98.1±5.8 | 100.4±9.9 |
| Treatment dose | 25 mg/day | 25 mg/day | 12.5 mg/day 25 mg/day |
at physician discretion |
12.5 mg/day 25 mg/day |
25 mg/day |
| Period of treatment | 4 weeks | 4 weeks | 8 weeks (timepoint at 4 weeks) |
6 months | 8 weeks (timepoint at 2 weeks) |
8 weeks (timepoint at 4 weeks) |
| Run-in period | 4 weeks placebo |
4 weeks | Never treated | 2 weeks | ~ 31 days | Never treated |
BMI, body mass index; SBP, systolic blood pressure; DBP, diastolic blood pressure
Numeric characteristics were presented as mean ± SD; categorical variables were presented as number
The mean BP level of four placebo treatment periods was used as the baseline ("pretreatment") level
GWAS Meta-Analysis
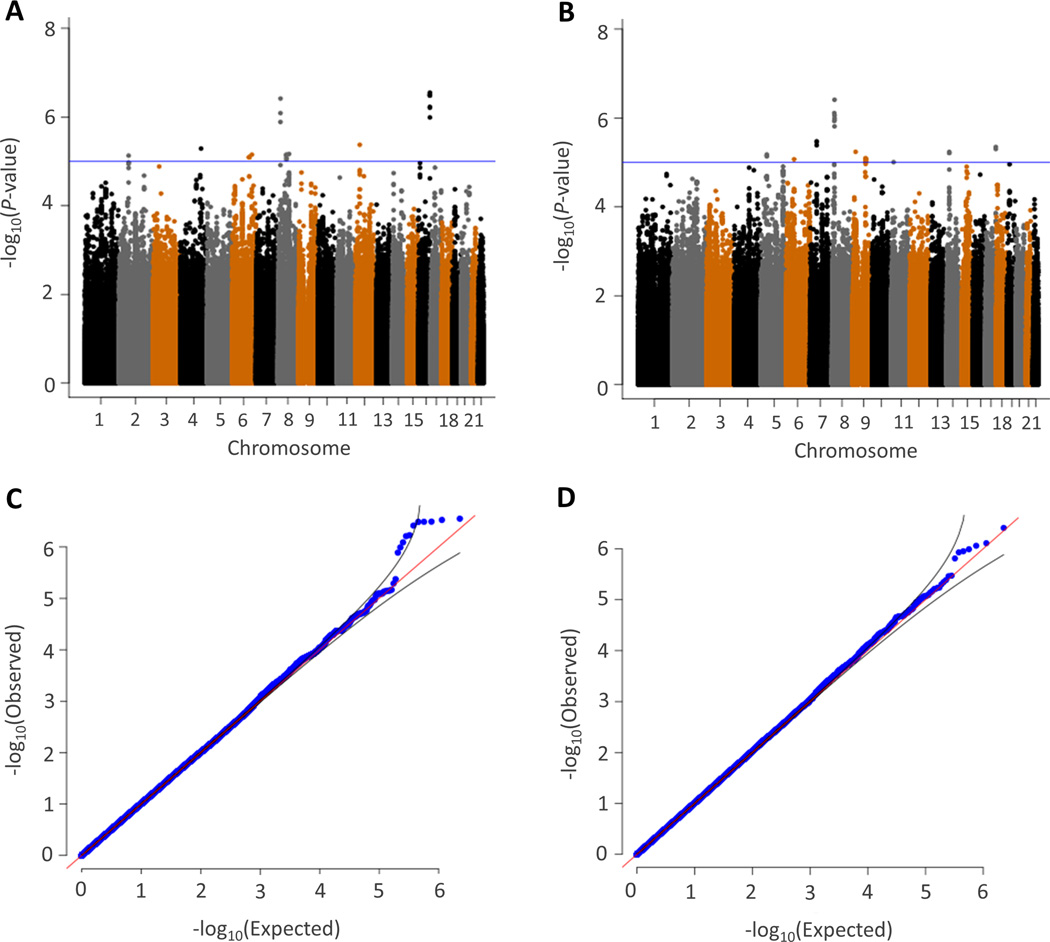
Manhattan and q-q plots for the meta-analysis of SBP and DBP response to HCTZ are shown in Figure 1. Although no SNP achieved Bonferroni corrected genome-wide significance (P< 5×10−8), the q-q plots indicated that the SNPs with P<10−5 deviated above the straight line, indicating that there is a suggestion of relationships between SNPs and BP response. Thus we considered SNPs with P<10−5 as suggestive. Accordingly, there were 10 SNPs with suggestive associations with SBP response and there were 11 SNPs with suggestive associations with DBP response (Table 2). These SNPs were in high LD (r2>0.80) with 34 additional SNPs according to HapMap reference (Tagged SNPs, Table 2).
Figure 1.
Manhattan (a, b) and quantile-quantile (c, d) plots from meta-analysis of genome-wide association for systolic (a, c) and diastolic (b, d) blood pressure response to hydrochlorothiazide. Each dot signifies a SNP. In the Manhattan plots, genomic coordinates are displayed along the X-axis with the negative logarithm of the association P-value displayed on the Y-axis. The blue line refers to −log10(P)=5. The quantile-quantile plots compare the observed results (−log10(P), X axis) versus the theoretically expected values (Y axis).
Table 2.
Meta-analysis results for blood pressure response to hydrochlorothiazide with P<10−5
| Trait | Marker | Chr | Position (bp) |
Gene/Region | Function | Coded/ other alleles |
Coded allele Freq |
Beta | P | Tagged SNPs (r2>0.80) |
|---|---|---|---|---|---|---|---|---|---|---|
| SBP | ||||||||||
| rs7565329 | 2 | 67959340 | ETAA1/C1D | Intergenic | T/C | 0.09 | −1.7 | 7.42×10−6 | ||
| rs12642634 | 4 | 147205077 | ZNF827/LSM6 | Intergenic | A/G | 0.78 | 1.1 | 5.15×10−6 | ||
| rs11750990 | 6 | 121977301 | GJA1/HSF2 | Intergenic | A/G | 0.96 | 2.44 | 8.11×10−6 | rs11755230, rs11755743, rs11752212, rs11754717 |
|
| rs2876449 | 6 | 141099634 | CITED2/NMBR | Intergenic | T/G | 0.03 | −2.8 | 7.06×10−6 | ||
| rs11784910 | 8 | 17119626 | ZDHHC2 | Intronic | A/T | 0.95 | 2.31 | 3.85×10−7 | rs11775427, rs11786857 |
|
| rs2925663 | 8 | 59424423 | FAM110B/UBXN2B | Intergenic | T/C | 0.12 | −1.4 | 7.11×10−6 | rs747401, rs733665, rs2970756, rs2925666 |
|
| rs13253998 | 8 | 78697117 | PEX2/PKIA | Intergenic | T/G | 0.51 | −0.9 | 6.80×10−6 | ||
| rs7960884 | 12 | 31251907 | FAM60A | Intergenic | A/G | 0.27 | 1.09 | 4.23×10−6 | ||
| rs16962897 | 16 | 82766581 | DNAAF1 | Intronic | A/G | 0.12 | −1.7 | 3.25×10−7 | rs1056616, rs2288025, rs1056612, rs3743642, rs4150162 |
|
| rs4150161 | 16 | 82771737 | TAF1C | Intronic | T/C | 0.86 | 1.53 | 1.02×10−6 | ||
| DBP | ||||||||||
| rs822127 | 5 | 41582716 | PLCXD3/OXTC1 | Intergenic | A/G | 0.37 | −0.6 | 6.64×10−6 | rs1495757, rs4957159 |
|
| rs9370524 | 6 | 56327712 | COL21A1 | Intronic | A/G | 0.36 | 0.58 | 8.51×10−6 | ||
| rs10951933 | 7 | 47873739 | PKD1L1 | Intronic | T/G | 0.2 | 0.75 | 3.37×10−6 | rs10951934, rs17131904 |
|
| rs2198596 | 8 | 15045137 | SGCZ | Intronic | C/G | 0.91 | −1.3 | 7.76×10−7 | rs17575278 | |
| rs13256445 | 8 | 17106584 | ZDHHC2 | Intronic | T/C | 0.85 | 0.96 | 1.02×10−6 | rs13278086 | |
| rs11784910 | 8 | 17119626 | ZDHHC2 | Intronic | A/T | 0.95 | 1.48 | 3.89×10−7 | rs11775427, rs11786857 |
|
| rs17755650 | 9 | 21770037 | MTAP | Intergenic | T/C | 0.9 | 1.17 | 5.78×10−6 | ||
| rs12685849 | 9 | 93570641 | ROR2 | Intronic | A/G | 0.04 | −1.7 | 9.98×10−6 | rs12685213, rs3935544, rs10992090, rs3802377, rs3802378, rs16907776, rs10992095 |
|
| rs4757718 | 11 | 18761510 | PTPN5 | Intronic | A/G | 0.67 | −0.6 | 9.87×10−6 | ||
| rs177848 | 14 | 37173757 | TTC6/FOXA1 | Intronic/5'- Flanking |
A/C | 0.59 | −.6 | 5.81×10−6 | rs1998125, rs177829 |
|
| rs11657217 | 17 | 75323934 | ENPP7 | Coding | C/G | 0.69 | −0.7 | 4.50×10−6 | rs8081537 |
SNPs are ranked by chromosome (Chr) and position based on hg18 (NCBI 36) assembly. Tagged SNPs are in high LD (r2>0.80) with markers. P refer to the meta-analysis results of GWAS from the 6 ICAPS cohorts.
Trans-ethnic replication
We then assessed the association of suggestive meta-analysis loci (P<10−5) and neighboring regions in two black ancestry samples from GERA-1 and PEAR-1. Samples characteristics are detailed in the Supplement and in Table S1. GERA-1 and PEAR-1 black participants had similar mean age and BMI. Pre-treatment DBP was higher in GERA-1 group than in PEAR-1 (P=0.004) while pre-treatment SBP was not significantly different between the cohorts (P=0.069).
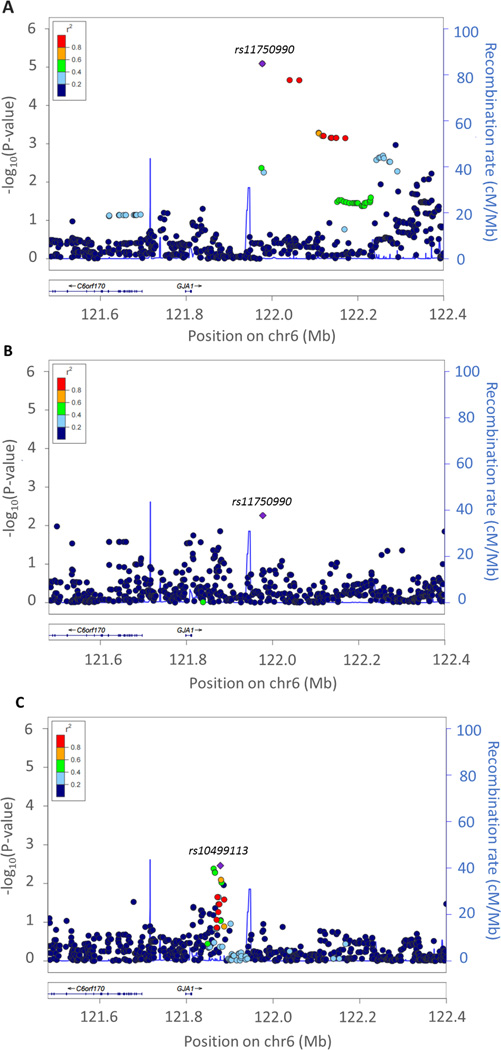
In blacks, we identified two regions of interest on chromosome 6 and 14, neighboring the identified suggestive SNPs in whites, for SBP and DBP response to HCTZ.
With regard to SBP associations, we identified a signal of interest located in the 3’-flanking region of gap junction protein alpha1 gene (GJA1) on chromosome 6q22.31, where rs11750990 was associated with SBP in the meta-analysis of white participants (P=8.11×10−6 and beta=2.44 mmHg per A allele) (Figure 2a, and Table 2). This variant was nominally associated in the meta-analysis of PEAR1 and GERA1 black individuals (P=5–46×10−3) with opposite beta coefficient (beta=−6.17) per A allele (Figure 2b). In the same region, a peak of suggestive significance was observed in the GERA-1 sample with a different SNP, rs10499113, associated with SBP response (P=3.46×10−3 and beta=−3.14 per C allele) (Figure 2c).
Figure 2.
Local regional plot for chromosome 6q22.31. a) Meta-analysis results for white samples b) Replication results in meta-analysis of PEAR-1 and GERA-1 black samples c) Replication results in GERA-1 black sample. Each dot signifies a SNP. Genomic coordinates are displayed along the X-axis with the negative logarithm of the association P-value displayed on the Y-axis. SNPs are colored based on their r2 with the top signal SNP which has the lowest P-value in the region.
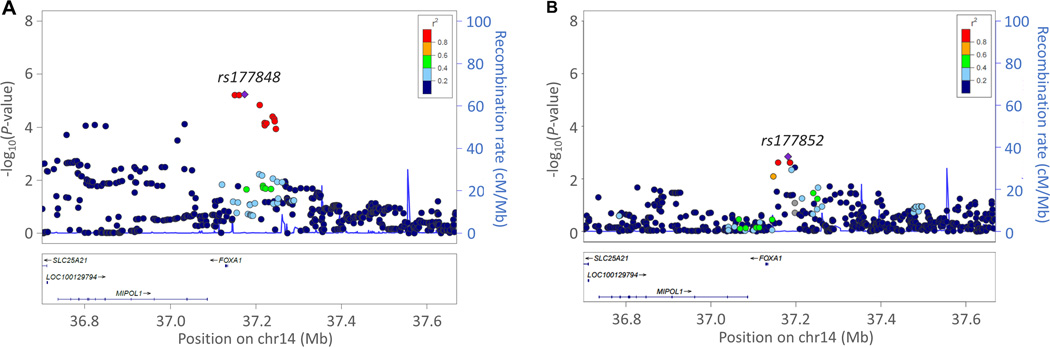
With regard to DBP associations, we identified a signal of interest in the 5’-flanking region of forkhead box A1 gene (FOXA1) on chromosome 14q21.1 (Figure 3). In the meta-analysis of whites, rs177848 was associated with DBP response (P=5.8×10−6 and beta=−0.60 per A allele) (Table 2 and Figure 3a). Rs177852, 7.9 kb upstream of rs177848, was associated with DBP response in the PEAR-1 black cohort (P=1.43×10−3 and beta=−2.95 per C allele) (Figure 3b).
Figure 3.
Local regional plot for chromosome 14q21. a) Meta-analysis results for white samples b) Replication results in PEAR1 black sample. Each dot signifies a SNP. Genomic coordinates are displayed along the X-axis with the negative logarithm of the association P-value displayed on the Y-axis. SNPs are colored based on their r2 with the top signal SNP which has the lowest P-value in the region.
Gene-based Meta-Analysis
Results of the gene-based analysis are shown in Table S2. Applying a Bonferroni-corrected significance threshold for gene-based analysis (P<2.28×10−4), we identified the hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 1 gene (HSD3B1) on chromosome 1p12 as significantly associated with DBP response (P= 2.80×10−5) and SBP response (P= 7.5 ×10−5) to HCTZ (Table S2).
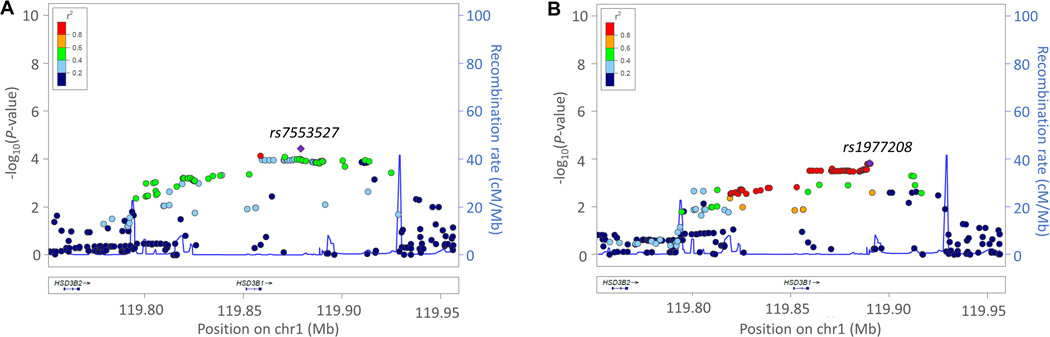
In the GWAS meta-analysis results for HSD3B1, we observed a cluster of SNPs with P≤10−4 associated with BP response to HCTZ (Figure 4, Table S3). For SBP response, we identified the 3’-flanking SNP rs7553527 (Beta= −0.86, P= 3.67×10−5 per C allele) in high LD with the coding SNP rs6203 and the 3’-flanking SNP rs10754403 (Beta= −0.83, P= 8.07×10−5 per G allele), which is in LD with 58 other SNPs within the 3’ to 5’ flanking region (Table S3, Figure 4a). The coding SNP rs6203 had a P=2.32X10−3 for DBP response (Table S3, Figure 4b).
Figure 4.
Local regional plot for HSD3B1 gene. a) SBP and b) DBP meta-analysis results for white samples. Each dot signifies a SNP. Genomic coordinates are displayed along the X-axis with the negative logarithm of the association P-value displayed on the Y-axis. SNPs are colored based on their r2 with the top signal SNP which has the lowest P-value in the region.
While gene-based analysis for HSD3B1 in blacks was not significant it is important to consider that: 1) the LD pattern of the HSD3B1 locus is different comparing CEU and YRI HapMap populations (Figure S1); and 2) rs7553527 and rs6203 are not present in HapMap data for the YRI population.
DISCUSSION
This paper describes the largest genome-wide meta-analysis of loci influencing the antihypertensive response to HCTZ monotherapy and includes a total of 1739 white individuals from six independent cohorts from ICAPS. Using single-SNP GWAS, we identified two suggestive regulatory regions on chromosomes 6 and 14, potentially linked to genes relevant for cardiovascular and kidney function. These signals were nominally validated in two independent black cohorts. Through a complementary candidate gene-based approach, we identified HSD3B1, at a significance level taking Bonferroni correction into account, which was not detected using the single-SNP GWAS.
In the genome-wide meta-analysis, no SNP reached the genome wide significance level, which is not surprising given the total sample size included in the meta-analysis. In fact, despite being the largest HCTZ monotherapy meta-analysis in whites (1739 individuals), we calculated that a sample size ranging from 2860 to 5571 samples would be required to achieve 80% power at P=5×10−8, assuming an expected effect of at least 2 mmHg and an allele frequency ranging from 0.15 to 0.45.33 The sample size of the present study was powered to find variants with effects ≥ 3 mmHg and frequencies ≥ 0.2 whereas, for the suggestive SNPs identified, the average effect observed was approximately 1.3 mmHg.
The suggestive SNPs, as well as SNPs in neighboring regions, that were identified in the meta-analysis were tested in two samples of different ancestry.
The first interesting region for SBP response is located in the 3’-flanking region of GJA1. GJA1 encodes the Connexin43 (Cx43), the predominant gap junction protein in myocardial and aortic smooth muscle cells with involvement in the regulation of cell-to-cell communication and elasticity and contractility of the vascular wall.34 The expression of Cx43 was observed to be increased in aortic wall34 and muscular artery35 of hypertensive rats and was decreased following exposure to the combination of hydralazine-HCTZ and candesartan.35 According to the HaploReg database36, the suggestive 3’-flanking region of GJA1 contains eQTLs, transcription factor binding sites and histone marks. SNP rs10499113 is reported to be an eQTL for GJA1 in skin-sun exposed tissue with C allele carriers exhibiting increased gene expression compared to the GG carriers. In the GERA-1 black cohort, the C allele was associated with greater SBP response (effect size= −3.1 mmHg and P=3.5×10−3). Additionally, rs10499113 is in high LD with another eQTL for GJA1, rs2104334 (not present in the HapMap reference). According to ChiP-Seq experiments from ENCODE Project Consortium 2011, in HUVEC cells, rs2104334 maps within a probable peak of binding of GATA2 transcription factor, 35 base-pairs upstream the GATA2 consensus binding motif.
Moreover, in aorta, rs2104334 co-localizes with H3K4me3 histone modifications that mark active promoters in chromatin regions and with H3K4me1 and H3K27ac histone marks associated with enhancers. SNP rs11750990, which we identified to be associated with SBP response in the GWAS meta-analysis of whites, co-localizes with H3K4me1 and H3K27ac.37
The 5’-flanking region of FOXA1 was associated with DBP response to HCTZ in the GWAS meta-analysis and nominally validated with different SNPs in the PEAR-1 black cohort. According to Haploreg annotation36, rs177852, identified to be associated with HCTZ response in PEAR-1 black cohorts, is related to FOXA1 expression in the brain cortex.38 FOXA1, is also expressed in the collecting duct of the kidney.39,40 Its putative binding sites were found in the promoters of several genes expressed in the urothelium of the renal pelvis including genes encoding the vasopressin receptor, several subunits of the Na/K ATPase and E-cadherin.39–41 Foxa1 has also been identified as a vasopressin induced gene in a differentiated mouse clonal cortical collecting duct cell line.42 Furthermore, Foxa1-deficient mice develop nephrogenic diabetes insipidus with a defect in renal water reabsorption.40
For all of the above-mentioned variants, we observed a greater effect size in blacks compared to whites. This could be related to the greater antihypertensive efficacy of HCTZ in blacks due to their greater volume expansion, salt sensitivity and lower renin activity, compared to white hypertensives.43
Our gene-based meta-analysis provided a very interesting, biologically plausible and statistically significant signal that would have remained indistinguishable from random noise with the traditional single-SNP GWAS approach. Gene-based tests can highlight regions that display substantial allelic heterogeneity, defined as the presence of multiple alleles that act through one gene to influence a trait. Furthermore, gene-based tests can increase statistical power by combining single variants from GWAS into a gene-based score, which substantially reduces the burden of multiple testing.29
With this approach, we identified HSD3B1. HSD3B1 is expressed as 3β-hydroxysteroid dehydrogenase (3-beta-HSD) with a crucial role in the biosynthesis of hormonal steroids including aldosterone.44 HSD3B1, markedly overexpressed in the hypothalamus of Milan hypertensive rats, is involved in endogenous ouabain (EO) synthesis in an adrenal medullary–derived cell line (PC12).45 Hypertensive patients have elevated circulating EO levels which are positively correlated with higher BP, higher plasma Na concentrations and increased proximal tubular reabsorption.46,47 EO is also higher in patients with kidney failure48, myocardial infarction49 and congestive heart failure.50 Multiple studies have described the association of genetic variants in HSD3B1 with hypertension or BP variation. The CC genotype at rs6203 was associated with hypertension51 and higher BP.51–54 This association was reported as stronger in males as also confirmed by our single GWAS data (Table S4). Additionally, rs3765945 and rs1047303 have been significantly associated with SBP.53 The T-C haplotype, established by rs3088283-rs1047303, correlated with significantly higher level of aldosterone and BP54 and the G-C-C haplotype of rs2236780-rs3765945-rs6203 was related to left ventricular diastolic function.55
In conclusion, the following can be gathered from our study: (1) this is the largest genome-wide meta-analysis of BP response to HCTZ conducted to date; (2) while ours is the largest HCTZ meta-analysis conducted to date, our sample size still lacked sufficient power; (3) using the single-SNP approach with validation in blacks, we identified two suggestive regions linked to the regulation of GJA1 and FOXA1; and (4) the gene-based approach, never applied before to pharmacogenomics of antihypertensive drugs to our knowledge, highlights HSD3B1 as a susceptibility gene of BP response to HCTZ. This gene was not identified in the single-SNP analysis due to high allelic heterogeneity.
These data pave the way for future research on new pathways and drug targets in hypertension toward better-personalized therapeutic approaches.
Perspectives
Hypertension is a major risk factor for global disease burden and is also the most common chronic disease for which medications are prescribed. Pharmacogenomics, may represent a useful tool in the future to select antihypertensive therapy with the greatest efficacy, based on individual’s genetic profile. This study performed the largest pharmacogenomic genome-wide meta-analysis of BP response to HCTZ in hypertensive cohorts from ICAPS applying both SNP-based and gene-based approaches. Three new biologically plausible loci linked to hypertension and blood pressure regulation were identified as markers of BP response to HCTZ. Further investigations of the associated regions may enhance the current understanding of personalized antihypertensive treatment.
Supplementary Material
Novelty and Significance.
What Is New?
this is the largest pharmacogenomics study of BP response to HCTZ.
the GWAS SNP-based approach identified two novel loci of BP response to HCTZ linked to the regulation of GJA1 and FOXA1.
the gene-based approach, never before applied to pharmacogenomics of antihypertensive drugs, highlighted HSD3B1 gene as new marker of BP response to HCTZ.
What Is Relevant?
The identified variants can be considered new biologically plausible loci associated with hypertension and blood pressure regulation.
Summary
By amassing all the available pharmacogenomic studies of BP response to HCTZ, and using two different analysis approaches, we identified three novel loci influencing BP response to HCTZ. These data open the way for future research on new pathways and drug targets in hypertension toward better personalized therapeutic approaches.
Acknowledgments
Sources of funding
GENRES was supported by the Sigrid Juselius Foundation and the Finnish Foundation for Cardiovascular Research. HCTZ-Milan and PHSS studies were supported by the HYPERGENES project (FP7-HEALTH-F4-2007-201550), InterOmics (PB05 MIUR-CNR Italian Flagship Project) and The ‘Associazione per lo sviluppo della ricerca sull’ipertensione arteriosa e sulle malattie cardiovascolari – ONLUS’. PEAR was supported by the National Institute of Health Pharmacogenetics Research Network grant U01-GM074492 and the National Center for Advancing Translational Sciences under the award number UL1 TR000064 (University of Florida); UL1 TR000454 (Emory University) and UL1 TR000135 (Mayo Clinic). PEAR was also supported by funds from the Mayo Foundation. CL and PM are funded by the Italian Ministry of Health Grant #RF-2011-02347356. SP is funded by the MRC (MR/M016560/1, The AIM-HY Study) and the British Heart Foundation (PG/12/85/29925, CS/16/1/31878). AFD has funding from the Scottish Ecosystem for Precision Medicine.
Footnotes
Conflicts of interest/Disclosures
None.
References
- 1.Kearney PM, Whelton M, Reynolds K, Muntner P, Whelton PK, He J. Global burden of hypertension: analysis of worldwide data. Lancet. 2005;365:217–223. doi: 10.1016/S0140-6736(05)17741-1. [DOI] [PubMed] [Google Scholar]
- 2.Go AS, Mozaffarian D, Roger VL, et al. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129:e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakris G, Sarafidis P, Agarwal R, Ruilope L. Review of blood pressure control rates and outcomes. J Am Soc Hypertens. 2014;8:127–141. doi: 10.1016/j.jash.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 4.James PA, Oparil S, Carter BL, et al. Evidence-Based Guideline for the Management of High Blood Pressure in Adults. JAMA. 2013;1097:1–14. doi: 10.1001/jama.2013.284427. [DOI] [PubMed] [Google Scholar]
- 5.Turner ST, Schwartz GL, Chapman aB, Hall WD, Boerwinkle E. Antihypertensive pharmacogenetics: getting the right drug into the right patient. J Hypertens. 2001;19:1–11. doi: 10.1097/00004872-200101000-00001. [DOI] [PubMed] [Google Scholar]
- 6.Cooper-DeHoff RM, Johnson JA. Hypertension pharmacogenomics: in search of personalized treatment approaches. Nat Rev Nephrol. 2016;12:110–122. doi: 10.1038/nrneph.2015.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li Y, Zhou Y, Yang P, Niu JQ, Wu Y, Zhao DD, Wu SL. Interaction of ACE and CYP11B2 genes on blood pressure response to hydrochlorothiazide in Han Chinese hypertensive patients. Clin Exp Hypertens. 2011;33:141–146. doi: 10.3109/10641963.2010.531838. [DOI] [PubMed] [Google Scholar]
- 8.Sciarrone MT, Stella P, Barlassina C, Manunta P, Lanzani C, Bianchi G, Cusi D. ACE and alpha-adducin polymorphism as markers of individual response to diuretic therapy. Hypertension. 2003;41:398–403. doi: 10.1161/01.HYP.0000057010.27011.2C. [DOI] [PubMed] [Google Scholar]
- 9.Svensson-Färbom P, Wahlstrand B, Almgren P, Dahlberg J, Fava C, Kjeldsen S, Hedner T, Melander O. A functional variant of the NEDD4L gene is associated with beneficial treatment response with β-blockers and diuretics in hypertensive patients. J Hypertens. 2011;29:388–395. doi: 10.1097/HJH.0b013e3283410390. [DOI] [PubMed] [Google Scholar]
- 10.McDonough CW, Burbage SE, Duarte JD, Gong Y, Langaee TY, Turner ST, Gums JG, Chapman AB, Bailey KR, Beitelshees AL, Boerwinkle E, Pepine CJ, Cooper-DeHoff RM, Johnson JA. Association of variants in NEDD4L with blood pressure response and adverse cardiovascular outcomes in hypertensive patients treated with thiazide diuretics. J Hypertens. 2013;31:698–704. doi: 10.1097/HJH.0b013e32835e2a71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gong Y, McDonough CW, Wang Z, Hou W, Cooper-DeHoff RM, Langaee TY, Beitelshees AL, Chapman AB, Gums JG, Bailey KR, Boerwinkle E, Turner ST, Johnson JA. Hypertension susceptibility loci and blood pressure response to antihypertensives: results from the pharmacogenomic evaluation of antihypertensive responses study. Circ Cardiovasc Genet. 2012;5:686–691. doi: 10.1161/CIRCGENETICS.112.964080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turner ST, Bailey KR, Fridley BL, Chapman AB, Schwartz GL, Chai HS, Sicotte H, Kocher J-P, Rodin AS, Boerwinkle E. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension. 2008;52:359–365. doi: 10.1161/HYPERTENSIONAHA.107.104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chittani M, Zaninello R, Lanzani C, et al. TET2 and CSMD1 genes affect SBP response to hydrochlorothiazide in never-treated essential hypertensives. J Hypertens. 2015;33:1301–1309. doi: 10.1097/HJH.0000000000000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner ST, Boerwinkle E, Connell JRO, et al. Genomic Association Analysis of Common Variants Influencing Antihypertensive Response to Hydrochlorothiazide. Hypertension. 2013;62:391–397. doi: 10.1161/HYPERTENSIONAHA.111.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hiltunen TP, Donner KM, Sarin A-P, et al. Pharmacogenomics of hypertension: a genome-wide, placebo-controlled cross-over study, using four classes of antihypertensive drugs. J Am Heart Assoc. 2015;4:e001521–e001521. doi: 10.1161/JAHA.114.001521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fontana V, Luizon MR, Sandrim VC. An update on the pharmacogenetics of treating hypertension. J Hum Hypertens. 2015;29:283–291. doi: 10.1038/jhh.2014.76. [DOI] [PubMed] [Google Scholar]
- 17.Lupoli S, Salvi E, Barcella M, Barlassina C. Pharmacogenomics considerations in the control of hypertension. Pharmacogenomics. 2015;16:1951–1964. doi: 10.2217/pgs.15.131. [DOI] [PubMed] [Google Scholar]
- 18.Shahin MH, Johnson JA. Mechanisms and pharmacogenetic signals underlying thiazide diuretics blood pressure response. Curr Opin Pharmacol. 2016;27:31–37. doi: 10.1016/j.coph.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hiltunen TP, Suonsyrjä T, Hannila-Handelberg T, Paavonen KJ, Miettinen HE, Strandberg T, Tikkanen I, Tilvis R, Pentikäinen PJ, Virolainen J, Kontula K. Predictors of antihypertensive drug responses: Initial data from a placebo-controlled, randomized, cross-over study with four antihypertensive drugs (The GENRES Study) Am J Hypertens. 2007;20:311–318. doi: 10.1016/j.amjhyper.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Chapman AB, Schwartz GL, Boerwinkle E, Turner ST. Predictors of antihypertensive response to a standard dose of hydrochlorothiazide for essential hypertension. Kidney Int. 2002;61:1047–1055. doi: 10.1046/j.1523-1755.2002.00200.x. [DOI] [PubMed] [Google Scholar]
- 21.The Nordic Diltiazem Study (NORDIL) A prospective intervention trial of calcium antagonist therapy in hypertension. Blood Press. 1993;2:312–321. doi: 10.3109/08037059309077174. [DOI] [PubMed] [Google Scholar]
- 22.Johnson JA, Boerwinkle E, Zineh I, Chapman AB, Bailey K, Cooper-DeHoff RM, Gums J, Curry RW, Gong Y, Beitelshees AL, Schwartz G, Turner ST. Pharmacogenomics of antihypertensive drugs: Rationale and design of the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) study. Am Heart J. 2009;157:442–449. doi: 10.1016/j.ahj.2008.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner ST, Schwartz GL, Chapman AB, Beitelshees AL, Gums JG, Cooper-Dehoff RM, Boerwinkle E, Johnson JA, Bailey KR. Power to identify a genetic predictor of antihypertensive drug response using different methods to measure blood pressure response. J Transl Med. 2012;10:47. doi: 10.1186/1479-5876-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 26.Winkler TW, Day FR, Croteau-Chonka DC, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9:1192–1212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDonough CW, Gillis NK, Alsultan A, et al. Atenolol Induced HDL-C Change in the Pharmacogenomic Evaluation of Antihypertensive Responses (PEAR) Study. PLoS One. 2013;8:1–10. doi: 10.1371/journal.pone.0076984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu JZ, McRae AF, Nyholt DR, Medland SE, Wray NR, Brown KM, Hayward NK, Montgomery GW, Visscher PM, Martin NG, MacGregor S. A versatile gene-based test for genome-wide association studies. Am J Hum Genet. 2010;87:139–145. doi: 10.1016/j.ajhg.2010.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hägg S, Ganna A, Van Der Laan SW, et al. Gene-based meta-analysis of genome-wide association studies implicates new loci involved in obesity. Hum Mol Genet. 2015;24:6849–6860. doi: 10.1093/hmg/ddv379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dewey M. metap: meta-analysis of significance values. [Accessed July 25, 2016]; https://CRAN.R-project.org/package=metap. R package version 0.7. Published April 07, 2016. [Google Scholar]
- 32.Padmanabhan S, Caulfield M, Dominiczak AF. Genetic and Molecular Aspects of Hypertension. Circ Res. 2015;116:937–959. doi: 10.1161/CIRCRESAHA.116.303647. [DOI] [PubMed] [Google Scholar]
- 33.Maranville JC, Cox NJ. Pharmacogenomic variants have larger effect sizes than genetic variants associated with other dichotomous complex traits. Pharmacogenomics J. 2016;16:388–392. doi: 10.1038/tpj.2015.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haefliger JA, Castillo E, Waeber G, Bergonzelli GE, Aubert JF, Sutter E, Nicod P, Waeber B, Meda P. Hypertension increases connexin43 in a tissue-specific manner. Circulation. 1997;95:1007–1014. doi: 10.1161/01.cir.95.4.1007. [DOI] [PubMed] [Google Scholar]
- 35.Kansui Y, Fujii K, Nakamura K, Goto K, Oniki H, Abe I, Shibata Y, Iida M. Angiotensin II receptor blockade corrects altered expression of gap junctions in vascular endothelial cells from hypertensive rats. Am J Physiol Heart Circ Physiol. 2004;287:H216–H224. doi: 10.1152/ajpheart.00915.2003. [DOI] [PubMed] [Google Scholar]
- 36.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–D934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hon GC, Hawkins RD, Ren B. Predictive chromatin signatures in the mammalian genome. Hum Mol Genet. 2009;18:12–14. doi: 10.1093/hmg/ddp409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol. 2008;6:2869–2879. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peterson S, Clevidence E, Costa H. Hepatocyte Nuclear Factor-3d Recognition by Cell-specific Factor-I, and Autoactivation. Cell Growth Differ. 1997;8:69–82. [PubMed] [Google Scholar]
- 40.Behr R, Brestelli J, Fulmer JT, Miyawaki N, Kleyman TR, Kaestner KH. Mild nephrogenic diabetes insipidus caused by Foxa1 deficiency. J Biol Chem. 2004;279:41936–41941. doi: 10.1074/jbc.M403354200. [DOI] [PubMed] [Google Scholar]
- 41.Overdier DG, Ye H, Peterson RS, Clevidence DE, Costa RH. The winged helix transcriptional activator HFH-3 is expressed in the distal tubules of embryonic and adult mouse kidney. J Biol Chem. 1997;272:13725–13730. doi: 10.1074/jbc.272.21.13725. [DOI] [PubMed] [Google Scholar]
- 42.Robert-Nicoud M, Flahaut M, Elalouf JM, Nicod M, Salinas M, Bens M, Doucet A, Wincker P, Artiguenave F, Horisberger JD, Vandewalle A, Rossier BC, Firsov D. Transcriptome of a mouse kidney cortical collecting duct cell line: effects of aldosterone and vasopressin. Proc Natl Acad Sci U S A. 2001;98:2712–2716. doi: 10.1073/pnas.051603198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams SF, Nicholas SB, Vaziri ND, Norris KC. African Americans, hypertension and the renin angiotensin system. World J Cardiol. 2014;6:878. doi: 10.4330/wjc.v6.i9.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mason JI, Keeney DS, Bird IM, Rainey WE, Morohashi K, Leers-Sucheta S, Melner MH. The regulation of 3 beta-hydroxysteroid dehydrogenase expression. Steroids. 1997;62:164–168. doi: 10.1016/s0039-128x(96)00176-6. [DOI] [PubMed] [Google Scholar]
- 45.Murrell JR, Randall JD, Rosoff J, Zhao J, Jensen RV, Gullans SR, Haupert GT. Endogenous ouabain: upregulation of steroidogenic genes in hypertensive hypothalamus but not adrenal. Circulation. 2005;112:1301–1308. doi: 10.1161/CIRCULATIONAHA.105.554071. [DOI] [PubMed] [Google Scholar]
- 46.Manunta P, Maillard M, Tantardini C, Simonini M, Lanzani C, Citterio L, Stella P, Casamassima N, Burnier M, Hamlyn JM, Bianchi G. Relationships among endogenous ouabain, alpha-adducin polymorphisms and renal sodium handling in primary hypertension. J Hypertens. 2008;26:914–920. doi: 10.1097/HJH.0b013e3282f5315f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tentori S, Messaggio E, Brioni E, Casamassima N, Simonini M, Zagato L, Hamlyn JM, Manunta P, Lanzani C. Endogenous ouabain and aldosterone are coelevated in the circulation of patients with essential hypertension. J Hypertens. 2016;34:2074–2080. doi: 10.1097/HJH.0000000000001042. [DOI] [PubMed] [Google Scholar]
- 48.Manunta P, Hamlyn JM, Simonini M, Messaggio E, Lanzani C, Bracale M, Argiolas G, Casamassima N, Brioni E, Glorioso N, Bianchi G. Endogenous ouabain and the renin-angiotensin-aldosterone system: distinct effects on Na handling and blood pressure in human hypertension. J Hypertens. 2011;29(2):349–356. doi: 10.1097/HJH.0b013e32833ea821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Goto A, Yamada K, Hazama H, Uehara Y, Atarashi K, Hirata Y, Kimura K, Omata M. Ouabainlike compound in hypertension associated with ectopic corticotropin syndrome. Hypertension. 1996;28:421–425. doi: 10.1161/01.hyp.28.3.421. [DOI] [PubMed] [Google Scholar]
- 50.Hamlyn JM, Blaustein MP, Bova S, DuCharme DW, Harris DW, Mandel F, Mathews WR, Ludens JH. Identification and characterization of a ouabain-like compound from human plasma. Proc Natl Acad Sci U S A. 1991;88:6259–6263. doi: 10.1073/pnas.88.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosmond R, Chagnon M, Bouchard C, Björntorp P. Polymorphism in exon 4 of the human 3 beta-hydroxysteroid dehydrogenase type I gene (HSD3B1) and blood pressure. Biochem Biophys Res Commun. 2002;293:629–632. doi: 10.1016/S0006-291X(02)00234-6. [DOI] [PubMed] [Google Scholar]
- 52.Speirs HJL, Katyk K, Kumar NN, Benjafield AV, Wang WYS, Morris BJ. Association of G-protein-coupled receptor kinase 4 haplotypes, but not HSD3B1 or PTP1B polymorphisms, with essential hypertension. J Hypertens. 2004;22:931–936. doi: 10.1097/00004872-200405000-00014. [DOI] [PubMed] [Google Scholar]
- 53.Tripodi G, Citterio L, Kouznetsova T, Lanzani C, Florio M, Modica R, Messaggio E, Hamlyn JM, Zagato L, Bianchi G, Staessen JA, Manunta P. Steroid biosynthesis and renal excretion in human essential hypertension: association with blood pressure and endogenous ouabain. Am J Hypertens. 2009;22:357–363. doi: 10.1038/ajh.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shimodaira M, Nakayama T, Sato N, Aoi N, Sato M, Izumi Y, Soma M, Matsumoto K. Association of HSD3B1 and HSD3B2 gene polymorphisms with essential hypertension, aldosterone level, and left ventricular structure. Eur J Endocrinol. 2010;163:671–680. doi: 10.1530/EJE-10-0428. [DOI] [PubMed] [Google Scholar]
- 55.Jin Y, Kuznetsova T, Citterio L, Thijs L, Messaggio E, Casamassima N, Manunta P, Fagard R, Bianchi G, Staessen JA. Left ventricular structure and function in relation to steroid biosynthesis genes in a white population. Am J Hypertens. 2012;25:986–993. doi: 10.1038/ajh.2012.69. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.