Abstract
MicroRNAs (miRNAs) regulate gene expression by binding mRNA transcripts and inhibiting translation and/or inducing degradation of the associated transcripts. Expression levels of miRNAs have been shown to be altered in response to environmental toxicants, thus impacting cellular function and influencing disease risk. Transcription factors (TFs) are known to be altered in response to environmental toxicants and play a critical role in the regulation of miRNA expression. To date, environmentally-responsive TFs that are important for regulating miRNAs remain understudied. In a state-of-the-art analysis, we utilized in silico bioinformatic analysis to characterize potential transcriptional regulators of environmentally-responsive miRNAs. Using the miRStart database, genomic sequences of promoter regions for all available human miRNAs (n=847) were identified and promoter regions were defined as −1000/+500 base pairs from the transcription start site. Subsequently, the promoter region sequences of environmentally-responsive miRNAs (n=128) were analyzed using enrichment analysis to determine overrepresented TF binding sites (TFBS). While most (56/73) TFs differed across environmental contaminants, a set of 17 TFs was enriched for promoter binding among miRNAs responsive to numerous environmental contaminants. Of these, one TF was common to miRNAs altered by the majority of environmental contaminants, namely SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily A, member 3 (SMARCA3). These identified TFs represent candidate common transcriptional regulators of miRNAs perturbed by environmental toxicants.
Keywords: Transcription factors, microRNA, miRNA, promoters, environmental toxicants
1. Introduction
MicroRNAs (miRNAs) are single-stranded RNA molecules of ~22 nucleotides in length that regulate gene expression by binding to messenger RNA (mRNA) and inhibiting translation or degrading the transcript (Roush and Slack, 2008; Bartel, 2009). miRNAs function in many fundamental biological processes including development (Zhao et al., 2007), differentiation (Esquela-Kerscher and Slack, 2006), metabolism (Xu et al., 2003), and apoptosis (Xu et al., 2003; Xu et al., 2004). The expression levels of miRNAs are altered in many diseases, including neurodegenerative, oncogenic, cardiovascular, metabolic, and auto-inflammatory diseases (Wang and Sen, 2011; Rottiers and Naar, 2012; Satoh, 2012; Wang et al., 2012; Papaconstantinou et al., 2013; Dong et al., 2014). Additionally, expression levels of miRNAs in tissues and in circulation can be diagnostic, as well as prognostic, and predictive of patient therapeutic response (Slaby et al., 2010; Preis et al., 2011; Peck et al., 2015).
Importantly, miRNAs have been shown to be responsive to numerous environmental agents, indicating that they are possible biomarkers of exposure (Vrijens et al., 2015). Environmental contaminants/exposures shown to alter miRNA expression include air pollution, aluminum, arsenic, bisphenol A (BPA), diethyl phthalate (DEP), dichloro-diphenyl-trichloroethane (DDT), formaldehyde, polycyclic aromatic hydrocarbon (PAH), particulate matter (PM) and nanoparticles (NP), perfluorooctanoic acid (PFOA) and perfluorooctane sulfonate (PFOS), smoking, hexahydro-1,3,5-trinitro-1,3,5-triazine (RDX), and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Kappil and Chen, 2014; Vrijens et al., 2015). As miRNAs regulate critical biological pathways in the cell, these changes may partially contribute to environmentally-induced disease.
At present, the transcriptional control of miRNAs is understudied. Studies have identified transcription start sites (TSSs) of pri-miRNAs (Barski et al., 2007; Marson et al., 2008; Ozsolak et al., 2008; Corcoran et al., 2009; Stitzel et al., 2010). The TSSs can be 50k nucleotides or more upstream of the mature miRNA sequence (Chien et al., 2011). These TSSs help define promoter regions where transcription factor (TF) binding occurs to facilitate pri-miRNA transcription. With increasing knowledge about the location of the miRNA promoter regions, several TFs have been implicated in miRNA transcription in both normal physiologic processes and pathologic conditions. For example, tumor promoter 53 (p53) is known to regulate the expression of miR-24 (Yamakuchi and Lowenstein, 2009), Thus, TFs may be regulators of environmental contaminant-induced expression of miRNAs. To date, there has been little consideration of the role of TFs as regulators of miRNAs in response to environmental toxicants. The majority of studies have looked only at associations between environmental contaminants and miRNA responses without consideration for transcriptional control.
In the present study, we examined the relationship among miRNAs responsive to environmental contaminants and characterized enriched sequences for transcription factor binding within their promoter regions. This was achieved by compiling all promoter region sequences for human miRNAs, generating a database of all miRNAs known to be modulated by environmental contaminants and sequence-based analysis of the promoter regions for overrepresented transcription factor binding sites. The findings inform the extent to which particular miRNAs are responsive to different contaminants and also reveal common TFs that may be responsible for the miRNA response to various toxicants.
2. Materials and methods
2.1 Establishing genomic locations of miRNA transcription start sites
The identification of the miRNA transcription start sites (TSS) was performed using the miRStart microRNA transcription start site database (Chien et al., 2011). The miRStart database was developed by integrating three experimental datasets including transcription start site (TSS) sequence libraries, high-throughput sequencing analysis of gene initiation using H3K4me3 chromatin signatures, and cap analysis of gene expression (CAGE) tags (Chien et al., 2011). Using the miRStart database, 847 miRNA TSSs and promoter coordinates were systematically established by analyzing these three experimental data sets enabled through a machine-learningbased Support Vector Machine (SVM) (Chien et al., 2011).
2.2 Establishment of miRNA promoter location and sequences
Based upon prior evidence (Cooper et al., 2006), the promoter region for each miRNA was defined as being located 1,000 base pairs (bp) upstream and 500 bp downstream from the TSS (a total of 1,500 bp). Once the miRNA TSS locations were established, batch browser extensible display (BED) format files with the predicted promoter location were generated for groups of miRNAs altered in expression upon exposure to environmental contaminants. Each miRNA promoter coordinate consisted of six fields as follows: chromosome number, initiating chromosome location, terminating chromosome location, miRNA name, arbitrary Id, and, positive or negative strand, (e.g. chr18, 56112494, 56113994, hsa-mir-122, 48, and +). DNA coordinates were queried in Galaxy (https://usegalaxy.org/) and miRNA promoter DNA sequences were obtained through UCSC Genome Browser (https://genome.ucsc.edu/cgibin/hgGateway).
2.3 Developing a database of environmentally-altered miRNAs
To characterize TF-regulated miRNA expression in response to environmental contaminants, we identified a total of 128 different studies, across four species that studied changes in miRNA expression in response to one of 13 different environmental contaminants. The studies 13 different environmental contaminants included: air pollution, aluminum, arsenic, BPA and mixtures, DEP and mixtures, DDT, formaldehyde, PAH, particulate matter and nano particles, PFOA and PFOS, smoking, RDX and TCDD. These data were also collected from four different species: Homo sapiens, Mus musculus, Rattus norvegicus, and Macaca mulatta. From this, a database of 128 unique environmentally-responsive miRNAs was established.
2.4 Cross-species analysis of miRNA homology
As multiple species were considered in this analysis, a homology analysis was conducted to determine the conservation of miRNAs between species. To conduct this analysis any miRNA that was included from a non-human species was compared to its human homolog using miRViewer (Kiezun et al., 2012) where the lowest observed homology was found to be 0.89. Importantly, five miRNAs were identified that had no human homologs and thus were not included for downstream analysis.
2.5 Analysis of miRNA promoter DNA sequences for enriched transcription factor binding sites
The miRNA promoter batch files were analyzed using the Genomatix software suite common transcription factor sites analysis platform (https://www.genomatix.de). Each batch file was analyzed for TFBS enrichment with individual matrices, 0.95 core similarity, and TF sites common to a ≥ 85% of the miRNA batch file. The p-value for the enrichment represents the probability of obtaining an equal or greater number of sequences with a match in a randomly drawn sample of the same size as the input sequence set. As TFBS enrichment analysis would not be effective on miRNA promoter batch files containing <5 different miRNAs, these batch files were excluded from analysis. This resulted in ten miRNA promoter batch files being analyzed for TFBS enrichment, including air pollution, arsenic, BPA & mixtures, DDT, formaldehyde, PAH, PM & NP, PFOA & PFOS, smoking, and TCDD.
3. Results
3.1 Genomic locations of miRNA transcriptional start sites and promoter locations
Transcription start site coordinates for 847 miRNAs were established using miRStart. This database relies on three lines of evidence when determining promoter coordinates, specifically CAGE tags, TSS Sequence tags and H3K4me3 enrichment (Chien et al., 2011). Subsequently, the promoter locations were defined as 1,000 bp up-stream and 500 bp downstream of the annotated TSS (Chien et al., 2011) for a total of 1,500 bp. The promoter locations for the miRNAs are provided in Supplementary Table 1.
3.2 Compilation of environmentally-responsive miRNAs
A set of 128 environmentally-responsive miRNAs was compiled from studies representing exposure to 13 different environmental contaminants (Supplementary Table 2). These studies included the assessment of miRNA expression across various tissues from human, mouse, and rat models as well as cell lines. For any study that derived miRNA results from a non-human species, a homology analysis was performed to the human miRNA. These results are summarized in Supplementary Table 2.
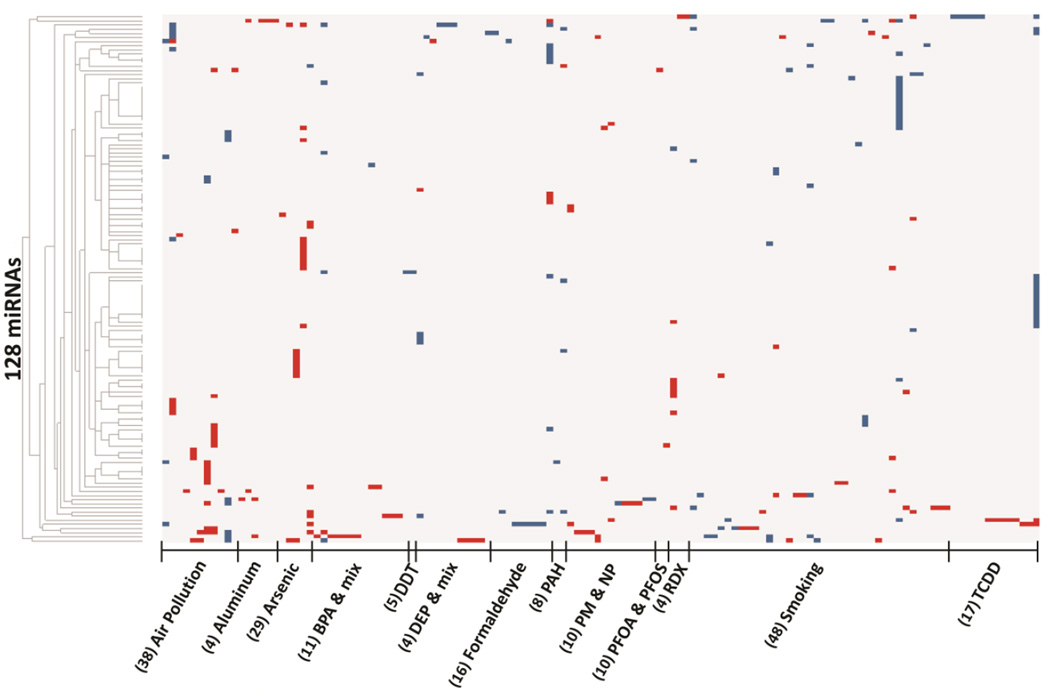
A heat-map displays the altered expression of the miRNAs in response to the 13 different environmental contaminants (Fig. 1). It is apparent that the expression levels of the altered miRNAs are highly variable across studies and contaminants, possibly reflecting the diverse tissues studied. Further examination revealed that the majority of the miRNAs (n=75, 59%) were altered in response to single contaminants, while a subset (n=53, 41%) were commonly altered in response to two or more contaminants. The most commonly altered miRNA was miR-21 which was significantly modulated in 15 studies in relation to exposure to six contaminants (Table 1, Supplementary Table 2). Other miRNAs that were altered in expression across multiple studies and contaminants include miR-146, miR-181a, miR-125b, and let-7e (Supplementary Table 2).
Fig. 1.
Heat map of the expression patterns of 128 environmentally-regulated miRNAs responsive to 13 different environmental contaminants. Each column represents a single study (n=128). Red represents increased expression of miRNAs, blue represents decreased expression of miRNAs. (N) represents the number of miRNAs observed to be altered in expression in relationship to the environmental contaminant.
Table 1.
Expression changes of miR-21 associated with environmental contaminant exposure.
| miR-21 Expression |
Pollutant | Tissue | Homology | Source |
|---|---|---|---|---|
| Increased | Air pollution Metal-rich PM | Blood leukocytes | Bollati et al. 2010 |
|
| Increased | Smoking (in vitro) | Human squamous carcinoma cells |
Zhang et al. 2014 |
|
| Increased | Air pollution (in vivo) 300 µg PM2.5/m3 DEP |
Peripheral blood | Yamamoto et al. 2013 |
|
| Decreased | Air pollution PM2.5, black carbon, organic carbon, sulfate |
Leukocytes | Fossati et al. 2014 |
|
| Increased | Arsenic | HUVEC cells | Li et al. 2012b |
|
| Decreased | BPA | MCF-7 cells | Tilghman et al. 2012 |
|
| Increased | DEP, metal-rich PM | Breast cancer | Ji et al 2007 | |
| Increased | DEP, metal-rich PM | Glioblastoma | Raitoharju et al. 2011 |
|
| Increased | DEP, metal-rich PM | Neo-intimal lesions | van Rooij et al. 2007 |
|
| Increased | DEP, metal-rich PM | Cardiac hypertrophy, atherosclerosis |
Volinia et al. 2006 |
|
| Increased | Nano particles 0.268 or 0.162 mg carbon black |
Mouse Lung | 0.99 | Bourdon et al. 2012 |
| Decreased | Smoking | Plasma from type 2 diabetic indivdiuals |
Zampetaki et al. 2010 |
|
| Decreased | Smoking | Placenta | Maccani et al. 2010 |
|
| Increased | Smoking | Gastric tissue | Stánitz et al. 2013 |
|
| Increased | Arsenic | Blood samples | Kong et al. 2012 |
3.3 Enrichment of TFBSs observed within promoter regions of environmentally-regulated miRNA
Using the database of all currently available Homo sapiens miRNA promoter coordinates (n=847), we characterized TF regulation of the environmentally-responsive miRNAs using TFBS over-representation analysis. Of the 128 miRNAs, only five were excluded based on no known homology to Homo sapiens. The miRNA promoter regions were analyzed for enrichment of TFBSs to predict common TF regulators of miRNA expression in response to different environmental contaminants. TFBS enrichment analysis is not effective on a limited number of promoter sequences, thus the analysis was only carried out for studies that identified >5 altered miRNAs. Therefore, three contaminants (aluminum, DEP and mixtures, and RDX) were excluded from analysis leaving ten for inclusion (air pollution, arsenic, BPA, DDT, formaldehyde, PAH, PM, NP, PFOA, PFOS, smoking, and TCDD). As a result of this exclusion, the final number of studies analyzed for TFBS was reduced from 128 to 108 and the number of miRNAs was reduced from 128 to 121.
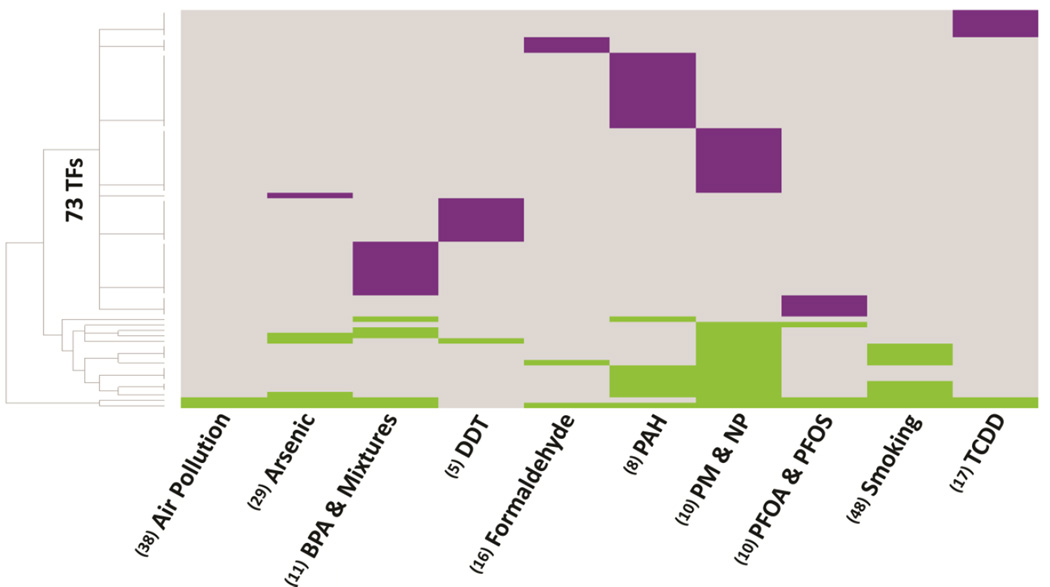
Sequence-based analysis of the miRNA promoter regions identified 73 unique TFs that were significantly (P < 0.05) enriched within miRNA promoter regions (Fig. 2, Supplementary Table 3). The majority of the TFs (n=56/73, 76.7%) were predicted to regulate miRNAs responsive to single contaminants. The remaining 17 TFs displayed TFBS enrichment across miRNAs responsive to at least two environmental contaminants. (Supplementary Table 4). Notable TFs with a high frequency of enrichment within miRNA promoter regions across contaminants in this category are SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 3 (SMARCA3), which was enriched among miRNAs altered by nine contaminants, and an alternative splicing variant of Forkhead Box P1 (FOXP1), activated in embryonic stem cells (FOXP1_ES), which was enriched among miRNAs responsive to seven contaminants (Supplementary Table 5).
Fig. 2.
Heat map of 73 TFs with enriched binding sites within promoters of environmentally-regulated miRNAs. A total of 56 TFs were observed to be enriched within a single miRNA promoter region, suggesting TF regulation of the miRNA that is specific for single environmental contaminants (displayed in purple). The remaining TFs (n=17) were observed to be enriched within multiple miRNA promoter regions, suggesting a TF regulation of the miRNAs common to two or more environmental contaminants. TF mediated regulation of the miRNAs that are common to environmental contaminants are represented in green. (N) represents the number of miRNAs known to be regulated by the environmental contaminant.
3.4 TFs as regulators of environmentally-responsive miRNAs
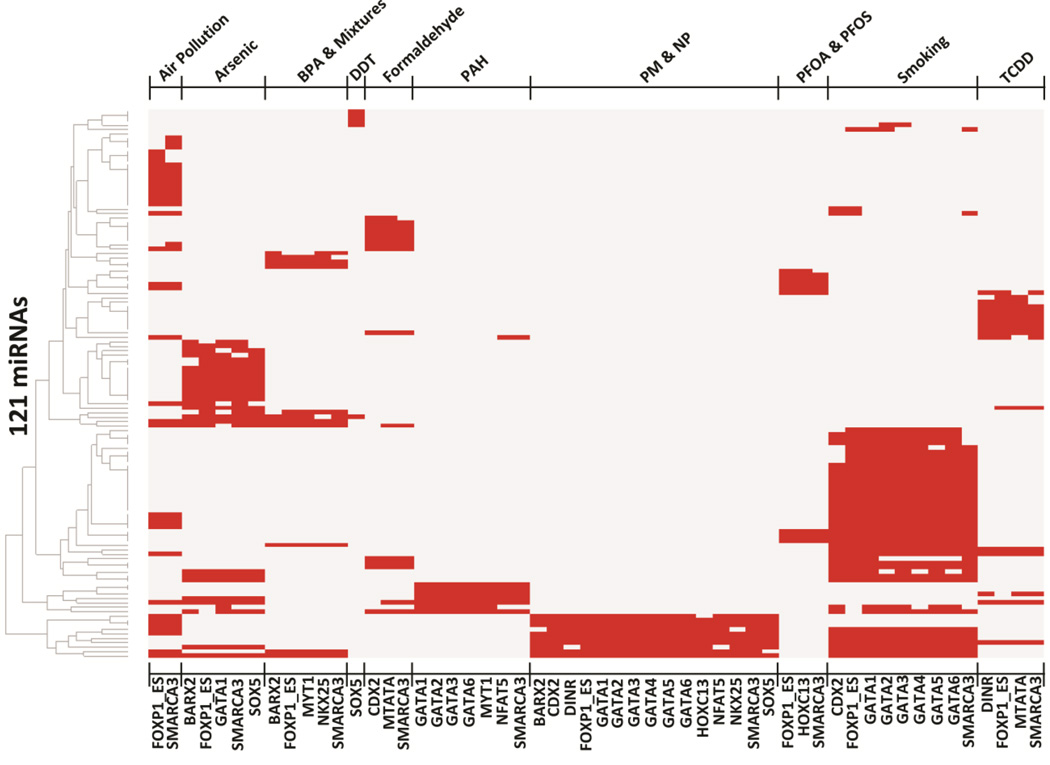
We next characterized the patterns of miRNA regulation for the 17 TFs that were common to two or more contaminants (Fig. 3). We show the relationship of the 17 TFs and their 121 target environmentally–responsive miRNAs (Fig. 3, Supplementary Table 5). As a specific example, there are 38 miRNAs responsive to air pollution and 29 miRNAs responsive to arsenic, with only five miRNAs shared between these two contaminants, namely hsa-mir-21, hsa-mir-26b hsa-mir-126, hsa-mir-181a-1, and hsa-mir-222 (Supplementary Table 5). The TFBS analysis demonstrated that the promoter regions of 31/38 of the air pollution-associated miRNAs and 25/29 of the arsenic-associated miRNAs contain binding sites for SMARCA3 and FOXP1_ES, respectively (Supplementary Table 5). The results illustrate the novel finding that distinct miRNAs that are responsive to various environmental contaminants may in fact be regulated by common transcription factors.
Fig. 3.
Heat-map of the 121 environmentally-regulated miRNAs that are enriched for common TFs (n=17) responsive to environmental contaminants.
Discussion
Expression levels of miRNAs are altered in response to environmental toxicants, as well as in multiple pathological conditions. As a result, these small RNAs have emerged as potential biomarkers of environmental exposure, as well as disease. While miRNAs are transcriptionally regulated by TFs, the specific TFs that mediate the altered expression of miRNAs to various environmental contaminants are currently understudied. In the present study, an in silico approach was used to systematically analyze the promoter sequences of environmentally-regulated miRNAs in order to characterize enriched TFBSs. This resulted in the identification of a set of 17 TFs, including SMARCA3, and FOXP1_ES, with binding sites enriched among the promoters of miRNAs with expression altered in response to two or more environmental agents. These data highlight specific TFs as common transcriptional regulators of environmentally-modulated miRNAs.
Of the currently known environmentally-responsive miRNAs, many (~60%) were changed only in a single study and only in relationship to single contaminants. The remaining miRNAs (~40%) were responsive to two or more environmental contaminants. Notably, miR-21, the most commonly dysregulated miRNA, has been shown to have altered expression in response to six different contaminants, including air pollution, arsenic, BPA and mixtures, DEP and mixtures, nano particles, and smoking. As an example of the complexity of environmental induction of miRNAs, miR-21 displayed increased expression in relation to arsenic, nanoparticles, and DEP metal-rich PM. In contrast, it displayed decreased expression in relation to BPA. Depending upon the study, miR-21 displayed either increased or decreased expression in relation to smoking and air pollution. MiR-21 is known to play a critical role in various diseases including cancer, cardiac damage, and inflammation (Wang et al., 2014; Duygu and Da Costa Martins, 2015; Sheedy, 2015). The establishment of common miRNAs, such as miR-21, as responders to a diverse number of environmental contaminants suggests a potential non-specific cellular response to environmental insult.
When the miRNA promoter regions were analyzed for TF binding sites, a total of 17 TFs were determined to be potential common regulators in relation to different contaminants. The two most common TFs were SMARCA3 and FOXP1_ES. The TF SMARCA3, also known as helicase-like transcription factor (HLTF), plays a role in preventing genomic instability after genotoxic stress (Motegi et al., 2008; Qing et al., 2011). Prolonged stalling of DNA replication due to DNA damage from stressors such as ionizing radiation, UV radiation, chemicals, and biological reactive metabolites can lead to apoptosis (Motegi et al., 2008). Cells have evolved to prevent apoptosis via post-replication repair mechanisms without removal of DNA damage and SMARCA3 has been shown to be important in this process (Motegi et al., 2008). The TF FOXP1_ES is an alternatively spliced form of the forkhead family transcription factor (FOXP1) specifically expressed in embryonic stem cells and is important in the transcription of genes required for stem cell pluripotency (Gabut et al., 2011). The enrichment of common TF binding sites within the promoter regions of miRNAs dysregulated in relationship to different environmental contaminants suggests a potential common mechanism of miRNA regulation in response to environmental insult.
In summary, this is the first study to apply sequence-based bioinformatic analysis to miRNA promoter regions to characterize TFs as transcriptional regulators in relation to various environmental contaminants. The results narrow the range of potential TFs that regulate environmentally-regulated miRNAs from thousands to a priority subset of specific TFs. It is important to note that the compiled miRNAs come from studies performed across different tissues and cell types, likely impacting differences in the expression of miRNAs across the studies. To address this, future work could characterize cell type-specific TFBS enrichment. Moving forward, the results of this study suggest that investigations on miRNA dysregulation in response to environmental contaminants should include analysis of TF regulation. To this end, we have provided the sequence locations of human miRNAs promoters required for such analysis. Additionally, future studies could examine miRNA expression in cells proficient or deficient for the specified transcription factors. The TFs characterized in this research represent potential regulators of miRNAs in response to numerous environmental contaminants. These TFs may ultimately be utilized to diagnose, treat, and prevent pathological conditions associated with environmental toxicants.
Supplementary Material
Highlights.
The submitted manuscript focuses on applying a bioinformatic in silico approach to genomic DNA sequences proximal to transcription start sites (TSS) of miRNAs as a unique method for predicting potential TFs that regulate miRNA expression. Specifically, we set out to identify transcription factor binding sites (TFBS) located within DNA promoter regions of miRNAs that are known to be dysregulated by environmental exposures.
A total of 13 environmental contaminants/stressors have been reported to influence the expression of miRNAs. These contaminants include; air pollution, aluminum, arsenic, BPA, DEP, DDT, formaldehyde, PAH, particulate matter and nano particles, PFOA and PFOS, smoking, RDX, and TCDD. Analyzing these, we developed a database of promoter sequences of 847 miRNAs. This database was queried for enriched TFBS among those miRNAs altered in relation to specific environmental contaminants (n=128) resulting in the identification of 73 TFs with high significance.
Predicted binding sites for the majority of the 73 TFs (n=56) were enriched within a single miRNA promoter region, suggesting a TF-mediated regulation of the miRNA that is specific for a single environmental contaminant. Predicted binding sites for the rest of the 73 (n=17) TFs were enriched within multiple miRNA promoter regions, suggesting TF-mediated regulation of miRNAs that are common for two or more environmental contaminants. Notable TFs common to multiple environmental contaminants included SMARCA3, which was enriched among miRNAs altered by nine contaminants and FOXP1_ES, which was common to seven contaminants.
Taken together, this study identifies specific transcription factors as potential master regulators of environmentally-mediated microRNA expression that may ultimately be utilized to diagnose, treat, and prevent pathological diseases associated with environmental exposures
Acknowledgments
The authors would like to thank Lisa Smeester for her comments on this manuscript. This research was supported by the National Institute of Environmental Health Sciences (T32-ES007018 and P42-ES005948).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-Resolution Profiling Of Histone Methylations In The Human Genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bartel DP. Micrornas: Target Recognition And Regulatory Functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien CH, Sun YM, Chang WC, Chiang-Hsieh PY, Lee TY, Tsai WC, Horng JT, Tsou AP, Huang HD. Identifying Transcriptional Start Sites Of Human Micrornas Based On High-Throughput Sequencing Data. Nucleic Acids Research. 2011;39:9345–9356. doi: 10.1093/nar/gkr604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper SJ, Trinklein ND, Anton ED, Nguyen L, Myers RM. Comprehensive Analysis Of Transcriptional Promoter Structure And Function In 1% Of The Human Genome. Genome Research. 2006;16:1–10. doi: 10.1101/gr.4222606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features Of Mammalian Microrna Promoters Emerge From Polymerase Ii Chromatin Immunoprecipitation Data. Plos One. 2009;4:E5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Yu J, Ng SS. Microrna Dysregulation As A Prognostic Biomarker In Colorectal Cancer. Cancer Management And Research. 2014;6:405–422. doi: 10.2147/CMAR.S35164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duygu B, Da Costa Martins PA. Mir-21: A Star Player In Cardiac Hypertrophy. Cardiovascular Research. 2015;105:235–237. doi: 10.1093/cvr/cvv026. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - Micrornas With A Role In Cancer. Nature Reviews. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Gabut M, Samavarchi-Tehrani P, Wang X, Slobodeniuc V, O'hanlon D, Sung HK, Alvarez M, Talukder S, Pan Q, Mazzoni EO, Nedelec S, Wichterle H, Woltjen K, Hughes TR, Zandstra PW, Nagy A, Wrana JL, Blencowe BJ. An Alternative Splicing Switch Regulates Embryonic Stem Cell Pluripotency And Reprogramming. Cell. 2011;147:132–146. doi: 10.1016/j.cell.2011.08.023. [DOI] [PubMed] [Google Scholar]
- Kappil M, Chen J. Environmental Exposures In Utero And Microrna. Current Opinion In Pediatrics. 2014;26:243–251. doi: 10.1097/MOP.0000000000000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiezun A, Artzi S, Modai S, Volk N, Isakov O, Shomron N. Mirviewer: A Multispecies Microrna Homologous Viewer. Bmc Research Notes. 2012;5:92. doi: 10.1186/1756-0500-5-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting Microrna Genes To The Core Transcriptional Regulatory Circuitry Of Embryonic Stem Cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi A, Liaw HJ, Lee KY, Roest HP, Maas A, Wu X, Moinova H, Markowitz SD, Ding H, Hoeijmakers JH, Myung K. Polyubiquitination Of Proliferating Cell Nuclear Antigen By Hltf And Shprh Prevents Genomic Instability From Stalled Replication Forks. Proceedings Of The National Academy Of Sciences Of The United States Of America. 2008;105:12411–12416. doi: 10.1073/pnas.0805685105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin Structure Analyses Identify Mirna Promoters. Genes & Development. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaconstantinou I, Stamatis K, Tzathas C, Vassiliou I, Giokas G, Gazouli M. The Role Of Variations Within Microrna In Inflammatory Bowel Disease. European Journal Of Gastroenterology & Hepatology. 2013;25:399–403. doi: 10.1097/MEG.0b013e32835c34ea. [DOI] [PubMed] [Google Scholar]
- Peck BC, Weiser M, Lee SE, Gipson GR, Iyer VB, Sartor RB, Herfarth HH, Long MD, Hansen JJ, Isaacs KL, Trembath DG, Rahbar R, Sadiq TS, Furey TS, Sethupathy P, Sheikh SZ. Micrornas Classify Different Disease Behavior Phenotypes Of Crohn's Disease And May Have Prognostic Utility. Inflammatory Bowel Diseases. 2015;21:2178–2187. doi: 10.1097/MIB.0000000000000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preis M, Gardner TB, Gordon SR, Pipas JM, Mackenzie TA, Klein EE, Longnecker DS, Gutmann EJ, Sempere LF, Korc M. Microrna-10b Expression Correlates With Response To Neoadjuvant Therapy And Survival In Pancreatic Ductal Adenocarcinoma. Clinical Cancer Research : An Official Journal Of The American Association For Cancer Research. 2011;17:5812–5821. doi: 10.1158/1078-0432.CCR-11-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qing P, Han L, Bin L, Yan L, Ping WX. Usp7 Regulates The Stability And Function Of Hltf Through Deubiquitination. Journal Of Cellular Biochemistry. 2011;112:3856–3862. doi: 10.1002/jcb.23317. [DOI] [PubMed] [Google Scholar]
- Rottiers V, Naar AM. Micrornas In Metabolism And Metabolic Disorders. Nature Reviews. Molecular Cell Biology. 2012;13:239–250. doi: 10.1038/nrm3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roush S, Slack FJ. The Let-7 Family Of Micrornas. Trends In Cell Biology. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- Satoh J. Molecular Network Of Microrna Targets In Alzheimer's Disease Brains. Experimental Neurology. 2012;235:436–446. doi: 10.1016/j.expneurol.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Sheedy FJ. Turning 21: Induction Of Mir-21 As A Key Switch In The Inflammatory Response. Frontiers In Immunology. 2015;6:19. doi: 10.3389/fimmu.2015.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slaby O, Lakomy R, Fadrus P, Hrstka R, Kren L, Lzicarova E, Smrcka M, Svoboda M, Dolezalova H, Novakova J, Valik D, Vyzula R, Michalek J. Microrna-181 Family Predicts Response To Concomitant Chemoradiotherapy With Temozolomide In Glioblastoma Patients. Neoplasma. 2010;57:264–269. doi: 10.4149/neo_2010_03_264. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Sethupathy P, Pearson DS, Chines PS, Song L, Erdos MR, Welch R, Parker SC, Boyle AP, Scott LJ, Program NCS, Margulies EH, Boehnke M, Furey TS, Crawford GE, Collins FS. Global Epigenomic Analysis Of Primary Human Pancreatic Islets Provides Insights Into Type 2 Diabetes Susceptibility Loci. Cell Metabolism. 2010;12:443–455. doi: 10.1016/j.cmet.2010.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijens K, Bollati V, Nawrot TS. Micrornas As Potential Signatures Of Environmental Exposure Or Effect: A Systematic Review. Environmental Health Perspectives. 2015;123:399–411. doi: 10.1289/ehp.1408459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sen S. Microrna Functional Network In Pancreatic Cancer: From Biology To Biomarkers Of Disease. Journal Of Biosciences. 2011;36:481–491. doi: 10.1007/s12038-011-9083-4. [DOI] [PubMed] [Google Scholar]
- Wang LL, Huang Y, Wang G, Chen SD. The Potential Role Of Microrna-146 In Alzheimer's Disease: Biomarker Or Therapeutic Target? Medical Hypotheses. 2012;78:398–401. doi: 10.1016/j.mehy.2011.11.019. [DOI] [PubMed] [Google Scholar]
- Wang Y, Gao X, Wei F, Zhang X, Yu J, Zhao H, Sun Q, Yan F, Yan C, Li H, Ren X. Diagnostic And Prognostic Value Of Circulating Mir-21 For Cancer: A Systematic Review And Meta-Analysis. Gene. 2014;533:389–397. doi: 10.1016/j.gene.2013.09.038. [DOI] [PubMed] [Google Scholar]
- Xu P, Guo M, Hay BA. Micrornas And The Regulation Of Cell Death. Trends In Genetics : Tig. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila Microrna Mir-14 Suppresses Cell Death And Is Required For Normal Fat Metabolism. Current Biology : Cb. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- Yamakuchi M, Lowenstein CJ. Mir-34, Sirt1 And P53: The Feedback Loop. Cell Cycle. 2009;8:712–715. doi: 10.4161/cc.8.5.7753. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, Von Drehle M, Muth AN, Tsuchihashi T, Mcmanus MT, Schwartz RJ, Srivastava D. Dysregulation Of Cardiogenesis, Cardiac Conduction, And Cell Cycle In Mice Lacking Mirna-1–2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.