Abstract
Background
The callipyge mutation is located within an imprinted gene cluster on ovine chromosome 18. The callipyge trait exhibits polar overdominant inheritance due to the fact that only heterozygotes inheriting a mutant paternal allele (paternal heterozygotes) have a phenotype of muscle hypertrophy, reduced fat and a more compact skeleton. The mutation is a single A to G transition in an intergenic region that results in the increased expression of several genes within the imprinted cluster without changing their parent-of-origin allele-specific expression.
Results
There was a significant effect of genotype (p < 0.0001) on the transcript abundance of DLK1, PEG11, and MEG8 in the muscles of lambs with the callipyge allele. DLK1 and PEG11 transcript levels were elevated in the hypertrophied muscles of paternal heterozygous animals relative to animals of the other three genotypes. The PEG11 locus produces a single 6.5 kb transcript and two smaller antisense strand transcripts, referred to as PEG11AS, in skeletal muscle. PEG11AS transcripts were detectable over a 5.5 kb region beginning 1.2 kb upstream of the PEG11 start codon and spanning the entire open reading frame. Analysis of PEG11 expression by quantitative PCR shows a 200-fold induction in the hypertrophied muscles of paternal heterozygous animals and a 13-fold induction in homozygous callipyge animals. PEG11 transcripts were 14-fold more abundant than PEG11AS transcripts in the gluteus medius of paternal heterozygous animals. PEG11AS transcripts were expressed at higher levels than PEG11 transcripts in the gluteus medius of animals of the other three genotypes.
Conclusions
The effect of the callipyge mutation has been to alter the expression of DLK1, GTL2, PEG11 and MEG8 in the hypertrophied skeletal muscles. Transcript abundance of DLK1 and PEG11 was highest in paternal heterozygous animals and exhibited polar overdominant gene expression patterns; therefore, both genes are candidates for causing skeletal muscle hypertrophy. There was unique relationship of PEG11 and PEG11AS transcript abundance in the paternal heterozygous animals that suggests a RNA interference mechanism may have a role in PEG11 gene regulation and polar overdominance in callipyge sheep.
Background
The mutation responsible for the callipyge trait is located within an imprinted gene cluster on the distal end of ovine chromosome 18 [1-3]. The callipyge phenotype is associated with an altered carcass composition including a 30–40% increase in muscle mass, a 6–7% decrease in carcass fat, decreased organ weights, and a more compact skeleton, all without a net affect on animal growth [4-7]. There is a pronounced hypertrophy of muscles in the loin and pelvic limbs and a lesser degree of hypertrophy in muscles of the thoracic limbs [6-8]. The callipyge phenotype is inherited in a non-Mendelian mode termed polar overdominance [9,10] in which only animals that inherit a normal allele (wild type; +) from the dam and the mutant callipyge allele from the sire (CLPGPat) exhibit the callipyge phenotype. Maternal heterozygotes (CLPGMat/+Pat) and callipyge allele homozygotes (CLPGMat/CLPGPat) have muscling and carcass compositions that are similar to normal sheep (wild type homozygotes; +/+).
A physical contig spanning the region containing the callipyge mutation was constructed using overlapping ovine bacterial artificial chromosomes [11] and 215 kb of sequence was obtained from the contig [3]. Comparisons of the ovine sequence to the human genome sequence and to expressed sequence databases indicated the presence of at least six transcribed genes with allele-specific expression [3] (Figure 1). The gene order along the contig was found to be Delta-like 1 (DLK1), DLK associated transcript (DAT), gene-trap locus 2 (GTL2), paternal expressed gene 11 (PEG11 / PEG11AS) and maternal expressed gene 8 (MEG8). The same conserved gene order was also found as an imprinted domain on human chromosome 14 and mouse chromosome 12 [12-15]. The DLK1 locus (also referred to as PREF-1, Zog-1 and pG2) encodes a transmembrane protein that contains epidermal-growth factor repeats [16-19]. Cleavage of the extracellular domain of DLK1 produces the circulating protein fetal antigen-1 [20]. DAT is a short non-coding RNA that has been proposed to be a cleavage product of extended DLK1 transcripts [21]. Both DLK1and DAT are expressed from the paternal allele [3,13-15]. GTL2 (also referred to as MEG3) and MEG8 genes express non-coding RNA from the maternal allele [3,13-15]. The PEG11 gene contains an intronless open reading frame of 1333 amino acids in sheep [3]. The human and mouse orthologues known as retrotransposon-like 1 (RTL1/rtl1) encode 1358 and 1745 amino acids, respectively. RNA transcripts were detected from the opposite strand of the same gene, referred to as PEG11AS (formerly known as antiPEG11) [3]. In the mouse, two maternally expressed microRNAs have been identified with perfect complementarity to mouse Rtl1 [22].
Figure 1.
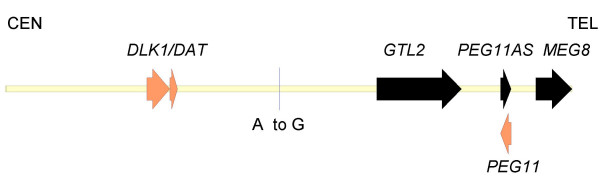
The callipyge region of ovine chromosome 18. A diagram of the callipyge region [3] based on GenBank accession No. AF345168 is shown. Six known transcripts are indicated along with the direction of transcription (arrows). Transcripts expressed from the paternal allele are shown as orange arrows and those expressed from the maternal allele are shown as black arrows [3]. A blue line indicates the position of the causative mutation. Alleles with an A are wild type (+) and the mutant callipyge allele (CLPG) has a G at this position [23, 24].
The causative mutation for callipyge is a single base transition of A (wild type; +) to G (CLPG) in the intergenic region located between DLK1 and GTL2 [23] (Figure 1). This mutation has been shown to be 100% concordant with all animals of the +Mat/CLPGPat genotype based on haplotype analysis [23]. Analysis of sheep from 19 different breeds as well as 13 mammalian species revealed a highly conserved 12 base sequence that includes the single nucleotide polymorphism [24]. The G polymorphism is unique to direct descendents of the first known callipyge animal, a ram named "Solid Gold". This animal was mosaic for the mutation [24], providing strong evidence that this single nucleotide polymorphism is the causative mutation. Initial results indicate that the mutation alters the expression of several of the genes within the imprinted cluster [25,26] when they are inherited in cis without altering their parent-of-origin-specific expression [25].
In this study, we analyzed the expression of five genes within the callipyge cluster in the muscles of lambs of all four genotypes. Quantitative analysis of gene expression using a series of orthogonal contrasts showed that DLK1, PEG11 and MEG8 exhibited a polar overdominant pattern of gene expression. The expression of PEG11 and PEG11AS transcripts in the muscles of paternal heterozygous callipyge lambs (+Mat/CLPGPat) was different from the other three genotypes. A sense/antisense interaction of PEG11 and PEG11AS, such as an RNA interference mechanism, would be consistent with a trans interaction between reciprocally imprinted genes that has been previously proposed as a mechanism for polar overdominance [27,28].
Results
Northern blot analysis
Muscle samples were collected from 12- and 8-week-old lambs, when muscle hypertrophy is well established in animals with the callipyge phenotype. Total RNA was extracted from three muscles that undergo hypertrophy including longissimus dorsi (loin), semimembranosus, and gluteus medius (pelvic limb), and one muscle that does not undergo hypertrophy, the supraspinatus, (thoracic limb). Strand specific probes were used to analyze PEG11 and PEG11AS expression. Hybridization of longissimus dorsi, semimembranosus and gluteus medius northern blots with a PEG11 probe indicated expression of a 6.5 kb PEG11 transcript in paternal heterozygotes (+Mat/CLPGPat) that was not readily detectable in the other three genotypes (Figure 2). Two smaller PEG11AS transcripts of 1.7 kb and 0.8 kb were detected in the two genotypes with maternally inherited callipyge alleles (CLPGMat/+Pat and CLPGMat/CLPGPat) by a probe from the complementary strand. PEG11 and PEG11AS transcripts were not detected in the supraspinatus. The expression of DLK1 was detectable at various levels in each of the four muscles and all four genotypes, although DLK1 appeared to be up-regulated in the loin and pelvic limb muscles of +Mat/CLPGPat and CLPGMat/CLPGPat animals (Figure 2). GTL2 transcripts of around 2.4 kb were evident in the CLPGMat/+Pat and CLPGMat/CLPGPat genotypes. A distinct 1.9 kb GTL2 transcript was consistently detected in the +Mat/CLPGPat genotype and no GTL2 transcripts were detected in the +/+ genotype. A 1.8 kb MEG8 transcript was expressed to a lesser degree than GTL2 in the CLPGMat/+Pat and CLPGMat/CLPGPat genotypes and was also detectable in the +Mat/CLPGPat genotype. The expression pattern of MEG8 was the least consistent in the northern blot analysis of individuals of the four genotypes.
Figure 2.
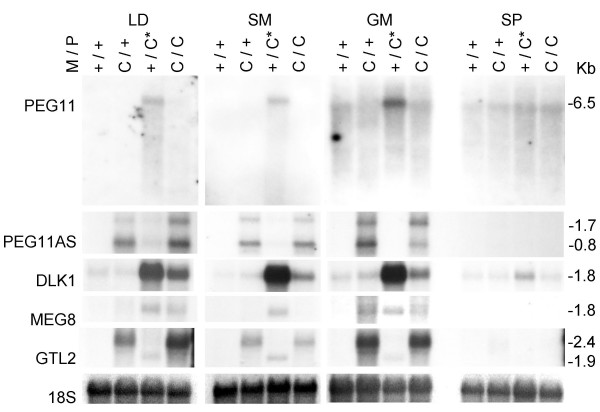
Northern blot analysis of skeletal muscle RNA from 12-week-old lambs. Expression of five genes from the callipyge region is shown for the four possible genotypes in 12-week-old lambs. Each lane contains total RNA from an individual animal. The genotypes are given with the maternal allele first followed by the paternal allele. The callipyge allele (CLPG) has been abbreviated to C and the paternal heterozygote that has the callipyge phenotype is indicated with an asterisk (*). Strand specific probes are indicated on the left and transcript sizes are indicated on the right. The skeletal muscles shown are the longissimus dorsi (LD), semimembranosus (SM), gluteus medius (GM) and the supraspinatus (SP). The blots were hybridized with an 18S RNA probe to show the equivalence of RNA loading and transfer.
PEG11/PEG11AS
Ribonuclease protection assays were performed using five riboprobes to map the PEG11 and PEG11AS transcripts to the contig sequence (Figure 3A). The results for riboprobe P confirm the expression of PEG11 transcripts in the gluteus medius of +Mat/CLPGPat animals and showed a very low level of PEG11 in the gluteus medius of CLPGMat/CLPGPat animals. PEG11AS transcripts were readily detectable in animals of the +/+, CLPGMat/+Pat and CLPGMat/CLPGPat genotypes but were slightly above background levels in the +Mat/CLPGPat animals (Figure 3B). Two other PEG11 probes from the open reading frame (E) and from the 3'UTR (C) showed equivalent results to riboprobe P in the gluteus medius (Figure 3B). The same expression pattern was seen for PEG11 and PEG11AS transcripts in the supraspinatus, but signal from the protected riboprobes was just above background level.
Figure 3.
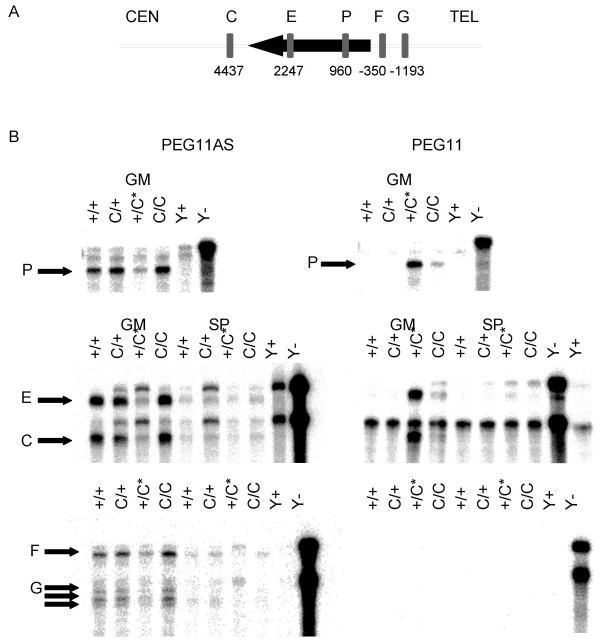
Mapping of PEG11 and PEG11AS transcripts by ribonuclease protection assay. A) A diagram of the PEG11 open reading frame (black arrow) and the position of the five riboprobes are shown. The PEG11 open reading frame extends from base 191859 to 187858 on the complementary strand of AF354168. B) Ribonuclease protection assays using PEG11AS (left column) and PEG11 (right column) are shown using total RNA from the gluteus medius (GM) and the supraspinatus (SP) muscles for each of the four possible genotypes. The genotypes are given with the maternal allele first followed by the paternal allele. The CLPG allele has been abbreviated to C and the paternal heterozygote that demonstrates the callipyge phenotype is indicated with an asterisk (*). Yeast RNA with RNase treatment (Y+) and without RNase treatment (Y-) are shown as controls. A black arrow indicates the RNA fragments protected by each of the five RNA probes. Some full-length probes for C and E that could not be completely eliminated by RNase digestion are present in both muscle RNA samples and the yeast RNA control (Y+).
Two probes from upstream of the PEG11 coding sequence (riboprobes F and G) did not detect any PEG11 transcripts, indicating that transcription of the 6.5 kb PEG11 was initiated within 350 bp of the start codon of the open reading frame. Riboprobe F detected PEG11AS transcripts in the gluteus medius and supraspinatus of all four genotypes, with the lowest expression in the paternal heterozygous (+Mat/CLPGPat) animals. The PEG11AS riboprobe G shows three protected fragments, suggesting variable splice junctions or transcription termination sites for PEG11AS transcripts.
Quantitative analysis of the effect of genotype on gene expression
Gene expression was measured in the gluteus medius and supraspinatus of 8 week-old animals (Figure 4) using quantitative PCR. The expression of glyceraldehyde-3-phosphate dehydrogenase was not significantly different across the four genotypes in the gluteus medius and supraspinatus (Table 1), indicating that equivalent amounts of RNA were used for cDNA synthesis and quantitative PCR. The effect of the callipyge mutation on genotype-specific expression of DLK1 and PEG11 in gluteus medius was the same (Table 1) although the magnitude of the response was greater for PEG11 than DLK1 (Figure 4A and 4B). The paternal heterozygous (+Mat/CLPGPat) animals had the highest transcript abundance (p < 0.05), followed by CLPGMat/CLPGPat animals, which had significantly greater transcript abundance (p < 0.05) than CLPGMat/+Pat or +/+ animals. The mRNA abundance of DLK1 in the gluteus medius was 6-fold and 2.5-fold greater in +Mat/CLPGPat lambs and CLPGMat/CLPGPat lambs respectively, relative to normal lambs (+/+; Figure 4A). PEG11 mRNA abundance in the gluteus medius was 200-fold and 13-fold greater in +Mat/CLPGPat lambs and CLPGMat/CLPGPat lambs respectively, relative to normal lambs (+/+; Figure 4B).
Figure 4.
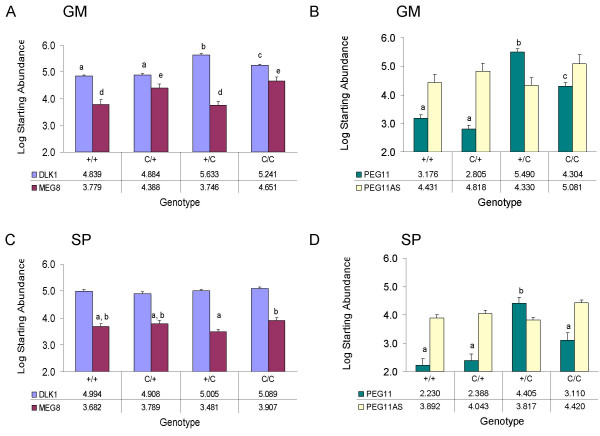
Quantitative reverse transcriptase PCR analysis of transcript abundance in the gluteus medius and supraspinatus muscles. Least square means and standard errors for transcript abundance by genotype are shown for the gluteus medius (GM), and supraspinatus (SP). The genotypes are given with the maternal allele first followed by the paternal allele and the CLPG allele has been abbreviated to C. Quantification of DLK1 and MEG8 transcripts (A, C) was based on random primed cDNA synthesis using 213 ng of total RNA. Quantification of PEG11 and PEG11AS (B, D) transcripts was based on gene-specific priming of cDNA synthesis using 400 ng of total RNA. Different superscripts indicate significant differences (p < 0.05) between genotypic means for a given mRNA transcript and muscle. Numerical values for the log starting abundance are given for each mRNA transcript in a table below the genotypes.
Table 1.
Statistical analysis of gene expression by quantitative PCR
| Orthogonal Contrast P-Values | |||||
| Muscle | Gene | Effect of Genotype (p-values) |
Additive | Maternal Dominance | Paternal Polar Overdominance |
| GM | DLK1 | 0.0001 | 0.0001 | 0.0261 | 0.0001 |
| MEG8 | 0.0005 | 0.0009 | 0.4083 | 0.0047 | |
| PEG11 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | |
| PEG11AS | 0.2622 | ||||
| G3PD | 0.1393 | ||||
| SP | DLK1 | 0.1865 | |||
| MEG8 | 0.0325 | 0.1398 | 0.9665 | 0.0094 | |
| PEG11 | 0.0001 | 0.0179 | 0.3315 | 0.0001 | |
| PEG11AS | 0.1438 | ||||
| G3PD | 0.3626 | ||||
The effect of the callipyge mutation on DLK1 and PEG11 expression in the supraspinatus was different (Table 1). No differences in DLK1 transcript abundance in the supraspinatus were found among the four genotypes (Figure 4C). Although not statistically analyzed, the level of DLK1 expression was similar between the gluteus medius and supraspinatus (Figure 4A and 4C). PEG11 expression in the supraspinatus had a different genotype-specific pattern than in the gluteus medius. PEG11 expression was elevated 150-fold in +Mat/CLPGPat animals (p < 0.05) but was not significantly changed in the other three genotypes (Figure 4D). The transcript abundance of PEG11 in supraspinatus was generally much lower than in gluteus medius of the same genotype. PEG11 expression in the supraspinatus was 12-fold lower than in the gluteus medius of paternal heterozygous animals (Figure 4B and 4D).
Maternal inheritance of the callipyge allele significantly altered the expression of MEG8 but not PEG11AS in the gluteus medius (Table 1). Transcript abundance of MEG8 in gluteus medius was 6-fold greater (p < 0.05) in the CLPGMat/+Pat and CLPGMat/CLPGPat animals relative to the +Mat/CLPGPat and +/+ animals (Figure 4A). Expression of MEG8 in the supraspinatus was also affected by the callipyge mutation but to a lesser degree (Figure 4C). The homozygous callipyge animals had significantly higher MEG8 transcript abundance than paternal heterozygotes, but those two genotypes were not significantly different from maternal heterozygotes and homozygous wild type lambs.
Orthogonal contrasts were used to analyze different models of gene action for the genes that had a significant effect for genotype (Table 1). The polar overdominance contrast was significant for transcript abundances of DLK1, PEG11 and MEG8 in gluteus medius and for PEG11 and MEG8 in the supraspinatus. The additive contrasts were also significant for DLK1, PEG11 and MEG8 in gluteus medius but were only significant for PEG11 in supraspinatus. The maternal dominance contrast was significant for DLK1 and PEG11 gluteus medius.
Discussion
Our results show a clear pattern of increased expression for genes within the imprinted callipyge cluster in muscles that become hypertrophied, whereas expression of these genes was either reduced or absent in a muscle that does not become hypertrophied. The increased gene expression occurred when the mutation was inherited in cis and was dependent on each gene's imprinting status, consistent with previous reports [25,26]. These results support the hypothesis that the mutation has disrupted a long range control element [27]. Two paternally expressed genes, DLK1 and PEG11, had significantly increased transcript abundance when a callipyge allele was inherited from the sire. One maternally expressed gene, MEG8, showed significantly increased transcript abundance when the callipyge allele was inherited from the dam. The changes in gene expression were sustained until 12 weeks of age, when the differences in growth and body composition between callipyge and normal lamb are established and are subsequently maintained [7].
Northern blot analysis suggests that expression of GTL2 and PEG11AS was increased by maternal inheritance of the callipyge allele. Quantitative analysis was not done for GTL2 in this study due to the expression of numerous alternatively spliced transcripts that have been reported for mice and sheep [26,29]. The 2.4 kb GTL2 mRNA seen in this study consisted of a heterogeneous population of alternatively spliced mRNAs. Similarly, two PEG11AS transcripts were detected over a 5.6 kb area extending from beyond the 5' end of the PEG11 transcript to the 3'UTR. The 1.7 and 0.8 kb PEG11AS transcripts detected by northern blot analysis using probe P would not be protected by probes C or G unless the PEG11AS transcripts undergo intron splicing or there are other transcripts that were not detected in the northern blots. Therefore, the effect of the callipyge mutation on GTL2 and PEG11AS will require a more extensive analysis to fully elucidate their expression patterns in the four genotypes and determine their role in the callipyge model.
Due to their paternal allele-specific expression, the DLK1 and PEG11 genes are both candidates for an effector gene that is responsible for the skeletal muscle hypertrophy exhibited by paternal heterozygous (+Mat/CLPGPat) animals. In this study, both genes showed a polar overdominant expression pattern in that paternal heterozygotes had significantly higher levels of gene expression than the other three genotypes. The major differences between DLK1 and PEG11 were the magnitude and the muscle specificity of the up-regulation. DLK1 was readily detectable in all muscles and genotypes by northern blot, but the up-regulation in paternal heterozygous animals was restricted to muscles of the loin and pelvic limb. The quantitative results showed a 6-fold increase in DLK1 transcript abundance in the gluteus medius and no change of DLK1 abundance in the supraspinatus. This pattern of gene expression is consistent with studies on individual muscle growth that show significant muscle hypertrophy in the gluteus medius but not in the supraspinatus [6,8]. DLK1 transcripts were significantly increased (2.5-fold) in the gluteus medius of CLPGMat/CLPGPat animals, which do not exhibit muscle hypertrophy. The lack of a phenotype in CLPGMat/CLPGPat animals could be due to a threshold effect that requires more than a 2.5-fold increase in DLK1 transcript abundance to change muscle growth.
The expression of PEG11 was very low in the gluteus medius and supraspinatus of normal sheep and was induced in both muscles of callipyge lambs. Northern blot analysis, ribonuclease protection assay and quantitative PCR results all show that the expression of PEG11 and PEG11AS transcripts was much lower in the supraspinatus than the other muscles. The high level of expression of PEG11 in the gluteus medius relative to the supraspinatus of callipyge lambs indicates that PEG11 could also be the gene responsible for muscle hypertrophy if there is a threshold level required to change muscle growth. The PEG11 gene has a long intronless open reading frame but it is not known if a protein is produced or what function it may have.
The overdominant nature of the callipyge phenotype through the lack of muscle hypertrophy in animals with the CLPGMat/CLPGPat genotype is one of the more intriguing aspects of the trait and has led to a hypothesis of trans effects by other reciprocally imprinted genes in the callipyge region [25,27]. If the PEG11 gene has a direct role in muscle hypertrophy, either alone or in concert with DLK1, then PEG11AS may have a role in a trans effect on PEG11 expression. In +Mat/CLPGPat animals, PEG11 transcripts were 14-fold and 4-fold more abundant than PEG11AS transcripts in the gluteus medius and supraspinatus respectively. PEG11AS transcripts were more abundant than PEG11 transcripts in the other three genotypes for both muscles. Therefore, the relative abundance of PEG11 transcripts to PEG11AS transcripts was unique in paternal heterozygous animals. MicroRNAs are a central component of RNA interference mechanisms. In the mouse, two antisense microRNA have been identified for the orthologous rtl1 locus [22]. RNA interference mechanisms have been shown both to repress transcription by inducing heterochromatin formation [30-32] and to cause post-transcriptional silencing through nuclear retention or targeted degradation [33-35]. MicroRNAs may be produced from post-transcriptional processing of PEG11AS RNA and be involved in normal regulation of the locus and in generating overdominance [28]. Expression of PEG11AS may normally cause repression of the paternal PEG11 locus since very little PEG11 mRNA was detectable in the muscles of normal animals. In the animals with a paternally inherited callipyge allele, (+Mat/CLPGPat and CLPGMat/CLPGPat), the normal repression of the PEG11 locus has been disrupted by the mutation in a putative long range control element [27]. PEG11 transcripts only accumulate in significant excess of PEG11AS in the paternal heterozygous animals, whereas in the CLPGMat/CLPGPat animals the PEG11AS transcripts remain in excess and may prevent the accumulation of the threshold level of PEG11 mRNA required to produce a muscle hypertrophy phenotype. Although the expression of PEG11AS was not affected by genotype in quantitative PCR, the northern blots and transcript mapping indicate there are multiple transcripts that are likely to undergo different intron splicing. Further analysis of PEG11AS expression will be necessary to determine its role in the PEG11 locus regulation.
Conclusions
The effect of the callipyge mutation has been to increase transcript abundance of four genes, DLK1, GTL2, PEG11 and MEG8, within the imprinted cluster in skeletal muscles that become hypertrophied. The increase in transcript abundance was consistent with each gene's parental allele-specific expression. The DLK1 and PEG11 genes were both expressed at their highest levels in paternal heterozygous animals and exhibited polar overdominant gene expression patterns. Therefore, both genes are candidates for causing muscle hypertrophy. DLK1 expression was only elevated in muscles that undergo hypertrophy, so its muscle-specific increase was consistent with the callipyge phenotype. PEG11 was 12-fold more abundant in hypertrophied muscle than non-hypertrophied muscle and only paternal heterozygous animals had PEG11 transcript levels in excess of PEG11AS transcript levels. The unique relationship of PEG11 and PEG11AS in paternal heterozygous animals suggests that an RNA interference mechanism may have a role in regulating the PEG11 locus and polar overdominance in callipyge sheep.
Methods
Sample collection
A series of planned matings were conducted to produce the four possible callipyge genotypes. The genotypes of all lambs were verified using the single nucleotide polymorphism [23,24] and several markers that flank the callipyge region on chromosome 18. Lambs were slaughtered in accordance with humane practices approved by the Utah State University Institutional Animal Care and Use Committee. Samples were collected from the longissimus dorsi, semimembranosus, gluteus medius and supraspinatus, and preserved in RNAlater (Ambion Inc., Woodlands, TX USA). The tissue samples were homogenized in 4 M guanidinium thiocyanate, 25 mM sodium citrate, 50 mM EDTA, 1% sodium-N-lauroyl-sarcosine and total RNA was sedimented by ultracentrifugation of the homogenate on a cushion of 5.7 M CsCl, 50 mM EDTA [36]. Purified RNA was quantified by spectrophotometry and the use of a constant mass of RNA for each quantitative assay was based on absorbance at 260 nm. Total RNA was treated with DNase I using DNA free™ reagents (Ambion Inc.) to remove trace genomic DNA prior to use in the ribonuclease protection assay and quantitative PCR.
Northern blot analysis
Northern blots were prepared using denaturing formaldehyde gel electrophoresis (NorthernMax™; Ambion Inc) of 10 μg of total RNA and transferred to positively charged nylon membranes using standard methods [37]. Primer sequences used to amplify probes from the callipyge region are given in Charlier et al. [25], and the PCR products were verified by DNA sequencing. Strand specific DNA probes were synthesized by 40 cycles of asymmetric PCR with Strip-EZ™ nucleotides (Ambion Inc.) and 50 μCi of α-[32P]-dATP (Amersham-Pharmacia, Piscataway, NJ USA). Unincorporated nucleotides were removed by spin column chromatography (BioSpin P30; Bio-Rad Inc., Hercules, CA USA). The probes were hybridized to the membranes without denaturation using Ultrahyb™ (Ambion Inc.) at 42°C overnight. After hybridization, the membranes were washed in 2X SSC (0.3 M sodium chloride, 0.03 M sodium citrate)/0.5% SDS followed by 3 washes in 1X SSC/ 0.1% SDS at 65°C for 30 min and a final high stringency wash in 0.1X SSC/ 0.1%SDS at 65°C for 30 min. The northern blots were exposed to Kodak XAR autoradiography film for 18 to 72 h at -80°C. After autoradiography, the probe was degraded and removed from the membranes using Strip-EZ™ reagents (Ambion Inc.).
Ribonuclease protection assay
Templates for synthesizing strand specific RNA probes for the ribonuclease protection assay (RPA) were generated from PCR products (Table 2) by ligation of double stranded oligonucleotides containing T7 or SP6 promoter sequences and re-amplification with an adapter and gene specific primer (Lig'n Scribe™, Ambion Inc.). Labeled RNA probes were synthesized using MAXIscript™ reagents (Ambion, Inc.) and 50 μCi of α-[32P]-UTP (Amersham-Pharmacia). The RPA were conducted using RPA III™ reagents (Ambion Inc.) and standard urea/acrylamide gel electrophoresis methods [37]. Dried gels were exposed to phosphorimaging screens and images were collected using a Cyclone Storage Phosphorimager (Packard Instrument Co., Meriden, CT USA).
Table 2.
Primer sequences for ribonuclease protection assay probes and quantitative PCR
| Sizea | Tm | Locationb | |||
| Primer | Sequence | bp | °C | ||
| PEG11/PEG11AS | |||||
| C | F | AGGAACACCGCTGTGGAGGTAGAA | 135 | 59 | 4337 |
| R | ACAGCAGAGGCAGCCAAGCA | ||||
| E | F | GGTGACGCCCGTCTGCAAGT | 180 | 61 | 2427 |
| R | GTGGAACGGTTCGCCGACAT | ||||
| Pc | F | ACAGCTCAACAGTGGAGGTCATG | 199 | 55 | 996 |
| R | ATCAGCTGGCAGAGCACGATGAAC | ||||
| F | F | TTCCCCCATGGCTGTGAGAAAT | 175 | 58 | -390 |
| R | CCTCTGTGACCTTCTGGTGACCAA | ||||
| G | F | AGGCTGAATTGACAGAGATGT | 124 | 49 | -1193 |
| R | GTTAAATGGCTCAAGAACGA | ||||
| DLK1 | F | CCCGTCCTCTTGCTCCTGCT | 116 | 58 | |
| R | GGCTGGCACCTGCACACACT | ||||
| MEG8 | F | CCCAGGGAGTGTGAGGCTCTTCT | 100 | 56 | |
| R | GGACCCACGGCTGACCTGTT | ||||
| G3PD | F | TGAGTGTCGCTGTTGAAGT | 150 | 58 | |
| R | CCTGCCAAGTATGATGAGAT | ||||
aSize of the PCR product and protected fragment for RPA bLocation for the PCR product is relative to the first base of the PEG11 open reading frame. cCharlier et al. [3]
Quantitative PCR
Complementary DNA was synthesized from 3.2 μg of total RNA using random hexamer priming and MMLV reverse transcriptase reagents (Invitrogen, Carlsbad, CA USA) with RNase inhibitor supplementation (Superase Inhibitor, Ambion Inc). The cDNA samples were diluted with water and aliquoted so that the quantification was based on 213 ng of total RNA for analysis of glyceraldehyde-3-phosphate dehydrogenase, DLK1 and MEG8 transcripts. PEG11 and PEG11AS transcripts were measured using gene-specific priming of cDNA synthesis from 1.6 μg of total RNA. The cDNA was diluted with water and aliquoted for quantification based on 400 ng of total RNA. Each cDNA sample was amplified in triplicate using the SYBR Green Jump Start™ system (Sigma-Aldridge, St. Louis, MO USA). Quantification standards were composed of aliquots of plasmids containing target PCR products in 10-fold serial dilutions ranging from 108 to 102 molecules. The standards were used to calculate a regression of threshold cycle on molecule copy number to determine a log value of starting abundance for each of the cDNA samples based on their threshold cycle. The PCR reactions were run for 40 cycles in an iCycler Real-Time PCR Detection System (Bio-Rad Inc.). The log value of starting abundance for each gene was analyzed by analysis of variance using the PROC MIXED procedure of SAS [38]. The analysis model included genotype as a fixed effect and animal within genotype as a random effect. The number of animals representing each genotype (6 to 8) is given in Table 3. Orthogonal contrasts were used to evaluate different models of gene action if the effect of genotype on log value of starting abundance was significant. Initially, traditional additive, dominance and reciprocal heterozygote effects were evaluated (see Table 3 for contrasts). If the reciprocal heterozygote effect was significant (p < 0.05), a second set of orthogonal contrasts was used to test additive, maternal dominance and polar overdominance effects as previously described by Freking et al. [10] (Table 3).
Table 3.
Statistical methods for genetic models
| Orthogonal Contrast Values by Genotype | ||||
| Genetic Model | +/+ | CLPGMat/+Pat | +Mat/CLPGPat | CLPGMat/CLPGPat |
| Additive | -1 | 0 | 0 | 1 |
| Dominance | -1 | 1 | 1 | -1 |
| Reciprocal Heterozygote | 0 | -1 | 1 | 0 |
| Maternal Dominance | -1 | 2 | 0 | -1 |
| Paternal Polar Overdominance | -1 | -1 | 3 | -1 |
| Number of Animals | 6 | 7 | 8 | 8 |
Authors' contributions
CB participated in planning the study and developing the experimental design, conducted the northern blot analysis and ribonuclease protection assays, and wrote the drafts of the manuscript. LK and AP isolated RNA and performed the quantitative PCR assays. TH collected the muscle samples and genotyped the animals used in this study. DM participated in experimental design and performed the statistical analysis. NC participated in planning the study, set up a series of matings to generate all four genotypes and provided the experimental animals. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgements
This project was supported by funds from the USDA/NRI (2002-35205-11602) and the Purdue University Agricultural Research Program (paper No. 17430). The authors acknowledge Michel Georges and Carole Charlier, University of Liège, Belgium, for providing DNA sequence and PCR primer data and thank Christine Bynum, Anne Danielsson and David Forrester for their assistance with this experiment.
Contributor Information
Christopher A Bidwell, Email: cbidwell@purdue.edu.
Lauren N Kramer, Email: kramerl@purdue.edu.
Allison C Perkins, Email: aperkins@purdue.edu.
Tracy S Hadfield, Email: tracy.hadfield.shay@cc.usu.edu.
Diane E Moody, Email: moodyd@purdue.edu.
Noelle E Cockett, Email: noelle.cockett@cc.usu.edu.
References
- Cockett NE, Jackson SP, Shay TL, Nielsen D, Moore SS, Steele MR, Barendse W, Green RD, Georges M. Chromosomal localization of the callipyge gene in sheep (Ovis aries) using bovine DNA markers. Proc Natl Acad Sci USA. 1994;91:3019–3023. doi: 10.1073/pnas.91.8.3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrug SC, Freking BA, Rexroad CE, III, Leymaster KA, Kappes SM, Smith TPL. Comparative mapping of the ovine clpg locus. Mamm Genome. 2000;11:871–876. doi: 10.1007/s003350010150. [DOI] [PubMed] [Google Scholar]
- Charlier C, Segers K, Wagenaar D, Karim L, Berghmans S, Jaillon O, Shay T, Weissenbach J, Cockett N, Gyapay G, Georges M. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome Res. 2001;11:850–862. doi: 10.1101/gr.172701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Green RD, Miller MF. Phenotypic characterization of rambouillet sheep expressing the callipyge gene: I. Inheritance of the condition and production characteristics. J Anim Sci. 1997;75:14–18. doi: 10.2527/1997.75114x. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Miller MF, Green RD. Phenotypic characterization of rambouillet sheep expressing the callipyge gene: II. Carcass characteristics and retail yield. J Anim Sci. 1997;75:125–132. doi: 10.2527/1997.751125x. [DOI] [PubMed] [Google Scholar]
- Jackson SP, Miller MF, Green RD. Phenotypic characterization of rambouillet sheep expression the callipyge gene: III. Muscle weights and muscle weight distribution. J Anim Sci. 1997;75:133–138. doi: 10.2527/1997.751133x. [DOI] [PubMed] [Google Scholar]
- Freking BA, Keele JW, Nielsen MK, Leymaster KA. Evaluation of the ovine callipyge locus: II. Genotypic effects on growth, slaughter, and carcass traits. J Anim Sci. 1998;76:2549–2559. doi: 10.2527/1998.76102549x. [DOI] [PubMed] [Google Scholar]
- Koohmaraie M, Shackelford SD, Wheeler TL, Lonergan SM, Doumit ME. A muscle hypertrophy condition in lamb (callipyge): characterization of effects on muscle growth and meat quality traits. J Anim Sci. 1995;73:3596–3607. doi: 10.2527/1995.73123596x. [DOI] [PubMed] [Google Scholar]
- Cockett NE, Jackson SP, Shay TL, Farnir F, Berghmans S, Snowder GD, Nielsen DM, Georges M. Polar overdominance at the ovine callipyge locus. Science. 1996;273:236–238. doi: 10.1126/science.273.5272.236. [DOI] [PubMed] [Google Scholar]
- Freking BA, Keele JW, Beattie CW, Kappes SM, Smith TP, Sonstegard TS, Nielsen MK, Leymaster KA. Evaluation of the ovine callipyge locus: I. Relative chromosomal position and gene action. J Anim Sci. 1998;76:2062–2071. doi: 10.2527/1998.7682062x. [DOI] [PubMed] [Google Scholar]
- Segers K, Vaiman D, Berghmans S, Shay T, Meyers S, Beever J, Cockett N, Georges M, Charlier C. Construction and characterization of an ovine BAC contig spanning the callipyge locus. Anim Genet. 2000;31:352–359. doi: 10.1046/j.1365-2052.2000.00676.x. [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko-Ishino T, Ishino F. Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells. 2000;5:211–220. doi: 10.1046/j.1365-2443.2000.00320.x. [DOI] [PubMed] [Google Scholar]
- Schmidt JV, Matteson PG, Jones BK, Guan XJ, Tilghman SM. The Dlk1 and Gtl2 genes are linked and reciprocally imprinted. Genes Dev. 2000;14:1997–2002. [PMC free article] [PubMed] [Google Scholar]
- Takada S, Tevendale M, Baker J, Georgiades P, Campbell E, Freeman T, Johnson MH, Paulsen M, Ferguson-Smith AC. Delta-like and Gtl2 are reciprocally expressed, differentially methylated linked imprinted genes on mouse chromosome 12. Curr Biol. 2000;10:1135–1138. doi: 10.1016/S0960-9822(00)00704-1. [DOI] [PubMed] [Google Scholar]
- Wylie AA, Murphy SK, Orton TC, Jirtle RL. Novel imprinted DLK1/GTL2 domain on human chromosome 14 contains motifs that mimic those implicated in IGF2/H19 regulation. Genome Res. 2000;10:1711–1718. doi: 10.1101/gr.161600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laborda J, Sausville E, Hoffman T, Notario V. Dlk, a putative mammalian homeotic gene differentially expressed in small cell lung carcinoma and neuroendocrine tumor cell line. J Biol Chem. 1993;268:3817–3820. [PubMed] [Google Scholar]
- Smas CM, Sul HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-L. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Takemori H, Halder SK, Nonaka Y, Osamu H. Implication of Zog protein (zona glomerulosa-specific protein) in zone development of the adrenal cortex. Endocr Res. 1998;24:515–520. doi: 10.3109/07435809809032640. [DOI] [PubMed] [Google Scholar]
- Helman LJ, Sack N, Plon SE, Israel MA. The sequence of an adrenal specific human cDNA, pG2. Nucleic Acids Res. 1990;18:685. doi: 10.1093/nar/18.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen C, Krogh T, Hojrup P, Clausen P, Skjodt K, Larsson L, Enghild J, Teisner B. Protein structure of fetal antigen 1 (FA1). A novel circulating human epidermal-growth-factor-like protein expressed in neuroendocrine tumors and its relation to the gene products of dlk and pG2. Eur J Biochem. 1994;225:83–92. doi: 10.1111/j.1432-1033.1994.00083.x. [DOI] [PubMed] [Google Scholar]
- Paulsen M, Takada S, Youngson NA, Benchaib M, Charlier C, Segers K, Georges M, Ferguson-Smith AC. Comparative sequence analysis of the imprinted Dlk1-Gtl2 locus in three mammalian species reveals highly conserved genomic elements and refines comparison with the Igf2-H19 region. Genome Res. 2001;11:2085–2094. doi: 10.1101/gr.206901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitz H, Youngson N, Lin S-P, Dalbert S, Paulsen M, Bachellerie J-P, Ferguson-Smith AC, Cavaillé J. Imprinted microRNA genes transcribed antisense to a reciprocally imprinted retrotransposon-like gene. Nat Genet. 2003;34:261–262. doi: 10.1038/ng1171. [DOI] [PubMed] [Google Scholar]
- Freking BA, Murphy SK, Wylie AA, Rhodes SJ, Keele JW, Leymaster KA, Jirtle RL, Smith TPL. Identification of the single base change causing the callipyge muscle hypertrophy phenotype, the only known example of polar overdominance in mammals. Genome Res. 2002;12:1496–1506. doi: 10.1101/gr.571002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit M, Segers K, Carrascosa LG, Shay T, Baraldi F, Gyapay G, Snowder G, Georges M, Cockett N, Charlier C. Mosaicism of Solid Gold supports the causality of a noncoding A-to-G transition in the determinism of the callipyge phenotype. Genetics. 2003;163:453–456. doi: 10.1093/genetics/163.1.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlier C, Segers K, Karim L, Shay T, Gyapay G, Cockett N, Georges M. The callipyge mutation enhances the expression of coregulated imprinted genes in cis without affecting their imprinting status. Nat Genet. 2001;27:367–369. doi: 10.1038/86856. [DOI] [PubMed] [Google Scholar]
- Bidwell CA, Shay TL, Georges M, Beever JE, Berghmans S, Cockett NE. Differential expression of the GTL2 gene within the callipyge region of ovine chromosome 18. Anim Genet. 2001;32:248–256. doi: 10.1046/j.1365-2052.2001.00776.x. [DOI] [PubMed] [Google Scholar]
- Georges M, Charlier C, Cockett N. The callipyge locus: evidence for the trans interaction of reciprocally imprinted genes. Trends Genet. 2003;19:248–252. doi: 10.1016/S0168-9525(03)00082-9. [DOI] [PubMed] [Google Scholar]
- Lin S-P, Youngson N, Takada S, Seitz A, Reik W, Paulsen M, Cavaille J, Ferguson-Smith AC. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 2. Nat Genet. 2003;35:97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- Schuster-Gossler K, Bilinski P, Sado T, Ferguson-Smith A, Gossler A. The mouse Gtl2 gene is differentially expressed during embryonic development, encodes multiple alternatively spliced transcripts, and may act as an RNA. Dev Dyn. 1998;212:214–228. doi: 10.1002/(SICI)1097-0177(199806)212:2<214::AID-AJA6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SIS, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Hall IM, Shankaranarayana GD, Noma K-i, Ayoub N, Cohen A, Grewal SIS. Establishment and maintenance of a heterochromatin domain. Science. 2002;297:2232–2237. doi: 10.1126/science.1076466. [DOI] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SIS, Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004;303:672–676. doi: 10.1126/science.1093686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ. RNA interference. Nature. 2002;481:246–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Cerutti H. RNA interference: traveling in the cell and gaining functions? Trends Genet. 2003;19:39–46. doi: 10.1016/S0168-9525(02)00010-0. [DOI] [PubMed] [Google Scholar]
- Shi Y. Mammalian RNAi for the masses. Trends Genet. 2003;19:9–12. doi: 10.1016/S0168-9525(02)00005-7. [DOI] [PubMed] [Google Scholar]
- Chirgwin JM, Przybyla AE, MacDonald RJ, Rutter WJ. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry. 1979;24:5294–5299. doi: 10.1021/bi00591a005. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual, 3. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- SAS SAS Manual. In Cary, NC: SAS Institute, Inc. 2001.


