Abstract
Background
Ideal management of sexually transmitted infections (STI) may require risk markers for pathology or vaccine development. Previously, we identified common genetic variants associated with chlamydial pelvic inflammatory disease (PID) and reduced fecundity. As this explains only a proportion of the long-term morbidity risk, we utilized whole-exome sequencing to identify biological pathways that may be associated with STI-related infertility.
Methods
We obtained stored DNA from 43 non-Hispanic black women with PID from the PID Evaluation and Clinical Health Study. Infertility was assessed at a mean of 84 months. Principal component analysis revealed no population stratification. Potential covariates did not significantly differ between groups. Sequencing kernel association test (SKAT) was used to examine associations between aggregates of variants on a single gene and infertility. The results from the SKAT test were used to choose “focus genes” (p-value <0.01; n=150) for subsequent Ingenuity Pathway Analysis (IPA) to identify “gene sets” that are enriched in biologically relevant pathways.
Results
Pathway analysis revealed that focus genes were enriched in canonical pathways including, Interleukin-1 (IL-1) signaling, P2Y Purinergic Receptor signaling, and Bone Morphogenic Protein (BMP) signaling.
Conclusions
Focus genes were enriched in pathways that impact innate and adaptive immunity, protein kinase A activity, cellular growth and DNA repair. These may alter host resistance or immunopathology following infection. Targeted sequencing of biological pathways identified in this study may provide insight into STI-related infertility.
Keywords: Chlamydia trachomatis, Genetics, Infertility, Neisseria gonorrhoeae, Pelvic inflammatory disease
Introduction
Pelvic inflammatory disease (PID) is the infection and inflammation of the female upper genital tract and can lead to serious complications including tubal factor infertility and ectopic pregnancy (1). In a landmark Scandinavian cohort study of 2500 women with clinical PID, approximately 16% of women with salpingitis became infertile versus 2.7% of controls (1). Furthermore, tubal factor infertility roughly doubles with each PID episode reaching as high as 40% after three or more episodes (1).
Chlamydia trachomatis and Neisseria gonorrhoeae are well established causes of PID and routine screening for these pathogens has been implemented worldwide with the goal of interrupting progression or reducing transmission to prevent long term reproductive morbidity. However, both microbes remain a significant public health burden. Management of sexually transmitted infections (STI) may be improved by identifying biomarkers that can be used to predict pathology or can be used for vaccine development (2). However, a factor complicating control efforts is the range of clinical outcomes observed following bacterial STIs (2). The reasons for this variability are not clear because the mechanisms underlying the immunopathogenesis of C. trachomatis and N. gonorrhoeae have yet to be elucidated fully.
Studies focused on identifying bacterial, clinical, and environmental factors associated with late complications following C. trachomatis infection have failed to identify prognostic or diagnostic markers associated with reproductive morbidity (3). We have shown that N. gonorrhoeae coinfection increases risk for ascending as well as incident chlamydial infection (4), indicating that coinfection likely contributes to risk for upper genital tract disease. Host genetics also influences disease progression among infected women by modulating immune responses (5). A study of 64 Gambian twin pairs estimated that host genetics accounts for approximately 39% of variation in C. trachomatis outcome (6). We previously reported that single nucleotide polymorphisms in Toll-like receptor (TLR) 1 and 4 genes, innate inflammatory receptors, are associated with C. trachomatis infection, upper genital tract infection with C. trachomatis and/or N. gonorrhoeae and reduced pregnancy rates among African American women with PID (7). This suggests that some women may be predisposed to disproportionate and potentially damaging inflammatory responses following innate immune recognition of STIs.
Variants identified via candidate gene studies explain a small fraction of individual variation for long-term morbidity risk and focus only on known pathophysiological pathways. High throughput technologies such as next generation sequencing (8) support broader, unbiased detection of genetic variation. Compared to whole-genome sequencing, whole-exome sequencing (WES) is a more cost effective approach to increase our understanding of many complex diseases (9). WES may be particularly useful to reveal novel and rare coding variants across the genome to support inferences regarding biologically relevant pathways. WES has yet to be applied to the identification of host genetic factors that influence progression of upper genital tract infection to infertility. However, the ability of WES to identify a single variant association with disease is limited by the need for large samples sizes and the challenge of finding adequately characterized patient samples (10). The objective of this discovery phase study was to utilize novel methods such as variant pooling tests and pathway analyses to obtain useful biological information from WES. Pathway based tests are more powerful than single-variant association tests and may be more appropriate given that a complex disease such as PID is unlikely to have a single causal variant but could be modulated by multiple causal genes within a biological pathway (10). We anticipate that these analyses will support development of mechanistic hypotheses related to the pathophysiology of infertility following PID in women with C. trachomatis infection with or without N. gonorrhoeae coinfection. We expect that biologically relevant pathways identified by this study will be the basis of a larger follow-up study using targeted sequencing to improve understanding of STI-driven infertility.
Materials and Methods
Patient population
Study participants were selected from a large cohort of individuals who participated in the PID Evaluation and Clinical Health (PEACH) study, a randomized clinical trial that compared inpatient and outpatient treatment for preventing long-term complications among 831 women with clinically suspected PID (11). Briefly, between March 1996 and February 1999 women aged 14–37 years were recruited from 7 primary and 6 secondary sites throughout the United States. Eligible women had a history of pelvic discomfort for less than 30 days, findings of pelvic organ tenderness (uterine or adnexal) on bimanual examination, and leukorrhea and/or mucopurulent cervicitis and/or untreated but documented gonococcal or chlamydial cervicitis. Women with suspected PID, and who provided informed consent, were eligible to participate in the treatment trial, approved by the University of Pittsburgh Institutional Review Board.
The PEACH study collected blood from 692 of the 831 women enrolled for measurement of erythrocyte sedimentation rate and any residual sample was frozen and archived for subsequent studies. Our previous genetic study focused on 290 PEACH participants with DNA available for analysis (7). Among these women, 70% were African American, 18% were Caucasian, 8% were Hispanic and 4% were of other races, which reflected the distribution of the overall study. A total of 94 of 205 non-Hispanic African American women had C. trachomatis. This investigation included 43 of these women who had stored DNA remaining [15 of 43 (35%) were coinfected with N. gonorrhoeae]. This group was selected because narrowing our focus to a single racial/ethnic group reduced population stratification which can lead to erroneous results. In addition, African American women have the highest risk for chlamydia and PID. The Texas A&M University Institutional Review Board approved this study.
Data collection
Women enrolled in PEACH were followed for a median of 84 months and extensive data on sexually transmitted infections and reproductive morbidities were obtained. An extensive interview, administered at enrollment, collected demographic and clinical information regarding reason for visit, pain history, history of PID/sexually transmitted infections, sexual and contraceptive practices, reproductive decisions, douching, pregnancies, medical and gynecological history and lifestyle habits. A gynecological examination was also performed. Vaginal smears were gram stained for diagnosis of bacterial vaginosis by Nugent criteria. Endometrial biopsy specimens were obtained for histology. Endometritis was based on a modification of the criteria proposed by Kiviat et al (12) and defined as the presence of at least five neutrophils in the endometrial surface epithelium in the absence of menstrual endometrium and/or at least two plasma cells in the endometrial stroma. Cervical and endometrial swabs were also stored and later used to test for M. genitalium.
Reproductive outcomes monitored during follow-up for the PEACH study included pregnancy, infertility, live birth, recurrent PID, and chronic pelvic pain. In PEACH, infertility was assessed among sexually active women reporting no birth control or methods considered being unreliable, including withdrawal, rhythm method, vasectomy, or using the following methods rarely or occasionally – diaphragm, condoms, spermicidal foam/cream/jelly/suppositories, or cervical cap. Women were determined to be infertile if they did not conceive (positive urine or blood test, or doctor’s diagnosis of pregnancy) during follow-up (≥ 12 months).
Whole-exome sequencing and bioinformatics
An estimated 2 μg of stored DNA was shipped to Genomics and Bioinformatics Services, Texas A&M AgriLife for WES. Libraries were constructed using TruSeq Exome Enrichment kits (Illumina) and sequenced on the Illumina HiSeq 2500 platform as paired end 100bp reads. Quality control for the raw reads was archived using Fastq-mcf Toolkit (REF) to filter, trim, and correct reads. Reads were aligned by comparison with the standard reference genome GRch38. PCR duplications were removed and reads were realigned around potential indel regions using standard GATK version 2.6.5 procedures (13). Variants were filtered for quality control if there was strand bias (Fisher score >60), low coverage (<5 reads), low quality score (<50) or read depth <8×.
Statistical analyses
Logistic regression with Firth’s penalized likelihood approach was used to examine associations between baseline variables and infertility to identify potential covariates. No significant associations between potential covariates and infertility were detected so unadjusted models are presented. Principal component analysis was conducted but did not identify any significant population stratification (PLINK V 9).
Sequencing Kernel Association Test
To examine associations between the aggregate effects of variants on a single gene and infertility, we used the sequencing kernel association test (SKAT) (9). This is a variance-component test within a random effects model that evaluates the distribution of genetic effects of multiple variants on a single gene (i.e. examining cumulative variant gene-disease associations) and can account for bi-directionality of variant effect sizes within a gene (9). Thus, SKAT calculates p-values where the outcome is infertility and the genetic independent variable is not a single variant but rather an aggregate of variants from a single gene. We ranked genes according to p-value and selected a set of “focus genes” with p-values <0.01 (n=150 genes) for subsequent pathway analysis. All analyses were conducted with R V3.2.2.
Ingenuity Pathway Analysis (IPA)
We used Ingenuity Pathway Analysis (IPA) (Redwood City, CA, USA) (14), to understand biological relationships, if present, between the 150 focus genes identified via WES. Pathway analyses have greater power to identify biologically relevant genes compared to single variant or gene-disease association tests (10). Canonical biochemical or signaling pathways are based on the data accumulated in the IPA Knowledge Base (KB). Significant associations between a gene set and a canonical pathway are determined using the ratio of the number of focus genes mapping to a canonical pathway divided by the total number of genes from the IPA KB that map to that pathway. IPA ranks focus genes by significance where the p-value (Fisher’s exact test) determines the probability that a set of focus genes (gene sets) is enriched for a specific canonical pathway more than would be expected by chance alone.
Results
Among the subset of PEACH participants assessed via WES (n=43), a total of 13 women were classified as infertile at follow-up, while the remainder (n=30) were fertile. Compared to fertile women, infertile women were more likely to be >35 years of age (23.1% vs. 10.0%), have 12 + years of education (38.6% vs. 16.7%), report a previous history of chlamydia (50.0% vs. 33.3%), and to have N. gonorrhoeae co-infection (66.7% vs. 37.5%), although none of these differences reached statistical significance. History of PID, history of gonorrhea, histologic endometritis, M. genitalium co-infection, BV, and smoking status did not significantly differ between groups (Table 1).
Table 1.
Comparison of baseline characteristics between fertile and infertile women
| Fertile (n=30) | Infertile (n=13) | Odds ratio 95% confidence interval |
|
|---|---|---|---|
| Age | |||
| <25 | 12(40.0) | 6(46.2) | Reference |
| 25–34 | 15(50.0) | 4(30.8) | 0.5 (0.1–2.3) |
| 35+ | 3(10.0) | 3(23.1) | 1.9 (0.3–12.5) |
|
| |||
| Education | |||
| < 12 years | 18(60.0) | 5(38.5) | 0.3 (0.06–1.4) |
| 12 years | 7(23.3) | 3(23.1) | 0.5 (0.07–2.7) |
| 12 + years | 5(16.7) | 5(38.6) | Reference |
|
| |||
| History of PID | |||
| No | 24(80.0) | 11(84.6) | Reference |
| Yes | 11(20.0) | 2(15.8) | 0.8(0.1–4.4) |
|
| |||
| History of chlamydia | |||
| No | 20(66.7) | 6(50.0) | Reference |
| Yes | 10(33.3) | 6(50.0) | 1.9 (0.5–7.5) |
|
| |||
| History of gonorrhea | |||
| No | 21(70.0) | 9(30.0) | Reference |
| Yes | 11(84.6) | 2(15.4) | 0.9 (0.2–3.9) |
|
| |||
| Smoker | |||
| No | 16(53.3) | 8(61.5) | Reference |
| Yes | 14(46.7) | 5(38.5) | 0.7 (0.2–2.7) |
|
| |||
| Drug use | |||
| No | 17(56.7) | 6(46.2) | Reference |
| Yes | 13(43.3) | 7(53.9) | 1.5 (0.4–5.4) |
|
| |||
| Histologic Endometritis | |||
| No | 7(25.9) | 3(33.3) | Reference |
| Yes | 20(74.1) | 6(66.7) | 0.7 (0.1–3.4) |
|
| |||
| Gram stain results | |||
| Normal | 5(16.7) | 4(36.4) | Reference |
| Intermediate | 6(20.0) | 2(18.2) | 0.5 (0.1–3.5) |
| BV | 19(63.3) | 5(45.5) | 0.3 (0.1–1.8) |
|
| |||
| N. gonorrhoeae co-infection | |||
| No | 15(62.5) | 3(33.3) | Reference |
| Yes | 9(37.5) | 6(66.7) | 3.0 (0.6–14.6) |
|
| |||
| M. genitalium co-infection | |||
| No | 18(85.7) | 8(14.3) | Reference |
| Yes | 3(72.7) | 3(27.3) | 2.2 (0.4–13.2) |
Sequencing Kernel Association Test
We identified a total of 324,913 single nucleotide polymorphisms (SNPs) by WES. After removing missing data and unspecified gene locations, a total of 85,315 SNPs linked to 17,873 genes were included in the SKAT test. Variants associated with infertility were found for 668 genes (p<0.05), with 150 focus genes being identified with p-values <0.01. Table 2 shows a select subset of these genes with p-values <0.0005 (determined by SKAT: BMP3, POLR2J3, C1RL, NME4, POFUT2, TMEM70, and ASS1). These genes have functions related to cellular growth (BMP3); RNA polymerase activity (POLRJ2); complement activation, immune system regulation and response (C1RL), ATP binding or transport (NME4, TREM70); metabolic responses (POFUT2); and cellular response to IFN-γ (ASS1). However, none of the focus genes would remain significant at the Bonferroni threshold of 2.8E-06.
Table 2.
Select focus genes with ap-values <0.0005 from the sequencing kernel association test
| GENE | Chromosome | bMolecular Function | bBiological Function |
|---|---|---|---|
| BMP3 Bone morphogenetic protein 3 |
4 | BMP receptor binding; cytokine activity; growth factor activity; transforming growth factor beta receptor binding | cell-cell signaling; cell development; cell differentiation; growth |
| POLR2J3 Polymerase (RNA) II subunit J |
7 | DNA binding; protein dimerization activity; RNA polymerase II activity | transcription from RNA polymerase II promoter |
| C1RL Complement C1r subcomponent like |
12 | hydrolase activity; peptidase activity; serine-type endopeptidase activity; serine-type peptidase activity | complement activation, immune system process; innate immune response; proteolysis |
| NME4 Nucleoside diphosphate kinase 4 |
16 | ATP binding; calcium ion binding; kinase activity; lipid binding; metal ion binding; nucleoside; protein binding | CTP biosynthetic process; GTP biosynthetic process; lipid transport; nucleobase-containing small molecule |
| POFUT2 Protein O-fucosyltransferase 2 |
21 | fucosyltransferase activity; peptide-O-fucosyltransferase activity; transferase activity; transferase activity, transferring glycosyl groups | carbohydrate metabolic process; cellular protein metabolic process; fucose metabolic process; fucosylation; |
| TMEM70 Transmembrane protein 70 |
8 | Unknown | mitochondrial proton-transporting ATP synthase complex assembly |
| ASS1 Argininosuccinate synthase 1 |
9 | amino acid binding; argininosuccinate synthase activity; ATP binding; | acute-phase response; aging; cellular response to interferon-gamma; |
P-values determined by SKAT test and based on associations between aggregates of variants on a single gene and infertility
Function determined by IPA analysis
Genes would not be significant after correction for multiple comparisons (Bonferroni p-value =2.8E-06).
IPA canonical pathways
Figures 1–3 show the top three canonical pathways enriched with respect to focus genes. These canonical pathways included IL-1 signaling pathways (Figure 1), P2Y purinergic receptor (Figure 2) and BMP signaling (Figure 3). Genes enriched in these canonical pathways have several relevant biological functions including innate immune signaling, G-protein coupled receptor signaling, protein kinase A activity, platelet aggregation, and cell growth and differentiation.
Figure 1.
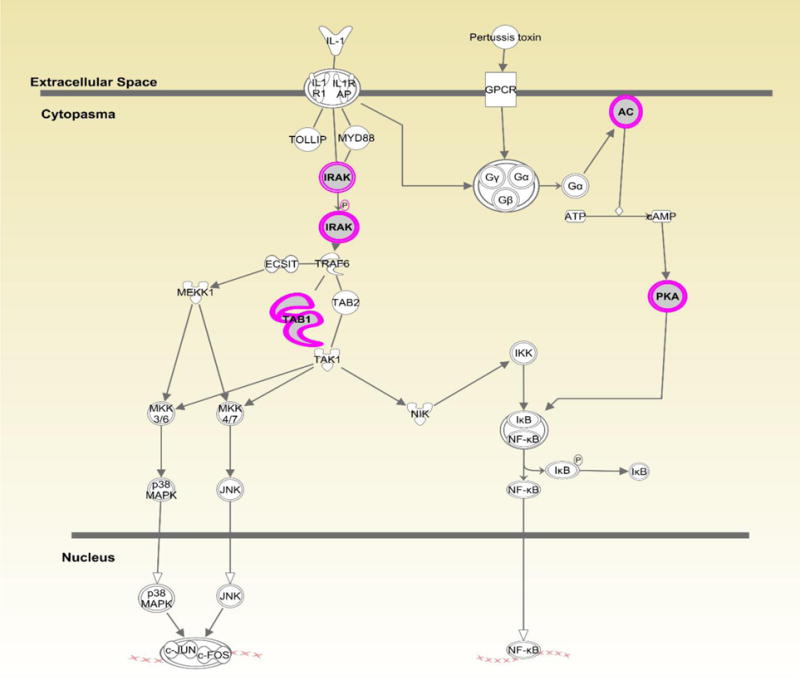
Displays canonical IL-1 signaling pathway identified through Ingenuity Pathway Analysis (IPA). Genes highlighted in pink represent focus genes. ADCYL [(AC) adenylate cyclase)] is involved in biological processes including activation of protein kinase A activity and G-protein coupled receptor signaling. PRKACB [(PKA) protein kinase cAMP-activated catalytic subunit beta)] is involved in activation of protein kinase A and G-protein coupled receptor signaling. TAB1(TGF-beta activated kinase 1/MAP3K7 binding protein 1) is involved in regulation of MAPK and NFκB activation, signaling through MyD88, and TGFβ signaling. IRAK (interleukin 1 receptor associated kinase 1) is involved in TLR signaling via MyD88, activation of protein kinase A, and G-protein coupled receptor signaling.
Figure 3.
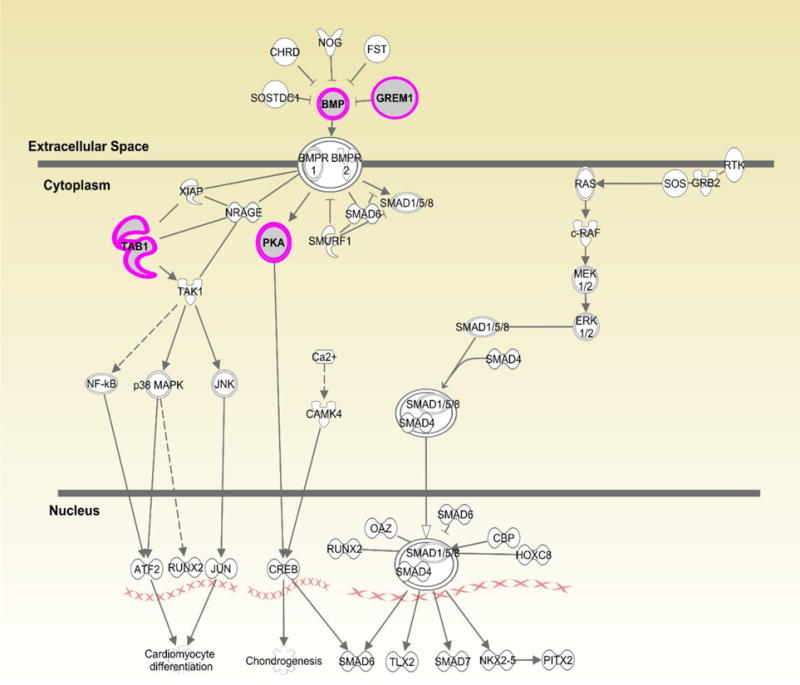
Displays canonical BMP pathway identified through Ingenuity Pathway Analysis (IPA). Genes highlighted in pink represent focus genes. BMP3 (Bone morphogenetic protein 3) is involved in biological processes including cell differentiation and growth, and regulation of apoptosis. GREM1(gremlin 1, DAN family BMP antagonist) is an antagonist of BMP and is involved in angiogenesis, apoptosis, and regulation of NFκB activation. PRKACB [(PKA) protein kinase cAMP-activated catalytic subunit beta)] is involved in G-protein coupled receptor signaling and activation of protein kinase A activity. TAB1(TGF-beta activated kinase 1/MAP3K7 binding protein 1) is involved in regulation of TGFβ signaling, signaling through MyD88, and MAPK and NFκB activation.
Figure 2.
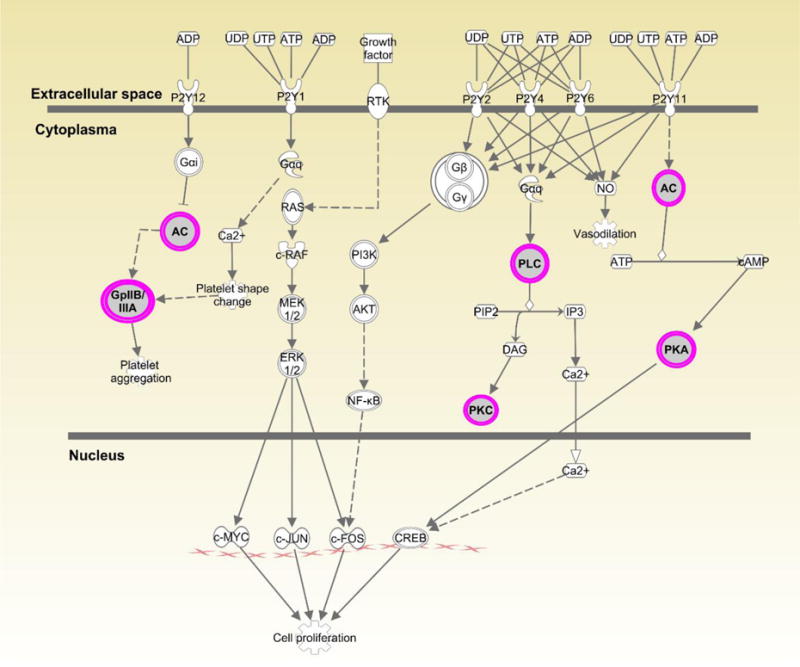
Displays canonical P2Y Purinergic Receptor Signaling pathway identified through Ingenuity Pathway Analysis (IPA). Genes highlighted in pink represent focus genes. ADCYL [(AC) adenylate cyclase)] is involved in biological processes including activation of protein kinase A activity and G-protein coupled receptor signaling. ITGA2B [(GpIIB/IIA) integrin subunit alpha 2b)] is involved in cell adhesion, regulation of leukocyte migration and platelet aggregation. PLCD [(PLC) phospholipase C delta 1)] is involved in angiogenesis and is a regulator of cell proliferation. PRKACB [(PKA) protein kinase cAMP-activated catalytic subunit beta)] is involved in activation of protein kinase A activity and G-protein coupled receptor signaling. PRKCH [(PKC) protein kinase C eta)] is involved in cell differentiation and proliferation, apoptosis, and NFκB activity.
Discussion
We used WES to examine the aggregated effects of multiple variants on a single gene to identify potential associations with infertility following PID. Focus genes displayed in Table 2 represented associations with infertility that had the lowest corresponding p-values. These genes impact BMP signaling that orchestrates tissue architecture throughout the body, immune function and complement (C1RL, ASS1), metabolism (POFUT2), RNA polymerase (POLR2J3) and ATP binding or synthesis (NME4, TMEM70). The relationship between some of these biological functions and STIs is described below. It is less clear how POFUT2 or POLR2J3 function would impact infertility. POFUT2 is linked to cancer (15) and there is a growing appreciation that changes in immune cell function, particularly T cell proliferation and T cell effector signaling pathways are dependent on metabolic state (16). There is no information on POLR2J3 in the literature. However, none of these focus genes have been directly linked to STIs, and associations were not significant after correction for multiple comparisons.
The top 150 focus genes after SKAT testing were included in Ingenuity Pathway Analyses to identify biologically relevant canonical pathways common among the focus genes. Several focus genes (N=4) were enriched in the canonical IL1 signaling pathway (Fig. 1). IL-1 is involved in tissue destruction in human Fallopian tubes following C. trachomatis infection (17). Specifically, IL1-receptor antagonist (IL-1RA) eliminates tissue damage induced by C. trachomatis in human Fallopian tube organ culture. The mouse model of chlamydial genital tract disease reveals IL-1 is a key mediator of oviduct damage. Thus, variants in this pathway may exacerbate tissue damage following C. trachomatis infection (18). One focus gene in this pathway, IRAK2, is involved in TLR function and is upregulated following C. trachomatis infection (19). Indeed, we have previously linked the TLR1 gene and TLR4 gene to C. trachomatis and upper genital tract pathology (7). Other focus genes in this pathway have not been directly linked to STIs. However, TAB1 dependent activation of MAPK is involved in T-cell senescence which could alter proliferation and adaptive immunity (20). Our results are consistent with the hypothesis that altered function in immune pathways may be involved in C. trachomatis pathogenesis.
Some focus genes within the IL-1 signaling pathway (ADCYL and PRKACB) were also enriched in the P2Y Purinergic Receptor Signaling pathway (Fig. 2). However, there is little information on the possible role of purinergic receptor signaling in STIs. The purinergic receptor P2X7R has been shown to inhibit C. trachomatis infection in epithelial cells via ATP-mediation (21). Purinergic receptors have been implicated in immune cell trafficking (22) and in the development of fibrosis through the binding of ATP and ADP (23). These receptors may also contribute to T cell activation (24).
Lastly, focus genes were enriched in the BMP canonical pathway (Fig. 3) that impacts TGF-β signaling, cell growth and apoptosis. Gene variants in this pathway could alter the immune response to N. gonorrhoeae as well as C. trachomatis. Mouse models have shown that N. gonorrhoeae induces TGF-β in order to suppress the host immune response and blockade of TGF-β improves host defense against this pathogen (25). Genes that regulate apoptosis can be manipulated by pathogens to increase the probability of replication and survival. Both N. gonorrhoeae and C. trachomatis have been shown to modulate apoptosis pathways to avoid host defense mechanisms (26, 27). BMP genes and antagonist GREM1 are involved in inflammation and tissue damage (28). Although there is no direct link to bacterial STIs, BMP3 is involved in Hepatitis C virus-induced cirrhosis (29). Thus, it is possible that variants involved in this pathway that affect cellular repair or growth may lead to tissue damage following persistent infection.
Our study utilized data with long term follow-up that enabled us to examine infertility following clinically suspected PID. We did not have access to laparoscopic findings and relied on self-report to determine infertility. Thus, misclassification is possible. However, PEACH is one of only a few studies that have assessed reproductive morbidity following clinical PID. There is no widely accepted approach for the analysis of WES data or for the selection of candidate genes in the discovery phase (9). The use of the SKAT test allowed us to capture regions that may have a high proportion of causal or non-causal variants with bi-directionality. However, given our sample size this test was not powerful enough to detect significant associations after correction for multiple comparisons. Methods of correction for multiple comparisons for sequencing studies are not established and existing methods including Bonferroni and FDR are suggested to be suboptimal (30). Thus, relying on strict multiple comparison correction in exome sequencing studies may lead to ruling out potentially important associations. Overall, pathway analyses are a more powerful approach to identify biologically relevant pathways that may be related to disease (10). The limitation of these analyses is that correction for control of type I error is still not well developed (10).
This was the first study to utilize whole-exome sequencing in order to identify novel biological pathways that may be associated with STI-related infertility. The IL-1 signaling pathway is of interest as variations in this pathway may enhance disease following infection. Pathways that impact tissue repair and growth (BMP pathway) may be involved in the formation of scarring or enhanced cellular damage following infection. Our study identified biologically relevant pathways that may serve as targets for a larger study utilizing targeted sequencing. Other avenues of research should include examining some of the identified novel pathways of interest (i.e. BMP signaling and Purinergic Receptor) in animal models. As STI’s remain a major public health burden, the use of genomics in STI research may enhance our understanding of pathogenesis. This could lead to the identification of biomarkers for prediction of tissue damage and perhaps contribute towards vaccine development. Biomarkers of upper genital tract infection might also be useful for monitoring efficacy in vaccine trials.
Acknowledgments
We would like to thank Dr. Charles Johnson and the staff at Genomics & Bioinformatics, Texas A&M AgriLife for the sequencing support.
Funding source: This work was supported by the American Sexually Transmitted Disease Association Developmental Grant for B.T., the National Institute for Allergy and Infectious Diseases [AI084024] for T.D, and [U19AI113170] for X.Z., and the Agency for Healthcare Research and Quality grant [HS08358-05] for R.N.
Footnotes
Conflict of interest: The authors have no conflict of interest to disclose.
References
- 1.Westrom L, Joesoef R, Reynolds G, Hagdu A, Thompson SE. Pelvic inflammatory disease and fertility. A cohort study of 1,844 women with laparoscopically verified disease and 657 control women with normal laparoscopic results. Sexually transmitted diseases. 1992;19(4):185–92. [PubMed] [Google Scholar]
- 2.Brunham RC, Rappuoli R. Chlamydia trachomatis control requires a vaccine. Vaccine. 2013;31(15):1892–7. doi: 10.1016/j.vaccine.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.den Hartog JE, Morre SA, Land JA. Chlamydia trachomatis-associated tubal factor subfertility: Immunogenetic aspects and serological screening. Human reproduction update. 2006;12(6):719–30. doi: 10.1093/humupd/dml030. [DOI] [PubMed] [Google Scholar]
- 4.Russell AN, Zheng X, O’Connell CM, et al. Analysis of Factors Driving Incident and Ascending Infection and the Role of Serum Antibody in Chlamydia trachomatis Genital Tract Infection. J Infect Dis. 2016;213(4):523–31. doi: 10.1093/infdis/jiv438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stamm WE. Chlamydia trachomatis infections of the adult. In: Holmes KK, Sparling FP, Stamm WE, editors. Sexually Transmitted Diseases. 4. New York: McGraw Hill; 2008. pp. 575–93. [Google Scholar]
- 6.Bailey RL, Natividad-Sancho A, Fowler A, et al. Host genetic contribution to the cellular immune response to Chlamydia trachomatis: Heritability estimate from a Gambian twin study. Drugs Today (Barc) 2009;45(Suppl B):45–50. [PubMed] [Google Scholar]
- 7.Taylor BD, Darville T, Ferrell RE, Kammerer CM, Ness RB, Haggerty CL. Variants in toll-like receptor 1 and 4 genes are associated with Chlamydia trachomatis among women with pelvic inflammatory disease. The Journal of infectious diseases. 2012;205(4):603–9. doi: 10.1093/infdis/jir822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pabinger S, Dander A, Fischer M, et al. A survey of tools for variant analysis of next-generation genome sequencing data. Briefings in bioinformatics. 2014;15(2):256–78. doi: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Abecasis GR, Boehnke M, Lin X. Rare-variant association analysis: study designs and statistical tests. Am J Hum Genet. 2014;95(1):5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu G, Zhi D. Pathway-based approaches for sequencing-based genome-wide association studies. Genet Epidemiol. 2013;37(5):478–94. doi: 10.1002/gepi.21728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ness RB, Soper DE, Holley RL, et al. Effectiveness of inpatient and outpatient treatment strategies for women with pelvic inflammatory disease: results from the Pelvic Inflammatory Disease Evaluation and Clinical Health (PEACH) Randomized Trial. Am J Obstet Gynecol. 2002;186(5):929–37. doi: 10.1067/mob.2002.121625. [DOI] [PubMed] [Google Scholar]
- 12.Kiviat NB, Wolner-Hanssen P, Eschenbach DA, et al. Endometrial histopathology in patients with culture-proved upper genital tract infection and laparoscopically diagnosed acute salpingitis. Am J Surg Pathol. 1990;14(2):167–75. doi: 10.1097/00000478-199002000-00008. [DOI] [PubMed] [Google Scholar]
- 13.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngwa JS, Manning AK, Grimsby JL, Lu C, Zhuang WV, Destefano AL. Pathway analysis following association study. BMC Proc. 2011;5(Suppl 9):S18. doi: 10.1186/1753-6561-5-S9-S18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong S, Nutt CL, Betensky RA, et al. Histology-based expression profiling yields novel prognostic markers in human glioblastoma. J Neuropathol Exp Neurol. 2005;64(11):948–55. doi: 10.1097/01.jnen.0000186940.14779.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pearce EL, Poffenberger MC, Chang CH, Jones RG. Fueling immunity: insights into metabolism and lymphocyte function. Science. 2013;342(6155):1242454. doi: 10.1126/science.1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hvid M, Baczynska A, Deleuran B, et al. Interleukin-1 is the initiator of Fallopian tube destruction during Chlamydia trachomatis infection. Cell Microbiol. 2007;9(12):2795–803. doi: 10.1111/j.1462-5822.2007.00996.x. [DOI] [PubMed] [Google Scholar]
- 18.Prantner D, Darville T, Sikes JD, et al. Critical role for interleukin-1beta (IL-1beta) during Chlamydia muridarum genital infection and bacterial replication-independent secretion of IL-1beta in mouse macrophages. Infect Immun. 2009;77(12):5334–46. doi: 10.1128/IAI.00883-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agrawal T, Bhengraj AR, Vats V, Salhan S, Mittal A. Expression of TLR 2, TLR 4 and iNOS in cervical monocytes of Chlamydia trachomatis-infected women and their role in host immune response. Am J Reprod Immunol. 2011;66(6):534–43. doi: 10.1111/j.1600-0897.2011.01064.x. [DOI] [PubMed] [Google Scholar]
- 20.Lanna A, Henson SM, Escors D, Akbar AN. The kinase p38 activated by the metabolic regulator AMPK and scaffold TAB1 drives the senescence of human T cells. Nat Immunol. 2014;15(10):965–72. doi: 10.1038/ni.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darville T, Welter-Stahl L, Cruz C, Sater AA, Andrews CW, Jr, Ojcius DM. Effect of the purinergic receptor P2X7 on Chlamydia infection in cervical epithelial cells and vaginally infected mice. J Immunol. 2007;179(6):3707–14. doi: 10.4049/jimmunol.179.6.3707. [DOI] [PubMed] [Google Scholar]
- 22.Ferrari D, McNamee EN, Idzko M, Gambari R, Eltzschig HK. Purinergic Signaling During Immune Cell Trafficking. Trends Immunol. 2016;37(6):399–411. doi: 10.1016/j.it.2016.04.004. [DOI] [PubMed] [Google Scholar]
- 23.Lu D, Insel PA. Cellular mechanisms of tissue fibrosis. 6. Purinergic signaling and response in fibroblasts and tissue fibrosis. Am J Physiol Cell Physiol. 2014;306(9):C779–88. doi: 10.1152/ajpcell.00381.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsukimoto M, Tokunaga A, Harada H, Kojima S. Blockade of murine T cell activation by antagonists of P2Y6 and P2X7 receptors. Biochem Biophys Res Commun. 2009;384(4):512–8. doi: 10.1016/j.bbrc.2009.05.011. [DOI] [PubMed] [Google Scholar]
- 25.Liu Y, Russell MW. Diversion of the immune response to Neisseria gonorrhoeae from Th17 to Th1/Th2 by treatment with anti-transforming growth factor beta antibody generates immunological memory and protective immunity. MBio. 2011;2(3):e00095–11. doi: 10.1128/mBio.00095-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nudel K, Massari P, Genco CA. Neisseria gonorrhoeae Modulates Cell Death in Human Endocervical Epithelial Cells through Export of Exosome-Associated cIAP2. Infect Immun. 2015;83(9):3410–7. doi: 10.1128/IAI.00732-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kontchou CW, Tzivelekidis T, Gentle IE, Hacker G. Infection of epithelial cells with Chlamydia trachomatis inhibits TNF-induced apoptosis at the level of receptor internalisation while leaving non-apoptotic TNF-signalling intact. Cell Microbiol. 2016 doi: 10.1111/cmi.12598. [DOI] [PubMed] [Google Scholar]
- 28.Chang K, Weiss D, Suo J, et al. Bone morphogenic protein antagonists are coexpressed with bone morphogenic protein 4 in endothelial cells exposed to unstable flow in vitro in mouse aortas and in human coronary arteries: role of bone morphogenic protein antagonists in inflammation and atherosclerosis. Circulation. 2007;116(11):1258–66. doi: 10.1161/CIRCULATIONAHA.106.683227. [DOI] [PubMed] [Google Scholar]
- 29.Huang XX, McCaughan GW, Shackel NA, Gorrell MD. Up-regulation of proproliferative genes and the ligand/receptor pair placental growth factor and vascular endothelial growth factor receptor 1 in hepatitis C cirrhosis. Liver Int. 2007;27(7):960–8. doi: 10.1111/j.1478-3231.2007.01542.x. [DOI] [PubMed] [Google Scholar]
- 30.Jeng XJ, Daye ZJ, Lu W, Tzeng JY. Rare Variants Association Analysis in Large-Scale Sequencing Studies at the Single Locus Level. PLoS Comput Biol. 2016;12(6):e1004993. doi: 10.1371/journal.pcbi.1004993. [DOI] [PMC free article] [PubMed] [Google Scholar]


