Summary
Phylogenetic analysis reveals AMPA, kainate and NMDA receptor families in insect genomes, suggesting conserved functional properties corresponding to their vertebrate counterparts. However, heterologous expression of the Drosophila kainate receptor DKaiR1D and the AMPA receptor DGluR1A revealed novel ligand selectivity at odds with the classification used for vertebrate glutamate receptor ion channels (iGluRs). DKaiR1D forms a rapidly activating and desensitizing receptor that is inhibited by both NMDA and the NMDA receptor antagonist AP5; crystallization of the KaiR1D ligand-binding domain reveals that these ligands stabilize open cleft conformations explaining their action as antagonists. Surprisingly, the AMPA receptor DGluR1A shows weak activation by its namesake agonist AMPA and also by quisqualate. Crystallization of the DGluR1A ligand-binding domain reveals amino acid exchanges that interfere with binding of these ligands. The unexpected ligand binding profiles of insect iGluRs allows classical tools to be used in novel approaches for the study of synaptic regulation.
Keywords: Drosophila, Glutamate Receptors, NMDA, AP5, Crystal Structures
Introduction
Glutamate is the major excitatory neurotransmitter in the vertebrate central nervous system; its actions are mediated largely via three classes of ionotropic glutamate receptors (iGluRs) named AMPA, kainate and NMDA receptors (Traynelis et al., 2010). The classification of iGluRs into AMPA, kainate and NMDA receptors was based on the efforts of medicinal chemists who identified subtype selective heterocyclic amino acids such as AMPA, kainate and quisqualate (Krogsgaard-Larsen et al., 1980; Watkins and Evans, 1981), and amino acid analogs such as NMDA (Curtis and Watkins, 1960) and 2(R)-amino-5-phosphonopentanoic acid (D-AP5) that act as agonists and antagonists (Evans et al., 1982). This work was so successful that the selective action of NMDA and D-AP5 formed the corner stone on which the role of NMDA receptors in synaptic plasticity was established (Collingridge and Bliss, 1995).
Subsequent cloning of insect iGluRs, which revealed sequence similarity with their vertebrate AMPA, kainate and NMDA receptor counterparts, suggests that the same series of ligands can be used to investigate their role in CNS function. However, with the exception of the neuromuscular junction (NMJ) of larval Drosophila (Jan and Jan, 1976) and the NMJ of adult locusts (Cull-Candy, 1976), the small size and inaccessibility of insect neurons has to date challenged characterization of the functional properties of native insect iGluRs. Sequence analysis of the Drosophila genome identified fourteen iGluR genes that resemble vertebrate AMPA, kainate and NMDA receptors (Littleton and Ganetzky, 2000). Transcript profiling revealed that nine of these iGluRs are expressed in the brain, with five expressed at the neuromuscular junction (Graveley et al., 2011). Very little is known about the structure and functional properties of Drosophila iGluRs, and only recently was a functional reconstitution achieved for recombinant Drosophila NMJ iGluRs (Han et al., 2015). As a result, iGluRs are understudied in model organisms like Drosophila for which powerful genetic techniques have otherwise yielded numerous insights into the molecular neurobiology of synapse development and function (Charng et al., 2014; Thomas and Sigrist, 2012; Yamamoto et al., 2014).
Four presumptive Drosophila kainate receptors (Clumsy, DKaiR1C, DKaiR1D, and CG11155) are functionally required for spectral preference behavior and are thought to mediate excitatory synaptic transmission from the second-order neuron Dm8 to the third-order neuron Tm5c (Karuppudurai et al., 2014). The eye-enriched kainate receptor (EKAR) is expressed in photoreceptors, receiving feedback glutamatergic signals from amacrine cells (Hu et al., 2015), but so far, in vitro reconstitution has not been achieved for any of these presumptive kainate receptors. Instead, functional analysis of their role in CNS glutamatergic circuits relies solely on chronic inactivation using genetic mutants and RNAi-mediated knockdown (Hu et al., 2015). In this study, we combined electrophysiological, biochemical and crystallographic analyses to determine receptor activity and ligand specificity of a Drosophila kainate receptor DKaiR1D and a Drosophila AMPA receptor DGluR1A. We found that DKaiR1D forms functional homomeric channels in HEK cells and oocytes with pharmacological properties distinct from vertebrate and Drosophila muscle iGluRs. Crystal structures of DKaiR1D ligand-binding dimer complexes with glutamate, NMDA and AP5, revealed that only glutamate triggers domain closure, and that NMDA and AP5 are antagonists. DGluR1A receptors respond weakly to AMPA and quisqualate; the crystal structure of DGluR1A revealed that the binding of these ligands is hindered by steric occlusion. Thus, despite structural and sequence similarity between insect and vertebrate iGluRs, insect iGluRs do not conform to the pharmacology-based classification of vertebrate iGluRs. However, the agonist/antagonist binding properties of insect iGluRs we report here provide a new approach for acute inactivation/activation in vivo and for dissecting their functions in complex neural circuits.
RESULTS
Phylogenetic Analysis of Insect iGluR Families
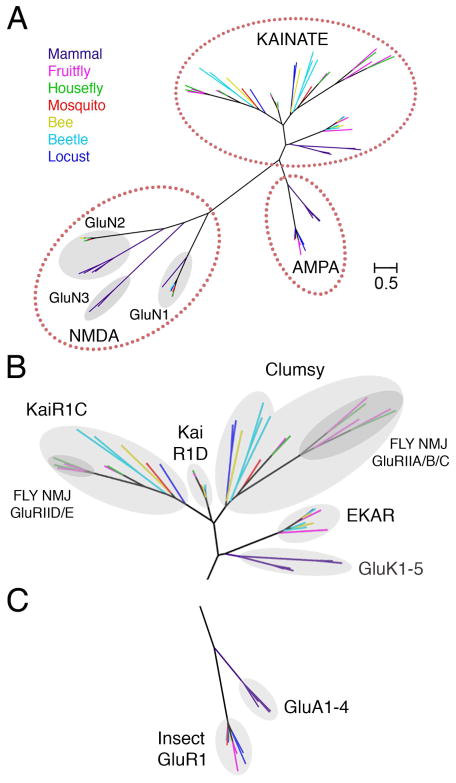
A maximum likelihood phylogenetic analysis of 64 iGluR sequences culled from Fruit fly (Drosophila melanogaster), Housefly (Musca domestica), Mosquito (Anopheles gambiae), Bee (Apis mellifera), Locust (Locusta migratoria) and Beetle (Tribolium castaneum) genomes revealed branching into three major families, together with their vertebrate AMPA, kainate and NMDA receptor counterparts (Figure 1A and Figure S1). Notably, the kainate receptor branch has undergone a large expansion in insects (Figure 1B and Figure S1) with separate branches for 4 families named KaiR1C, KaiR1D, Clumsy and EKAR, with the fly NMJ GluRIIA, GluRIIB and GluRIIC subunits forming a branch within the clumsy clade, and the GluRIID and GluRIIE subunits forming a branch within the KaiR1C clade. By contrast, AMPA (Figure 1C and Figure S1) and NMDA receptor GluN1 and GluN2 families are largely conserved in insects and vertebrates (Figure 1A and Figure S1).
Figure 1. Phylogenetic Analysis of Insect iGluRs.
(A) Maximum likelihood topology tree for diverse insect and mammalian iGluRs reveals conserved clades for AMPA, kainate and NMDA receptors.
(B) An expanded view of the kainate receptor clade reveals multiple insect iGluR subtypes corresponding to KaiR1C, KaiR1D, Clumsy and EKAR families, with the fly NMJ GluRII subunits forming sub-branches in the KaiR1C and Clumsy families.
(C) An expanded view of the AMPA receptor clade showing separate branches for the insect and mammalian AMPA receptor subunits.
To gain insight into the molecular function of insect CNS iGluRs we took a dual approach of screening individual Drosophila AMPA and kainate receptor subtypes for functional activity using heterologous expression systems, and in addition prepared ligand-binding domain (LBD) constructs for structural and biochemical analysis. Of the six Drosophila CNS iGluRs selected for functional analysis only the kainate receptor DKaiR1D and the AMPA receptor DGluR1A produced glutamate activated currents when expressed in HEK293 cells or Xenopus oocytes; coexpression of additional kainate receptor species with DKaiR1D did not reveal evidence for formation of heteromeric receptor assemblies with emergent functional properties. Likewise, of the six Drosophila iGluR LBDs selected, only the kainate receptor DKaiR1D and the AMPA receptor DGluR1A yielded crystal structures.
DKaiR1D forms Rapidly Desensitizing Ca2+ Permeable Receptors
Rapid application of 10 mM glutamate for 1 ms to outside-out patches from HEK cells transfected with DKaiR1D revealed rapid activation (Figure 2A), 10–90% rise time 217 ± 6 μs (n = 9), and fast deactivation, rate constant 2523 ± 218 s−1 (n = 9); longer applications of glutamate revealed profound desensitization, 94 ± 1.5 % (n = 9), with the decay well fit by the sum of two exponentials, kfast 1317 ± 105 s−1, kslow 107 ± 22 s−1, Afast 90 ± 2.5 % (n = 9); the extent of desensitization varied from cell to cell (Figure S2). Recovery from desensitization, measured using a twin pulse protocol (Figure 2B), was complete within a second, and described by the sum of two exponentials of rate constants kfast 122 s−1, kslow 9.9 ± 22 s−1, Afast 88%. Because expression of DKaiR1D was poor in HEK cells, with 69/103 patches yielding currents too small for reliable analysis (< 10 pA), we switched to expression in Xenopus oocytes to further characterize DKaiR1D after finding that the lectin Concanavalin A could be used to attenuate desensitization.
Figure 2. Functional characterization of DKaiR1D.
(A) Superimposed responses to 10 mM glutamate applied for 1 and 100 ms to an outside-out patch from a HEK cell transfected with DKaiR1D; black lines show the average of 10 responses; red lines show decay of the responses fit with the sum of one or two exponential functions; open tip junction currents measured at the end of the experiments are shown at the top.
(B) Kinetics of recovery from desensitization measured using a twin pulse protocol fit with the sum of two exponentials of rate constants 388 and 9.9 s−1 (right panel).
(C) Responses to 10 mM glutamate at −60 mV for DKaiRID wild type (left) or DKaiRID Q604R (right) recorded from Xenopus oocytes with either 2 mM Ca2+, 50 μM Ca2+, or 0.8 mM Ba2+ in the extracellular solution, as indicated.
(D) Leak subtracted current-voltage plots for wild type DKaiRID (left) or the Q604R mutant right (right) responses to 10 mM glutamate recorded with 0.8 mM extracellular Ba2+.
(E) Block of responses to 3 mM glutamate at -60 mV by 3 μM philanthotoxin-433 for wild type DKaiRID, showing slow recovery. Philanthotoxin-433 did not block responses for the Q604R mutant (right).
When expressed in Xenopus oocytes DKaiR1D yielded robust responses to 10 mM glutamate after application of 0.6 mg/ml Concanavalin A for 4 mins. As observed previously for Drosophila NMJ iGluRs (Han et al., 2015), the response to glutamate recorded with 2 mM extracellular Ca2+ showed a large transient inward current, which was abolished when Ca2+ was replaced by Ba2+, or when the extracellular Ca2+ concentration was reduced to 50 μM (Figure 2C). This characteristic pattern is due to activation of endogenous TMEM16A chloride channels by glutamate triggered influx of Ca2+ (Boton et al., 1989; Schroeder et al., 2008), indicating that DKaiR1D forms Ca2+ permeable ion channels. Mutation of Gln 604 to Arg, a position in the pore loop equivalent to the Q/R site which undergoes RNA editing in vertebrate AMPA and kainate receptors (Higuchi et al., 1993; Köhler et al., 1993) abolished these transient responses (Figure 2C), indicating a loss of Ca2+ permeability as occurs in edited vertebrate AMPA and kainate receptors (Hume et al., 1991; Köhler et al., 1993).
All subsequent experiments were performed using Ba2+ to prevent activation of TMEM16A. Under these conditions, analysis of current-voltage plots for DKaiR1D responses revealed pronounced inward rectification which was abolished for the DKaiR1D Q604R mutant (Figure 2D and Figure S2), exactly as found for Ca2+ permeable vertebrate AMPA and kainate receptors which undergo RNA editing at the equivalent position, indicating that current flow through DKaiR1D is blocked by cytoplasmic polyamines, with relief from block on strong depolarization (Bähring et al., 1997; Bowie and Mayer, 1995). Also similar to Ca2+ permeable vertebrate kainate receptors (Bähring and Mayer, 1998), wild type but not DKaiR1D Q604R mutant responses to glutamate were blocked by extracellular philanthotoxin, with several minutes required for full recovery from block (Figure 2E).
Ligand Selectivity of DKaiR1D Activation
Consistent with classification as a kainate receptor, glutamate, kainate and quisqualate, but not AMPA, aspartate or NMDA produced activation of DKaiR1D (Figure 3A); all ligands were applied at a concentration of 10 mM, and on average the amplitude of responses kainate and quisqualate were 0.37 ± 0.03 and 0.78 ± 0.03 of those to glutamate (n = 7). Concentration response curves for glutamate (Figure 3B) and kainate (Figure 3C) revealed that glutamate is a very low affinity agonist, EC50 10.7 ± 0.5 mM (n = 8), while kainate acts as a potent, but weak partial agonist, EC50 39 ± 1 μM (n = 8), producing only 16% of the response to glutamate at saturating concentrations. The low sensitivity of DKaiR1D to glutamate resembles that found previously for Drosophila NMJ iGluRs (Han et al., 2015), but neither type A nor type B NMJ receptors respond to kainate.
Figure 3. Unique DKaiR1D Ligand Binding Profile.
(A) Upper panel, DKaiRID responses to different ligands (all 10 mM) recorded from a Xenopus oocyte at -60 mV; lower panel, peak current responses normalized to the glutamate response, presented as mean ± SEM (n=7).
(B) Upper panel, responses to glutamate at -60 mV at the indicated concentrations (in mM) with Na+ gluconate used to maintain a constant extracellular sodium concentration; lower panel, concentration-response plot fit with the Hill equation; data points show the mean for 8 cells, normalized to the peak current recorded in individual cells; error bars show ± SD.
(C) Upper panel, responses to kainate at -60 mV at the indicated concentrations (in mM); lower panel, concentration-response plot fit with the Hill equation; data points show the mean for 8 cells, normalized to the peak current recorded in individual cells; error bars show ± SD.
(D) Proteolysis protection assays for digestion of DKaiR1D S1S2 for 60 min by trypsin at a 1:20 ratio, with the indicated ligands added at a concentration of 1 mM (MW: 29.3 kDa molecular weight marker; UC: 10 μg uncut DKaiR1D S1S2).
(E) Stereoview of the DKaiR1D LBD glutamate complex; the cartoon representation is shaded gold and cyan for domains 1 and 2 respectively; glutamate, three trapped water molecules, and the side chains with they interact are drawn as sticks, with transparent shading for the surface of the LBD cavity, and hydrogen bonds and salt bridges drawn as dashed lines.
To further explore the ligand binding profile of DKaiR1D, and as a complement to electrophysiological experiments, we performed proteolysis protection assays using the purified DKaiR1D ligand-binding domain expressed as a soluble protein. These experiments revealed that, as expected, glutamate, kainate and quisqualate prevented digestion by trypsin, while AMPA was inactive, and the non-selective vertebrate AMPA and kainate receptor antagonist CNQX produced weak protection. Unexpectedly, NMDA, the GluN2 subunit antagonist AP5, and to a lesser extent aspartate were also protective (Figure 3D), even though in electrophysiological experiments NMDA and aspartate did not activate DKaiR1D ion channel currents (Figure 3A).
To investigate the molecular basis for this unexpected result we crystallized the DKaiR1D ligand-binding domain complex with glutamate and solved its structure to a resolution of 1.85 Å by X-ray diffraction (Figure 3E and Table S1). Our goal was to use this structure as a template to dock other ligands and thus resolve the unique ligand binding profile of DKaiR1D. The DKaiR1D ligand-binding domain crystallized as a dimer assembly in which the two subunits adopted essentially identical conformations, 0.39 Å RMSD for least squares superposition using 263 CA atom coordinates. Notably, although the structure revealed a two domain clam shell assembly similar to that of vertebrate kainate receptors, RMSD 0.50/0.44 Å for separate superposition of domain 1 and domain 2 on the GluK2 glutamate complex (Chaudhry et al., 2009b; Mayer, 2005), the DKaiR1D ligand-binding domains adopted an intermediate extent of domain closure. After superposition using domain 1 coordinates, rotations of 8.15° and 8.31° for protomers A and B respectively were required to superimpose domain 2 on the GluK2 glutamate complex. Despite this more open conformation, the volume of the DKaiR1D LBD cavity, 215 ± 1.5 and 206 ± 1.0 Å3 for protomers A and B respectively, was smaller than that for GluK2, and contained only three trapped water molecules, versus five for GluK2. The bound glutamate ligand is coordinated via interactions that closely mimic those found in vertebrate kainate receptors (Figure 3E) as described in detail in supplementary information (Figure S3A). Docking experiments using the DKaiR1D glutamate complex as a template revealed that the ligand N-methyl group of NMDA makes a bad steric contact with Glu723, and that the NMDA β-COOH group collides Ser672 at the tip of α-helix F in domain 2. Likewise, both stereoisomers of AP5 made bad steric contacts with multiple domain 2 residues indicating that the conformation of the NMDA and AP5 complexes that underlies proteolysis protection must differ from that stabilized by glutamate.
NMDA and AP5 are DKaiR1D Antagonists
Since neither NMDA (Figure 3A) nor AP5 (Figure S5) produced DKaiR1D activation we considered the possibility that they might act as antagonists. This proved to be the case, and surprisingly NMDA was a much more effective antagonist than CNQX (Figure 4A); in addition, both the D- and L-isomers of AP5 also produced inhibition of DKaiR1D responses to glutamate (Figure 4B and Figure S5). At a concentration of 10 mM, NMDA produced 79.2 ± 2.2% (n = 6) block of responses to 10 mM glutamate, while CNQX produced 34.3 ± 3.0% block (n = 6). D-AP5 produced 16.6 ± 1.0% block (n = 5) of responses to 10 mM glutamate and 22.7 ± 0.8% block (n = 8) of responses to 1 mM glutamate. L-AP5 produced 11.7 ± 1.2% block (n = 5) of responses to 10 mM glutamate and 35.7 ± 1.4% block (n = 8) of responses to 1 mM glutamate. Block by NMDA was concentration dependent, and the resulting concentration inhibition plot yielded an IC50 of 0.9 mM (Figure 4C).
Figure 4. NMDA and AP5 are DKaiR1D Antagonists.
(A) DKaiRID responses to 10 mM glutamate at -60 mV inhibited by 10 mM CNQX (left) and 10 mM NMDA (right).
(B) DKaiRID responses to 1 mM glutamate at -60 mV inhibited by 10 mM D-AP5 (left) and 10 mM L-AP5 (right).
(C) Concentration-dependent inhibition of responses to 10 mM glutamate at -60 mV by 0.3, 1, 3 and 10 mM NMDA (left), and concentration inhibition plot for block of DKaiRID responses to 10 mM glutamate by NMDA fit with the Hill equation (right); data points show mean ± SD for 10 cells.
Open Cleft Conformations of DKaiR1D NMDA and AP5 Complexes
To gain further insight into how NMDA and AP5 act as antagonists we solved crystal structures of their DKaiR1D LBD complexes at resolutions of 1.28 and 1.75 Å respectively (Table S1). Both complexes crystallized as dimer assemblies (Figure 5), but with strikingly different conformations from the DKaiR1D glutamate complex. In the both the NMDA and AP5 complex dimer assemblies, consistent with their action as antagonists in electrophysiological experiments, the two protomers adopt open cleft conformations such that the separation of the lower lobes, measured using Ile651 CA atom positions at the base of domain 2, proximal to the linker leading to the M3 helix in the ion channel, decreases from 36.6 Å in the glutamate complex (Figure 5A) to 25.7 Å and 19.8 Å in the NMDA (Figure 5B and Movie S1) and AP5 (Figure 5C and Movie S2) complexes respectively.
Figure 5. Crystal Structures of DKaiR1D LBD NMDA and AP5 Complexes.
(A) Crystal structure of the DKaiR1D LBD glutamate complex dimer assembly with domains 1 and 2 colored gold and teal for subunit A, and pale yellow and pale blue for subunit B; α-helix J in each subunit is drawn as a transparent cylinder, with vectors through the helix illustrating the relative tilt of the two subunits in the dimer assembly.
(B) The DKaiR1D LBD NMDA complex dimer assembly drawn as described above.
(C) The DKaiR1D LBD AP5 complex dimer assembly colored as described above.
(D) Domain 2 rotation angles relative to the appropriate closed cleft glutamate complexes for the indicated antagonist complexes for DKaiR1D, GluK1 and GluN2D. (E) The DKaiR1D B subunit LBD NMDA complex with 2mFo-DFc electron density maps contoured at 2σ for the ligand and solvent molecules; the cartoon representation is shaded gold and cyan for domains 1 and 2 respectively; side chains are drawn as sticks, water molecules as red spheres, and hydrogen bonds and salt bridges drawn as dashed lines. (F) The DKaiR1D A subunit LBD D-AP5 complex with an mFo-DFc electron density map contoured at 3 σ for the ligand and 2mFo-DFc electron density maps for solvent molecules contoured at 1.2 σ and colored as described above.
In the NMDA but not AP5 complex the two subunits in the dimer assembly pivot away from the 2-fold axis of molecular symmetry, such the angle between α-helix J in the two subunits increases, and the separation between the upper lobes of the two subunits, measured as the distance between the CA atoms of Pro758 immediately preceding the N-terminus of α-helix J increases from 16.9 Å to 21.2 Å (Figure 5B). These movements are largely mediated by rigid body movements of domains 1 and 2, with RMSD values of 0.47/0.59 Å and 0.46/0.82 Å for individual superposition of domains 1 and 2 of the A and B subunits of the NMDA dimer assembly on the glutamate complex. Rigid body movements also account for the larger conformational change found for the AP5 complex, for which RMSD values of 0.38/0.88 and 0.35/0.82 Å were measured for individual superposition of domains 1 and 2 of the A and B subunits of the AP5 dimer assembly on the glutamate complex dimer assembly.
As noted above, although both subunits in the NMDA and AP5 complex dimer assemblies adopted open cleft conformations, the extent of opening relative to the glutamate complex differed for individual subunits in the dimer assembly. For the DKaiR1D NMDA complex, after superposition using domain 1 coordinates, rotations of 19° and 25° for the A and B subunits were required to superimpose domain 2 on the closed cleft DKaiR1D glutamate complex (Figure 5D and Figures S3B and C). By contrast, for vertebrate NMDA receptors NMDA produces the same extent of cleft closure for GluN2D as glutamate and acts as an agonist (Vance et al., 2011). For the DKAiR1D AP5 complex, after superposition using domain 1 coordinates, rotations of 30° and 23° for the A and B subunits were required to superimpose domain 2 on the closed cleft DKaiR1D glutamate complex (Figure 5D and Figures S3D and E). The 30° opening observed for the B subunit of the AP5 complex (Figure 5D) is similar to the opening observed for several vertebrate GluK1 antagonist structures (Alushin et al., 2011; Mayer et al., 2006), and much larger than for vertebrate GluN2A NMDA receptors (Figure 5D) for which D-AP5 produces only 15° opening (Jespersen et al., 2014).
Electron density maps for the DKaiR1D complex with NMDA were of exceptionally high quality (Figure 5E and Figures S3B and C) and revealed that coordination of the ligand α-COOH and α-NH2 groups is similar to that for glutamate (Figure S3A), with the exception that due to cleft opening, the hydrogen bond with the Ser672 main chain amide in α-helix F is lost; also, the Glu723 side chain rotates 82° about χ1 so that it no longer binds the ligand α-NH2 group, which is instead connected to a hydrogen bonded network of solvent molecules (Figure 5E and Figures S3B and S3C). The interaction of the NMDA β-COOH group with DKAiR1D domain 2 differs for subunits A and B, due to their different extent of opening, and also differs from interactions made by the glutamate γ-COOH group. In chain A (19° opening) the β-COOH group is bound by the side chain OH group and main chain amide of Ser672, and by a network of water molecules linked to the Asn704 side chain (Figure S3B). In chain B (25° opening) the ligand rotates 9° and 13° about χ1 and χ2 such that the β-COOH group points towards domain 1 where it is bound by the side chain OH group of Thr503 (Figure S3C), while additional water molecules are recruited into the hydrogen bonded solvent network linking the β-COOH group with domain 2.
The DKaiR1D complex with AP5 was crystallized using racemic ligand and, consistent with the results of electrophysiological experiments, separate refinements with D-AP5 and L-AP5 revealed that both enantiomers could be accommodated in Fo-Fc electron density maps, and that their interactions with the LBD were nearly identical (Figures S3D and S3E); thus we used a mixed occupancy refinement strategy with both ligands present, but to facilitate illustration show electron density maps for the results of separate refinements for the two enantiomers. This revealed that the AP5 δ-phosphonate group is bound in a pocket formed by the N-terminus of α-helices F and H (Figure 5F). In chain A (30° opening) the δ-phosphonate group forms hydrogen bonds with the main chain amide and side chain OH group of Ser672, and with a network of water molecules connected to the side chain of Asn704. In chain A (23° opening) these contacts are lost due to rotation of domain 2, and only a single water molecule links the δ-phosphonate group to the main chain amide of Glu723. Notably, the side chain of Glu723 adopts different rotamers in the A and B subunits of the AP5 complex, such that in chain A, which adopts a more open conformation, Glu723 forms a salt bridge with Lys747 in domain 1 (Figure S3D), while in chain B, which adopts a more closed conformation, Glu723 forms a salt bridge with the AP5 α-NH2 group (Figure S3E).
Sodium Binding sites in the DKaiR1D Dimer assembly
A defining feature of vertebrate kainate receptors is their modulation by Na+ and Cl− ions (Bowie, 2002; Paternain et al., 2003). Structural and biochemical studies reveal that these ions bind to sites in the LBD dimer assembly and stabilize the active conformation that supports ion channel gating by glutamate (Chaudhry et al., 2009a; Plested and Mayer, 2007; Plested et al., 2008). In the subunit B of the DKaiR1D glutamate complex dimer assembly we observed a Na+ ion bound in exactly the same location as found in vertebrate GluK1 and GluK2 LBD crystal structures, with identical coordination by a conserved Glu (n+4) Asp motif, in which the side chain and main chain carbonyl group of Glu509, the main chain carbonyl group of Val512 and the side chain of Asp513 form the cation binding site (Figure 6A). Different from vertebrate GluK1 and GluK2 LBD crystal structures, the Na+ ion has six-fold coordination, with two water molecules completing the ion coordination shell, while in the GluK1 and GluK2 complexes an Ile or Met side chain, which is replaced by Pro755 in DKaiR1D, blocks one of the ion coordination sites. In subunit A of the DKaiR1D dimer assembly the Na+ binding site is plugged by the Lys715 side chain of an adjacent protein molecule, just as found previously for vertebrate GluK1 complexes with kainate (Plested et al., 2008); in the DKaiR1D NMDA and AP5 complex dimer assemblies which crystallized in a different space group, both Na+ binding sites are plugged by Lys715 from adjacent molecules in the crystal lattice.
Figure 6. DKaiR1D has an Allosteric Binding Site for Na+.
(A) Stereoview of the DKaiR1D LBD dimer assembly viewed from the top, with subunits A and B colored pale yellow and gold respectively; side chains are drawn in stick representation, and water molecules shown as red spheres; a 2mFo-DFc electron density map contoured at 3σ shows the bound Na+ ion; bonds connecting the Na+ ion to the protein and water molecules have characteristic lengths of 2.2 – 2.6 Å and were refined without restraints. Note that there is no Cl− ion trapped in the dimer interface, which is filled with water molecules instead.
(B) Amino acid sequence alignment for LBD segments forming Na+ and Cl− ion binding sites in mammalian kainate receptors, represented by GluK1, and the corresponding segments for Drosophila iGluRs in the kainate receptor clade. The secondary structure assignment is for DKaiR1D, with the Na+ binding site residues indicated by * in yellow circles, and residues forming or controlling the conformation of Cl− binding site shown by * in green circles.
(C) Responses to 10 mM glutamate recorded from an outside-out patch from a HEK cell transfected with DKaiR1D with either NaCl or CsCl in the extracellular solution; black lines show the average of 10 responses; red lines show fits with the sum of two exponential functions; open tip junction currents measured at the end of the experiments are shown at the top. The bar plot shows the mean amplitude of responses from 5 patches and the weighted rate constant of desensitization, calculated as (kfast * Afast) + (kslow * Aslow).
Different from vertebrate kainate receptors, there was no Cl− ion trapped in the dimer interface of the DKaiR1D dimer assembly, suggesting loss of the anion-binding site. An amino acid sequence alignment of regions forming the Na+ and Cl− ion binding sites in vertebrate kainate receptors with Drosophila iGluRs in the kainate receptor clade, reveals loss of the critical Lys side chain that coordinates the bound anion in GluK1 and GluK2, which is replaced by Thr516 in DKaiR1D, and by Ile and Pro in DKaiR1C and clumsy, respectively (Figure 6B). Although both EKAR subunits harbor a Lys at this position, additional amino acid sequence differences, including the substitution of Asn or Ser for the Thr residue that forms hydrogen bonds with water molecules bound to the Cl− ion in GluK1 and GluK2, and in DKAiR1D substitution of Thr761 for the Asp residue that positions Arg side chains at the N-terminus of α-helix J to support binding of a Cl− ion, further contribute to loss of the anion binding site in Drosophila kainate receptors. Consistent with this, the top of the DKaiR1D dimer assembly crystal structure lacked the electrostatic profile required for anion binding and was filled with water molecules (Figure 6A). The amino acid sequence alignment of Drosophila kainate receptors also revealed that only DKaiR1D has all of the conserved residues necessary to form a Na+ binding site in the upper lobe of the LBD dimer assembly, and that in all of the other subunits, different combinations of amino acid exchanges destroy both the Na+ and Cl− ion binding sites; conversely, the Na+ ion binding site is conserved in all KaiR1D subunits from diverse insect species (Figure S4).
In the DKaiR1D dimer assembly the bound Na+ ions are coordinated exclusively by residues in a single subunit, and there is no intermolecular cross link connecting these sites, a role played by the bound Cl− ion and the Lys500 side chain in GluK2, raising the question of whether Na+ ions have any functional role in DKaiR1D. To test this we performed ion substitution experiments with CsCl (Figure 6B). This revealed that DKaiR1D responses to glutamate are not strongly modulated by Na+; on average the peak amplitude of the glutamate response in 150 mM CsCl was 88.1 ± 2.8% of that measured in 150 mM NaCl (mean ± SD, 5 patches, all with two exchanges between NaCl and CsCl). By contrast, for GluK2 the amplitude of glutamate responses in CsCl is only 6–12% of that in NaCl (Bowie, 2002; Plested et al., 2008). Consistent with this, the exchange of CsCl for NaCl did not produce a large acceleration in the rate of onset of desensitization for DKaiR1D, kfast 1504 ± 90 s−1, kslow 165 ± 27 s−1, Afast 90 ± 1.0 % for responses measured in NaCl, and kfast 1884 ± 99 s−1, kslow 192 ± 28 s−1, Afast 88 ± 1.2 % for responses measured in CsCl (n = 10). By contrast, for GluK2 the rate of desensitization in 150 mM CsCl is 6–12-fold faster than in 150 mM NaCl (Bowie, 2002; Plested et al., 2008). Consistent with the absence of a bound Cl− ion in the DKaiR1D LBD crystal structure the exchange of NaMeSO3 for NaCl did not produce large changes in the rate of onset of desensitization, kfast 1970 ± 88 s−1, kslow 190 ± 31 s−1, Afast 92 ± 0.8 % for responses measured in NaCl, and kfast 1985 ± 68 s−1, kslow 236 ± 39 s−1, Afast 90 ± 1.9 % for responses measured in NaMeSO3 (n = 6), while for GluK2 this increases the rate of onset of desensitization 7-fold (Plested and Mayer, 2007).
Functional characterization of DGluR1A Reveals Weak Activation by AMPA
The Drosophila genome encodes two iGluRs with sequence similarity to vertebrate AMPA receptors, DGluR1A and DGluR1B (Figure 1 and Figures S1 and S6). Prior studies on DGluR1A reported activation by kainate, EC50 75 μM (Ultsch et al., 1992), and that coexpression with an auxiliary stargazin subunit from the TARP family is required for activation by glutamate (Walker et al., 2006). In our experiments, DGluR1A did not generate functional ion channels when expressed alone in Xenopus oocytes, but did respond to both glutamate and kainate when coexpressed with either Drosophila or Apis stargazin (Figure 7A). As found previously (Walker et al., 2006), DGluR1A responses to glutamate exhibit very slow activation with Drosophila stargazin, while with Apis stargazin (Stg1) responses were faster, and thus we used Apis Stg1, which has 39% amino acid identity with Drosophila stargazin, in all subsequent experiments. Current-voltage plots for DGluR1A revealed strong biphasic rectification (Figure 7B), consistent with inhibition of DGluR1A by philanthotoxin (Ultsch et al., 1992). In combination with amino acid sequence alignments that reveal a Gln residue at the Q/R site in the pore loop, these results suggest that DGluR1A forms Ca2+ permeable ion channels. However, different from vertebrate AMPA receptors, which show reduced block by cytoplasmic polyamines when coexpressed with stargazin (Soto et al., 2007), outward current flow remained strongly inhibited at positive membrane potentials.
Figure 7. Functional characterization of DGluR1A.
(A) Stargazin is required for functional reconstitution of DGluR1A. Representative traces show responses to 10 mM glutamate for oocytes injected with the indicated cRNAs for DGluR1A, Apis Stg1, DGluR1A plus Apis Stg1, and DGluR1A plus Drosophila Stg1.
(B) Current-voltage plot for DGluR1A responses to glutamate recorded from an oocyte co-injected with DGluR1A and Apis STG1 shows inward rectification.
(C) Responses to different ligands (all 3 mM) recorded from a Xenopus oocyte at -60 mV co-injected with DGluR1A and Apis STG1.
(D) Peak current responses at -60 mV for the indicated agonists normalized to the glutamate response, presented as mean ± SD (n=4).
(E) Concentration-response plot for glutamate-activated currents for DGluR1A co-expressed with Apis Stg1 fit with the Hill equation; data points show the mean for 6 cells, normalized to the peak current recorded in individual cells; error bars show ± SD.
(F) Concentration-response plot for kainate-activated currents for DGluR1A co-expressed with Apis Stg1 fit with the Hill equation; data points show the mean for 11 cells, normalized to the peak current recorded in individual cells; error bars show ± SD.
We next screened DGluR1A for activation by a panel of agonists, all applied at 3 mM (Figures 7C), and found that responses to kainate were 4.1 ± 0.3 (n=4) times larger than those to glutamate, with no activation by NMDA, weak activation by aspartate, 28 ± 8% of the response to glutamate recorded in the same cell and surprisingly, weak activation by AMPA, 40 ± 4% and quisqualate, 41 ± 3%, all n = 4 (Figure 7D). Concentration response analysis revealed that glutamate and kainate were of similar potency, EC50 for glutamate 136 ± 8 μM (n = 6, Figure 7E), EC50 for kainate 125 ± 4 μM (n = 8, Figure 7F). The 4-fold larger amplitude of the response to kainate parallels the effect of stargazin on vertebrate AMPA receptors (Levchenko-Lambert et al., 2011; Tomita et al., 2005).
Ligand binding profile and crystal structure of DGluR1A
The results of proteolysis assays using the DGluR1A ligand-binding domain expressed as a soluble protein revealed that at 1 mM, glutamate, kainate, quisqualate and CNQX, but not AMPA, NMDA, aspartate or AP5 were protective (Figure 8A), largely in agreement with the results for activation of ion channel gating by these ligands. The lack of protection by AMPA and its weak action as an agonist is surprising given that the LBDs of DGluR1A and vertebrate AMPA receptors share 70–71% sequence similarity (54–55% sequence identity). To investigate the unusual ligand binding profile of DGluR1A we solved the crystal structure of the glutamate complex at a resolution of 1.6 Å and compared it to the structure of the GluA2 glutamate complex (Table S1 and Figures 8B and S6A). This revealed that the structures of the DGluR1A and GluA2 LBDs are nearly identical, with similar extents of cleft closure, RMSD 0.69 Å for 249 CA atoms. The volumes of the cavity in which glutamate binds, 197 ± 0.3 Å3 for DGluR1A and 231 ± 0.2 Å3 for GluA2 are also similar, and the glutamate ligand makes identical interactions with the side chains of Arg573, Thr762 and Glu811 in the two receptors (Figure 8B and Figure S7).
Figure 8. DGluR1A Ligand Binding Profile and Crystal Structure.
(A) Proteolysis protection assays for digestion of DGluR1A S1S2 by trypsin at a 1:20 ratio, with the indicated ligands added at a concentration of 1 mM (MW: 29.3 kDa molecular weight marker; UC: 10 μg uncut DGluR1A S1S2; Trypsin).
(B) DGluR1A LBD glutamate complex; the cartoon representation is shaded gold and cyan for domains 1 and 2 respectively; glutamate, three trapped water molecules, and the side chains with they interact are drawn as sticks, with transparent shading for the surface of the LBD cavity, and hydrogen bonds and salt bridges drawn as dashed lines.
(C) Amino acid sequence alignment for α-Helix H showing conservation of the exchange of a Thr residue in mammalian AMPA receptors for a Tyr residue in insect receptors.
What differs between DGluR1A and GluA2 is the shape of the ligand binding cavity, and the number and location of water molecules trapped in the cavity, three for DGluR1A and four for GluA2. Only one of these water molecules adopts a conserved position in DGluR1A and GluA2, with W1 forming a hydrogen bond contact linking the glutamate ligand γ-COOH group with a main chain amide at the N-terminus of α-helix F (Figure 8B and Figure S7A and B). In domain 2 of DGluR1A, at the N-terminus of α-helix H, Tyr792 projects into the ligand-binding pocket, with the side chain OH group forming a charge assisted hydrogen bond contact with the glutamate ligand γ-COOH group; in GluA2 there is a Thr at this position (Figure 8B and Figure S7B). The Tyr side chain sculpts the domain 2 surface of the ligand-binding cavity, displacing three water molecules bound at this location in the GluA2 complex. An amino acid sequence alignment for seven AMPA receptor subunits from diverse insect species revealed that these all harbor a Tyr residue at the N-terminus of α-helix H, while all vertebrate AMPA receptors contain a Thr residue at the structurally equivalent position (Figure 8C). Amino acid sequence alignments also revealed that the binding site for cyclothiazide (Partin et al., 1995; Sun et al., 2002), a specific allosteric modulator of vertebrate AMPA receptors, is blocked by the exchange of Lys/Thr for the Ser residue found in vertebrate flip splice variant AMPA receptors (Figure S6) and thus cyclothiazide cannot be used to block desensitization of insect AMPA receptors.
When AMPA is docked in the DGluR1A LBD crystal structure, the isoxazole ring makes multiple bad contacts with the Tyr792 side chain (Figure S7C), and thus AMPA cannot bind to DGluR1A in its closed cleft conformation; likewise, when quisqualate was docked, the oxadiazolidin ring also made bad contacts with the Tyr792 side chain, suggesting that AMPA and quisqualate likely act as partial agonists in which the LBD adopts an intermediate extent of domain closure (Jin et al., 2003). We next crystallized the DGluR1A Y792T mutant glutamate complex (Table S1) and found that the volume of ligand binding cavity increased to 337 ± 0.1 Å3 allowing the binding of four additional water molecules (Figure S8A); as expected, docking experiments revealed relief from steric hindrance for both AMPA and quisqualate. However, both proteolysis protection assays (Figure S8B) and measurement of receptor activity (Figure S8C) revealed that quisqualate, but not AMPA, binds strongly and produces full receptor activation. Closer inspection of the Y792T mutant structure revealed that while quisqualate is able to bind via contacts like those found in vertebrate receptors, the isoxazole ring of AMPA is left ‘hanging in space’ and due to its different conformation fails to make good contacts with domain 2 residues, highlighting the complexity of predicting ligand specificity despite the availability of high resolution crystal structures.
DISCUSSION
In this study, we combine phylogenetic, electrophysiological and crystallographic analyses to characterize a Drosophila kainate receptor, DKaiR1D, and a Drosophila AMPA receptor, DGluR1A. We found that DGluR1A and DKaiR1D, similar to vertebrate GluA1-4 AMPA and GluK1-3 kainate receptor subunits, form homomeric calcium-permeable channels. Based on sequence alignments and the lack of RNA-editing of Drosophila iGluRs mRNA at their Q/R sites (St Laurent et al., 2013), it is likely that most insect iGluRs are calcium permeable and that they are inhibited by endogenous cytoplasmic polyamines and by spider venom polyamine toxins. We note that homomeric DKaiR1D has a very fast desensitization rate, while for DGluR1A we have not yet been able to achieve sufficient expression to allow recording from outside-out patches with rapid perfusion. Structural analyses revealed that DKaiR1D LBD dimers contain conserved Na+ ion binding sites characteristic of vertebrate kainate receptors, but these appear to not strongly modulate the activation or desensitization of KaiR1D, perhaps because the Cl− binding site found in vertebrate kainate receptors is absent in insect kainate receptors (Plested and Mayer, 2007; Plested et al., 2008). Sequence analysis revealed that this separation of Na+ and Cl− binding sites in KaiR1D subunits occurs in all insect species examined. Structure-aided sequence analysis also reveals that in the other three groups of fly kainate receptors, different combinations of amino acid substitutions destroy or significantly weaken both the Na+ and Cl−, binding sites. Thus, the allosteric modulation by both anions and cations that is characteristic of vertebrate kainate receptors is uncoupled in insect kainate receptors and for the majority of cases both ion binding sites are eliminated.
Expansion of the kainate receptor family in insects
Previous phylogenetic studies suggest that most bilateria, including insects, worms and vertebrates, have three major classes of cation-selective iGluRs, corresponding to vertebrate AMPA, kainate and NMDA receptors (Alberstein et al., 2015; Brockie and Maricq, 2003; Croset et al., 2010). Our analysis reveals that in insects the kainate receptor family is expanded into four groups, while a prior phylogenetic analysis revealed that in Mollusca the AMPA receptor family is expanded (Alberstein et al., 2015). At the neuromuscular junction of Drosophila (Jan and Jan, 1976) and the locust Schistocerca gregaria (Cull-Candy, 1976; Cull-Candy and Usherwood, 1973) iGluRs have been extensively studied, in part serving as a surrogate model for CNS iGluRs. Interestingly, we found that late in evolution, in higher Diptera, the five Drosophila NMJ iGluR subunits, GluRIIA-E, were derived from two separate kainate receptor subtypes, KaiR1C and Clumsy. Thus, despite their unique obligate heterotetrameric subunit stoichiometry (Marrus et al., 2004; Qin et al., 2005) and insensitivity to kainate (Han et al., 2015), fly NMJ iGluRs evolved from ancestral kainate-sensitive iGluRs and it is likely that in other insect species iGluRs related to KaiR1C and Clumsy may function in both the CNS and NMJ.
Novel pharmacology of Drosophila ionotropic glutamate receptors
Although phylogenetic analysis supports classification of Drosophila and other insect iGluRs into the familiar AMPA, kainate and NMDA receptor families, our results reveal unexpected differences in their ligand binding properties. The most dramatic change was the conversion of NMDA from an agonist for vertebrate NMDA receptors to an antagonist for Drosophila KaiR1D, a kainate receptor which is also inhibited by both isomers of AP5, while D-AP5 but not L-AP5 acts as a potent vertebrate NMDA receptor antagonist (Evans et al., 1982). The crystal structures we solved for the DKaiR1D LBD establish that NMDA and AP5 inhibit activation of DKaiR1D by stabilizing an open cleft conformation, similar to the action of competitive antagonists for vertebrate iGluRs from each of the three major families (Alushin et al., 2011; Armstrong and Gouaux, 2000; Furukawa and Gouaux, 2003; Jespersen et al., 2014). In addition, NMDA triggered separation of the upper lobes of the DKaiR1D LBD dimer assembly, a conformational change that occurs during desensitization of vertebrate AMPA and kainate receptors (Armstrong et al., 2006; Durr et al., 2014; Meyerson et al., 2014), may in addition contribute to the inhibitory action of NMDA on DKaiR1D.
AMPA receptors were initially identified by and named in response to their activation by quisqualic acid (Watkins and Evans, 1981; Watkins et al., 1990), a glutamate bioisostere that is non selective and which activates all of the major vertebrate iGluR subtypes, in addition to acting as a potent agonist for G protein coupled glutamate receptors (Sugiyama et al., 1987). Prior to the cloning of GluA1-4 subunits, the so called quisqualate receptors were renamed AMPA receptors, following the synthesis of AMPA and the discovery that it was a highly selective agonist, without activity at kainate, NMDA or G protein coupled glutamate receptors (Krogsgaard-Larsen et al., 1980; Watkins et al., 1990). These serendipitous events in the history of the development of selective ligands for iGluR subtypes were strongly reinforced when a large family of vertebrate iGluR subunits were cloned, and it was discovered that these encoded discrete families of iGluR subtypes, each with high sequence identity, the ligand binding properties of which corresponded to the familiar AMPA, kainate and NMDA receptor subtypes (Hollmann and Heinemann, 1994; Nakanishi, 1992; Seeburg, 1993). Our experiments reveal an unexpected breakdown of the classification scheme for Drosophila and most likely other insect species iGluRs.
Combining genetics and pharmacology for synaptic circuit dissection in Drosophila
With the plethora of genetic tools and advanced connectome analyses, Drosophila has emerged as a key model organism for studying the circuit basis of behavior. It is now evident that like vertebrates, glutamatergic synapses are abundantly utilized in fly CNS circuits (Daniels et al., 2008). Our functional and structural analyses revealed that Drosophila iGluRs have agonist and antagonist selectivity very different from those of vertebrates indicating that sequence and structural homology does not confer conserved pharmacological properties. However, the unique pharmacology of Drosophila iGluRs reported here has proven of use to reveal the role of KaiR1D in presynaptic homeostasis (Kiragasi and Dickman personal communication). We envision that appropriate use of pharmacological tools in combination with powerful fly genetics will greatly aid studies of complex neural circuits in Drosophila.
EXPERIMENTAL PROCEDURES
Molecular biology
The DKaiRID coding sequence was PCR-amplified from the full-length cDNA and cloned into pRK5-IRES-EGFP and pT7TS expression vectors for expression in HEK293 cells and Xenopus oocytes respectively. Site directed mutagenesis was performed to change a CAG codon to CGG to make KaiRID Q604R. HA tagged Drosophila HA::GluRIA and Drosophila STG1 (gifts from Andres Villu Maricq, Department of Biology, University of Utah, Salt Lake City, UT) were cloned into pSGEM, with the HA tag removed and three residues mutated (L61M, N301S, P317A) to match the published sequence of Drosophila melanogaster GluRIA (Flybase ID CG8442). A synthetic gene for Apis STG1 based on the sequence of A. mellifera first-strand cDNA (GenBank accession nos. DQ015968) was also cloned into pSGEM. All sequences were confirmed by DNA sequencing. pT7TS-KaiRID and pSGEM-based constructs were linearized with EcoRI and PacI, respectively. cRNAs were synthesized using T7 polymerase (Ambion, mMessage mMachine T7 transcription kit).
Functional analysis
Electrophysiological experiments using outside-out patches from transfected HEK293 cells and from Xenopus oocytes injected with cRNA were performed using standard techniques, as described in supplementary information.
Phylogenetic sequence analysis
Phylogenetic analysis using amino acid sequence alignments for 64 iGluR sequences culled from Fruitfly (Drosophila melanogaster), Housefly (Musca domestica), Mosquito (Anopheles gambiae), Bee (Apis mellifera), Locust (Locusta migratoria) and Beetle (Tribolium castaneum) genomes and 16 rat (Rattus norvegicus) iGluR cDNA sequences, was performed with MEGA6 and Maximum-Likelihood (ML) tree reconstruction, using the Le-Gascuel amino acid substitution model and 10 distinct gamma distributed rates and invariant sites (Le and Gascuel, 2008). The ML heuristic search was performed with the Nearest-Neighbor-Interchange method, and the initial tree was selected by applying the Neighbor Joining method to a matrix of pairwise distances estimated using the JTT method. The accuracy of the tree was tested with bootstrapping using 100 replicates. Subsequent formatting of the tree was performed with MEGA and FigTree (http://tree.bio.ed.ac.uk/software/figtree).
Crystallography
Synthetic genes in pET22b with codon optimization for expression in Escherichia coli were designed for the LBDs of the following Drosophila iGluRs, with a GT dipeptide connecting the S1 and S2 sequences: DKaiR1C (R407-K525 and P648-S790); DKaiR1D (A411-K522 and P650-R788); Clumsy (Q410-K530 and S652-E796); CG9935 (S400-L512 and P638-T778); and CG11155 (S420-V537 and P660-N800). Only the DKaiR1D and DGluR1A S1S2 ligand binding cores were successfully overexpressed, purified, and crystallized, essentially as previously described for other iGluR LBDs (Alberstein et al., 2015; Han et al., 2015; Mayer, 2005). Following crystallization, as described in supplementary information, X-ray diffraction data sets were collected at the Advanced Photon Source (SER-CAT) 22-ID beamline, indexed and scaled using HKL2000 (Otwinowski and Minor, 2001), and structures solved by molecular replacement as described in supplementary information.
Supplementary Material
Acknowledgments
We thank Carla Glasser, Chun-Yuan Ting, and Bo Shi for technical support, and Rich Grey for performing the phylogenetic analysis. Synchrotron diffraction data was collected at Southeast Regional Collaborative Access Team (SER-CAT) 22-ID beamline at the Advanced Photon Source, Argonne National Laboratory. Use of the Advanced Photon Source was supported by the U. S. Department of Energy, Office of Science, Office of Basic Energy Sciences, under Contract No. W-31-109-Eng-38. This work was supported by the intramural research program of NICHD, NIH, DHHS (ES, CHL and MLM).
Footnotes
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited with the Protein Data Bank; accession codes are DKaiR1D Glu (5DTB); DKaiR1D NMDA (5EHM); DKaiR1D AP5 (5EHS); DGluR1A Glu (5DT6); DGluR1A Y792T Glu (5ICT).
AUTHOR CONTRIBUTIONS
Y.L. performed TEVC recordings. T-H.H. performed HEK cell recordings. P.D. and M.L.M. performed biochemical and crystallographic experiments. All authors analyzed data and contributed ideas to the manuscript. M.S., C-H.L. and M.L.M. supervised the project. M.L.M. and C-H.L. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alberstein R, Grey R, Zimmet A, Simmons DK, Mayer ML. A family of novel glycine activated ion channel subunits encoded by ctenophore glutamate receptor genes. Proceedings of the National Academy of Sciences of the United States of America. 2015 doi: 10.1073/pnas.1513771112. pii: 201513771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alushin GM, Jane D, Mayer ML. Binding site and ligand flexibility revealed by high resolution crystal structures of GluK1 competitive antagonists. Neuropharmacology. 2011;60:126–134. doi: 10.1016/j.neuropharm.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong N, Gouaux E. Mechanisms for activation and antagonism of an AMPA-sensitive glutamate receptor: Crystal structures of the GluR2 ligand binding core. Neuron. 2000;28:165–181. doi: 10.1016/s0896-6273(00)00094-5. [DOI] [PubMed] [Google Scholar]
- Armstrong N, Jasti J, Beich-Frandsen M, Gouaux E. Measurement of conformational changes accompanying desensitization in an ionotropic glutamate receptor. Cell. 2006;127:85–97. doi: 10.1016/j.cell.2006.08.037. [DOI] [PubMed] [Google Scholar]
- Bähring R, Bowie D, Benveniste M, Mayer ML. Permeation and block of rat GluR6 glutamate receptor channels by internal and external polyamines. J Physiol (Lond) 1997;502:575–589. doi: 10.1111/j.1469-7793.1997.575bj.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähring R, Mayer ML. An analysis of philanthotoxin block for recombinant rat GluR6(Q) glutamate receptor channels. J Physiol (Lond) 1998;509:635–650. doi: 10.1111/j.1469-7793.1998.635bm.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boton R, Dascal N, Gillo B, Lass Y. Two calcium-activated chloride conductances in Xenopus laevis oocytes permeabilized with the ionophore A23187. J Physiol (Lond) 1989;408:511–534. doi: 10.1113/jphysiol.1989.sp017473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D. External anions and cations distinguish between AMPA and kainate receptor gating mechanisms. The Journal of physiology. 2002;539:725–733. doi: 10.1113/jphysiol.2001.013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie D, Mayer ML. Inward rectification of both AMPA and kainate subtype glutamate receptors generated by polyamine-mediated ion channel block. Neuron. 1995;15:453–462. doi: 10.1016/0896-6273(95)90049-7. [DOI] [PubMed] [Google Scholar]
- Brockie PJ, Maricq AV. Ionotropic glutamate receptors in Caenorhabditis elegans. Neurosignals. 2003;12:108–125. doi: 10.1159/000072159. [DOI] [PubMed] [Google Scholar]
- Charng WL, Yamamoto S, Bellen HJ. Shared mechanisms between Drosophila peripheral nervous system development and human neurodegenerative diseases. Curr Opin Neurobiol. 2014;27:158–164. doi: 10.1016/j.conb.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry C, Plested AJ, Schuck P, Mayer ML. Energetics of glutamate receptor ligand binding domain dimer assembly are modulated by allosteric ions. Proceedings of the National Academy of Sciences of the United States of America. 2009a;106:12329–12334. doi: 10.1073/pnas.0904175106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry C, Weston MC, Schuck P, Rosenmund C, Mayer ML. Stability of ligand-binding domain dimer assembly controls kainate receptor desensitization. EMBO J. 2009b;28:1518–1530. doi: 10.1038/emboj.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge GL, Bliss TV. Memories of NMDA receptors and LTP. Trends Neurosci. 1995;18:54–56. [PubMed] [Google Scholar]
- Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, Gibson TJ, Benton R. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 2010;6:e1001064. doi: 10.1371/journal.pgen.1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy SG. Two types of extrajunctional L-glutamate receptors in locust muscle fibres. The Journal of physiology. 1976;255:449–464. doi: 10.1113/jphysiol.1976.sp011289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cull-Candy SG, Usherwood PN. Two populations of L-glutamate receptors on locust muscle fibres. Nat New Biol. 1973;246:62–64. doi: 10.1038/newbio246062a0. [DOI] [PubMed] [Google Scholar]
- Curtis DR, Watkins JC. The excitation and depression of spinal neurones by structurally related amino acids. J Neurochem. 1960;6:117–141. doi: 10.1111/j.1471-4159.1960.tb13458.x. [DOI] [PubMed] [Google Scholar]
- Daniels RW, Gelfand MV, Collins CA, DiAntonio A. Visualizing glutamatergic cell bodies and synapses in Drosophila larval and adult CNS. J Comp Neurol. 2008;508:131–152. doi: 10.1002/cne.21670. [DOI] [PubMed] [Google Scholar]
- Durr KL, Chen L, Stein RA, De Zorzi R, Folea IM, Walz T, McHaourab HS, Gouaux E. Structure and dynamics of AMPA receptor GluA2 in resting, pre-open, and desensitized states. Cell. 2014;158:778–792. doi: 10.1016/j.cell.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans RH, Francis AA, Jones AW, Smith DA, Watkins JC. The effects of a series of omega-phosphonic alpha-carboxylic amino acids on electrically evoked and excitant amino acid-induced responses in isolated spinal cord preparations. Br J Pharmacol. 1982;75:65–75. doi: 10.1111/j.1476-5381.1982.tb08758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa H, Gouaux E. Mechanisms of activation, Inhibition and Specificity: Crystal Structures of NR1 Ligand-Binding Core. EMBO J. 2003;22:1–13. doi: 10.1093/emboj/cdg303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, et al. The developmental transcriptome of Drosophila melanogaster. Nature. 2011;471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han TH, Dharkar P, Mayer ML, Serpe M. Functional reconstitution of Drosophila melanogaster NMJ glutamate receptors. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:6182–6187. doi: 10.1073/pnas.1500458112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Single FN, Köhler M, Sommer B, Sprengel R, Seeburg PH. RNA editing of AMPA receptor subunit GluR-B: a base-paired intron-exon structure determines position and efficiency. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- Hollmann M, Heinemann S. Cloned glutamate receptors. Ann Rev Neurosci. 1994;17:31–108. doi: 10.1146/annurev.ne.17.030194.000335. [DOI] [PubMed] [Google Scholar]
- Hu W, Wang T, Wang X, Han J. Ih channels control feedback regulation from amacrine cells to photoreceptors. PLoS Biol. 2015;13:e1002115. doi: 10.1371/journal.pbio.1002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume RI, Dingledine R, Heinemann SF. Identification of a site in glutamate receptor subunits that controls calcium permeability. Science. 1991;253:1028–1031. doi: 10.1126/science.1653450. [DOI] [PubMed] [Google Scholar]
- Jan LY, Jan YN. L-glutamate as an excitatory transmitter at the Drosophila larval neuromuscular junction. The Journal of physiology. 1976;262:215–236. doi: 10.1113/jphysiol.1976.sp011593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen A, Tajima N, Fernandez-Cuervo G, Garnier-Amblard EC, Furukawa H. Structural insights into competitive antagonism in NMDA receptors. Neuron. 2014;81:366–378. doi: 10.1016/j.neuron.2013.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R, Banke TG, Mayer ML, Traynelis SF, Gouaux E. Structural basis for partial agonist action at ionotropic glutamate receptors. Nature Neurosci. 2003;6:803–810. doi: 10.1038/nn1091. [DOI] [PubMed] [Google Scholar]
- Karuppudurai T, Lin TY, Ting CY, Pursley R, Melnattur KV, Diao F, White BH, Macpherson LJ, Gallio M, Pohida T, Lee CH. A hard-wired glutamatergic circuit pools and relays UV signals to mediate spectral preference in Drosophila. Neuron. 2014;81:603–615. doi: 10.1016/j.neuron.2013.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler M, Burnashev N, Sakmann B, Seeburg PH. Determinants of Ca2+ permeability in both TM1 and TM2 of high affinity kainate receptor channels: diversity by RNA editing. Neuron. 1993;10:491–500. doi: 10.1016/0896-6273(93)90336-p. [DOI] [PubMed] [Google Scholar]
- Krogsgaard-Larsen P, Honore T, Hansen JJ, Curtis DR, Lodge D. New class of glutamate agonist structurally related to ibotenic acid. Nature. 1980;284:64–66. doi: 10.1038/284064a0. [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Molecular biology and evolution. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- Levchenko-Lambert Y, Turetsky DM, Patneau DK. Not all desensitizations are created equal: physiological evidence that AMPA receptor desensitization differs for kainate and glutamate. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2011;31:9359–9367. doi: 10.1523/JNEUROSCI.6761-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littleton JT, Ganetzky B. Ion channels and synaptic organization: analysis of the Drosophila genome. Neuron. 2000;26:35–43. doi: 10.1016/s0896-6273(00)81135-6. [DOI] [PubMed] [Google Scholar]
- Marrus SB, Portman SL, Allen MJ, Moffat KG, DiAntonio A. Differential localization of glutamate receptor subunits at the Drosophila neuromuscular junction. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2004;24:1406–1415. doi: 10.1523/JNEUROSCI.1575-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer ML. Crystal Structures of the GluR5 and GluR6 Ligand Binding Cores: Molecular Mechanisms Underlying Kainate Receptor Selectivity. Neuron. 2005;45:539–552. doi: 10.1016/j.neuron.2005.01.031. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Ghosal A, Dolman NP, Jane DE. Crystal structures of the kainate receptor GluR5 ligand binding core dimer with novel GluR5-selective antagonists. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2006;26:2852–2861. doi: 10.1523/JNEUROSCI.0123-06.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson JR, Kumar J, Chittori S, Rao P, Pierson J, Bartesaghi A, Mayer ML, Subramaniam S. Structural mechanism of glutamate receptor activation and desensitization. Nature. 2014;514:328–334. doi: 10.1038/nature13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S. Molecular diversity of glutamate receptors and implications for brain function. Science. 1992;258:597–603. doi: 10.1126/science.1329206. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Denzo and Scalepack. International Tables for Crystallography. 2001;F:226–235. [Google Scholar]
- Partin KM, Bowie D, Mayer ML. Structural determinants of allosteric regulation in alternatively spliced AMPA receptors. Neuron. 1995;14:833–843. doi: 10.1016/0896-6273(95)90227-9. [DOI] [PubMed] [Google Scholar]
- Paternain AV, Cohen A, Stern-Bach Y, Lerma J. A role for extracellular Na+ in the channel gating of native and recombinant kainate receptors. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2003;23:8641–8648. doi: 10.1523/JNEUROSCI.23-25-08641.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plested AJ, Mayer ML. Structure and mechanism of kainate receptor modulation by anions. Neuron. 2007;53:829–841. doi: 10.1016/j.neuron.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Plested AJ, Vijayan R, Biggin PC, Mayer ML. Molecular basis of kainate receptor modulation by sodium. Neuron. 2008;58:720–735. doi: 10.1016/j.neuron.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Schwarz T, Kittel RJ, Schmid A, Rasse TM, Kappei D, Ponimaskin E, Heckmann M, Sigrist SJ. Four different subunits are essential for expressing the synaptic glutamate receptor at neuromuscular junctions of Drosophila. The Journal of neuroscience: the official journal of the Society for Neuroscience. 2005;25:3209–3218. doi: 10.1523/JNEUROSCI.4194-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008;134:1019–1029. doi: 10.1016/j.cell.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeburg P. The molecular biology of mammalian glutamate receptor channels. Trends Neurosci. 1993;16:359–365. doi: 10.1016/0166-2236(93)90093-2. [DOI] [PubMed] [Google Scholar]
- Soto D, Coombs ID, Kelly L, Farrant M, Cull-Candy SG. Stargazin attenuates intracellular polyamine block of calcium-permeable AMPA receptors. Nat Neurosci. 2007;10:1260–1267. doi: 10.1038/nn1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Laurent G, Tackett MR, Nechkin S, Shtokalo D, Antonets D, Savva YA, Maloney R, Kapranov P, Lawrence CE, Reenan RA. Genome-wide analysis of A-to-I RNA editing by single-molecule sequencing in Drosophila. Nat Struct Mol Biol. 2013;20:1333–1339. doi: 10.1038/nsmb.2675. [DOI] [PubMed] [Google Scholar]
- Sugiyama H, Ito I, Hirono C. A new type of glutamate receptor linked to inositol phospholipid metabolism. Nature. 1987;325:531–533. doi: 10.1038/325531a0. [DOI] [PubMed] [Google Scholar]
- Sun Y, Olson R, Horning M, Armstrong N, Mayer M, Gouaux E. Mechanism of glutamate receptor desensitization. Nature. 2002;417:245–253. doi: 10.1038/417245a. [DOI] [PubMed] [Google Scholar]
- Thomas U, Sigrist SJ. Glutamate receptors in synaptic assembly and plasticity: case studies on fly NMJs. Advances in experimental medicine and biology. 2012;970:3–28. doi: 10.1007/978-3-7091-0932-8_1. [DOI] [PubMed] [Google Scholar]
- Tomita S, Adesnik H, Sekiguchi M, Zhang W, Wada K, Howe JR, Nicoll RA, Bredt DS. Stargazin modulates AMPA receptor gating and trafficking by distinct domains. Nature. 2005;435:1052–1058. doi: 10.1038/nature03624. [DOI] [PubMed] [Google Scholar]
- Traynelis SF, Wollmuth LP, McBain CJ, Menniti FS, Vance KM, Ogden KK, Hansen KB, Yuan H, Myers SJ, Dingledine R. Glutamate receptor ion channels: structure, regulation, and function. Pharmacol Rev. 2010;62:405–496. doi: 10.1124/pr.109.002451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ultsch A, Schuster CM, Laube B, Schloss P, Schmitt B, Betz H. Glutamate receptors of Drosophila melanogaster: cloning of a kainate-selective subunit expressed in the central nervous system. Proc Natl Acad Sci USA. 1992;89:10484–10488. doi: 10.1073/pnas.89.21.10484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KM, Simorowski N, Traynelis SF, Furukawa H. Ligand-specific deactivation time course of GluN1/GluN2D NMDA receptors. Nat Commun. 2011;2:294. doi: 10.1038/ncomms1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CS, Brockie PJ, Madsen DM, Francis MM, Zheng Y, Koduri S, Mellem JE, Strutz-Seebohm N, Maricq AV. Reconstitution of invertebrate glutamate receptor function depends on stargazin-like proteins. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10781–10786. doi: 10.1073/pnas.0604482103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JC, Evans RH. Excitatory amino acid transmitters. Ann Rev Pharmacol Toxicol. 1981;21:165–204. doi: 10.1146/annurev.pa.21.040181.001121. [DOI] [PubMed] [Google Scholar]
- Watkins JC, Krogsgaard-Larsen P, Honoré T. Structure-activity relationships in the development of excitatory amino acid receptor agonists and competitive antagonists. Trends Pharmacol Sci. 1990;11:25–33. doi: 10.1016/0165-6147(90)90038-a. [DOI] [PubMed] [Google Scholar]
- Yamamoto S, Jaiswal M, Charng WL, Gambin T, Karaca E, Mirzaa G, Wiszniewski W, Sandoval H, Haelterman NA, Xiong B, et al. A drosophila genetic resource of mutants to study mechanisms underlying human genetic diseases. Cell. 2014;159:200–214. doi: 10.1016/j.cell.2014.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.