Summary
Central to homologous recombination in eukaryotes is the RAD51 recombinase, which forms helical nucleoprotein filaments on single-stranded DNA (ssDNA) and catalyzes strand invasion with homologous duplex DNA. Various regulatory proteins assist this reaction including the RAD51 paralogs. We recently discovered that a RAD51 paralog complex from C. elegans, RFS-1/RIP-1, functions predominantly downstream of filament assembly by binding and remodeling RAD-51-ssDNA filaments to a conformation more proficient for strand exchange. Here, we demonstrate that RFS-1/RIP-1 acts by shutting down RAD-51 dissociation from ssDNA. Using stopped-flow experiments, we show that RFS-1/RIP-1 confers this dramatic stabilization by capping the 5′ end of RAD-51-ssDNA filaments. Filament end capping propagates a stabilizing effect with a 5′→3′ polarity approximately 40 nucleotides along individual filaments. Finally, we discover that filament capping and stabilization are dependent on nucleotide binding, but not hydrolysis by RFS-1/RIP-1. These data define the mechanism of RAD51 filament remodeling by RAD51 paralogs.
Keywords: homologous recombination, DNA repair, genome stability, Rad51, Rad51 paralogs, filaments
Graphical Abstract
Highlights
-
•
A nematode RAD51 paralog complex binds the 5′ end of RAD51 pre-synaptic filaments
-
•
Filament binding drives pre-synaptic complex remodeling with 5′→3′ polarity
-
•
Pre-synaptic complex remodeling propagates up to 40 nucleotides from the 5′ end
-
•
Filament binding is dependent on a nucleotide co-factor, but not ATP hydrolysis
RAD51 paralogs promote homologous recombination by remodeling RAD51-ssDNA filaments. Here, Taylor et al. demonstrate that RAD51 paralogs mediate remodeling by specifically binding the 5′ end of RAD51 filaments in a nucleotide-dependent manner. This propagates a stabilizing effect with 5′→3′ polarity distal to their binding site, slowing RAD51 dissociation from DNA.
Introduction
Homologous recombination (HR) is a highly conserved mechanism for the repair of DNA double-strand breaks and stalled replication forks. The central reactions of HR are catalyzed by the RAD51 recombinase when it is assembled on single-stranded (ss)DNA as helical RAD-51-ssDNA nucleoprotein filaments, which search for and invade homologous double-stranded (ds)DNA to form joint molecules. Repair DNA synthesis from the 3′ end of the invading strand then copies the correct sequence information from the intact duplex, before resolution of the joint molecules completes repair (Chapman et al., 2012, San Filippo et al., 2008). Failure to efficiently execute HR repair is associated with genome instability and cancer development, as well as the severe congenital disorder Fanconi anemia (Krejci et al., 2012).
HR is positively regulated by various RAD51 accessory factors to ensure its timely completion (San Filippo et al., 2008). HR mediators, such as BRCA2, promote RAD51 filament assembly on ssDNA coated with the ssDNA binding protein RPA (Jensen et al., 2010, Liu et al., 2010, Shahid et al., 2014, Thorslund et al., 2010), while RAD54 promotes RAD51 dissociation from dsDNA after strand invasion (Solinger et al., 2002). These activities ultimately allow these proteins to stimulate strand exchange by RAD51 in vitro (Jensen et al., 2010, Solinger et al., 2002, Thorslund et al., 2010) and promote HR and DNA damage resistance in vivo (Johnson and Jasin, 2001).
Another class of proteins that stimulate HR is the RAD51 paralogs (Chun et al., 2013, French et al., 2002, Johnson et al., 1999, Pierce et al., 1999, Rattray and Symington, 1995, Takata et al., 2001). Although these proteins share significant sequence and/or structural homology with RAD51, they do not exhibit intrinsic recombinase activity, but can stimulate strand exchange by RAD51 (Gaines et al., 2015, Genois et al., 2015, Sigurdsson et al., 2001, Sung, 1997, Taylor et al., 2015). For many years, the mechanism of action of this group of HR regulators was unknown due to the biochemical intractability of recombinant RAD51 paralogs. Recently, we discovered a heterodimeric RAD51 paralog complex from C. elegans, RFS-1/RIP-1, which stimulates the strand exchange activity of C. elegans RAD-51 and promotes its accumulation/stabilization at stalled replication forks (Taylor et al., 2015, Ward et al., 2007). RFS-1/RIP-1 promotes HR by binding to the pre-synaptic filament and converting it to a stabilized conformation in which the ssDNA is more accessible to degradation by nucleases and the overall flexibility of the filament is increased. This represents a previously unknown HR stimulatory mechanism, which we named filament “remodeling”. RFS-1 mutants defective in remodeling fail to stimulate RAD-51 strand invasion activity (Taylor et al., 2015), defining remodeling as a critical mechanism of HR stimulation by RFS-1/RIP-1.
In this study, we sought to better understand the mechanism by which RAD51 paralogs execute RAD51 filament remodeling and stabilization. We present evidence that RFS-1/RIP-1 binds to the 5′ end of individual RAD-51-ssDNA filaments and mediates remodeling in a 5′→3′ direction, in a manner dependent on ATP binding, but not hydrolysis by the complex. This blocks the turnover of RAD-51 from ssDNA, thus stabilizing the pre-synaptic filament in an active state. Together, these data define the mechanism of RAD51 filament remodeling and HR enhancement by RAD51 paralogs.
Results
RFS-1/RIP-1 Shuts down RAD-51 Dissociation from ssDNA
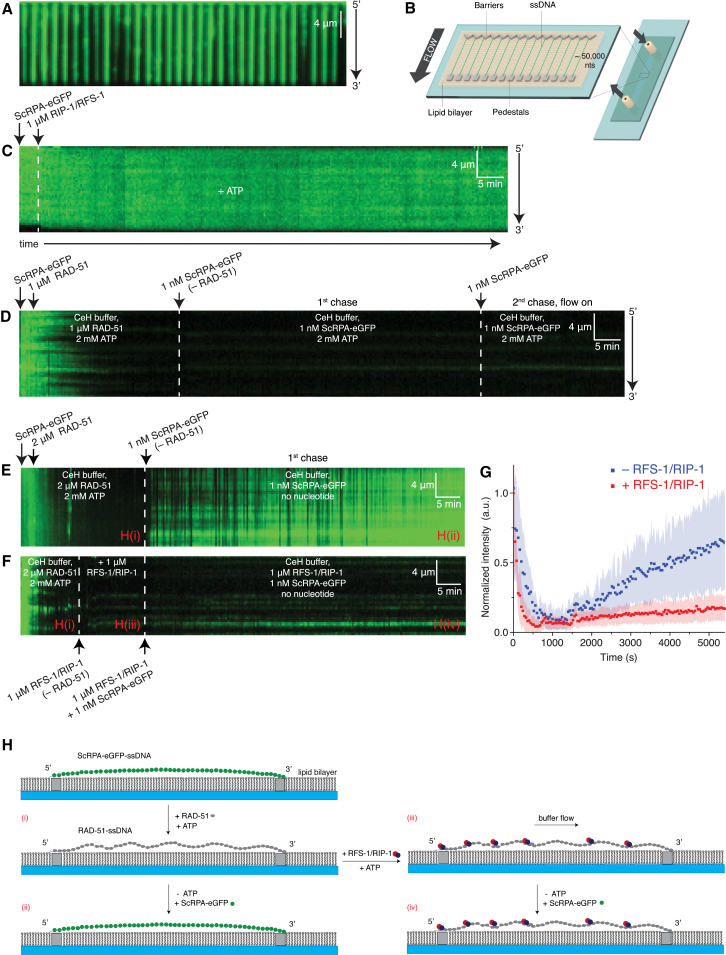
To investigate the mechanism by which RAD51 paralogs stabilize pre-synaptic filaments, we monitored the stability of RAD-51 filaments assembled on long (approximately 50 kb) ssDNA curtains using the rebinding of fluorescently labeled yeast RPA to naked ssDNA (ScRPA-eGFP, hereafter abbreviated to RPA) (Figures 1A and 1B) (Gibb et al., 2014a, Qi et al., 2015). RPA binds ssDNA with high affinity (Wold, 1997), and a molar of excess RFS-1/RIP-1 (1 μM) was unable to outcompete RPA binding to ssDNA curtains (Figure 1C), consistent with the low ssDNA affinity of RFS-1/RIP-1 (Taylor et al., 2015). Like yeast Rad51 (Qi et al., 2015), 1 μM RAD-51 effectively displaced RPA from the ssDNA, as observed by the loss of eGFP fluorescence signal from ssDNA curtains (Figure 1D). The rate of RPA displacement was increased at 2 μM RAD-51 (Figure 1E), which was used for subsequent experiments to ensure rapid and complete filament formation. Similar to yeast Rad51 (Gibb et al., 2014b, Qi et al., 2015), RAD-51-ssDNA filaments remained stable when free RAD-51 was removed and replaced by RPA as long as ATP was maintained in the buffer (Figures 1E and 1F). In contrast, transfer into buffer without nucleotide co-factors drove the rapid disassembly of RAD-51 filaments and re-establishment of the fluorescent RPA signal (Figures 1E and 1G).
Figure 1.
RFS-1/RIP-1 Shuts Down RAD-51-ssDNA Filament Dissociation
(A) Wide-field image of extended ScRPA-eGFP-ssDNA filaments, each anchored at both ends.
(B) Schematic depiction of (A) showing underlying ssDNA curtains anchored to the flow cell.
(C) Kymogram of a single filament over time in the absence of buffer flow. The ScRPA-eGFP complex is unperturbed by 1 μM RIP-1/RFS-1 (pre-incubated with 2 mM ATP for 10 min), as evident by the persistent GFP signal.
(D) RAD-51 is protected from replacement by ScRPA-eGFP in the presence of ATP both with and without flow, as indicated by the lack of ScRPA-eGFP binding.
(E) After RAD-51 filament assembly, free protein and ATP were washed out of the solution and the filament was allowed to disassemble in buffer lacking nucleotide and containing 1 nM ScRPA-eGFP, demonstrating the dependence of RAD-51 on ATP for stability. ScRPA-eGFP binding serves as a proxy for RAD51 dissociation.
(F) RIP-1/RFS-1 (pre-incubated with 2 mM ATP for 10 min) is able to bypass the ATP requirement for RAD-51 filament stability. The RIP-1/RFS-1 is first incubated with the RAD-51 filament in the presence of ATP for 10 min before ATP was washed away from the flow cell. The flow cell was chased with RFS-1/RIP-1 and 1 nM ScRPA-eGFP to monitor for disassembly.
(G) Quantitation of the intensity of the ScRPA-eGFP signal over the time course of the experiment for the data in (E) and (F).
(H) Schematic depiction of RAD-51-ssDNA filament stabilization by RFS-1/RIP-1 in this assay. The images labeled (i) –(iv) represent the indicated observations in (E) and (F).
Next, RAD-51 filaments formed in the presence of ATP were incubated with 1 μM RFS-1/RIP-1, while maintaining ATP in the buffer. Buffer with RPA, but without ATP, was then flowed onto the curtains while maintaining 1 μM RFS-1/RIP-1 in free solution to maintain its association from filaments. In contrast to RAD-51 alone, we observed a striking stabilization of the filaments induced by RFS-1/RIP-1, which persisted for over an hour (Figures 1F and 1G). This result demonstrates that RFS-1/RIP-1 shuts down RAD-51 turnover from ssDNA even when ATP is removed from the buffer (Figure 1H). This also suggests that RFS-1/RIP-1-mediated filament remodeling locks RAD-51 onto ssDNA by reducing its dissociation rate (koff). We next sought to determine the mechanism of action of RFS-1/RIP-1 in filament stabilization.
RFS-1/RIP-1 Binds the 5′ Ends of RAD-51-ssDNA Filaments
We previously employed immuno-gold labeling with electron microscopy to show that RFS-1/RIP-1 preferentially binds to the ends of RAD-51-ssDNA filaments compared to the filament body, which was only ever observed at one end and never both (Taylor et al., 2015). ssDNA molecules are intrinsically 5′→3′ polar with respect to the sugar-phosphate backbone, and crystal structures of the yeast Rad51 (Conway et al., 2004) and bacterial RecA (homolog of Rad51) (Chen et al., 2008) filaments assembled on ssDNA have also revealed a structural polarity in the filament with respect to the underlying DNA. We therefore considered the possibility that RFS-1/RIP-1 recognizes the intrinsic polarity of the ssDNA and/or RAD-51-ssDNA filament and binds at an interface exposed on one end, but not the other.
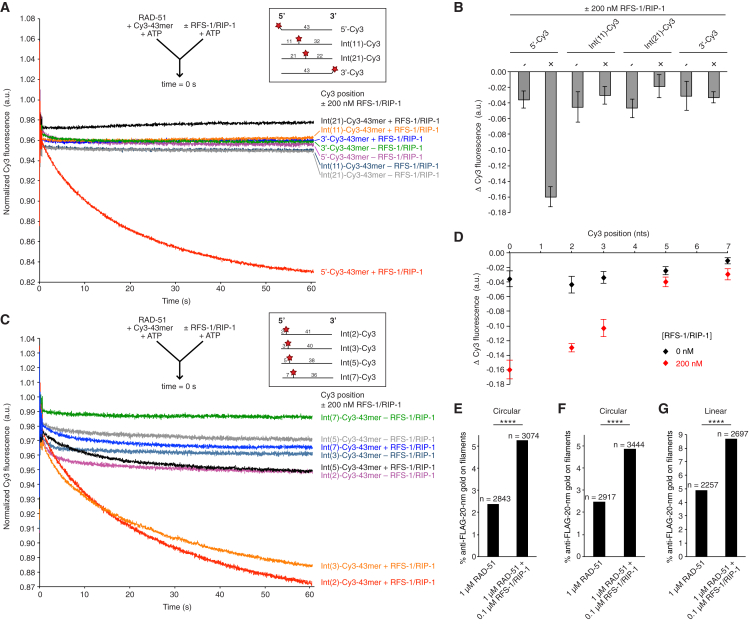
Intriguingly, RFS-1/RIP-1 drives a reduction in the fluorescence of RAD-51-ssDNA filaments formed on a 5′-Cy3-labeled 43-mer oligonucleotide in a stopped-flow system, representing an alteration in the biophysical properties of the RAD-51-ssDNA filament (Taylor et al., 2015). We reasoned this fluorescence reduction may reflect the binding of RFS-1/RIP-1 at the 5′ end, and that if the end binding observed in electron microscopy was random with respect to the underlying DNA polarity, then an equivalent fluorescence reduction should be observed on an oligonucleotide Cy3-labeled at the 3′ end. However, mixing RFS-1/RIP-1 with RAD-51-ssDNA filaments formed on DNA labeled with Cy3 at the 3′ end in stopped flow conferred no such reduction in fluorescence (Figures 2A and 2B), suggesting RFS-1/RIP-1 specifically binds the 5′ end. We also monitored the effect of RFS-1/RIP-1 on the fluorescence of RAD-51 filaments formed on internally Cy3-labeled oligonucleotides, in which the Cy3 was placed after the 11th or 22nd nucleotide (Int(11)-Cy3 and Int(22)-Cy3, respectively) from the 5′ end. The fluorescence of these filaments was also unaffected by RFS-1/RIP-1 (Figures 2A and 2B), confirming that RFS-1/RIP-1 is likely to predominantly engage with the filament end compared with the filament body. We verified that all four oligonucleotides exhibit an increase in Cy3 fluorescence upon mixing with RAD-51 in stopped flow, corresponding to RAD-51 nucleation on DNA (Figures S1A and S1B), and directly confirmed RAD-51 filament formation occurred with similar efficiency on all four oligonucleotides by electrophoretic mobility shift assay (EMSA) (Figure S1D). Intriguingly, we observed that the rate of ssDNA binding was reduced for RAD-51 binding near the 3′ label, but was faster on the other three constructs (Figure S1C). These data are consistent with RAD-51 filament extension displaying either a unidirectional polarity for the 5′→3′ direction or bidirectional extension that occurs faster in the 3′→5′ direction than 5′→3′. Notably, similar differences in the rate of fluorescence increase when monitoring 5′ and 3′ Cy3 labels have also been observed for the assembly of yeast Rad51 on DNA in ensemble stopped-flow (Antony et al., 2009) and single molecule protein-induced fluorescence enhancement (PIFE) studies (Hwang and Myong, 2014, Qiu et al., 2013). Our results therefore support a conserved preference in directionality of filament extension on naked ssDNA between yeast Rad51 and nematode RAD-51. However, the kinetic profiles of filament formation for both proteins display several phases suggesting further complexities in this process (Antony et al., 2009).
Figure 2.
RFS-1/RIP-1 Binds the 5′ End of Individual RAD-51-ssDNA Filaments
(A and C) Average normalized Cy3-43-mer fluorescence profiles plotted as a function of time. The arrow indicates the components of the two syringes rapidly mixed at the 0 s time point in a stopped-flow instrument. The schematics of the different Cy3 label positions are shown in the inset. The RAD-51-ssDNA filaments pre-formed with 1 μM RAD-51 + 15 nM Cy3-43-mer for 10 min were mixed with buffer ± 200 nM RFS-1/RIP-1. 5′-, Int(11)-, Int(22)-, and 3′-Cy3 constructs (n = 5–9) (A). Int(2)-, Int(3), Int(5)-, and Int(7)-Cy3 constructs (n = 5–8) (C).
(B) Graph of average Δ Cy3 fluorescence for the data presented in (A) (the error bars represent SD).
(D) Graph of Cy3 position-dependence of Δ Cy3 fluorescence in the presence of RFS-1/RIP-1 for the data presented in (A) (5′-Cy3) and (C) (other constructs) (the error bars represent SD).
(E) Quantification of % anti-FLAG-20-nm gold particle binding to RAD-51-ssDNA filaments ± RFS-1/RIP-1 formed on circular ssDNA.
(F) Quantification of an independent experimental repeat for the same experiment in (E).
(G) Quantification of % anti-FLAG-20-nm gold particle binding to RAD-51-ssDNA filaments ± RFS-1/RIP-1 formed on linearized ssDNA. ∗∗∗∗Two-tailed Chi-square test of independence, p < 0.0001. See also Figure S1.
The reduction in fluorescence induced by RFS-1/RIP-1 on RAD-51 filaments pre-formed on 5′ Cy3-labeled ssDNA could represent binding of RFS-1/RIP-1 to the 5′ end or the conformational change in the remodeled filament (Taylor et al., 2015). To investigate this, we first expanded the tested range of labeled oligonucleotides to those in which the Cy3 was placed after the second, third, fifth, or seventh nucleotide (Int(2)-Cy3, Int(3)-Cy3, Int(5)-Cy3, and Int(7)-Cy3, respectively) in the same stopped-flow experimental setup. We observed a clear length dependency in the magnitude of fluorescence reduction conferred by RFS-1/RIP-1 (Figures 2C and 2D). Significantly, there was a linear relationship between magnitude of fluorescence reduction and label position for the Cy3 labels between 0–5 nucleotides from the 5′ end (Figure 2D). This is consistent with the fluorescence reduction reflecting RFS-1/RIP-1 binding at the 5′ end, such that increasing distances away from the 5′ end renders the fluorophore less sensitive to changes in the environment induced by RFS-1/RIP-1 binding.
We previously demonstrated in stopped-flow experiments that at equilibrium, the final fluorescence of RAD-51-ssDNA in the presence of RFS-1/RIP-1 is intermediate to naked 5′ Cy3-43-mer ssDNA and RAD-51-ssDNA alone (Taylor et al., 2015). The mechanism of fluorescence reduction due to RFS-1/RIP-1 binding to the 5′ end of the filament may represent a modulation of the efficiency of RAD-51-induced PIFE, such that the overall equilibrium PIFE of 5′ Cy3-labeled ssDNA for the RAD-51/RFS-1/RIP-1 complex is lower than that for RAD-51 alone. Recent evidence suggests that Cy3 PIFE arises due to decreased photoisomerization of the dye to the non-fluorescent cis when sterically constrained by protein (Stennett et al., 2015). Indeed, our single molecule fluorescence resonance energy transfer (FRET) studies also established that RFS-1/RIP-1 converts RAD-51-ssDNA filaments to a more flexible conformation (Taylor et al., 2015). As such, the RFS-1/RIP-1-bound filament is expected to be less sterically constrained nearby the 5′ Cy3 dye, accounting for rapider photoisomerization and reduced PIFE. Furthermore, PIFE exhibits a strong and linear sensitivity to distance between the protein binding site and label, with significant effects induced in the 0–30 Å range (Hwang et al., 2011, Hwang and Myong, 2014). Given that the average rise per nucleotide along the axis of ssDNA within the RAD51 filaments is 5.1 Å (Yu et al., 2001), one would predict modulation of RAD-51 PIFE due to RFS-1/RIP-1 filament association at the 5′ end should be detectable up to 5–6 nucleotides (25.5–30.6 Å) from the 5′ end, but no further and decline linearly with distance, which is in very good agreement with our experimental data (Figure 2D). Notably, the ssDNA conformation in RecA-ssDNA filaments is non-uniform: for any given triplet, the first two nucleotides exhibit a rise similar to B-DNA, but the third is greatly stretched (Chen et al., 2008). This might be expected to result in a non-linear dependency between PIFE modulation and distance from the RFS-1/RIP-1 binding site within the 0–30 Å range. However, the register of one filament relative to another is likely to be random and therefore in the population of filaments monitored in stopped-flow measurements, this nuance is lost due to averaging.
RFS-1/RIP-1 Recognizes an Interface Exposed at the 5′ End of the Filament, but Not the 5′ End of the Underlying ssDNA
We next examined if RFS-1/RIP-1 binding to the 5′ end of the filament requires a free 5′ DNA end or if the filament end defined by the terminal RAD-51 protomer is sufficient. To test this, we analyzed RFS-1/RIP-1 binding to RAD-51 filaments formed on circular ssDNA, which lacks DNA ends, by immuno-gold labeling and electron microscopy. Filaments formed on circular ssDNA tended to clump together more on the grid (Figure S1E) compared to filaments on linear ssDNA (Figure S1F), consistent with the constrained path of closed ssDNA molecules. Using anti-FLAG-20-nm gold conjugates directed against the FLAG tag on RIP-1, we observed RFS-1/RIP-1 binding to filaments assembled on both linear and circular ssDNA (Figures 2E–2G and S1E–S1J). This suggests RFS-1/RIP-1 primarily recognizes the 5′ filament end rather than the underlying 5′ DNA end. Furthermore, the ssDNA curtains used to demonstrate RAD-51-ssDNA filament stabilization by RFS-1/RIP-1 (Figure 1) are anchored to the flow cell at both ends and are therefore effectively endless. Together, these observations suggest that the 5′ end of ssDNA is not a major determinant of the structure recognized by RFS-1/RIP-1. In addition, the likely substrate for RFS-1/RIP-1 in vivo is a RAD-51 filament loaded at a ssDNA gap or 3′ overhang ssDNA at resected double strand breaks, which lack naked 5′ ssDNA ends.
RFS-1/RIP-1 Propagates Remodeling and Stabilization of RAD-51-ssDNA Filaments beyond Immediate Proximity to the 5′ End
Rather than representing 5′ filament end binding, an alternative interpretation of the stopped-flow data in Figure 2 is that the fluorescence reduction induced by RFS-1/RIP-1 reflects the altered conformation of the remodeled RAD-51-ssDNA filament, with remodeling restricted to a short section of the filament extending 5–7 nucleotides from the 5′ end. We also previously demonstrated DNaseI sensitization by RFS-1/RIP-1 on filaments formed on 5′ fluorescently labeled oligonucleotides (Taylor et al., 2015), which could formally represent a localized conformational change at the 5′ filament end, since only cleavage products retaining the 5′ fluorescent label are detectable. Similarly, we previously only tested filament stabilization by RFS-1/RIP-1 using 5′ labeled oligonucleotides.
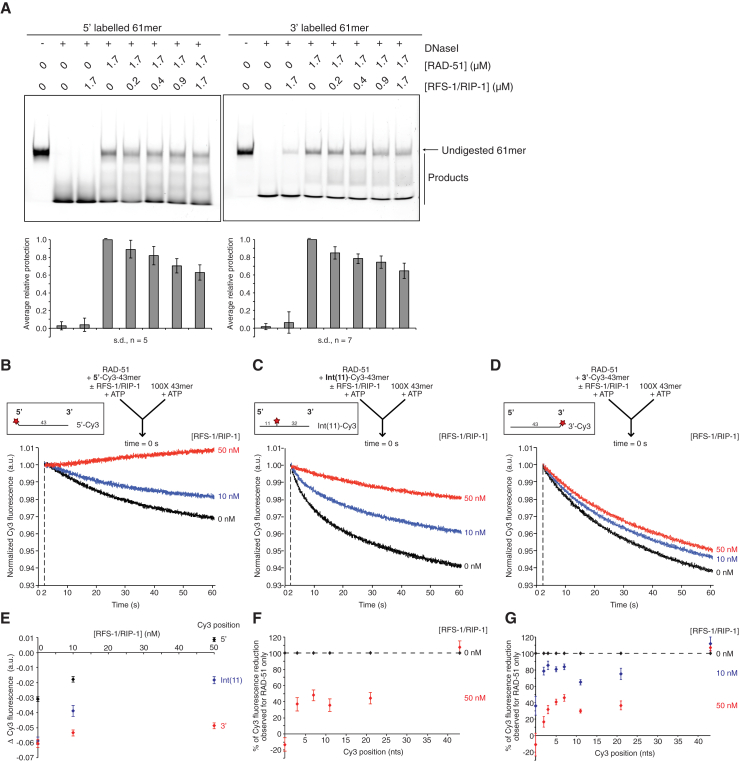
To distinguish between these possibilities, we sought to determine if these properties of remodeling propagate beyond this restricted region close to the 5′ filament end. We first performed nuclease protection assays on a 61-mer substrate fluorescently labeled at either the 5′ or 3′ end (Figure 3A). Interestingly, in the presence of RAD-51, both substrates were protected to a similar extent (Figure 3A), but the pattern of degradation products was different (Figure S2). Longer degradation products were enriched for the 5′ construct and shorter products enriched for the 3′ construct, suggesting DNaseI exhibits a degree of sequence preference in its cleavage of ssDNA, as for dsDNA (Herrera and Chaires, 1994), and that cleavage events nearer the 3′ end of this oligonucleotide predominate. Importantly, in the presence of RFS-1/RIP-1, ssDNA degradation and formation of all cleavage products, regardless of length, was enhanced on both constructs (Figures 3A and S2). This suggests that the DNaseI sensitization induced by RFS-1/RIP-1 reflects filament remodeling, rather than 5′ end binding, since the effect also propagates along the filament to cleavage sites closer to the 3′ end than the 5′ end.
Figure 3.
RFS-1/RIP-1 Induces Remodeling throughout RAD-51-ssDNA Filaments a Significant Distance beyond the 5′ End
(A) DNaseI protection assay on protein-DNA complexes formed by RAD-51 and RFS-1/RIP-1 on 5′ or 3′ fluorescently labeled 61-mer ssDNA. The products were resolved by native PAGE. The average extent of protection relative to RAD-51 only samples is quantified.
(B–D) Average normalized Cy3-43-mer fluorescence profiles plotted as a function of time. The arrow indicates the components of the two syringes rapidly mixed at the 0 s time point in a stopped-flow instrument. The RAD-51-ssDNA filaments pre-formed with 1 μM RAD-51 + 15 nM Cy3-43-mer and indicated concentrations of RFS-1/RIP-1 for 10 min were mixed with 100-fold excess unlabeled 43-mer (n = 9–11). 5′-Cy3 construct (B); Int(11)-Cy3 construct (C); and 3′-Cy3 construct (D).
(E) Graph of RFS-1/RIP-1 concentration-dependence of Δ Cy3 fluorescence for the data presented in (B)–(D).
(F) Graphs of Δ Cy3 fluorescence in the presence of RFS-1/RIP-1 as a % of Δ Cy3 fluorescence for RAD-51 only samples on constructs Cy3-labeled 5′, 3′, 3, 7, 11, or 21 nt from the 5′ end for the data presented in Figures S3A–S3F (n = 6–8; the error bars represent SD).
(G) Graphs of Δ Cy3 fluorescence in the presence of RFS-1/RIP-1 as a % of Δ Cy3 fluorescence for RAD-51 only samples on constructs Cy3-labeled 5′, 3′, 2, 3, 5, 7, 11 or 21 nt from the 5′ end for the data presented in Figures S3G-S3N (n = 6–8; the error bars represent SD).
See also Figures S2 and S3.
Second, we tested for propagation of RAD-51 filament stabilization. Initially, we compared the stability of filaments formed on the 5′-Cy3, Int(11)-Cy3, and 3′-Cy3 constructs used in Figure 2 to unlabeled competitor DNA (Figures 3B–3E). In the absence of RFS-1/RIP-1, the magnitude of fluorescence reduction induced by mixing RAD-51 filaments with 100-fold excess of unlabeled competitor DNA corresponding to RAD-51 dissociation was smallest for the 5′ Cy3 labeled DNA (Figure 3B) and larger, but roughly equivalent, for the Int(11)-Cy3 and 3′-Cy3 constructs (Figures 3C and 3D). This observation is consistent with the fact that the fluorophores in these positions fluoresce more intensely upon RAD-51 binding than Cy3 at the 5′ position (Figures S1A and S1B) and therefore would be expected to exhibit a larger fluorescence reduction to return to the intensity associated with naked DNA.
In the presence of RFS-1/RIP-1, strong, concentration-dependent stabilization against filament disruption by competitor DNA was observed for the 5′-Cy3 construct, with 50 nM RFS-1/RIP-1 sufficient to almost completely stabilize the filament as previously reported (Figure 3B) (Taylor et al., 2015). However, no significant stabilization was observed when monitoring the 3′ Cy3-labeled oligonucleotide (Figure 3D). In contrast, the Int(11)-Cy3 oligonucleotide exhibited a clear, concentration-dependent stabilization, but the extent of stabilization at both concentrations of RFS-1/RIP-1 was smaller than for the 5′-Cy3 construct (Figure 3C). Given that RFS-1/RIP-1 did not induce a fluorescence reduction in filaments formed on the Int(11)-Cy3 construct (Figure 2A), these data support the view that the fluorescence reduction reported in Figure 2 represents the binding of RFS-1/RIP-1 to the 5′ end, which only induces a change in the biophysical properties of the Cy3 label when it is in close proximity (approximately 5–7 nucleotides) to the 5′ end and not the remodeling of the filament. However, RFS-1/RIP-1 induces a remodeled conformation that propagates along the filament at least as far as 11 nucleotides from the 5′ end, since the filament exhibits partial stabilization at this position.
These results raised the possibility that filament remodeling and stabilization could be propagated through the filament from the 5′ end with a 5′→3′ polarity following RFS-1/RIP-1 binding, such that RAD-51 protomers nearer the 5′ end are more significantly stabilized. This model is supported by the fact that the magnitude of stabilization observed on the Int(11)-Cy3 construct is intermediate to that on the 5′-Cy3 construct, whereas the 3′-Cy3 construct exhibited no stabilization (Figure 3E). To monitor possible stabilization propagation further, we expanded the competition experiments to the other internally labeled constructs used in Figure 2. We also quantified the extent of stabilization observed with RFS-1/RIP-1 relative to RAD-51 alone (Figures 3F and 3G), rather than analyzing the absolute fluorescence reduction (Figure 3E). This normalization accounts for the inherent difference in the magnitude of Cy3 fluorescence changes associated with RAD-51 binding (Figures S1A and S1B) and dissociation (Figures 3B–3D and S3) from the different constructs, allowing exclusive analysis of the position-dependency of stabilization with respect to 5′ filament end binding of RFS-1/RIP-1. If the 5′ end propagation model is true, we predicted that there would be a direct relationship between Cy3 label position and the extent of filament stabilization by a fixed concentration of RFS-1/RIP-1, analogous to that observed in Figure 2D.
Strikingly, in two independent experiments (Figures 3F, 3G, and S3), we failed to detect a strong correlation between the extent of filament stabilization and Cy3 label position within the first 21 nucleotides. As before, 50 nM RFS-1/RIP-1 was sufficient to completely stabilize the filament when monitoring the 5′ end, whereas no stabilization was observed at the 3′ end. In contrast, all internal labels tested showed a concentration-dependent (Figures 3G and S3G–S3N), but intermediate stabilization effect, even as close as two nucleotides from the 5′ end (Figures 3F and 3G). These results may be explained by a model in which RFS-1/RIP-1 exhibits two distinct modes of filament stabilization. First, RFS-1/RIP-1 binding to the 5′ end may sterically impair RAD-51 dissociation from the DNA entirely, accounting for the dramatic stabilization exhibited by 50 nM RFS-1/RIP-1 at the 5′ end. Second, RFS-1/RIP-1-induced filament remodeling may also alter the biophysical properties of the filament, which slows the rate of RAD-51 dissociation from the DNA internally, and this propagates along the filament at least as far as 21 nucleotides from the 5′ end. This would account for the intermediate stabilization effect exhibited at Cy3 label positions from 2–21 nucleotides from the 5′ end.
RFS-1/RIP-1 Propagates Filament Stabilization from the 5′ End with 5′→3′ Polarity
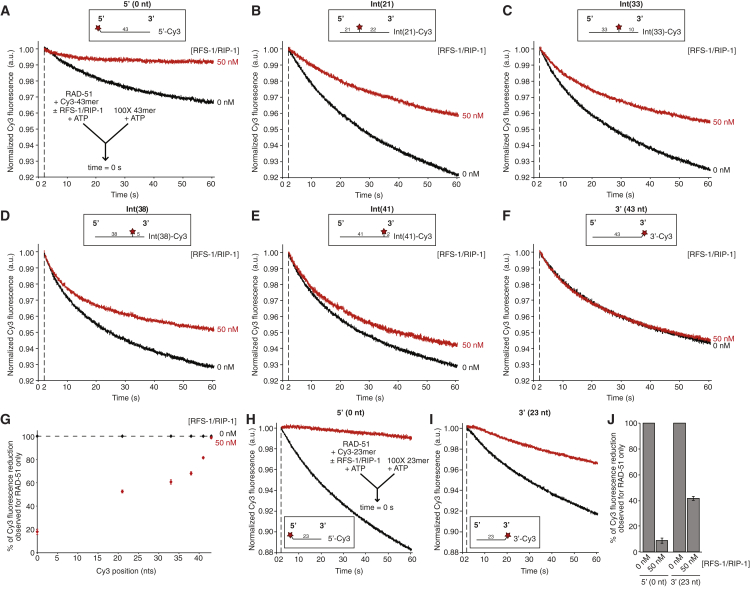
To further test the 5′ end propagation model, we tested if stabilization declined between 21 nucleotides and the 3′ end by expanding the range of oligonucleotides analyzed to include those labeled after the 33rd, 38th, or 41st nucleotide (Int(33)-Cy3, Int(38)-Cy3, and Int(41)-Cy3, respectively), as well as the 5′-Cy3, Int(21)-Cy3 and 3′-Cy3 constructs (Figures 4A–4G). We compared the stability of RAD-51 in the presence or absence of 50 nM RFS-1/RIP-1 on these constructs and observed a profound length-dependent decline in the magnitude of stabilization (Figure 4G). Specifically, a similar magnitude of stabilization was observed on the Int(21)-Cy3 construct in the presence of RFS-1/RIP-1 to previous experiments (Figures 3F, 3G, S3E, S3M, 4B, and 4G), with a fluorescence reduction of approximately 40%–50% of that for RAD-51 only. As the Cy3 dye position was moved toward the 3′ end, this fluorescence reduction relative to RAD-51 increased progressively (Int(33)-Cy3: 61%; Int(38)-Cy3: 68%; Int(41)-Cy3: 82%; and 3′-Cy3 [43 nucleotides]: 99%). Therefore, the cutoff point where significant stabilization was no longer detectable occurs approximately 40 nucleotides from the 5′ end (Figure 4G). These results demonstrate filament stabilization propagates a significant distance through the length of the filament from the 5′ end with 5′→3′ polarity.
Figure 4.
RFS-1/RIP-1 Binding Propagates a Stabilizing Effect along RAD-51-ssDNA Filaments with 5′→3′ Polarity
(A–F) Average normalized Cy3-43-mer fluorescence profiles plotted as a function of time. The RAD-51-ssDNA filaments pre-formed with 1 μM RAD-51 + 15 nM Cy3-43-mer ± 50 nM RFS-1/RIP-1 for 10 min were mixed with 100-fold excess unlabeled 43-mer. The label position is indicated at the top center of each profile. The data were pooled from three independent experiments (n = 19–22). (A) 5′-Cy3 construct. (B) Int(21)-Cy3 construct. (C) Int(33)-Cy3 construct. (D) Int(38)-Cy3 construct. (E) Int(41)-Cy3 construct. (F) 3′-Cy3 construct.
(G) Graph of Cy3 position-dependence of Δ Cy3 fluorescence in the presence of RFS-1/RIP-1 as a % of Δ Cy3 fluorescence for RAD-51 only samples for the data presented in (A)–(F) (the error bars represent SEM).
(H and I) Average normalized Cy3-23-mer fluorescence profiles plotted as a function of time. The RAD-51-ssDNA filaments were pre-formed with 750 nM RAD-51 + 15 nM Cy3-23-mer ± 50 nM RFS-1/RIP-1 for 10 min, then mixed with 100-fold excess unlabeled 23-mer. The label position is indicated at the top center of each profile. The data were pooled from two independent experiments (n = 12–16). (H) 5′-Cy3 construct. (I) 3′-Cy3 construct.
(J) Graph of Δ Cy3 fluorescence in the presence of RFS-1/RIP-1 as a % of Δ Cy3 fluorescence for RAD-51 only samples for the data presented in (H) and (I) (the error bars represent SEM).
Next, we wished to verify that the failure of RFS-1/RIP-1 to stabilize the filament at a 3′ end and confer weak stabilization at the Int(38)-Cy3 and Int(41)-Cy3 labels was due to termination of remodeling propagation rather than the influence of possible additional biophysical properties of the 3′ filament end that could drive faster RAD-51 turnover in this locality. To this end, we monitored filament stability on 5′ and 3′ Cy3-labeled 23-mer oligo(dT) constructs (Figures 4H–4J). As previously reported, RFS-1/RIP-1 almost completely stabilizes the 5′ end of this construct (Figure 4H) (Taylor et al., 2015). Notably, the same concentration (50 nM) of RFS-1/RIP-1 that was used in the above experiments on 43-mer constructs was also able to induce a striking stabilization at the 3′ end of the 23-mer construct (Figure 4I). The magnitude of fluorescence reduction relative to RAD-51 alone was 41.9% ± 1.4% (Figure 4J), which is comparable to that observed on the Int(21)-Cy3 43-mer construct (Figures 3F, 3G, and 4G), which is labeled at a similar position downstream of the 5′ end. Notably, this is in complete contrast to the effect observed at the 3′ end of the 43-mer (Figures 3D–3G, S3F, S3N, 4F, and 4G). This result reinforces the above evidence that stabilization propagates approximately 40 nucleotides and is not antagonized by unique properties of the 3′ filament end.
Together, these data demonstrate that RFS-1/RIP-1 binding at the 5′ filament end is able to induce a conformational change in the filament beyond the locality of its binding site, which manifests in DNaseI sensitization and stabilization against competitor DNA. The stopped-flow stability assays reveal in high resolution the magnitude and 5′→3′ polarity of the propagation of remodeling.
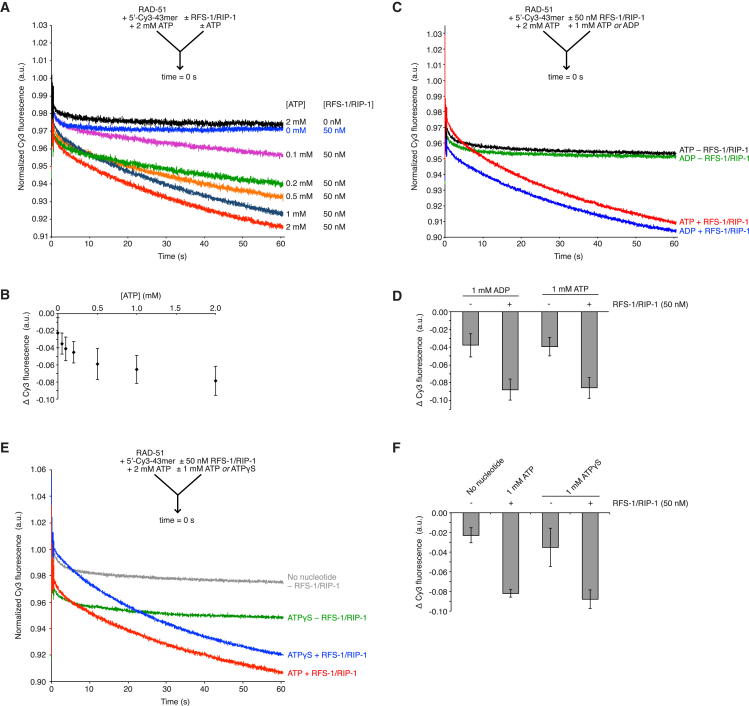
RFS-1/RIP-1 5′ End Filament Binding Is Dependent on Nucleotide Binding, but Not Hydrolysis
RFS-1/RIP-1 contains ATP binding Walker motifs (Walker et al., 1982), and the Walker A and B boxes of RFS-1 are required for RAD-51-ssDNA filament remodeling and stimulation of strand exchange activity (Taylor et al., 2015). We considered the possibility that a defect in ATP binding or hydrolysis underlies the failure of these mutants to mediate filament remodeling. To test this, we monitored the ability of wild-type RFS-1/RIP-1 pre-incubated with different concentrations of ATP to induce a fluorescence reduction in RAD-51 filaments pre-formed on 5′ Cy3 labeled ssDNA in the presence of 2 mM ATP. Strikingly, we observed a strong concentration-dependent effect of ATP on the magnitude of fluorescence reduction, with no fluorescence reduction seen when RFS-1/RIP-1 was not pre-incubated with ATP (Figures 5A and 5B), suggesting RFS-1/RIP-1 must be nucleotide-bound to associate with RAD-51-ssDNA filaments.
Figure 5.
Binding of RAD-51-ssDNA Filaments Requires ATP Binding, but Not Hydrolysis by RFS-1/RIP-1
(A) Average normalized Cy3-43-mer fluorescence profiles plotted as a function of time. The arrow indicates the components of the two syringes rapidly mixed at the 0 s time point in a stopped-flow instrument. The RAD-51-ssDNA filaments pre-formed with 1 μM RAD-51 + 15 nM Cy3-43-mer for 10 min were mixed with buffer with the indicated concentration of ATP ± 50 nM RFS-1/RIP-1 (n = 7–9).
(B) Graph of ATP-dependence of Δ Cy3 fluorescence in the presence of RFS-1/RIP-1 for the data presented in (A) (the error bars represent SD).
(C) Average normalized Cy3-43-mer fluorescence profiles plotted as a function of time. The RAD-51-ssDNA filaments pre-formed with 1 μM RAD-51 + 15 nM Cy3-43-mer for 10 min were mixed with buffer with the indicated nucleotide ± 50 nM RFS-1/RIP-1 (n = 8–9).
(D) Graph of average Δ Cy3 fluorescence for the data presented in (C) (the error bars represent SD).
(E) Average normalized Cy3-43-mer fluorescence profiles plotted as a function of time. The RAD-51-ssDNA filaments pre-formed with 1 μM RAD-51 + 15 nM Cy3-43-mer for 10 min were mixed with buffer with the indicated nucleotide ± 50 nM RFS-1/RIP-1 (n = 4–8).
(F) Graph of average Δ Cy3 fluorescence for the data presented in (E) (the error bars represent SD).
See also Figure S4.
The lack of response of RFS-1/RIP-1 not pre-incubated with ATP in this assay is surprising since RFS-1/RIP-1 encounters 1 mM ATP contributed from the RAD-51-ssDNA filament syringe upon mixing in stopped flow. In addition, the ATP concentration dependency of the filament response to RFS-1/RIP-1 occurs at relatively high ATP concentrations. To test if RFS-1/RIP-1 binds ATP weakly and slowly, we compared RAD-51 and RFS-1/RIP-1 binding to a fixed concentration of the fluorescent ATP analog TNP-ATP. RAD-51 has a high affinity for TNP-ATP (KD ∼3.5 μM), whereas RFS-1/RIP-1 exhibited a weak response and failed to achieve saturation within the testable concentration range (Figures S4A and S4B). We also monitored RAD-51 and RFS-1/RIP-1 binding to 0.5 μM MANT-ATP, an ATP analog that becomes fluorescent upon binding to protein, by stopped flow. Both proteins exhibited a concentration-dependent binding of MANT-ATP to the protein in the testable range (0–8 μM), and RAD-51 exhibited a much stronger MANT-ATP binding response than RFS-1/RIP-1 (Figure S4E). RAD-51 also binds MANT-ATP rapidly, causing a strong increase in fluorescence (Figure S4C), whereas RFS-1/RIP-1 binds much more slowly, causing a very small fluorescence response before the onset of MANT-ATP photobleaching (Figure S4D).
Together, these results indicate that RFS-1/RIP-1 has a low nucleotide affinity, which is in part due to a slow nucleotide binding rate (kon). Therefore, when RFS-1/RIP-1 mixes with 1 mM ATP from the RAD-51-ssDNA syringe (Figure 5A), its slow and weak ATP binding does not allow sufficient active complex to assemble within the 60 s timescale of the stopped-flow experiment to undergo detectable filament binding. We also validated that RFS-1/RIP-1 does not become inactivated by prolonged incubation in the absence of nucleotide, since the filament binding response is rescued by introducing ATP to the RFS-1/RIP-1 syringe after initial pre-incubation in the absence of nucleotide (Figures S4F and S4G).
To determine the nucleotide requirements of RFS-1/RIP-1 activity, we performed similar experiments in which RFS-1/RIP-1 was pre-incubated with ADP (Figures 5C and 5D), the product of ATP hydrolysis, or ATPγS (Figures 5E and 5F), a non-hydrolysable ATP analog. Both nucleotides induced an equivalent magnitude of fluorescence reduction in the stopped-flow assay compared to that observed in the presence of ATP (Δ Cy3 fluorescence = −0.08 to −0.09, 50 nM RFS-1/RIP-1) (Figures 5D and 5F), suggesting that RFS-1/RIP-1 requires nucleotide binding, but not hydrolysis for normal 5′ filament end association.
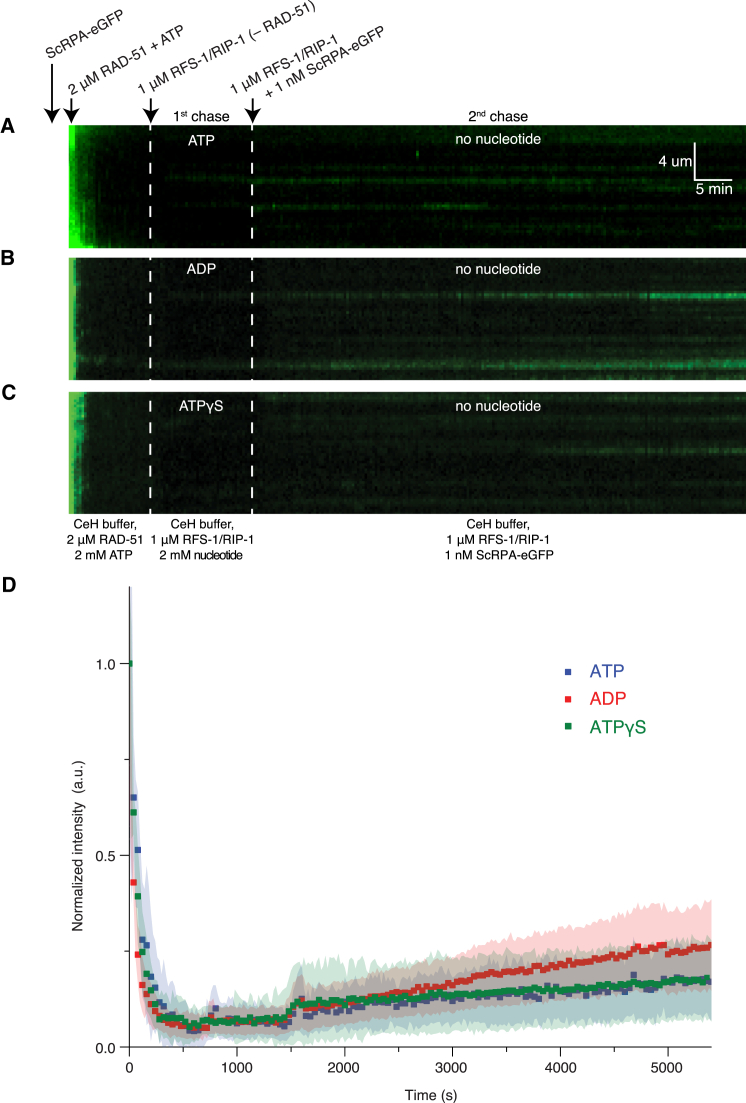
Next, we wanted to test if RAD-51-ssDNA filaments remodeled by ADP- or ATPγS-bound RFS-1/RIP-1 become as efficiently stabilized as those remodeled by ATP-bound RFS-1/RIP-1. To test this, we employed the ssDNA curtains system, which allows for multiple buffer exchanges. As before (Figure 1), RAD-51-ssDNA filaments were first assembled on ssDNA curtains in the presence of ATP, while RFS-1/RIP-1 was pre-incubated in buffer supplemented with ADP or ATPγS. After filament assembly, RAD-51 and ATP were removed and RFS-1/RIP-1 injected with ADP or ATPγS buffer and incubated for 10 min to bind filaments. Excess protein and nucleotide was washed away for 2 min with a buffer lacking nucleotide, before injecting RFS-1/RIP-1 and RPA in the same buffer to assess stability in the same way as in Figure 1. We observed RFS-1/RIP-1 incubated with either ADP (Figure 6B) or ATPγS (Figure 6C) are able to stabilize RAD-51 filaments as effectively as in the presence of ATP (Figures 6A and 6D).
Figure 6.
Nucleotide Hydrolysis by RFS-1/RIP-1 Is Dispensable for RAD-51-ssDNA Filament Stabilization
(A–C) Kymograms showing the rapid exchange of ScRPA-eGFP by 2 μM RAD-51 in the presence of 2 mM ATP and the subsequent stabilization on the RAD-51 filament by the RFS-1/RIP-1 complex when it is pre-incubated for 10 min with 2 mM ATP (A), ADP (B), or ATPγS (C). Free RAD-51 and ATP were washed out of the flow cell before the injection of RFS-1/RIP-1, which is pre-incubated with the indicated nucleotide before injection. The RIP-1/RFS-1 is incubated with RAD-51 filaments in the flow cell for 10 min before free nucleotide was washed away and more RIP-1/RFS-1 is flown in with buffer containing 1 nM ScRPA-eGFP. The buffer conditions inside the flow cell during the incubations are indicated below the kymograms. ScRPA-eGFP binding serves as a proxy for RAD51 dissociation.
(D) Quantitation of the intensity of the ScRPA-eGFP signal over the time course of the experiment for the data in (A)–(C). Data with ATP (blue trace) is reproduced from Figure 1G for comparison purposes to the other nucleotide analogs.
Together, these results demonstrate that nucleotide binding is necessary and sufficient to activate the 5′ filament end binding and remodeling activities of RFS-1/RIP-1, whereas nucleotide hydrolysis by the complex is dispensable for these functions. The Walker box mutants of RFS-1 also fail to induce a fluorescence reduction on pre-formed RAD-51-ssDNA filaments, which mimics the situation observed for nucleotide-free RFS-1/RIP-1 (Figures 5A and 5B). These data therefore suggest that the Walker box mutants of RFS-1 are likely to be defective for ATP binding, which could explain their overall failure to remodel RAD-51-ssDNA filaments (Taylor et al., 2015).
Discussion
Mechanism of RAD51 Filament Remodeling by RAD51 Paralogs
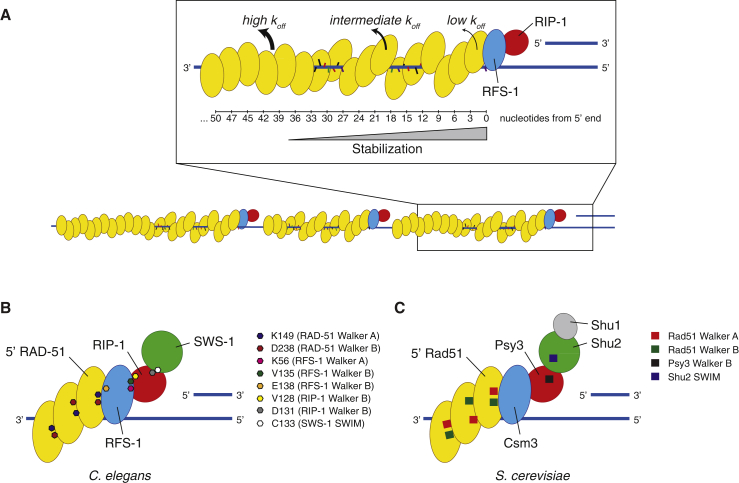
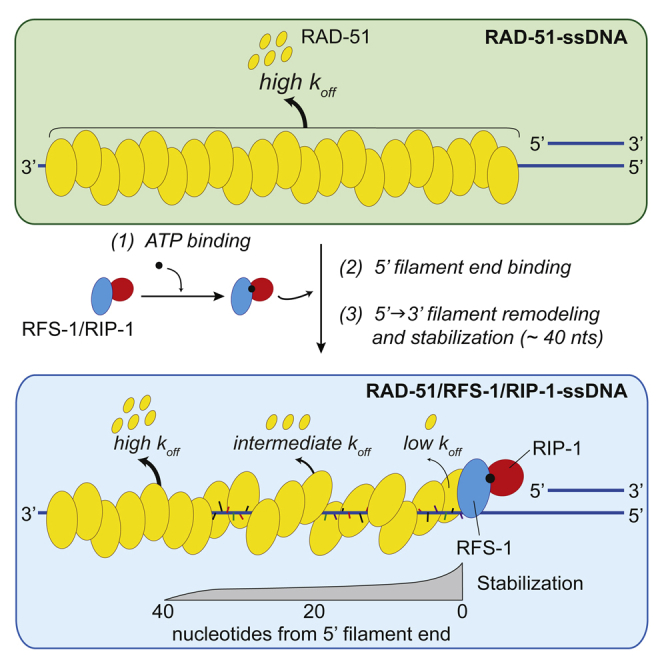
Our previous work revealed a function for RAD51 paralogs in remodeling RAD51 pre-synaptic filaments to a relaxed and stable structure, which is more proficient for strand invasion (Taylor et al., 2015). Despite these insights, important mechanistic questions remained unresolved, particularly in relation to the precise nature of filament stabilization, how the RAD51 paralogs engage with the pre-synaptic filament, and the role of nucleotide co-factors in this process. We establish here that RAD51 paralogs act by capping the 5′ end of the pre-synaptic filament in a nucleotide-dependent manner, which induces a conformational transition that propagates along the filament in a 5′→3′ direction. This extends up to 40 nucleotides away from the 5′ end of the filament and shuts down the dissociation of RAD-51 from ssDNA (Figure 7A).
Figure 7.
Model for the Proposed Mechanism of RAD51-ssDNA Filament Remodeling by RAD51 Paralogs
(A) RFS-1/RIP-1 binds the 5′ end of individual RAD-51-ssDNA filaments and propagates a stabilizing effect with 5′→3′ polarity up to 40 nucleotides from the 5′ end. In the context of a larger pre-synaptic complex, such as in ssDNA curtain experiments, RFS-1/RIP-1 may cap RAD-51-ssDNA filaments arising from separate nucleation events to allow stabilization of the entire assembly.
(B and C) Putative model for the interaction network between RAD51, RAD51 paralogs, and Shu complex proteins from C. elegans (B) and S. cerevisiae (C) in the context of the RAD51-ssDNA filament 5′ end. Hypothetical positions of critical residues for these interactions from the Walker boxes and SWIM domain of C. elegans RAD-51, RFS-1, RIP-1, and SWS-1 are indicated. The equivalent protein features identified in the S. cerevisiae proteins are also noted.
Molecular Basis of RFS-1/RIP-1 Binding to the 5′ Filament End
A major question arising from our work is how RFS-1/RIP-1 engages with the 5′ filament end. Interestingly, modeling of the crystal structure of Psy3-Csm2 from the budding yeast Shu complex, which contains divergent RAD51 paralogs, revealed that it could be specifically docked onto the 5′ end of the yeast RAD51 filament crystal structure (Conway et al., 2004, Sasanuma et al., 2013). In contrast, docking to the 3′ end was associated with steric clashes rendering such an association less plausible (Sasanuma et al., 2013). Our study provides experimental evidence that RAD51 paralogs recognize an intrinsic polarity within the RAD51 filament and bind specifically to the 5′ end. Given that RFS-1 mutants that abolish the ability of RFS-1/RIP-1 to drive the fluorescence reduction associated with 5′ filament end binding also fail to mediate remodeling and stabilization (Taylor et al., 2015), we propose that normal 5′ filament end binding is a pre-requisite for initiation of remodeling.
In the future, it will be important to establish tractable methods to obtain atomic resolution structural models of RAD51 paralogs bound to RAD51 filaments, to better understand the intricacies underlying filament capping and remodeling. Docking of the yeast Csm2-Psy3 heterodimer and Rad51-ssDNA filament crystal structures was achieved by modeling the complex as a direct extension of the filament (Sasanuma et al., 2013), consistent with the fact that the fundamental repetitive units of the yeast Rad51 filament are Rad51 homodimers, which assemble in tandem with a distinctive interface to that between the two monomers of each homodimer pair (Conway et al., 2004). Both protomer-protomer interfaces involve the ATP binding pockets of adjacent protomers and bound ATP co-factor (Conway et al., 2004), similar to that observed in the RecA-ssDNA crystal structure (Chen et al., 2008). Csm2-Psy3 may be a good model for RFS-1/RIP-1 since Psy3, like RIP-1, only contains a Walker B box (Martín et al., 2006), while Csm2, despite lacking sequence homology with RAD51, adopts a RAD51-like fold (Sasanuma et al., 2013, She et al., 2012, Tao et al., 2012). Accordingly, Csm2 is proposed to dock directly to the 5′ filament end, while Psy3 is distal (Sasanuma et al., 2013), consistent with the fact that RFS-1, but not RIP-1, interacts with RAD-51 in yeast two hybrid (Taylor et al., 2015). In addition, recent work identified a homolog of the SWIM domain-containing protein of the Shu complex (yeast Shu2 or human SWS1) (Liu et al., 2011, Martín et al., 2006) in C. elegans, SWS-1 (Godin et al., 2015, McClendon et al., 2016). SWS-1 interacts with RIP-1, but not RFS-1, in a manner dependent on the RIP-1 Walker B box aspartic acid residue 131 (McClendon et al., 2016). Notably, the same D131A mutation in RIP-1 does not dramatically interfere with the RFS-1/RIP-1 interaction in yeast two hybrid (Taylor et al., 2015), consistent with this residue exhibiting greater functional importance in maintaining a RIP-1/SWS-1 interface. SAXS data also indicate that Shu2 associates with Psy3 rather than Csm2 (She et al., 2012) and, by deduction, distal to the 5′ filament end. Together, these observations support a model in which RFS-1 directly contacts RAD-51 at the 5′ filament end and RIP-1 is positioned distally, allowing it to interact with SWS-1 (Figures 7B and 7C). How SWS-1 influences filament binding and remodeling remains to be tested.
Another key question is how do nucleotide co-factors contribute to 5′ filament end recognition? Since the RFS-1/RAD-51 interaction is highly sensitive to mutations in their Walker boxes (Taylor et al., 2015) and RFS-1/RIP-1 must be nucleotide-bound to mediate normal 5′ filament end binding, a nucleotide co-factor may sit at the proposed RAD-51/RFS-1 interaction interface, but nucleotide binding may also be required between RFS-1 and RIP-1 to allow them to adopt an active form. RFS-1/RIP-1 bound to ADP was equally efficient as ATP in 5′ filament end binding, raising the question of exactly which nucleotide binds each of these proposed interfaces physiologically.
EM experiments on circular ssDNA revealed filament binding is independent of a 5′ DNA end. This is supported by the fact filament binding by RFS-1/RIP-1 is detectable in stopped flow even when the native 5′ DNA end is modified by the Cy3 dye. Notably, RFS-1/RIP-1 also readily stabilizes the RAD-51 filaments in the ssDNA curtain assays, even though the long ssDNA molecules have no accessible 5′ ends. In these assays, it is likely that RFS-1/RIP-1 binds to the 5′ filament termini located between filament discontinuities (Figure 7A). RFS-1/RIP-1 may still contact the ssDNA backbone during filament capping. In EMSA, RFS-1/RIP-1 binds ssDNA very weakly, but strongly enriches equilibrium protein-ssDNA complex levels in the presence of RAD-51 (Taylor et al., 2015). Thus, RFS-1/RIP-1 could contact ssDNA with higher affinity in the context of a pre-established RAD-51 filament. Both RAD-51 and ssDNA contacts by RFS-1/RIP-1 could therefore be required for its filament capping activity.
Interplay between HR Regulators in Pre-synaptic Complex Regulation
In contrast to Psy2-Csm3, the crystal structure of a fragment of human BRCA2 fused to RAD51 can be modeled as binding the 3′ end of the yeast Rad51 filament (Pellegrini et al., 2002, Sasanuma et al., 2013). This is consistent with electron microscopy studies demonstrating 3′ filament end binding of full-length BRCA2, suggesting BRCA2 promotes filament growth with 3′→5′ polarity, at least on naked ssDNA (Shahid et al., 2014). Our data on spontaneous assembly of RAD-51 filaments are consistent with unidirectional filament extension in the 5′→3′ direction, as has also been interpreted for yeast Rad51 (Antony et al., 2009, Hwang and Myong, 2014, Qiu et al., 2013) or bidirectional assembly occurring more rapidly 3′→5′ than 5′→3′. BRCA2 may restrict filament extension polarity to 3′→5′, which importantly would leave the 5′ end of the extending filament free for binding by RAD51 paralogs.
Given the distinct filament binding polarity exhibited by BRCA2 and RFS-1/RIP-1, it will be interesting to analyze the interplay between the nematode BRCA2 ortholog, BRC-2, and RFS-1/RIP-1 during filament assembly and remodeling, since these proteins may synergize in these processes on naked and/or RPA-bound ssDNA. Indeed, it was recently shown that although the yeast RAD51 paralog complex Rad55-Rad57 and the Shu complex do not stimulate Rad51 binding to RPA-coated ssDNA, they significantly synergize with Rad52 in this process (Gaines et al., 2015). We have also observed that RFS-1/RIP-1 stimulates filament formation in the presence of ATP (Taylor et al., 2015), so it is possible this activity may synergize with BRC-2. In addition, by capping opposing ends of filaments, BRC-2 and RFS-1/RIP-1 may even exhibit a degree of allostery in mediating filament remodeling and stability. In the case of the β2-adrenergic and μ-opioid G protein coupled receptors, binding of agonists is not sufficient to drive the complete establishment of the active conformation of the receptor and full activation requires heterotrimeric G protein engagement (Manglik et al., 2015, Sounier et al., 2015). As such, anchoring of the 3′ filament end by BRC-2 could co-operatively enhance filament remodeling by RFS-1/RIP-1 to allow propagation beyond the 40 nucleotide threshold defined in this study.
Propagation of RAD51 Filament Remodeling and Stabilization
Another critical structural question is exactly how RFS-1/RIP-1 propagates a biophysical change along the filament to a point considerably distal to its binding site, manifested as stabilization and nuclease-sensitization of the filament beyond the region in immediate proximity to the 5′ filament end where RFS-1/RIP-1 binds. One possible mechanism is that the binding of RFS-1/RIP-1 induces a conformational change in the proximal RAD-51 protomer in the filament. This in turn could modulate the interface between this RAD-51 molecule and an adjacent, distal RAD-51 protomer, ultimately propagating remodeling with 5′→3′ polarity through allosteric communication between adjacent RAD-51 monomers within the filament, up to 40 nucleotides (13–14 RAD-51 protomers) from the end. Such long-range conformational propagation in response to a binding event by proteins and/or small molecule ligands is well established for signal transduction by membrane receptors, such as G protein coupled receptors upon agonist and heterotrimeric G protein binding (Manglik et al., 2015, Sounier et al., 2015). It is therefore conceivable that long ranging organizational changes could occur throughout RAD51 filaments in response to terminal binding events by RAD51 paralogs.
In conclusion, our study provides insights into the molecular mechanism of RAD51-ssDNA filament remodeling by RAD51 paralogs, paving the way for future structural studies of this process, and interplay with other HR mediators.
Experimental Procedures
All experiments were performed at 25°C. For stopped-flow experiments, equal volumes of the indicated components were rapidly mixed and Cy3 fluorescence monitored for 1 min. Raw data sets were normalized as follows: for RAD-51 ssDNA binding (Figure S1) and RFS-1/RIP-1 filament binding experiments (Figures 2 and 5), normalized to the starting value for Cy3 fluorescence; for competition experiments (Figures 3, 4, and S3), normalized to the value for Cy3 fluorescence at the 2.01998 s time point and truncated before this. ssDNA curtains were prepared by rolling circle amplification of an M13mp18 single-strand plasmid, tethered at both ends in a flow cell, and visualized by binding of ScRPA-eGFP. RAD-51 filaments were assembled by flushing out ScRPA-eGFP and flowing in RAD-51 in the presence of ATP. Filaments were equilibrated with buffer containing the indicated nucleotide before incubation with RFS-1/RIP-1. To assess filament stability, RAD-51 was removed and replaced with ScRPA-eGFP in the absence of ATP, while maintaining RFS-1/RIP-1 in the buffer. For quantification, the fluorescence intensity of individual kymograms was adjusted to the background and normalized to the starting value. Immuno-gold EM was performed by incubating proteins (1 μM RAD-51 and 0.1 μM RFS-1/RIP-1) and circular p8064 ssDNA or linearized PhiX ssDNA, then incubating with anti-FLAG antibody conjugated to 20-nm gold particles, staining with uranyl acetate, and imaging. For nuclease protection assays, protein-DNA complexes were assembled on 5′ or 3′ fluorescently labeled 61-mer ssDNA before challenging with DNaseI, deproteinizing, and resolving DNA products by PAGE. See also Supplemental Experimental Procedures online.
Author Contributions
M.R.G.T., M.S., L.K., and S.J.B. conceived the study. M.R.G.T. purified proteins. M.R.G.T. and M.S. performed stopped-flow, EMSA, and nuclease protection assays. M.S. performed nucleotide binding analyses. C.J.M. and E.C.G. performed ssDNA curtains experiments. R.C., M.R.G.T., and L.M.C. performed EM. T.T. and M.R.G.T. analyzed EM data. M.R.G.T. and S.J.B. wrote the manuscript.
Acknowledgments
We thank A. Alidoust and N. Patel for yeast fermentation. M.R.G.T. is supported by a Sir Henry Wellcome Postdoctoral Fellowship from the Wellcome Trust (110014/Z/15/Z). The S.J.B. laboratory is supported by the Francis Crick Institute, which receives its core funding from Cancer Research UK (FC0010048), the UK Medical Research Council (FC0010048), the Wellcome Trust (FC0010048), a European Research Council (ERC) Advanced Investigator grant (RecMitMei), and a Wellcome Trust Senior Investigator grant. The L.K. laboratory is supported by the Czech Science Foundation (GA13–26629S and GAP207/12/2323), the National Program of Sustainability II (MEYS CR, project no. LQ1605), the project FNUSA-ICRC no. CZ.1.05/1.1.00/02.0123 (OP VaVpI), and by project ICRC-ERA-HumanBridge (no. 316345) funded by the European Commission. The E.C.G. laboratory is funded by a grant from the NIH (1R35GM118026).
Published: November 17, 2016
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.molcel.2016.10.020.
Contributor Information
Lumir Krejci, Email: lkrejci@chemi.muni.cz.
Simon J. Boulton, Email: simon.boulton@crick.ac.uk.
Supplemental Information
References
- Antony E., Tomko E.J., Xiao Q., Krejci L., Lohman T.M., Ellenberger T. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol. Cell. 2009;35:105–115. doi: 10.1016/j.molcel.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman J.R., Taylor M.R.G., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. doi: 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Chen Z., Yang H., Pavletich N.P. Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature. 2008;453:489–494. doi: 10.1038/nature06971. [DOI] [PubMed] [Google Scholar]
- Chun J., Buechelmaier E.S., Powell S.N. Rad51 paralog complexes BCDX2 and CX3 act at different stages in the BRCA1-BRCA2-dependent homologous recombination pathway. Mol. Cell. Biol. 2013;33:387–395. doi: 10.1128/MCB.00465-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway A.B., Lynch T.W., Zhang Y., Fortin G.S., Fung C.W., Symington L.S., Rice P.A. Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 2004;11:791–796. doi: 10.1038/nsmb795. [DOI] [PubMed] [Google Scholar]
- French C.A., Masson J.-Y., Griffin C.S., O’Regan P., West S.C., Thacker J. Role of mammalian RAD51L2 (RAD51C) in recombination and genetic stability. J. Biol. Chem. 2002;277:19322–19330. doi: 10.1074/jbc.M201402200. [DOI] [PubMed] [Google Scholar]
- Gaines W.A., Godin S.K., Kabbinavar F.F., Rao T., VanDemark A.P., Sung P., Bernstein K.A. Promotion of presynaptic filament assembly by the ensemble of S. cerevisiae Rad51 paralogues with Rad52. Nat. Commun. 2015;6:7834. doi: 10.1038/ncomms8834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genois M.M., Plourde M., Éthier C., Roy G., Poirier G.G., Ouellette M., Masson J.Y. Roles of Rad51 paralogs for promoting homologous recombination in Leishmania infantum. Nucleic Acids Res. 2015;43:2701–2715. doi: 10.1093/nar/gkv118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb B., Ye L.F., Gergoudis S.C., Kwon Y., Niu H., Sung P., Greene E.C. Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS ONE. 2014;9:e87922. doi: 10.1371/journal.pone.0087922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb B., Ye L.F., Kwon Y., Niu H., Sung P., Greene E.C. Protein dynamics during presynaptic-complex assembly on individual single-stranded DNA molecules. Nat. Struct. Mol. Biol. 2014;21:893–900. doi: 10.1038/nsmb.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin S.K., Meslin C., Kabbinavar F., Bratton-Palmer D.S., Hornack C., Mihalevic M.J., Yoshida K., Sullivan M., Clark N.L., Bernstein K.A. Evolutionary and functional analysis of the invariant SWIM domain in the conserved Shu2/SWS1 protein family from Saccharomyces cerevisiae to Homo sapiens. Genetics. 2015;199:1023–1033. doi: 10.1534/genetics.114.173518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera J.E., Chaires J.B. Characterization of preferred deoxyribonuclease I cleavage sites. J. Mol. Biol. 1994;236:405–411. doi: 10.1006/jmbi.1994.1152. [DOI] [PubMed] [Google Scholar]
- Hwang H., Myong S. Protein induced fluorescence enhancement (PIFE) for probing protein-nucleic acid interactions. Chem. Soc. Rev. 2014;43:1221–1229. doi: 10.1039/c3cs60201j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H., Kim H., Myong S. Protein induced fluorescence enhancement as a single molecule assay with short distance sensitivity. Proc. Natl. Acad. Sci. USA. 2011;108:7414–7418. doi: 10.1073/pnas.1017672108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R.B., Carreira A., Kowalczykowski S.C. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.D., Jasin M. Double-strand-break-induced homologous recombination in mammalian cells. Biochem. Soc. Trans. 2001;29:196–201. doi: 10.1042/0300-5127:0290196. [DOI] [PubMed] [Google Scholar]
- Johnson R.D., Liu N., Jasin M. Mammalian XRCC2 promotes the repair of DNA double-strand breaks by homologous recombination. Nature. 1999;401:397–399. doi: 10.1038/43932. [DOI] [PubMed] [Google Scholar]
- Krejci L., Altmannova V., Spirek M., Zhao X. Homologous recombination and its regulation. Nucleic Acids Res. 2012;40:5795–5818. doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Doty T., Gibson B., Heyer W.-D. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T., Wan L., Wu Y., Chen J., Huang J. hSWS1·SWSAP1 is an evolutionarily conserved complex required for efficient homologous recombination repair. J. Biol. Chem. 2011;286:41758–41766. doi: 10.1074/jbc.M111.271080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manglik A., Kim T.H., Masureel M., Altenbach C., Yang Z., Hilger D., Lerch M.T., Kobilka T.S., Thian F.S., Hubbell W.L. Structural insights into the dynamic process of β2-adrenergic receptor signaling. Cell. 2015;161:1101–1111. doi: 10.1016/j.cell.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín V., Chahwan C., Gao H., Blais V., Wohlschlegel J., Yates J.R., 3rd, McGowan C.H., Russell P. Sws1 is a conserved regulator of homologous recombination in eukaryotic cells. EMBO J. 2006;25:2564–2574. doi: 10.1038/sj.emboj.7601141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClendon T.B., Sullivan M.R., Bernstein K.A., Yanowitz J.L. Promotion of homologous recombination by SWS-1 in complex with RAD-51 paralogs in Caenorhabditis elegans. Genetics. 2016;203:133–145. doi: 10.1534/genetics.115.185827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Yu D.S., Lo T., Anand S., Lee M., Blundell T.L., Venkitaraman A.R. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- Pierce A.J., Johnson R.D., Thompson L.H., Jasin M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 1999;13:2633–2638. doi: 10.1101/gad.13.20.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Z., Redding S., Lee J.Y., Gibb B., Kwon Y., Niu H., Gaines W.A., Sung P., Greene E.C. DNA sequence alignment by microhomology sampling during homologous recombination. Cell. 2015;160:856–869. doi: 10.1016/j.cell.2015.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Antony E., Doganay S., Koh H.R., Lohman T.M., Myong S. Srs2 prevents Rad51 filament formation by repetitive motion on DNA. Nat. Commun. 2013;4:2281. doi: 10.1038/ncomms3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rattray A.J., Symington L.S. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J., Sung P., Klein H. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sasanuma H., Tawaramoto M.S., Lao J.P., Hosaka H., Sanda E., Suzuki M., Yamashita E., Hunter N., Shinohara M., Nakagawa A., Shinohara A. A new protein complex promoting the assembly of Rad51 filaments. Nat. Commun. 2013;4:1676. doi: 10.1038/ncomms2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid T., Soroka J., Kong E.H., Malivert L., McIlwraith M.J., Pape T., West S.C., Zhang X. Structure and mechanism of action of the BRCA2 breast cancer tumor suppressor. Nat. Struct. Mol. Biol. 2014;21:962–968. doi: 10.1038/nsmb.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- She Z., Gao Z.Q., Liu Y., Wang W.J., Liu G.F., Shtykova E.V., Xu J.H., Dong Y.H. Structural and SAXS analysis of the budding yeast SHU-complex proteins. FEBS Lett. 2012;586:2306–2312. doi: 10.1016/j.febslet.2012.06.024. [DOI] [PubMed] [Google Scholar]
- Sigurdsson S., Van Komen S., Bussen W., Schild D., Albala J.S., Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev. 2001;15:3308–3318. doi: 10.1101/gad.935501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger J.A., Kiianitsa K., Heyer W.D. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol. Cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Sounier R., Mas C., Steyaert J., Laeremans T., Manglik A., Huang W., Kobilka B.K., Déméné H., Granier S. Propagation of conformational changes during μ-opioid receptor activation. Nature. 2015;524:375–378. doi: 10.1038/nature14680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stennett E.M., Ciuba M.A., Lin S., Levitus M. Demystifying PIFE: The photophysics behind the protein-induced fluorescence enhancement phenomenon in Cy3. J. Phys. Chem. Lett. 2015;6:1819–1823. doi: 10.1021/acs.jpclett.5b00613. [DOI] [PubMed] [Google Scholar]
- Sung P. Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev. 1997;11:1111–1121. doi: 10.1101/gad.11.9.1111. [DOI] [PubMed] [Google Scholar]
- Takata M., Sasaki M.S., Tachiiri S., Fukushima T., Sonoda E., Schild D., Thompson L.H., Takeda S. Chromosome instability and defective recombinational repair in knockout mutants of the five Rad51 paralogs. Mol. Cell. Biol. 2001;21:2858–2866. doi: 10.1128/MCB.21.8.2858-2866.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y., Li X., Liu Y., Ruan J., Qi S., Niu L., Teng M. Structural analysis of Shu proteins reveals a DNA binding role essential for resisting damage. J. Biol. Chem. 2012;287:20231–20239. doi: 10.1074/jbc.M111.334698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor M.R., Špírek M., Chaurasiya K.R., Ward J.D., Carzaniga R., Yu X., Egelman E.H., Collinson L.M., Rueda D., Krejci L., Boulton S.J. Rad51 paralogs remodel pre-synaptic Rad51 filaments to stimulate homologous recombination. Cell. 2015;162:271–286. doi: 10.1016/j.cell.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T., McIlwraith M.J., Compton S.A., Lekomtsev S., Petronczki M., Griffith J.D., West S.C. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat. Struct. Mol. Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker J.E., Saraste M., Runswick M.J., Gay N.J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J.D., Barber L.J., Petalcorin M.I., Yanowitz J., Boulton S.J. Replication blocking lesions present a unique substrate for homologous recombination. EMBO J. 2007;26:3384–3396. doi: 10.1038/sj.emboj.7601766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wold M.S. Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- Yu X., Jacobs S.A., West S.C., Ogawa T., Egelman E.H. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl. Acad. Sci. USA. 2001;98:8419–8424. doi: 10.1073/pnas.111005398. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.