Abstract
Background
The widely known terminology gap between health professionals and health consumers hinders effective information seeking for consumers.
Objective
The aim of this study was to better understand consumers’ usage of medical concepts by evaluating the coverage of concepts and semantic types of the Unified Medical Language System (UMLS) on diabetes-related postings in 2 types of social media: blogs and social question and answer (Q&A).
Methods
We collected 2 types of social media data: (1) a total of 3711 blogs tagged with “diabetes” on Tumblr posted between February and October 2015; and (2) a total of 58,422 questions and associated answers posted between 2009 and 2014 in the diabetes category of Yahoo! Answers. We analyzed the datasets using a widely adopted biomedical text processing framework Apache cTAKES and its extension YTEX. First, we applied the named entity recognition (NER) method implemented in YTEX to identify UMLS concepts in the datasets. We then analyzed the coverage and the popularity of concepts in the UMLS source vocabularies across the 2 datasets (ie, blogs and social Q&A). Further, we conducted a concept-level comparative coverage analysis between SNOMED Clinical Terms (SNOMED CT) and Open-Access Collaborative Consumer Health Vocabulary (OAC CHV)—the top 2 UMLS source vocabularies that have the most coverage on our datasets. We also analyzed the UMLS semantic types that were frequently observed in our datasets.
Results
We identified 2415 UMLS concepts from blog postings, 6452 UMLS concepts from social Q&A questions, and 10,378 UMLS concepts from the answers. The medical concepts identified in the blogs can be covered by 56 source vocabularies in the UMLS, while those in questions and answers can be covered by 58 source vocabularies. SNOMED CT was the dominant vocabulary in terms of coverage across all the datasets, ranging from 84.9% to 95.9%. It was followed by OAC CHV (between 73.5% and 80.0%) and Metathesaurus Names (MTH) (between 55.7% and 73.5%). All of the social media datasets shared frequent semantic types such as “Amino Acid, Peptide, or Protein,” “Body Part, Organ, or Organ Component,” and “Disease or Syndrome.”
Conclusions
Although the 3 social media datasets vary greatly in size, they exhibited similar conceptual coverage among UMLS source vocabularies and the identified concepts showed similar semantic type distributions. As such, concepts that are both frequently used by consumers and also found in professional vocabularies such as SNOMED CT can be suggested to OAC CHV to improve its coverage.
Keywords: controlled vocabulary, consumer health vocabulary, concept coverage
Introduction
Background
There is a widely known language gap between health consumers and health care professionals [1-3]. This gap may hinder effective communication between the 2 groups [4-7]; thus, impacting consumers’ health information seeking [3,8,9] and subsequent decision making regarding their health issues [10]. To assess the gap, Roberts and Demner-Fushman [11] used a variety of natural language processing (NLP) techniques to analyze the difference between health questions asked by consumers and health professionals in different online question and answer (Q&A) sites (eg, Yahoo! Answers, and WebMD). They found that consumer questions tend to contain more misspelled medical terms, have longer background information, and resemble open-domain language more closely than texts written by professionals. One major aspect of the gap is the difference in medical vocabulary used by consumers and health professionals. Zeng and colleagues [12] observed that when searching online health information, using only consumer terms leads to poor information retrieval results. Plovnick and Zeng [13] later reformulated consumers’ health queries with professional terminology and about 40% of reformulated queries yielded better search performance.
To bridge the vocabulary gap between health professionals and consumers, early researchers have collected and analyzed diverse textual data generated by consumers to identify medical terms used by consumers. Brennan and Aronson [14] used the MetaMap tool to extract salient concepts in nursing vocabularies from consumers’ email messages. Smith and collegues [15] also used MetaMap to successfully identify the Unified Medical Language System (UMLS) concepts used by consumers in their email messages submitted to University of Pittsburg Cancer Institute’s Cancer Information and Referral Service. These studies aimed to bridge the vocabulary gap between health professionals and consumers by identifying frequently-used consumer health terms that are relevant in developing consumer-oriented health information applications and linking free text to complex clinical knowledge resources. These ad hoc studies represent early efforts in bridging the vocabulary gap.
A controlled vocabulary is “an organized arrangement of words and phrases used to index content and/or to retrieve content through browsing or searching[16].” In an effort to formalize consumer vocabulary for various applications, a controlled vocabulary called Open-Access Collaborative Consumer Health Vocabulary (“OAC CHV,” “CHV” for short) was recently developed as a collection of expressions and concepts that are commonly used by ordinary health information users [17]. Moreover, CHV has been integrated in the largest medical terminological system–the UMLS, which has mapped terms from different source vocabularies with the same meaning into the same concept by the United States National Library of Medicine (NLM). As such, consumer terms are connected to their corresponding professional terms in professional vocabularies such as SNOMED Clinical Terms (SNOMED CT). With CHV in the UMLS, one can translate a sentence with consumer terms to a sentence with professional terms in an automated fashion.
Domain coverage—the extent to which a controlled vocabulary covers the intended domain—is one of the most desired properties for a controlled vocabulary [18]. The usability and the overall structure of a controlled vocabulary heavily rely upon its coverage [19]. Traditionally, controlled vocabulary development takes a top-down approach, which reflects a group of experts’ knowledge in the respective subject matter [20,21]. For the development of CHV, however, a bottom-up approach was taken, emphasizing 2 fundamental properties: (1) CHV should capture actual consumers’ terms and expressions that reflect their health information needs, and (2) the expressions should be familiar to and used by consumers [17].
To keep up with continuous evolution of medical knowledge, CHV needs to be updated and maintained by incorporating new, consumer-provided terms and expressions [17,22-24]. Existing studies have shown promising results in discovering consumer terms for CHV from social media, in particular. Vydiswaran et al [7] applied a pattern-based text mining approach to identify pairs of consumer and professional terms from Wikipedia. Hicks et al [25] analyzed consumer messages exchanged in Twitter in order to evaluate terms related to gender identification on intake forms. Doing-Harris and Zeng-Treitler [24] developed a computer assisted CHV update system, which can automatically identify prospective terms from social media. Identifying terms used by consumers in consumer-generated text in aggregate fashion can account for the variability of lay health language. These terms can be used to refine and enrich CHV [17].
Consumers, however, may also learn and use professional terms [17,24,26]. In this sense, medical terms that are familiar to consumers and are already established in other controlled vocabularies could be used to improve the coverage of CHV. Term reuse is a principle and best practice in ontology/terminology development as it promises to support the semantic interoperability and to reduce engineering costs [27]. Researchers have previously developed semi-automated methods to facilitate systematic term reuse. He et al [28] developed a topological-pattern-based method to identify terms from UMLS source vocabularies to enrich SNOMED CT [28,29] and National Cancer Institute Thesaurus (NCIt) [30].
However, this method cannot be directly applied to CHV, because it does not have hierarchical relationships (eg. parent-child relationship) that are necessary to construct topological patterns [28-30]. Recently, Chandar et al [31] introduced a similarity-based term recommendation method that represents n-grams extracted from the free-text eligibility criteria of clinical trials as a set of linguistic and contextual features. SNOMED CT terms are clustered with K-means clustering. The new terms are ordered by their distance to the nearest cluster centroid, representing their similarity to existing SNOMED CT terms. This method performed well on the corpus of free-text clinical study eligibility criteria, because they are mostly short and partial sentences written by health professionals with fruitful medical terms and little noise. It has yet to be tested on free-form consumer text that typically contains lengthy sentences and lay terms.
Most previous studies concerning CHV development concentrated on the identification of new terms used by consumers [17,22-24]. To the best of our knowledge, no prior studies have conducted in-depth assessment of the coverage and popularity of medical concepts in user-generated documents on social media. In this respect, there is a need to understand consumers’ use of terms in existing controlled vocabularies, and to perceive if there is the potential to improve CHV by incorporating health-related concepts used by consumers that are covered by professional vocabularies. In this study, therefore, we performed such an analysis in order to assess consumers’ use of medical concepts on social media postings pertaining to health concerns and to evaluate how many popular consumer terms have been included in the existing source vocabularies of the UMLS [32].
In this study, we focus on diabetes, which is recognized as one of the most important public health problems with escalating health concerns by the World Health Organization (WHO) [33]. Diabetes caused 1.5 million deaths in 2012 alone. It is known to cause disability and an array of serious health issues such as hypertension, nephropathy, and stroke [34]. Global diabetes cases skyrocketed from 108 million in 1980 to 422 million in 2014. The number of diabetes onset will likely reach 700 million by 2025 [35]. Diabetes and its complications not only impair population health but also impose substantial economic burdens on patients, their family, and the society [33].
In this study, we collected diabetes-related consumer-generated blog postings from Tumblr and diabetes-related questions and answers from Yahoo! Answers. We carried out text mining to identify UMLS concepts from our datasets. Thus, we formulated the 2 research questions (RQs): (1) To what degree do the concepts from UMLS source vocabularies cover the concepts used by consumers describing their diabetes-related concerns on health postings of social media, especially blogs and social Q&A? Which concepts do or do not overlap? (2) To what degree are the UMLS semantic types applicable to analyzing the concepts used by consumers when describing their diabetes-related concerns in social media, especially blogs and social Q&A? Which semantic types are observed?
In the first research question, we evaluated the coverage of all of the 178 English source vocabularies of the UMLS in our 2 datasets from Tumblr and Yahoo! Answers. In the second research question, we analyzed the semantic types of the UMLS concepts identified in our datasets.
The current study mainly investigated the overlap between consumer concepts from social media and professional concepts in the UMLS. Indeed, consumers often proactively seek and share online health information on social media [36,37]. Their use of professional terms could be sophisticated covering both laypersons’ expressions and medical terminologies. In fact, not only consumers but also health care professionals have actively participated in creating health postings in social media [38,39]. Their use of terms in social media, however, is likely to be more consumer/patient-centric for health education and promotion to the public. The comparative analysis of the concept coverage between consumers and professional vocabularies in social media may be helpful in understanding the scale of the phenomenon. The comparison will also help yield insights into the nature of the vocabulary gap, which will contribute to the consistent development of the CHV. The current study, in particular, could shed light on how much social media users use existing terms in UMLS source vocabularies on the web. At the same time, findings from the current study could inform the feasibility of leveraging existing UMLS source vocabularies to enrich the CHV.
The Unified Medical Language System
The UMLS, maintained by the NLM of the National Institutes of Health, is the largest biomedical terminological system. Its 2-level structure consists of Metathesaurus and Semantic Network. The UMLS Metathesaurus is “a large, multi-purpose, and multi-lingual thesaurus that contains millions of biomedical and health related concepts, their synonymous names, and their relationships” [40]. The UMLS Metathesaurus integrates more than 9.1 million terms from over 170 English source vocabularies into 3.1 million medical concepts (2015AA version). Besides English, the UMLS also contains source vocabularies in 20 other languages. The UMLS has integrated most of the well-designed and well-maintained medical terminologies such as SNOMED CT, the International Classification of Diseases 9th Revision, Clinical Modification (ICD-9-CM), NCIt, and RxNORM. SNOMED CT is the most comprehensive and precise clinical terminology in the world with over 310,000 active concepts [41]. ICD-9-CM is used primarily to encode the diagnoses and procedures for billing purposes [42]. RxNORM, on the other hand, normalizes names of all clinical drugs available on the US market and their links to many of the drug vocabularies commonly used in pharmacy management [43]. Most significantly, the terms with the same meaning are mapped to the same concept in the UMLS. Due to its native term mapping, the UMLS is a valuable resource for supporting interoperability and translation in biomedicine [32]. The NLM releases a new version of the UMLS twice a year.
The UMLS semantic types represent “a set of broad subject categories that provide a consistent categorization of all concepts represented in the UMLS Metathesaurus” [44]. Each concept in the UMLS is assigned 1 or more semantic types. In the 2015AA version of the UMLS, there are a total of 127 semantic types, describing concepts at the levels of entity and event. Entities include physical objects such as organism, anatomical structure, and substances. Events describe activities, phenomenon, and processes. For example, the semantic type “Disease or Syndrome” categorizes a set of concepts in the UMLS that indicate “a condition which alters or interferes with a normal process, state, or activity of an organism.”
Consumer Health Vocabularies and Their Use in Consumer-Oriented Health Applications
OAC CHV has been used in various health-related applications to improve patients’ access to health information. Zeng et al developed a translator specifically to convert texts in electronic health records to consumer-friendly text in patient health records by replacing UMLS terms to their corresponding OAC CHV terms [45]. Many UMLS concepts have one to one match with OAC CHV concepts. All the OAC CHV concepts have predefined consumer-friendly display names. Besides OAC CHV, other proprietary consumer health vocabularies have been developed. For example, Apelon has developed a CHV and has mapped their CHV terms to corresponding clinical concepts in SNOMED RT (an earlier version of SNOMED CT, developed by College of American Pathologists), ICD-9-CM, and Physician’s Current Procedural Terminology (CPT) administrative codes. The CHV of Apelon has been used in various applications, such as consumer health data entry, patient results reporting clinical note translation, and Web-based information retrieval [46]. Mayo Clinic also developed their own consumer health vocabulary, which has a rich content of disease concepts as well as genetic and non-genetic risk factors to diseases [8]. In this paper, we used OAC CHV because it is the only publicly available consumer health vocabulary that we have access to (through the UMLS).
Methods
Data Collection
2 types of social media were analyzed in the current study, namely blogs and social Q&A, as they allow consumers to generate and freely exchange health information in text format. Health-related blogs are one of the most popular social media venues for health information distribution. Bloggers typically describe their personal experiences with diseases along with their encounters with health care professionals [47]. Health care professionals also create blogs for sharing their medical knowledge and information with patients [48]. Blogs have also been widely used for health promotion and education as a collaborative tool for both consumers and health care professionals [49-51]. On the other hand, social Q&A is an online community-based Q&A service where people gain knowledge through raising questions and receiving answers from others who willingly share their knowledge, experiences, and opinions regarding a wide range of topics including health. Social Q&A is considered to be a knowledge-shaping sphere for laypeople [52]. Consumers are motivated to use social Q&A because their searches on web search engines with short queries that are not fully expressive often fail in retrieving useful information for their specific problems, while social Q&A allows them to ask questions in natural language and in full sentences [11]. For data collection, we used 2 datasets: (1) Tumblr, a popular blogging service; and (2) Yahoo! Answers, a social Q&A service in North America.
Tumblr and Yahoo! Answers were chosen for the current study due to their popularity and the convenience of using their Application Program Interfaces (APIs), which allowed us to collect data automatically from these sites. Also, both Tumblr and Yahoo! Answers do not limit the number of words in postings. As such, their users can elaborate their health concerns and information on postings with sufficient details, thereby providing us ample opportunities to extract and analyze relevant concepts from the postings.
Tumblr is one of the fastest-growing blog sites with nearly twenty-fold increase in the number of blogs from October 2012 to October 2015 [53]. It launched relatively late in the market compared to other sites such as WordPress and Blogger, but is recognized as one of the best blog sites due to its ease of setup, stylish interface design, and micro-blogging support [54,55]. It has over 227 million blogs and 37 million unique visitors as of February 2016 [53]. From Tumblr, we collected a total of 3711 English text blogs with a tag related to “diabetes” (eg, “diabetes,” “diabetes mellitus,” and “Type 2 diabetes”) posted between February and October 2015.
Yahoo! Answers is one of the most popular social Q&A sites with approximately 5.6 million visitors per month as of February 2016 [56]. From Yahoo! Answers, we garnered a total of 58,422 questions and associated answers between 2009 and 2014 in the diabetes category of Yahoo! Answers. During data analysis, we carried out text mining with questions and answers (specifically, best answers) separately, because the information in questions and answers could be different. Questions could capture health concerns and associated problems, while answers could mainly discuss information resources intended to solve the problems. It is important to note that 1 question may have more than one answer. In this study, we limited answers to the one selected as the best answer by the questioner. The data collected from Yahoo! Answers were separated into questions and answers in the subsequent analyses.
Units of Analysis
Once we collected text data from Tumblr and Yahoo! Answers, we mined the text data for “concepts,” a unit of understanding which represents a fundamental component of terminology [57] or unit of meaning in an ontology [31]. Concepts are different from “terms” in that a term refers to an entity or “physical object” written or spoken in text to represent a concept or thought [58]. In the UMLS, a term is described as a “word or collection of words comprising an expression,” which indicates a class of all lexical variants (eg, “eye,” “Eye,” “eyes”) [59]. The UMLS assigned each term an atom unique identifier (AUI) and grouped the terms with the same meaning into a concept with a concept unique identifier (CUI). We also analyzed the semantic types of the extracted concepts in order to understand the broad semantic categories of the terms that are frequently used by consumers.
Textual Data Processing
We used a widely adopted biomedical text processing framework Apache cTAKES™ [60] and its extension YTEX [61] to identify UMLS terms in our datasets. Apache cTAKES is designed as a natural language processing (NLP) system for extraction of information from the free-text data available in electronic medical records (EMRs). It provides an agile and flexible platform based on the Unstructured Information Management Architecture (UIMA) and a rich NLP library. YTEX, a module of cTAKES, provides Word Sense Disambiguation (WSD), data mining and feature engineering functionalities. We mainly used the WSD function of YTEX to recognize the most possible UMLS concept when a term in the free text can be matched to multiple ambiguous concepts. We used the 3.2.2 release of cTAKES and YTEX with the default workflow configuration named “Aggregate Plaintext UMLS Processor.”
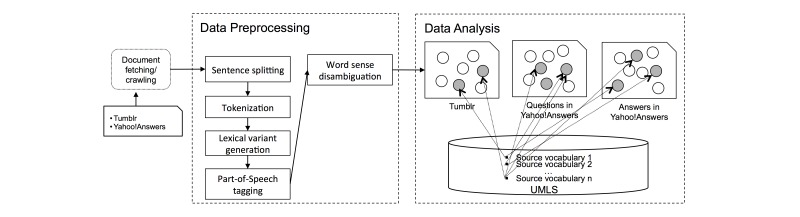
Figure 1 illustrates our overall analysis process. First, each document is a blog posting from Tumblr, a question or an answer from Yahoo! Answers. Each blog posting may consist of 1 or more sentences. Then, cTAKES detected and split each document into individual sentences using the sentence detector of OpenNLP [62,63], with the default configuration for English text. For each sentence, cTAKES performed tokenization with the default tokenizer of the OpenNLP, lexical variant generation using the lexical tool provided by the United States National Library of Medicine with the default configuration. Then, cTAKES performed Part-Of-Speech (POS) tagging using the POS tagger in OpenNLP with the information entropy-based model for English to generate the candidate terms for further processing. Then, YTEX matched the candidate terms with all the possible UMLS terms, which were preloaded from the MRCONSO table of the UMLS 2015AA release. We then stored the matching results to a MySQL database. For each candidate term, there may be 0, 1, or more matching UMLS terms with different semantics. To identify terms with reasonable semantics, we used YTEX to perform word sense disambiguation (WSD), in which the intrinsic information content (IC) measure is used as the semantic similarity metric with a window of 50 words as the context for WSD. The intrinsic information content is a measure of concept specificity computed from the structure of the taxonomy in a biomedical terminology and does not rely on the term frequency in the corpus. The details of the intrinsic IC measure can be found in Garla et al [64]. Finally, all the UMLS terms in each record were extracted with a UMLS CUI.
Figure 1.
Conceptual framework of the study. Dots refer to concepts extracted from the dataset and gray dots refer to concepts mapped to the concepts in one of the UMLS source vocabularies.
Concept Coverage Analysis
We first analyzed the basic characteristics of the overall concept coverage across our datasets collected from Tumblr and Yahoo! Answers: (1) blog postings from Tumblr, (2) questions in Yahoo! Answers, and (3) answers in Yahoo! Answers. We then analyzed the coverage of each source vocabulary in the UMLS across the datasets. SNOMED CT and CHV are the 2 vocabularies with the highest concept coverage in our datasets. Thus, we conducted a concept coverage analysis of SNOMED CT and CHV based on our datasets. We also analyzed the semantic types of the concepts identified from our datasets.
Results
Aggregate Characteristics of the Datasets
We identified 2415 UMLS concepts from blog postings, 6452 UMLS concepts from questions, and 10,378 UMLS concepts from answers. Table 1 shows the total number of documents and sentences in our datasets (ie, blog postings, questions, answers). These numbers were compared to the “# with UMLS concepts,” the unique number of documents and sentences containing the identified UMLS concepts. Note that we can only extract concepts that are presented in UMLS. Thus, the total number of concepts in our datasets (which can include concepts that are not in UMLS) is not provided in Table 1.
Table 1.
Basic characteristics of UMLS concept coverage in our datasets.
|
|
Tumblr | Yahoo! Answers | ||||
|
|
Blog postings | Questions | Answers | |||
|
|
Total # | # with UMLS concepts | Total # | # with UMLS concepts | Total # | # with UMLS concepts |
| Document | 3711 | 1388 (37.4%) | 58,422 | 51,850 (88.8%) | 58,422 | 51,550 (88.2%) |
| Sentence | 47,413 | 12,802 (27.0%) | 249,013 | 142,802 (57.3%) | 348,793 | 216,736 (62.1%) |
| Concepts | – | 2415 | – | 6452 | – | 10,378 |
There was a noticeable variation across the datasets. Over 80% of the documents from questions and answers contained 1 or more UMLS concepts whereas less than half of the documents from blogs did. Over half of the sentences from questions and answers contained at least 1 UMLS concept, while only 27% of those from blog posts contained at least 1 UMLS concept.
Coverage by the UMLS Source Vocabularies
The concepts in the blogs were covered by 56 UMLS source vocabularies, while those in questions and answers were covered by 58 source vocabularies. Table 2 illustrates the top 20 most covered UMLS source vocabularies (The full names and the version information of the source vocabularies can be found in the Multimedia Appendix 1 Table-A1). SNOMED CT was dominant across all our datasets, ranging from 84.9% to 95.9%. It was followed by CHV (between 73.5% and 80.0%) and MTH (between 55.7% and 73.5%). Other source vocabularies within the top 10 for all of our datasets are: NCIt, Medical Subject Headings (MeSH), Computer Retrieval of Information on Scientific Projects Thesaurus (CSP), Library of Congress Subject Headings Northwestern University subset (LCH NW), Logical Observation Identifier Names and Codes (LOINC), and National Drug File – Reference Terminology (NDFRT), although the ranking order varies slightly across different datasets. Multimedia Appendix 1 Table-A2 provides example concepts in the top 3 most covered source vocabularies.
Table 2.
Top 20 mostly covered UMLS source vocabularies.
|
|
Tumblr | Yahoo! Answers | ||||||||||
| Rank | Blogs (n=2415) | Questions (n=6452) | Answers (n=10,378) | |||||||||
| Source vocabulary | # of concepts | % | Source vocabulary | # of concepts | % | Source vocabulary | # of concepts | % | ||||
| 1 | SNOMED CT | 2315 | 95.9 | SNOMED CT | 5476 | 84.9 | SNOMED CT | 9032 | 87.0 | |||
| 2 | CHV | 1931 | 80.0 | CHV | 4928 | 76.4 | CHV | 7625 | 73.5 | |||
| 3 | MTH | 1774 | 73.5 | MTH | 3899 | 60.4 | MTH | 5780 | 55.7 | |||
| 4 | NCIt | 1156 | 47.9 | MeSH | 2957 | 45.8 | MeSH | 4796 | 46.2 | |||
| 5 | MeSH | 1130 | 46.8 | NCIt | 2917 | 45.2 | NCIt | 4485 | 43.2 | |||
| 6 | CSP | 812 | 33.6 | CSP | 1840 | 28.5 | NDFRT | 2999 | 28.9 | |||
| 7 | AOD | 775 | 32.1 | NDFRT | 1775 | 27.5 | CSP | 2839 | 27.4 | |||
| 8 | LCH_NW | 771 | 31.9 | LCH_NW | 1627 | 25.2 | LCH_NW | 2436 | 23.5 | |||
| 9 | LOINC | 697 | 28.9 | AOD | 1585 | 24.6 | AOD | 2335 | 22.5 | |||
| 10 | NDFRT | 659 | 27.3 | LOINC | 1510 | 23.4 | RXNORM | 2099 | 20.2 | |||
| 11 | LCH | 587 | 24.3 | RXNORM | 1421 | 22.0 | LOINC | 2081 | 20.1 | |||
| 12 | NCI_NCI-GLOSS | 475 | 19.7 | LCH | 1187 | 18.4 | LCH | 1730 | 16.7 | |||
| 13 | MEDLINEPLUS | 402 | 16.6 | NCI_NCI-GLOSS | 952 | 14.8 | NCI_FDA | 1387 | 13.4 | |||
| 14 | CST | 365 | 15.1 | NCI_FDA | 868 | 13.5 | DXP | 1322 | 12.7 | |||
| 15 | COSTAR | 362 | 15.0 | COSTAR | 835 | 12.9 | NCI_NCI-GLOSS | 1321 | 12.7 | |||
| 16 | NCI_FDA | 345 | 14.3 | DXP | 830 | 12.9 | COSTAR | 1257 | 12.1 | |||
| 17 | OMIM | 342 | 14.2 | CST | 794 | 12.3 | OMIM | 1234 | 11.9 | |||
| 18 | RXNORM | 338 | 14.0 | OMIM | 790 | 12.2 | CST | 1206 | 11.6 | |||
| 19 | DXP | 326 | 13.5 | MEDLINEPLUS | 721 | 11.2 | VANDF | 1117 | 10.8 | |||
| 20 | ICD9CM | 241 | 10.0 | VANDF | 644 | 10.0 | MTHSPL | 1033 | 10.0 | |||
There was significant overlap between the concepts from the top 2 source vocabularies, SNOMED CT and CHV⎯ 78.2% (1889/2415) concepts from blog postings, 70.0% (4518/6452) concepts in questions, and 68.4% (7095/10,378) concepts in answers. Table 3 shows the top 10 concepts. Note that we only show the preferred term of the concept in the UMLS throughout the paper. Diabetes-related concepts such as Blood, Sugars, Insulin, Glucose, and Diabetes mellitus were frequently mentioned (preferred names of a UMLS concept are denoted in italics). At the same time, it includes some general medical concepts such as disease, pharmaceutical preparations, and problem. Concepts related to glucose level in blood such as blood, sugars, glucose, and carbohydrates also appeared with high frequency.
Table 3.
Top 10 frequently observed concepts covered by both SNOMED CT and CHV.
| Rank | Tumblr | Yahoo! Answers | ||||
|
|
|
Questions | Answers | |||
| Concept | Freq. | Concept | Freq. | Concept | Freq. | |
| 1 | Blood (C0005767) | 816 | Blood (C0005767) | 30,654 | Blood (C0005767) | 54,689 |
| 2 | Pain (C0030193) | 798 | Sugars (C0242209) | 29,593 | Sugars (C0242209) | 49,207 |
| 3 | Insulin (C0021641) | 744 | Insulin (C0021641) | 10,816 | Insulin (C0021641) | 27,887 |
| 4 | Pharmaceutical preparations (C0013227) | 719 | Glucose (C0017725) | 7394 | Glucose (C0017725) | 26,420 |
| 5 | Sugars (C0242209) | 699 | Problem (C0033213) | 5111 | Pharmaceutical preparations (C0013227) | 11,571 |
| 6 | Disease (C0012634) | 617 | Water (C0043047) | 4781 | Diseases (C0012634) | 9733 |
| 7 | Problem (C0033213) | 568 | Pharmaceutical preparations (C0013227) | 4456 | Carbohydrates (C0007004) | 9517 |
| 8 | Diabetes mellitus (C0011849) | 501 | Hematologic tests (C0018941) | 3784 | Problem (C0033213) | 9248 |
| 9 | Teeth structure (C0040426) | 424 | Pain (C0030193) | 3625 | Water (C0043047) | 5994 |
| 10 | Operative surgery procedures (C0543467) | 375 | Urine (C0042036) | 2550 | Fasting (C0015663) | 5848 |
A few concepts were only covered by CHV: 1.7% (40/2415) concepts in blog postings, 6.3% (409/6452) concepts in questions, and 5.1% (529/10,378) concepts in answers. Table 4 shows the top 10 most frequently observed UMLS concepts covered by CHV but not SNOMED CT in our datasets.
Table 4.
Top 10 frequently observed concepts covered by CHV but not SNOMED CT.
|
|
Tumblr | Yahoo! Answers | |||||
| Rank |
|
|
Questions | Answers | |||
| Concept (CUI)a | Freq. | Concept (CUI) | Freq. | Concept (CUI) | Freq. | ||
| 1 | Cider vinegar (C0937941) | 54 | Stomach (C0038351) | 1050 | Lantus (C0876064) | 689 | |
| 2 | Apple cider vinegar (C1178459) | 54 | Lantus (C0876064) | 571 | Actos (C0875954) | 659 | |
| 3 | Lantus (C0876064) | 15 | Humalog (C0528249) | 260 | Avandia (C0875967) | 628 | |
| 4 | Gentle (C0720654) | 11 | NovoLog (C0939412) | 180 | HumaLog (C0528249) | 289 | |
| 5 | Corrective (C0719519) | 9 | Glucophage (C0591573) | 131 | NovoLog (C0939412) | 255 | |
| 6 | Botox (C0700702) | 9 | Levemir (C1314782) | 122 | Levemir (C1314782) | 184 | |
| 7 | RID (C0073361) | 6 | Actos (C0875954) | 95 | Glucophage (C1314782) | 161 | |
| 8 | HumaLog (C0528249) | 5 | Seroquel (C0287163) | 78 | Novolin (C0028467) | 112 | |
| 9 | Bead Dosage Form (C0991566) | 3 | Synthroid (C0728762) | 62 | Viagra (C0663448) | 105 | |
| 10 | Actos (C0875954) | 3 | Coumadin (C0699129) | 54 | Triphosphat (C0146894) | 77 | |
aCUI: concept unique identifier
All the concepts in Table 4 are about pharmacological substances or organic chemicals, except stomach found within questions. Three concepts regarding insulin therapy for diabetes, such as Lantus (ie, insulin glargine injection), Humalog (ie, insulin lispro injection), and Actos (ie, pioglitazone hydrochloride) in blog postings and questions/answers appeared with high frequency. Diabetes-treatment-related concepts, such as NovoLog and Glucophage, are more frequently observed in questions and answers than blog postings. In total, 9 out of the top 10 concepts in questions and answers were diabetes medications. Only 2 concepts, namely stomach in questions and Viagra in answers, are not directly related to diabetes treatment. On the other hand, some concepts in blogs were indirectly related to diabetes. For example, cider vinegar, apple cider vinegar, and Botox also frequently appeared.
There were also the concepts covered by SNOMED CT but not CHV: 17.6% (424/2415) concepts from blog postings, 957/6452 (14.8%) concepts in questions and 18.7% (1936/10,378) concepts in answers (See Table 5). Human body related concepts, such as back structure excluding neck, entire heart, entire pancreas, entire kidney, entire skin, and entire eye, were frequently used to describe their diabetes issues in blog postings or questions/answers. Three concepts, entire skin, symptoms and fatty acid glycerol esters were observed from all our datasets. Massage and training were frequently mentioned in blog postings, while injection procedure and protective cup were frequently mentioned in questions and answers but were not mentioned as frequently as in blog postings. As these concepts were frequently observed in social media, CHV should consider importing them to enrich its conceptual content.
Table 5.
Top 10 frequently observed concepts covered by SNOMED CT but not CHV.
|
|
Tumblr | Yahoo! Answers | ||||
| Rank |
|
|
Questions | Answers | ||
| Concept (CUI)a | Freq. | Concept (CUI) | Freq. | Concept (CUI) | Freq. | |
| 1 | Entire skin (C1278993) | 524 | Symptoms (C1457887) | 7690 | Symptoms (C1457887) | 12,727 |
| 2 | Symptoms (C1457887) | 393 | Fatty acid glycerol esters (C0015677) | 1789 | Fatty acid glycerol esters (C0015677) | 8727 |
| 3 | Back structure, excluding neck (C1995000) | 236 | Entire foot (C1281587) | 1647 | Entire cells (C1269647) | 6435 |
| 4 | Massage (C0024875) | 217 | Back structure, excluding neck (C1995000) | 1589 | Entire heart (C1281570) | 3204 |
| 5 | Fatty acid glycerol esters (C0015677) | 210 | Entire kidney (C1278978) | 1368 | Entire pancreas (C1278931) | 3003 |
| 6 | Training (C0220931) | 163 | Entire eye (C1280202) | 1210 | Entire skin (C1278993) | 2614 |
| 7 | Entire pancreas (C1278931) | 157 | Protective cup (C1533124) | 1159 | Protective cup (C1533124) | 2178 |
| 8 | Entire heart (C1281570) | 156 | Entire lower limb (C1269079) | 985 | Entire stomach (C1278920) | 1876 |
| 9 | Entire oral cavity (C1278910) | 138 | Entire hands (C1281583) | 969 | Injection procedure (C1533685) | 1561 |
| 10 | Entire spine (C1280065) | 137 | Entire skin (C1278993) | 912 | Entire bony skeleton (C1266909) | 1501 |
aCUI: concept unique identifier
Semantic Types of the Identified Concepts
Among 127 UMLS semantic types (STY), about half of them were identified in our datasets: 52 STYs (40.9%) in the blog postings, 59 STYs (46.5%) in the questions, and 54 STYs (42.5%) in the answers. In general, there was a significant overlap of STYs across our datasets with 52 shared STYs. Seven STYs, however, were identified in the questions only, including “Functional Concept,” “Intellectual Product,” “Laboratory Procedure,” “Organ or Tissue Function,” “Organism Attribute,” “Social Behavior,” and “Substance.” Two STYs, “Fully Formed Anatomical Structure” and “Cell or Molecular Dysfunction,” were not found in questions, but in both the answer dataset and the blog dataset. Table 6 shows the top 20 frequent semantic types of the identified UMLS concepts in the different datasets respectively.
Table 6.
Top 20 frequently observed semantic types of the identified concepts.
| Rank | Tumblr | Yahoo! Answers | ||||||||
| Blogs | Questions | Answers | ||||||||
| Semantic type | Concepta | Semantic type | Concept | Semantic type | Concept | |||||
| n (%) | Freq. | n (%) | Freq. | n (%) | Freq. | |||||
| 1 | Finding | 380 (15.7) |
5277 | Pharmacologic substance | 1240 (19.2) | 53,976 | Pharmacologic substance | 1995 (19.2) |
185,880 | |
| 2 | Pharmacologic substance | 307 (12.7) | 4413 | Organic chemical | 1006 (15.6) |
41,255 | Organic chemical | 1692 (16.3) |
123,509 | |
| 3 | Therapeutic or preventive procedure | 241 (10.0) |
3184 | Finding | 895 (13.9) |
30,458 | Disease or syndrome | 1511 (14.6) |
57,379 | |
| 4 | Disease or syndrome | 239 (9.9) |
2923 | Disease or syndrome | 743 (11.5) | 28,041 | Finding | 1302 (12.5) |
76,765 | |
| 5 | Organic chemical | 225 (9.3) |
2737 | Body part, organ, or organ component | 484 (7.5) |
27,172 | Body part, organ, or organ component | 666 (6.4) |
48,584 | |
| 6 | Body part, organ, or organ component | 208 (8.6) |
2566 | Sign or symptom | 338 (5.2) |
19,601 | Therapeutic or preventive procedure | 583 (5.6) |
16,555 | |
| 7 | Sign or symptom | 145 (6.0) | 2214 | Therapeutic or preventive procedure | 331 (5.1) |
16,372 | Amino acid, peptide, or protein | 495 (4.8) |
40,521 | |
| 8 | Medical device | 134 (5.5) |
1319 | Amino acid, peptide, or protein | 305 (4.7) |
13,178 | Sign or symptom | 436 (4.2) |
38,905 | |
| 9 | Amino acid, peptide, or protein | 70 (2.9) |
1112 | Medical device | 201 (3.1) |
12,862 | Medical device | 347 (3.3) |
20,391 | |
| 10 | Biologically active substance | 69 (2.9) |
1093 | Laboratory procedure | 180 (2.8) |
10,580 | Pathologic function | 292 (2.8) |
12,551 | |
aThe percentage was calculated based on the total number of unique identified UMLS concepts: blogs in Tumblr: n=2415, questions in Yahoo! Answers: n=6452, answers in Yahoo! Answers: n=10,378
When comparing the top 10 frequently observed STYs across the datasets, 9 out of 10 STYs (ie, “Finding,” “Pharmacologic Substance,” “Therapeutic or Preventive Procedure,” “Disease or Syndrome,” “Organic Chemical,” “Body Part, Organ, or Organ Component,” “Sign or Symptom,” “Medical Device,” and “Amino Acid, Peptide, or Protein”) commonly appeared across the datasets with minor differences in terms of frequency. “Laboratory Procedure” appeared frequently in questions but not in blogs and answers. “Pathologic Function” appeared frequently in answers but not in blogs and questions. Example concepts of the frequently observed STYs showed that laypeople tend to frequently use common concepts to describe their diabetes-related issues in social media. To illustrate, Sugars, Insulin, Glucose ranked in top 5 concepts of the STY “Pharmacologic Substance.” Similarly, the concepts such as Disease and Communicable Diseases appeared frequently among the concepts of the STY “Disease or Syndrome.” We provide the top 5 frequent concepts for the top 10 frequently observed semantic types in Multimedia Appendix 1 Table A3.
Discussion
Principal Findings
Previous studies [12-15] utilized user-generated documents including social media. However, they mainly used a single test bed based on the assumption that the selected test bed would properly reflect people’s medical concepts. Our study involved different types of social media which contains texts that laypeople generated for different purposes: questions are for expressing their health information seeking needs; blogs and answers are more likely for sharing their knowledge, experiences, and opinions to others. The current study investigated the terminology coverage in consumer-generated text in social media by identifying UMLS concepts and their semantic types. Our findings demonstrated that consumers use medical concepts not only from controlled vocabularies developed for consumers (ie, CHV) but also those for health professionals (eg, SNOMED CT). Our results are in line with prior observations that consumers use both lay and professional terms [24,26,65] and demonstrated that CHV can be enriched by other source vocabularies in the UMLS.
The UMLS concept usage in blogs and social Q&A was different in that the UMLS concepts appeared more frequently in the postings of social Q&A (almost 90% questions and answers) in comparison to blog postings (about 30%). Social Q&A users mainly discuss health-related issues (in the current study, diabetes-related issues) in their postings, because their participation in question asking and answering is purpose-driven. On the other hand, blog users often elaborate nonhealth related topics in their postings, although they tagged their postings with “diabetes.”
In spite of the differences of the overall UMLS concept coverage between blogs and social Q&A, we found that the UMLS concepts identified in different datasets can be covered by a similar number of UMLS source vocabularies. Two UMLS source vocabularies, ie, SNOMED CT and CHV, showed the best coverage. Social media users in our datasets may have advanced medical knowledge because they often use professional terms. CHV demonstrated the second largest coverage for all the datasets despite the fact that CHV has a much smaller number of concepts and terms than SNOMED CT (1:6 ratio). CHV was developed to incorporate consumer expressions presented in consumer-generated text data. Our findings showed that different social media platforms may play a similar role as consumer-generated documents for CHV enrichment, which confirmed the literature [66].
A comparison of the concept coverage between SNOMED CT and CHV in our datasets led us to examine the difference between the concept usages among blog and social media users. For example, cider vinegar, apple cider vinegar, massages, and training were frequently mentioned in blog postings, while they were not frequently mentioned in questions and answers. However, concepts pertaining to insulin therapy, such as Lantus, Humalog, and Actos, are frequently used in questions/answers. Consumers often inquire about a variety of insulin therapies in social Q&A, while blogs often include recipes specific to the use of vinegar, a popular ingredient in diabetes-controlling food. Botox and Viagra were often mentioned in blog postings and answers. They could be important for diabetic patients, although they may not be closely related to control diabetes. It would be interesting to further analyze the relationship of these terms to diabetes. An in-depth analysis of the identified concepts along with how they are used in the original postings could produce useful information for understanding consumers’ information needs and use.
According to our analysis, the percentage of unique concepts covered by CHV but not by SNOMED CT varied from 1.7% to 6.3%. In the blog dataset, where approximately 3000 blogs were analyzed, only 40 concepts were covered by CHV exclusively. On the other hand, in Yahoo! Answers, 409 concepts (6.3%) in questions and 529 concepts (5.1%) in answers were covered by CHV but not by SNOMED CT. These results indicate that the larger datasets would yield more lay concepts. The size of dataset also appeared to affect the diversity of semantics. The same set of 9 semantic types was observed frequently in all our datasets. “Finding,” “Pharmacologic Substance,” and “Disease and Syndrome” were among the top 4 most observed semantic types.
Differences were observed as well. Blogs might be better platforms for consumers to discuss organic chemical, pharmacologic substances, or therapeutic or preventive procedure for diabetes. Yet, concepts of organic chemical and pharmacologic substances were also frequently used in social Q&A. In social Q&A data, 7 semantic types that were not identified in blogs were observed, indicating that larger datasets may yield more diverse medical concepts.
Limitations
This study has a few limitations. First, the blog data in Tumblr and Yahoo! Answers data were collected in different time frames and are different in size, which might have affected the findings of this study. Smaller volumes of blog data used in this study may affect the diversity of the UMLS concepts identified. Even though blogging and question posting/answering are dynamic online activities for those living with chronic diseases, Tumblr and Yahoo! Answers may not represent all the health information users’ concept usage. The datasets could be expanded to include other types of social media such as diabetes-related discussion boards. The users of these online sources may be biased towards those with greater technological proficiency, such as those who are younger, more educated or those in a higher socioeconomic status who are more likely to seek health information on the Internet. This study may not reflect the experiences of those who are older adults, less educated or underprivileged [67]. Second, even though the automated NLP techniques that were employed in the current study were cost-effective, direct interaction with ordinary health information users would allow the researchers to capture more accurate meaning of medical concepts that these individuals commonly use to describe their health issues. Moreover, a qualitative approach such as content analysis also would help to identify contextual semantics of the concepts. Third, although the WSD function of YTEX is effective in selecting the most possible UMLS concepts for a term in free text, the same term may be matched to different ambiguous UMLS concepts. This is mainly due to the fact that the UMLS may contain unmapped synonymous concepts. Ideally, manual review by domain experts could be applied to further refine the automatic mapping results.
Conclusions
The current study examined the potential of social media as user-generated documents in which consumers’ medical concepts can be observed and leveraged for controlled vocabulary development for ordinary health information users. We selected and tested 2 social media venues, namely blogs and social Q&A. Our findings showed stronger similarities rather than differences in the controlled vocabulary usage. The size of a dataset may affect the number of concepts identified. However, the similarities in the source vocabularies, frequently used concepts, and semantic types of the concepts indicate that social media sites tend to reflect the common sense of laypeople. More importantly, we found that social media users not only employ consumer concepts in CHV but also concepts in professional vocabularies such as SNOMED CT. This indicates that CHV still has room for improvements by incorporating concepts from other UMLS source vocabularies. The focus of our study is not to identify a list of consumer medical concepts, but to test the feasibility of leveraging social media data to identify consumer concepts covered by existing UMLS source vocabularies. Ultimately, it would assist consumers’ health information searches online, narrowing the disparity between ordinary health information users and medical professionals. In future studies, we will employ automated approaches to identify and recommend new medical terms and concepts from social media to enrich CHV.
Acknowledgments
We would like to thank Dr Warren Allen for providing the computing resources for this work. This work was partially supported by an Amazon Web Services Education and Research Grant Award (PI: He). The work was also partially supported by National Center for Advancing Translational Sciences under the Clinical and Translational Science Award UL1TR001427 (PI: Nelson & Shenkman). The content is solely the responsibility of the authors and does not represent the official view of the National Institutes of Health.
Abbreviations
- APIs
Application Program Languages
- AUI
atom unique identifier
- CSP
Computer Retrieval of Information on Scientific Projects Thesaurus
- CUI
concept unique identifier
- IC
information content
- LCH NW
Library of Congress Subject Headings, Northwestern University subset
- LOINC
Logical Observation Identifier Names and Codes
- MeSH
Medical Subject Headings
- NCIt
National Cancer Institute Thesaurus
- NDFRT
National Drug File – Reference Terminology
- NER
named entity recognition
- NLM
National Library of Medicine
- NLP
natural language processing
- OAC CHV
Open-Access Collaborative Consumer Health Vocabulary
- POS
Part-Of-Speech
- Q&A
questions and answers
- SNOMED CT
SNOMED Clinical Terms
- STY
semantic type
- UIMA
Unstructured Information Management Architecture
- UMLS
Unified Medical Language System
- WSD
word sense disambiguation
Table A1. Full names of the UMLS source vocabularies in Table 2. Table A2. Top 10 frequently observed concepts in the top 3 most covered source vocabularies. Table A3. Top 5 most frequently concepts in the top 9 frequent semantic types.
Footnotes
Authors' Contributions: MP initiated the idea of this study. ZH led the conceptualization, design, and implementation of this study. MP collected and provided the blog data from Tumblr.com. SO collected and provided the social Q&A data from Yahoo! Answers. ZC performed the natural language processing on the datasets and structured the results in a relational database. MP performed the data analysis and drafted the initial version; ZH, SO, BJ extensively revised the draft critically and iteratively for important intellectual content. All authors contributed to the methodology development, results interpretation, edited the paper significantly, and gave final approval for the version to be published. ZH takes primary responsibility for the research reported here.
Conflicts of Interest: None declared.
References
- 1.Messai R, Simonet M, Bricon-Souf N, Mousseau M. Characterizing consumer health terminology in the breast cancer field. Stud Health Technol Inform. 2010;160(Pt 2):991–4. [PubMed] [Google Scholar]
- 2.Poikonen T, Vakkari P. Lay persons? and professionals? nutrition-related vocabularies and their matching to a general and a specific thesaurus. Journal of Information Science. 2009;35(2):232–43. [Google Scholar]
- 3.Smith CA, Wicks PJ. PatientsLikeMe: Consumer health vocabulary as a folksonomy. AMIA Annu Symp Proc. 2008:682–6. http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2656083&tool=pmcentrez&rendertype=abstract . [PMC free article] [PubMed] [Google Scholar]
- 4.Patrick TB, Monga HK, Sievert ME, Houston HJ, Longo DR. Evaluation of controlled vocabulary resources for development of a consumer entry vocabulary for diabetes. J Med Internet Res. 2001;3(3):E24. doi: 10.2196/jmir.3.3.e24. http://www.jmir.org/2001/3/e24/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zielstorff RD. Controlled vocabularies for consumer health. J Biomed Inform. 2003;36(4-5):326–33. doi: 10.1016/j.jbi.2003.09.015. http://linkinghub.elsevier.com/retrieve/pii/S1532046403000960 .S1532046403000960 [DOI] [PubMed] [Google Scholar]
- 6.Tse T, Soergel D. Exploring medical expressions used by consumers and the media: an emerging view of consumer health vocabularies. AMIA Annu Symp Proc. 2003:674–8. http://europepmc.org/abstract/MED/14728258 .D030002918 [PMC free article] [PubMed] [Google Scholar]
- 7.Vydiswaran VG, Mei Q, Hanauer DA, Zheng K. Mining consumer health vocabulary from community-generated text. AMIA Annu Symp Proc. 2014;2014:1150–9. http://europepmc.org/abstract/MED/25954426 . [PMC free article] [PubMed] [Google Scholar]
- 8.Seedorff M, Peterson KJ, Nelsen LA, Cocos C, McCormick JB, Chute CG, Pathak J. Incorporating expert terminology and disease risk factors into consumer health vocabularies. Pac Symp Biocomput. 2013:421–32. http://psb.stanford.edu/psb-online/proceedings/psb13/abstracts/2013_p421.html .9789814447973_0041 [PMC free article] [PubMed] [Google Scholar]
- 9.Gross T, Taylor A. What have we got to lose? The effect of controlled vocabulary on keyword searching results. College & Research Libraries. 2005;66(3):212–30. [Google Scholar]
- 10.Lewis D, Eysenbach G, Jimison Hb, Kukafka R, Stavri Pz. Consumer Health Informatics: Informing Consumers and Improving Health Care. New York, NY: Springer; 2005. Consumer health informatics; pp. 1–7. [Google Scholar]
- 11.Roberts K, Demner-Fushman D. Interactive use of online health resources: a comparison of consumer and professional questions. J Am Med Inform Assoc. 2016 Jul;23(4):802–11. doi: 10.1093/jamia/ocw024.ocw024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeng Q, Kogan S, Ash N, Greenes RA. Patient and clinician vocabulary: how different are they? Stud Health Technol Inform. 2001;84(Pt 1):399–403. [PubMed] [Google Scholar]
- 13.Plovnick RM, Zeng QT. Reformulation of consumer health queries with professional terminology: a pilot study. J Med Internet Res. 2004 Sep 03;6(3):e27. doi: 10.2196/jmir.6.3.e27. http://www.jmir.org/2004/3/e27/ v6e27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brennan PF, Aronson AR. Towards linking patients and clinical information: detecting UMLS concepts in e-mail. J Biomed Inform. 2003;36(4-5):334–41. doi: 10.1016/j.jbi.2003.09.017.S1532046403000984 [DOI] [PubMed] [Google Scholar]
- 15.Smith CA, Stavri PZ, Chapman WW. In their own words? A terminological analysis of e-mail to a cancer information service. Proc AMIA Symp. 2002:697–701. http://europepmc.org/abstract/MED/12463914 .D020002157 [PMC free article] [PubMed] [Google Scholar]
- 16.Harpring P. What Are Controlled Vocabularies? In: Baca M, editor. Introduction to Controlled Vocabularies: Terminology for Art, Architecture, and Other Cultural Works. Los Angeles, CA: Getty Publications; 2010. [Google Scholar]
- 17.Zeng QT, Tse T. Exploring and developing consumer health vocabularies. J Am Med Inform Assoc. 2006;13(1):24–9. doi: 10.1197/jamia.M1761. http://jamia.oxfordjournals.org/lookup/pmidlookup?view=long&pmid=16221948 .M1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cimino JJ. Desiderata for controlled medical vocabularies in the twenty-first century. Methods Inf Med. 1998 Nov;37(4-5):394–403. http://europepmc.org/abstract/MED/9865037 .98040394 [PMC free article] [PubMed] [Google Scholar]
- 19.Arts DG, Cornet R, de Jonge E, de Keizer NF. Methods for evaluation of medical terminological systems--a literature review and a case study. Methods Inf Med. 2005;44(5):616–25.05050616 [PubMed] [Google Scholar]
- 20.Greenberg J. Metadata and the World Wide Web. In: Drake M, editor. Encyclopedia of Library and Information Science. New York, NY: Marcel Deker, Inc; 2003. pp. 1876–1888. [Google Scholar]
- 21.Mathes A. [2016-11-02]. Folksonomies - cooperative classification and communication through shared metadata. 2004 Dec. http://www.adammathes.com/academic/computer-mediated-communication/folksonomies.html .
- 22.Kim S. An exploratory study of user-centered indexing of published biomedical images. J Med Libr Assoc. 2013 Jan;101(1):73–6. doi: 10.3163/1536-5050.101.1.011. http://europepmc.org/abstract/MED/23405049 .JMLA-D-12-00011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacLean DL, Heer J. Identifying medical terms in patient-authored text: a crowdsourcing-based approach. J Am Med Inform Assoc. 2013;20(6):1120–7. doi: 10.1136/amiajnl-2012-001110. http://jamia.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=23645553 .amiajnl-2012-001110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doing-Harris KM, Zeng-Treitler Q. Computer-assisted update of a consumer health vocabulary through mining of social network data. J Med Internet Res. 2011 May 17;13(2):e37. doi: 10.2196/jmir.1636. http://www.jmir.org/2011/2/e37/ v13i2e37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hicks A, Hogan WR, Rutherford M, Malin B, Xie M, Fellbaum C, Yin Z, Fabbri D, Hanna J, Bian J. Mining Twitter as a First Step toward Assessing the Adequacy of Gender Identification Terms on Intake Forms. AMIA Annu Symp Proc. 2015;2015:611–20. http://europepmc.org/abstract/MED/26958196 . [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis D, Brennan PF, McCray AT, Tuttle M, Bachman J. If We Build It, They Will Cometandardized Consumer Vocabularies. Studies in Health Technology and Informatics. 2001;84:1530. http://ebooks.iospress.nl/publication/19863 . [Google Scholar]
- 27.Kamdar MR, Tudorache T, Musen M. A Systematic Analysis of Term Reuse and Term Overlap across Biomedical Ontologies. Semantic Web – Interoperability, Usability, Applicability. 2016 doi: 10.3233/sw-160238. (forthcoming). http://www.semantic-web-journal.net/content/systematic-analysis-term-reuse-and-term-overlap-across-biomedical-ontologies-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.He Z, Geller J, Chen Y. A comparative analysis of the density of the SNOMED CT conceptual content for semantic harmonization. Artif Intell Med. 2015 May;64(1):29–40. doi: 10.1016/j.artmed.2015.03.002. http://europepmc.org/abstract/MED/25890688 .S0933-3657(15)00025-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Z, Geller J, Elhanan G. Categorizing the Relationships between Structurally Congruent Concepts from Pairs of Terminologies for Semantic Harmonization. AMIA Jt Summits Transl Sci Proc. 2014;2014:48–53. http://europepmc.org/abstract/MED/25717400 . [PMC free article] [PubMed] [Google Scholar]
- 30.He Z, Chen Y, de Coronado S, Piskorski K, Geller J. Topological-Pattern-based Recommendation of UMLS Concepts for National Cancer Institute Thesaurus. AMIA Annu Symp Proc. 2016 (forthcoming) [PMC free article] [PubMed] [Google Scholar]
- 31.Chandar P, Yaman A, Hoxha J, He Z, Weng C. Similarity-Based Recommendation of New Concepts to a Terminology. AMIA Annu Symp Proc. 2015;2015:386–95. http://europepmc.org/abstract/MED/26958170 . [PMC free article] [PubMed] [Google Scholar]
- 32.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004 Jan 1;32(Database issue):D267–70. doi: 10.1093/nar/gkh061. http://nar.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=14681409 .32/suppl_1/D267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. [2016-11-02]. Global Report on Diabetes. World Health Organization. http://apps.who.int/iris/bitstream/10665/204871/1/9789241565257_eng.pdf?ua=1 .
- 34.American Diabetes Association. [2016-11-02]. Diabetes complications. http://www.diabetes.org/living-with-diabetes/complications/
- 35.Krug EG. Trends in diabetes: sounding the alarm. Lancet. 2016 Apr 9;387(10027):1485–6. doi: 10.1016/S0140-6736(16)30163-5. http://linkinghub.elsevier.com/retrieve/pii/S0140-6736(16)30163-5 .S0140-6736(16)30163-5 [DOI] [PubMed] [Google Scholar]
- 36.Fox S. [2016-11-02]. The social life of health information. Pew Research Center. http://www.pewresearch.org/fact-tank/2014/01/15/the-social-life-of-health-information/
- 37.Andersen NB, Söderqvist T. [2016-11-02]. Social media and public health research. Copenhagen, Denmark: University of Copenhagen. 2012 Aug 20. http://www.museion.ku.dk/wp-content/uploads/FINAL-Social-Media-and-Public-Health-Research.pdf .
- 38.Oh S. The characteristics and motivations of health answerers for sharing information, knowledge, and experiences in online environments. J. Am. Soc. Inf. Sci. 2011 Nov 01;63(3):543–557. doi: 10.1002/asi.21676. [DOI] [Google Scholar]
- 39.Giustini D. How Web 2.0 is changing medicine. BMJ. 2006 Dec 23;333(7582):1283–4. doi: 10.1136/bmj.39062.555405.80. http://europepmc.org/abstract/MED/17185707 .333/7582/1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.U.S. National Library of Medicine. [2016-11-02]. Fact Sheet of the UMLS Metathesaurus. https://www.nlm.nih.gov/pubs/factsheets/umlsmeta.html .
- 41.U.S. National Library of Medicine. [2016-11-02]. SNOMED Clinical Terms. https://www.nlm.nih.gov/healthit/snomedct/index.html .
- 42.Finnegan R. ICD-9-CM coding for physician billing. J Am Med Rec Assoc. 1989 Feb;60(2):22–3. [PubMed] [Google Scholar]
- 43.Bennett CC. Utilizing RxNorm to support practical computing applications: capturing medication history in live electronic health records. J Biomed Inform. 2012 Aug;45(4):634–41. doi: 10.1016/j.jbi.2012.02.011. http://linkinghub.elsevier.com/retrieve/pii/S1532-0464(12)00041-X .S1532-0464(12)00041-X [DOI] [PubMed] [Google Scholar]
- 44.U.S. National Library of Medicine. [2016-07-13]. Fact Sheet of the UMLS Semantic Network. https://www.nlm.nih.gov/pubs/factsheets/umlssemn.html .
- 45.Zeng-Treitler Q, Goryachev S, Kim H, Keselman A, Rosendale D. Making texts in electronic health records comprehensible to consumers: a prototype translator. AMIA Annu Symp Proc. 2007:846–50. http://europepmc.org/abstract/MED/18693956 . [PMC free article] [PubMed] [Google Scholar]
- 46.Zielstorff Rd. Controlled vocabularies for consumer health. Journal of Biomedical Informatics. 2003 Aug;36(4-5):326–333. doi: 10.1016/j.jbi.2003.09.015. [DOI] [PubMed] [Google Scholar]
- 47.Miller EA, Pole A. Diagnosis blog: checking up on health blogs in the blogosphere. Am J Public Health. 2010 Aug;100(8):1514–9. doi: 10.2105/AJPH.2009.175125.AJPH.2009.175125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lagu T, Kaufman EJ, Asch DA, Armstrong K. Content of weblogs written by health professionals. J Gen Intern Med. 2008 Oct;23(10):1642–6. doi: 10.1007/s11606-008-0726-6. http://europepmc.org/abstract/MED/18649110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boulos Maged N Kamel. Maramba I, Wheeler S. Wikis, blogs and podcasts: a new generation of Web-based tools for virtual collaborative clinical practice and education. BMC Med Educ. 2006;6:41. doi: 10.1186/1472-6920-6-41. http://www.biomedcentral.com/1472-6920/6/41 .1472-6920-6-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oomen-Early J, Burke S. Entering the Blogosphere: Blogs as Teaching and Learning Tools in Health Education. International Electronic Journal of Health Education. 2007;10:186–196. http://eric.ed.gov/?id=EJ794207 . [Google Scholar]
- 51.Cobus Laura. Using blogs and wikis in a graduate public health course. Med Ref Serv Q. 2009;28(1):22–32. doi: 10.1080/02763860802615922.908112720 [DOI] [PubMed] [Google Scholar]
- 52.Shah C, Oh S, Oh J. Research agenda for social Q&A. Library & Information Science Research. 2009;31(4):205–9. doi: 10.1016/j.lisr.2009.07.006. [DOI] [Google Scholar]
- 53.Statista. [2016-02-06]. Statistics and facts about Tumblr. http://www.statista.com/topics/2463/tumblr/
- 54.DearBlogger: The Blogging Answers Community. [2016-11-02]. The Best Places to Start a Blog. http://www.dearblogger.org/blogger-or-wordpress-better .
- 55.Fitzpatrick J. Lifehacker. 2010. Jun 20, [2016-11-02]. Five Best Blogging Platforms. Lifehacker. 2010 Jun 20. http://lifehacker.com/5568092/five-best-blogging-platforms .
- 56.Quantcast. [2016-07-13]. Statistics of Yahoo! Answers. https://www.quantcast.com/answers.yahoo.com .
- 57.Temmerman R. Towards new ways of terminology description: the sociocognitive-approach. Amsterdam: J. Benjamins; 2000. Towards new ways of terminology description: The sociocognitive-approach. [Google Scholar]
- 58.Crystal D. Dictionary of Linguistics and Phonetics (The Language Library) Hoboken, NJ: Wiley-Blackwell; 2008. Jun, [Google Scholar]
- 59.U.S. National Library of Medicine. [2016-01-08]. UMLS Glossary. https://www.nlm.nih.gov/research/umls/new_users/glossary.html .
- 60.Apache Software Foundation. [2016-01-18]. cTAKES (clinical Text Analysis and Knowledge Extraction System). 2016 Jan 18. http://ctakes.apache.org .
- 61.Garla V, Lo RV, Dorey-Stein Z, Kidwai F, Scotch M, Womack J, Justice A, Brandt C. The Yale cTAKES extensions for document classification: architecture and application. J Am Med Inform Assoc. 2011;18(5):614–20. doi: 10.1136/amiajnl-2011-000093. http://jamia.oxfordjournals.org/cgi/pmidlookup?view=long&pmid=21622934 .amiajnl-2011-000093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Baldridge J. Apache Software Foundation. [2016-11-02]. The openNLP project. http://opennlp.apache.org/index .
- 63.Apache Software Foundation. [2016-11-02]. The OpenNLP Documentation. https://opennlp.apache.org/documentation.html .
- 64.Garla VN, Brandt C. Semantic similarity in the biomedical domain: an evaluation across knowledge sources. BMC Bioinformatics. 2012 Oct 10;13:261. doi: 10.1186/1471-2105-13-261. http://bmcbioinformatics.biomedcentral.com/articles/10.1186/1471-2105-13-261 .1471-2105-13-261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He Z, Park M, Chen Z. UMLS-Based Analysis of Medical Terminology Coverage for Tags in Diabetes-Related Blogs. Philadelphia, PA; iConference 2016; March 20-23, 2016; 2016. https://www.ideals.illinois.edu/handle/2142/89441 . [Google Scholar]
- 66.Sarasohn-Kahn J. [2016-11-02]. The Wisdom of Patients: Health Care Meets Online Social Media. California Health Care Foundation. 2008 Apr. http://www.chcf.org/publications/2008/04/the-wisdom-of-patients-health-care-meets-online-social-media .
- 67.Zhang Y. Beyond quality and accessibility: Source selection in consumer health information searching. J Assn Inf Sci Tec. 2014 Jan 07;65(5):911–927. doi: 10.1002/asi.23023. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table A1. Full names of the UMLS source vocabularies in Table 2. Table A2. Top 10 frequently observed concepts in the top 3 most covered source vocabularies. Table A3. Top 5 most frequently concepts in the top 9 frequent semantic types.