Abstract
Niemann–Pick type C (NPC) 1 protein plays important roles in moving cholesterol and other lipids out of late endosomes by means of vesicular trafficking, but it is not known whether NPC1 directly interacts with cholesterol. We performed photoaffinity labeling of intact cells expressing fluorescent protein (FP)-tagged NPC1 by using [3H]7,7-azocholestanol ([3H]AC). After immunoprecipitation, 3H-labled NPC1-GFP appeared as a single band. Including excess unlabeled sterol to the labeling reaction significantly diminished the labeling. Altering the NPC1 sterol-sensing domain (SSD) with loss-of-function mutations (P692S and Y635C) severely reduced the extent of labeling. To further demonstrate the specificity of labeling, we show that NPC2, a late endosomal/lysosomal protein that binds to cholesterol with high affinity, is labeled, whereas mutant NPC2 proteins inactive in binding cholesterol are not. Vamp7, an abundant late endosomal membrane protein without an SSD but with one transmembrane domain, cannot be labeled. Binding between [3H]AC and NPC1 does not require NPC2. Treating cells with either U-18666A, a compound that creates an NPC-like phenotype, or with bafilomycin A1, a compound that raises late endosomal pH, has no effect on labeling of NPC1-YFP, suggesting that both drugs affect processes other than NPC1 binding to cholesterol. We also developed a procedure to label the NPC1-YFP by [3H]AC in vitro and showed that cholesterol is more effective in protection against labeling than its analogs epicholesterol or 5-α-cholestan. Overall, the results demonstrate that there is direct binding between NPC1 and azocholestanol; the binding does not require NPC2 but requires a functional SSD within NPC1.
Niemann–Pick type C (NPC) disease is a fatal neurovisceral disorder characterized by clinically progressive hepatosplenomegaly and central nervous system neurodegeneration (reviewed in ref. 1). The disease involves the accumulation of unesterified cholesterol, sphingomyelin, glycolipids, and other lipids within the endosomal/lysosomal system, in cells of various tissues (1), and in neurons (2–4). Currently there is no cure for the disease. The disease can be caused by mutations in either of two genetic loci, NPC1 and NPC2. Mutations in NPC1 account for ≈95% of all of the NPC disease cases, whereas mutations at NPC2 account for the remaining ≈5% (1).
NPC1 has been extensively studied at the cellular and molecular levels. Chinese hamster ovary (CHO) cells with NPC1 mutations have been isolated, the CT60 and CT43 mutants from 25RA cells (5, 6) and other mutants from WT cells (7–9). The human NPC1 gene encodes a 1,278-aa (170–190 kDa) glycoprotein, with 13 putative transmembrane domains (TMD) that include a sterol-sensing domain (SSD) located between the third and seventh TMDs (10). The NPC2 gene encodes a soluble lysosomal protein (11) that can be secreted into the medium and binds cholesterol with high affinity (12). Single mutations within the putative cholesterol-binding pocket (13), including the F66A, V96F, and the Y100A mutants, disrupt the ability of the NPC2 to bind cholesterol in vitro (12).
SSDs consist of ≈180 aa organized in five consecutive membrane-spanning domains. NPC1 proteins with single mutations in the SSD, including the Y635C and the P692S mutants, are unable to rescue the cholesterol-trafficking defect present in CT60 cells (14, 15). The SSD is present in other membrane proteins including hydroxymethylglutaryl (HMG)-CoA reductase (16, 17), sterol regulatory element-binding protein cleavage-activating protein (SCAP) (18), PATCHED (19), and NPC1L1 (20). Thus, unraveling the mysteries of NPC1 will lead not only to greater knowledge of a devastating human disease, but also to an increased understanding of a family of proteins involved in diverse biological functions, including dietary uptake of cholesterol, mediation of cellular cholesterol homeostasis, and cell–cell signaling (21, 22).
The native NPC1 protein resides mainly in late endosomes and the related tubulovesicles (15, 23); it may interact transiently with lysosomes and the trans-Golgi network (24, 25). In mutant NPC1 cells, transport of cholesterol to the endosome/lysosome from low-density lipoprotein (LDL) (26, 27) or from endogenous biosynthesis (2, 28–30) is partially defective. It has been proposed that NPC1 may be involved in mediating the transport of multiple cargo, including cholesterol, fatty acids, and/or glycosphingolipids, in late endosomes/lysosomes (25, 31, 32). When expressed in Escherichia coli, the NPC1 protein exhibits lipid permease activity and transports fatty acids but not cholesterol (33). However, there is currently no evidence for NPC1 interacting directly with cholesterol or with other lipids. Photoaffinity labeling has been widely used to demonstrate direct binding between a macromolecule and a specific ligand (reviewed in ref. 34). We and other investigators have previously reported the syntheses and use of two similar 3H-labeled photolabile cholesterol analogs, [3H]6,6-azocholestanol (35) and [3H]7,7-azocholestanol ([3H]AC; ref. 36; Fig. 1A shows cholesterol and AC). When fed to intact CHO cells, [3H]AC is metabolized in manners very similar to [3H]cholesterol (36). After photolysis, [3H]AC cross-linked to a variety of proteins; one of them was caveolin-1, a protein known to bind cholesterol with high affinity (36).
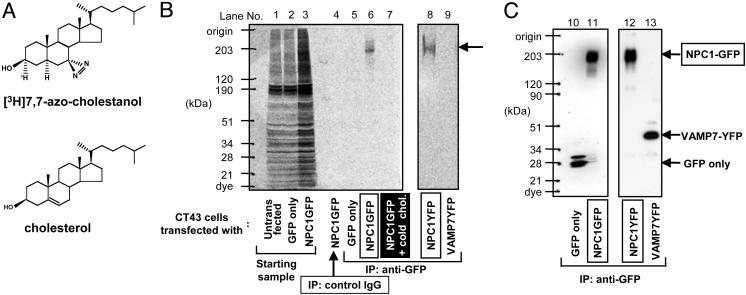
Fig. 1.
Identification of NPC1 protein as an azocholestanol-binding protein. (A) Chemical structures of 7,7-azo-5α-cholestan-3α[3H]-3β-ol ([3H]AC) and cholesterol. (B and C) Untransfected CT43 cells (lane 1) or CT43 cells transiently transfected with pREX-GFP (lanes 2, 5, and 10), pREX-NPC1-GFP (lanes 3, 4, 6, 7, and 11), pNPC1-EYFP (lanes 8 and 12), or pVAMP7-YFP (lanes 9 and 13) were photolabeled by using [3H]AC in the absence (lanes 1–6, 8, and 9) or presence (lane 7) of 60-fold excess of unlabeled cholesterol and processed for radioluminography (B) and Western blotting (C) according to procedures described in Materials and Methods. Antibodies used for IP were as follows: mouse monoclonal anti-GFP antibody (lanes 5–13) or control mouse IgG (lane 4). Antibodies used for Western blotting were as follows: rabbit anti-GFP antibodies (lanes 10–13). The labeled bands on the right are: (i) NPC1-GFP; (ii) VAMP7-YFP; (iii) GFP only. The results shown are representative of two experiments.
It is important to determine whether NPC1 binds sterols. To test the possibility that NPC1 may directly bind [3H]AC, we expressed fluorescent protein (FP)-tagged NPC1 protein in CHO cells or in human fibroblast cells. Intact cell labeling experiments using [3H]AC were used to assess binding to WT and mutant forms of NPC1, with the mutants providing important controls for specificity and information about SSD relevance. Our findings show that NPC1 does bind sterols and that this interaction requires an intact SSD.
Materials and Methods
Plasmids. pREX-NPC1-GFP and pREX-GFP were described in ref. 27. pNPC1-ECFP, pNPC1-EYFP, and plasmids containing various point mutations of pNPC1-FP were described in ref. 15. In addition, another point mutation, D945N, was produced. This mutation is found in people with severe NPC disease. pNPC2-myc-his and plasmids containing various point mutations of NPC2-myc-his were described in ref. 12. pVAMP7-YFP was from R. Advani (Stanford University, Stanford, CA).
Cell Culture and Transfection. The CT43 and CT60 mutant cell lines were isolated from mutagenized 25RA cells (5). Both mutants contain the same gain-of-function mutation in SCAP as the 25RA cells (18). In addition, the CT mutants contain a premature translational termination mutation near the 3′ end (for CT43), or near the 5′ end (for CT60) within the NPC1 coding sequence, producing nonfunctional, truncated NPC1 proteins (6). The CT60 cells stably expressing the NPC1-YFP protein (CT60-NPC1-YFP cells) have been described (15). CHO cell lines grown in 100-mm dishes were maintained in medium A (Ham's F-12, plus 10% FBS and 10 μg/ml gentamycin) as monolayers at 37°C with 5% CO2. The NPC2-deficient human fibroblast cell line expressing the SV40 large T antigen was from Dr. Yiannis A. Ioannou (37). Human fibroblasts were grown in the same condition as the CHO cells except that DMEM was used. For CT43 and 25RA cells, transfections with FuGENE 6 (Roche) were carried out. The cells were used for photolabeling within 2 days after transfection. NPC2-deficient human fibroblasts were transfected with 5 μg of pNPC1-EYFP and 15 μl of Lipofectamine 2000 (Invitrogen) in 8 ml of medium per dish, or cotransfected with 5 μg of pNPC1-EYFP, 5 μg of pNPC2-mychis, and 30 μl of Lipofectamine 2000 in 8 ml of medium per dish. Transfected human fibroblasts were used within 16 h after transfection.
Intact Cell Photolabeling. Transfected cells (3.0 × 106 cells unless specified) were grown in medium A for 48 h. Cells were detached by incubating with 10 mM EDTA in Hanks' buffer (unless specified otherwise) at room temperature for 5 min, then pelleted by brief centrifugation into 1.5-ml Eppendorf tubes on ice. After washing with Hanks' buffer, the cell pellets were preincubated with 74.6 μl of 5 mM methyl-β-cyclodextrin (CD; Sigma) for 15 min at 37°C. Next, 56.2 μl of the prewarmed [3H]AC/CD and 68 μl of CD with or without excess unlabeled sterol were added to the cell pellets. The [3H]AC/CD was prepared at 5 mM CD, with sterol/CD molar ratio at 1:64, and with [3H]AC at 34.0 μCi (1 Ci = 37 GBq) and 78.1 μM. The unlabeled sterol/CD complexes were prepared at 50 mM CD, with sterol/CD molar ratio at 1:16. CD with or without sterol was prepared in Hanks' buffer. The mixtures (3.0 × 106 cells per 300 μl final volume) were incubated for 1 h at 37°C, then ice-chilled and subjected to photolysis for 15 min on ice by using the photochemical reaction assembly (from Ace Glass; catalog no. 7825-34), with a 450-watt UV lamp at 366 nm. After photolysis, cells were washed with ice-chilled PBS and pelleted by a microcentrifuge. The pellets were lysed by using 1% Nonidet P-40 lysis buffer [50 mM Tris/100 mM NaCl/1% Nonidet P-40/1 mM EDTA/1 mM EGTA/protease inhibitors mixture (Sigma), pH 7.4]. The lysates were subjected to immunoprecipitations (IPs) as described (36), by using mouse monoclonal anti-GFP antibody (10 μg; Abcam1218), mouse monoclonal anti-c-myc antibody [10 μg; 9E10 (Roche)], or control mouse IgGs. The immunoprecipitates were subjected to 5–15% gradient SDS/PAGE, followed by radioluminography (exposure time, 5 days unless specified). In addition, a parallel set of samples was subjected to 5–15% SDS/PAGE, followed by immunoblotting with rabbit anti-GFP antibodies (1:10,000; Abcam290) or with rabbit anti-NPC2 antibodies (from P. Lobel, Robert Wood Johnson Medical School, New Brunswick, NJ) as indicated.
LysoTracker Staining in Live Cells and Fluorescence Microscopy. Cells (1.5 × 105 cells per well) were grown in medium A on glass coverslips in six-well plates. After washing with PBS, the cells were incubated in F-12 medium containing 150 nM LysoTracker Red (Molecular Probes) for 1 h at 37°C in 5% CO2 incubator. Afterward, the cells were washed two times with ice-chilled PBS containing 0.1% BSA, and two more times with ice-chilled PBS. The coverslips were quickly mounted with a drop of ProLong Antifade media (Molecular Probes) onto the glass slides. Samples were viewed and photographed by using a Leica TCS SP laser scanning confocal microscope. The images were processed by using leica confocal software (Leica, Deerfield, IL) and quantified by coollocalizer 1.1.2 (Cytolight Software, Washington, DC).
Preparation of the Membrane Fractions That Contain the NPC1-YFP Protein. All procedures were performed at 4°C. CT60-NPC1-YFP cells grown in medium A at 80% confluency, in fifteen 150-mm dishes, were scraped into the homogenization buffer (20 mM Hepes/250 mM sucrose/1 mM EDTA/protease inhibitors mixture, pH 7.3), and homogenized by using a stainless steel homogenizer (27). The postnuclear supernatants were collected after centrifugation (1,000 × g, 5 min) and were subjected to centrifugation (16,000 × g, 40 min) by using a Beckman model 70.1 Ti rotor. The pellets obtained were suspended in the resuspension buffer (50 mM Tris·HCl/150 mM NaCl/protease inhibitors mixture, pH 7.4), by using a glass homogenizer (Wheaton Scientific; 30 strokes), then repeatedly passed through a 25-gauge needle (10 times). The resuspended membrane fractions were used for the in vitro photolabeling experiments.
In Vitro Photolabeling. The freshly prepared membrane fractions (600 μg per sample) were preincubated in a total volume of 263 μl per sample, containing 60.0 μl of 50 mM CD or 60.0 μl of 1.5625 mM unlabeled sterol/50 mM CD (with sterol at 94.0 nmol), for 20 min at 30°C. CD solutions with or without sterols were prepared in the resuspension buffer. After preincubation, 37.0 μl of the prewarmed complex, consisting of 78.1 μM [3H]AC/5 mM CD (with [3H]AC at 34.0 μCi and 3.0 nmol), was added per sample. The mixtures (300 μl final volume per sample) were incubated for 1 h at 30°C, ice-chilled, and subjected to photolysis for 15 min on ice as described earlier. After photolysis, the membranes were pelleted by centrifugation (25,000 × g, 30 min) by using a Beckman model 70.1 Ti rotor. The pellets were solubilized by adding 300 μl per sample of 2% Nonidet P-40 lysis buffer (50 mM Tris/100 mM NaCl/2% Nonidet P-40/1 mM EDTA/1 mM EGTA/protease inhibitors mixture, pH 7.4) and vortexed. The solubilized membrane fractions underwent centrifugation (25,000 × g, 30 min). The supernatants were collected, and subjected to IP by using mouse monoclonal anti-GFP antibody (10 μg per sample), or control mouse IgGs (10 μg per sample). The immunoprecipitates were subjected to 4–14% gradient SDS/PAGE. To detect the labeled protein bands, a gel-slicing method (38) was used (instead of the radioluminography method). In brief, after electrophoresis, individual gel lanes were manually cut into 6-mm slices. The sliced gels were placed in glass vials and were dissolved in 0.5 ml per slice of 31% H2O2 by incubating at 70–80°C for 5–6 h. Afterward, 5.0 ml per vial of Ecoscint H (National Diagnosis) was added. After incubation for 12 h at room temperature in a dark room, the radioactivity levels were determined by scintillation counting. In addition, a parallel set of samples after SDS/PAGE was immunoblotted with rabbit anti-GFP antibodies.
Results
Labeling the NPC1 Protein with [3H]AC in Intact Cells. It is unknown whether the normal molecular function of NPC1 involves a direct interaction with cholesterol. To answer this question, we first showed that [3H]AC delivered to intact 25RA cells can be esterified and that the percentage of esterification is significantly reduced in the NPC1-deficient CT43 cells (Fig. 6, which is published as supporting information on the PNAS web site). Thus, the NPC1 protein functionally interacts with AC in intact cells. We next performed intact cell cross-linking experiments. GFP-tagged mouse NPC1 protein was produced in CT43 cells by transient transfection, followed by intact cell photoaffinity labeling using [3H]AC. The results show that, after IP, a single band with the same molecular mass as NPC1-GFP (200 kDa) is specifically photolabeled by [3H]AC (Fig. 1 B and C, lanes 6 and 11). The control experiment shows that GFP alone (28 kDa) is not photolabeled by [3H]AC (Fig. 1 B and C, lanes 5 and 10).
In separate experiments, we performed intact cell photoaffinity labeling by using [3H]AC in cells transiently transfected with the YFP-tagged late endosomal membrane protein Vamp7 (39). The result showed that Vamp7-YFP was not labeled by [3H]AC (Fig. 1B, lane 9). The expression level of the Vamp7-YFP is near that of NPC1-YFP (compare lanes 12 and 13 of Fig. 1C). Both NPC1 and Vamp7 are membrane proteins located mainly in late endosomes; Vamp7 contains one predicted transmembrane domain near its C terminus (39). Thus, the interaction between [3H]AC and NPC1 is not simply due to the overexpression of an integral membrane protein that resides within late-endosomal membranes.
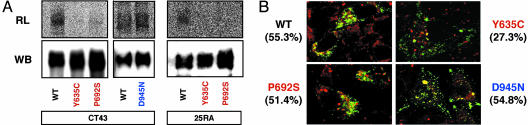
Role of the SSD Within NPC1 in Mediating the Binding Between NPC1 Protein and [3H]AC in Intact Cells. Further evidence of the specificity of the NPC1–sterol interaction comes from analyzing loss-of-function mutations in a specific domain within NPC1. We performed photolabeling experiments in CT43 cells expressing either WT NPC1 or one of three loss-of-function mutant NPC1 proteins [P692S and Y635C (15) and D945N]. Each of these three point mutations inactivates the NPC1 protein's ability to mobilize cholesterol from late endosomes (ref. 15; D.C.K., unpublished data). For the two NPC1 proteins that contain point mutations within the SSD [P692S and Y635C (15)], the results show much diminished labeling (Fig. 2A). In the third mutant (D945N), which contains a point mutation outside the SSD, the labeling is not diminished. These results suggest that the difference in labeling between the WT and the two SSD mutant NPC1 is not simply due to the loss of function of the two SSD mutants. To strengthen these results, in a separate experiment, we used 25RA cells instead of CT43 cells as the host for expression and compared the labeling of the WT and the two SSD mutants. The 25RA cells contain an active, endogenous NPC1, whereas the CT43 cells do not. We obtained the same result as in CT43 cells (Fig. 2 A Right). These data indicate that the difference in labeling between the WT and the two mutant SSD NPC1 proteins does not depend on whether the host cells contain a functional NPC1. Further experiments on protein localization were done to address the possibility that the different amounts of cross-linking with mutant vs. WT NPC1 proteins were due to changes in their subcellular localizations. WT NPC1 is localized mainly in late endosomes, whereas certain mutant NPC1s are mislocalized to the endoplasmic reticulum (ER) (15). Lyso-Tracker is a fluorescent dye that mainly stains the highly acidic subcellular organelles, i.e., the late endosomes/lysosomes, in live cells. We compared the degree of colocalization between various NPC1s and LysoTracker in CT43 cells. The results show that WT NPC1 and the P692S mutant protein significantly colocalize with LysoTracker (Fig. 2B Upper Left and Lower Left, 55% and 51%, respectively). The Y635C mutant colocalizes much less with LysoTracker (Fig. 2B Upper Right, 27%). The D945N mutant also significantly colocalizes with the LysoTracker (Fig. 2B Lower Right, 55%). Results similar to those shown in Fig. 2 were obtained when 25RA cells were used as the host for expression (results not shown). Thus, the large difference in [3H]AC labeling between the WT NPC1 and the P692S mutant NPC1 (Fig. 2 A) is unlikely to be accounted for by the small difference in their intracellular localizations (Fig. 2B). We conclude that mutations within the SSD specifically interrupt the binding between [3H]AC and NPC1 in intact cells.
Fig. 2.
Role of the SSD within NPC1 in mediating the binding between NPC1 protein and [3H]AC. (A) Effect of NPC1 mutations within the sterol-sensing domain on [3H]AC labeling. CT43 cells (3.0 × 106) or 25RA cells (6.0 × 106) transiently transfected with WT or various mutant pNPC1-FP as indicated were photolabeled by using [3H]AC and processed for radioluminography (RL) and Western blotting (WB) according to procedures as described in Materials and Methods. The results using CT43 cells are representative of four experiments. The results using 25RA cells are representative of two experiments. (B) Percentage colocalization of NPC1-FP with LysoTracker. CT43 or 25RA cells were plated in wells with coverslips, grown in medium A, transiently transfected with various NPC1-FP constructs as indicated, and then grown in medium A for 48 h. Afterward, cells were stained with LysoTracker red and viewed under a confocal microscope according to procedures described in Materials and Methods. Red signal, LysoTracker; green signal, NPC1-FP. The yellow signal indicates colocalization of the red and green signals. Values given are the percentage colocalization between LysoTracker and a given NPC1-FP as indicated. To estimate percentage colocalization for each construct, results from six different images were averaged and quantified by using coollocalizer 1.1.2 (Cytolight). Only the results using CT43 cells were shown. The results expressing various NPC1 constructs in 25RA cells as the host were very similar to those reported in CT43 cells.
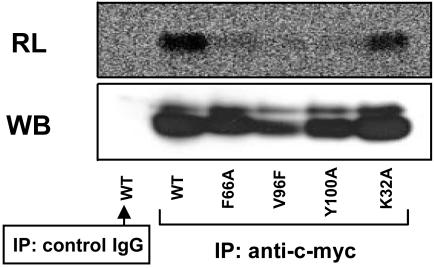
Role of NPC2 in Mediating the Binding Between NPC1 Protein and [3H]AC in Intact Cells. The other protein that is mutated in NPC disease, NPC2, is a soluble protein, mainly located within the lumen of late endosomes/lysosomes, that binds cholesterol with high affinity in vitro (12). We now ask two questions: (i) Can NPC2 also be labeled with [3H]AC in intact cells? (ii) Does the binding of [3H]AC to NPC1 require the presence of NPC2? Here, we show that NPC2 protein tagged with myc-his at the C terminus can be labeled by [3H]AC by using the same photolabeling procedure (Fig. 3, left lanes). For any of the three different point mutations of NPC2 that disrupts the cholesterol binding activity of NPC2 in vitro [F66A, V96F, Y100A (12)], its labeling by [3H]AC is much diminished (Fig. 3). These results further support the interpretation that the labeling seen by using our procedure is based on AC behaving as an analog of cholesterol.
Fig. 3.
Intact cell photolabeling of NPC2-myc-his with [3H]AC. CT43 cells transiently transfected with WT or with various mutant NPC2-myc-his constructs as indicated were photolabeled by using [3H]AC according to procedures described in Materials and Methods. The IPs were with mouse monoclonal anti-c-myc antibody or with control mouse IgG as indicated. RL, radioluminography; WB, Western blotting. The Western blotting was with rabbit polyclonal anti-NPC2 antibodies. Results are representative of two experiments.
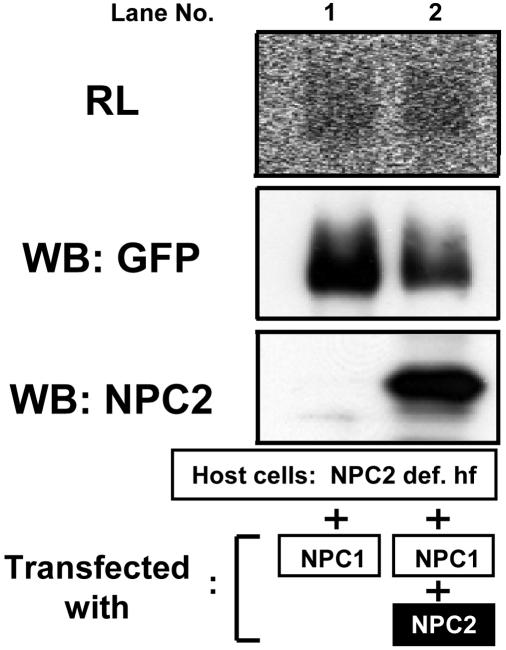
It has been proposed that NPC2 may be necessary to mediate the interaction between NPC1 and cholesterol (for example, ref. 40). To address this question, we expressed NPC1 alone or with NPC2 in a mutant human fibroblast cell line known to lack NPC2 (37) and then performed intact cell labeling with [3H]AC. The results show that the association between [3H]AC and NPC1 in cells with or without added NPC2 is approximately equal (Fig. 4). Thus, binding between AC and NPC1 does not require NPC2.
Fig. 4.
Intact cell photolabeling of NPC1 in the presence or absence of NPC2. NPC2-deficient human fibroblast (NPC2 def. hf) cells (8.0 × 106 cells) transiently transfected with pNPC1-YFP only (lane 1) or with pNPC1-YFP and pNPC2-myc-his (lane 2) were photolabeled by using [3H]AC. DMEM without phenol red, instead of Hanks' buffer, was used as the incubation buffer. The samples were processed for radioluminography (RL; exposure time, 7 days) and Western blotting (WB; with rabbit anti-GFP antibodies or with rabbit polyclonal anti-NPC2 antibodies as indicated). Lanes: 1, without pNPC2-mychis; 2, with pNPC2-myc-his. Results are representative of two experiments.
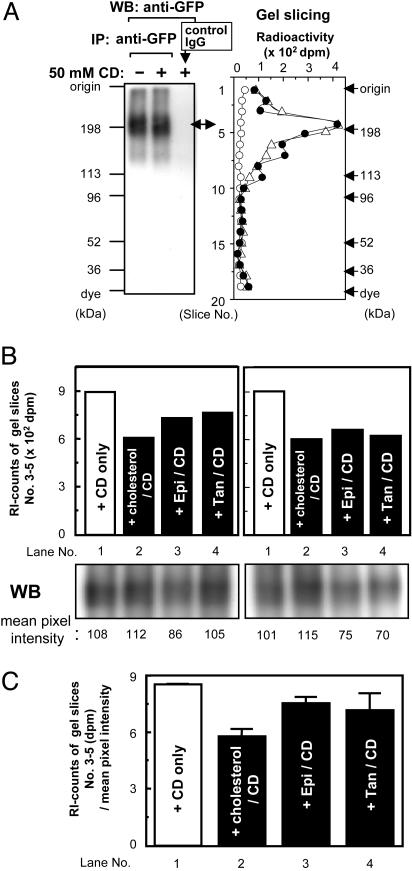
Labeling of NPC1-YFP by [3H]AC in Vitro and Protection Against Labeling by Various Unlabeled Steroids Added in Excess. To further test the specificity of labeling between NPC1 and [3H]AC, we prepared membrane fractions from cells that stably express the NPC1-YFP and performed photolabeling by [3H]AC in vitro. After labeling, the membranes were solubilized by detergent; the NPC1-YFP was then isolated by IP followed by SDS/PAGE. To evaluate the labeling more precisely, we quantitated the labeling by gel slicing. The results showed (Fig. 5A) that NPC1-YFP can be labeled by [3H]AC in vitro and that excess CD present in the labeling mixture does not significantly affect the extent of labeling. By using this procedure, we next tested the abilities of various unlabeled steroids, added in excess as a CD complex, to protect against labeling. The results showed that cholesterol is more effective in protection than its analog epi-cholesterol (a sterol with the OH-moiety at α orientation), or 5-α cholestan (a steroid without the OH-moiety at C-3). Thus, the NPC1 protein may contain a specific binding site(s) that recognizes the 3-β OH moiety of the steroid ring. We also noted that cholesterol present in 30-fold excess only partially inhibited the [3H]AC labeling (by 30%; results seen in two experiments). Thus, NPC1 may bind cholesterol with low affinity.
Fig. 5.
Photolabelings of NPC1-YFP in vitro and protections against labeling by various agents. (A) The membrane fractions from CT60-NPC1-YFP cells were preincubated in buffer, in the presence [center and right lanes in the Western blotting (WB; Left) panel; filled and open circles in the gel slicing (Right) panel] or absence (left lane in the WB panel; open triangles in the gel slicing panel) of 50 mM CD for 20 min at 30°C, then photolabeled with 5 mM CD complexed with [3H]AC, according to procedures described in Materials and Methods. The labeled samples were subjected to IP with mouse monoclonal anti-GFP antibody (left and center lanes in the WB panel; open triangles and filled circles in the gel slicing panel), or with control mouse IgGs (left lane in the WB panel; open circles in the gel slicing panel). Western blotting with anti-GFP antibodies or gel slicing was performed according to procedures described in Materials and Methods. Results shown are representative of two experiments. (B) The membrane fractions from CT60-NPC1-YFP cells were preincubated for 20 min at 30°C in buffer containing 50 mM CD only (lane 1; CD only), or 50 mM CD complexed with an unlabeled steroid as indicated (lanes 2–4). Lanes: 2, cholesterol (5α-cholesten-3β-ol); 3, epicholesterol (5α-cholesten-3α-ol); 4, 5α-cholestan. Afterward, 5 mM CD complexed with [3H]-AC was added, and the samples were photolabeled, according to procedures described in Materials and Methods. Raw radioactivity was the sum of the radioactivity of gel slices 3–5. Results from WB were quantified by using nih image 1.63; the values obtained are shown at the bottom of each lane. The assays were done in duplicate; the results of the duplicates are shown. (C) Normalized radioactivity. The values were obtained by dividing the raw radioactivities by the mean pixel intensities shown in B. The results shown were the averages of duplicate assays. Results shown in B and C are representative of two experiments.
Effects of Various Agents on Labeling Between NPC1 Protein and [3H]AC in Intact Cells. U-18666A is a hydrophobic compound that produces an NPC phenotype in treated cells (41). One mode of its action could be to compete with the association of cholesterol with NPC1. Adding U-18666A to CT60-NPC1-YFP cells for a short period (15 min) or for a long period (36 h) before cross-linking causes no significant alteration in the degree of labeling in intact cells (Fig. 7A, which is published as supporting information on the PNAS web site). Thus, the binding between AC and NPC1 is not altered by U-18666A. NPC1 seems to be a distant member of the RND permease family that functions through a proton-antiport mechanism (33). Thus, disrupting the proton gradient might interfere with labeling. To test this possibility, we found that treating cells with bafilomycin A1, a compound that interferes with vacuolar H+-ATPase and raises endosomal/lysosomal pH (42), caused no significant alteration in degree of labeling intact cells (Fig. 7B). Parallel control experiments showed that cells treated with bafilomycin A1 lost their ability to concentrate the fluorescent dye LysoTracker (result not shown). Thus, the binding between [3H]AC and NPC1 does not require an acidic pH environment.
Discussion
The results of our experiments show that the NPC1 protein can be labeled by a photoactivatable cholesterol analog in intact cells. The labeling, which implies direct binding between [3H]AC and NPC1, does not require NPC2. Binding does not need an acidic environment and is not restricted to cells that exhibit a NPC1 phenotype. Under our labeling conditions, AC also efficiently labels NPC2, a protein already known to bind cholesterol with high affinity in vitro. AC hardly labels various mutant NPC2s that are known to be inactive in binding cholesterol, nor does AC label Vamp7, an integral membrane protein that is abundantly expressed in the same compartments (late endosomes/lysosomes) as NPC1. We also showed that the NPC1-YFP present in membranes could be labeled by [3H]AC in vitro; cholesterol added in excess is more effective in protection against labeling than its analog epi-cholesterol or 5-α-cholestan. Cholesterol provided only partial protection against labeling in vitro. Taken together, our results strongly suggest that NPC1 is a low-affinity cholesterol-binding protein.
Two ideas about mechanism arise from the binding of sterol to NPC1. (i) NPC1 binds to cholesterol as its substrate and transports it at the late endosomal membrane; this possibility would be consistent with the current finding that NPC1 binds to cholesterol with low affinity. (ii) NPC1 participates in transporting various substances/complexes at the endosomal membrane; its activity is modulated by cholesterol bound as an allosteric regulator. These two possibilities are not mutually exclusive, and further investigations are required for resolution. If NPC1 transports cholesterol, it apparently does not use NPC2 bound to cholesterol as a donor for binding cholesterol. Additional results presented here indicate that a functional SSD is required for NPC1 to bind [3H]AC. To our knowledge, the current report provides the first evidence that a protein containing an SSD directly binds to a cholesterol analog. Several proteins, among others, contain SSDs: (i) HMG-CoA reductase, a rate-controlling enzyme in cholesterol biosynthesis (43); (ii) SCAP, a protein that forms a complex with the SRE binding proteins (SREBPs) and translocates from the endoplasmic reticulum (ER) to the Golgi in response to cholesterol deprivation in the ER (44); (iii) PATCHED, a tumor suppressor involved in the signal transduction cascade mediated by the cholesterol-modified morphogen Hedgehog (19); and (iv) NPC1L1, a protein recently reported to be involved in intestinal cholesterol absorption (20). These proteins all participate in various cholesterol-dependent regulatory events in some manner, and the SSDs present in these proteins play important roles in mediating the functions of these proteins (reviewed in ref. 22). On the other hand, whether binding occurs between sterol(s) and any of these SSD proteins and, if so, whether an SSD is required have not been tested. We have attempted to label two of these SSD proteins, the HMG-CoA reductase and PATCHED, each tagged with GFP, with [3H]AC by using the intact cell-labeling procedure described here. We were not able to detect significant labeling between [3H]AC and either one of these two proteins (data not shown), but the expression levels of HMG-CoA reductase-GFP and PATCHED-GFP in CHO cells were much lower than those of NPC1-GFP (<5%). Thus, a much better expression system will probably be needed to attempt photolabeling of AC with SSD proteins that are in low abundance in intact cells.
In NPC1/NPC1 homozygous mutant cells, abnormal accumulation of various sphingolipids (i.e., sphingomyelin, glucosylceramide, galactosylceramide, and the gangliosides GM2 and GM3) is observed in addition to alterations in cholesterol trafficking (reviewed in refs. 45–48). NPC1 protein may also bind other lipids in addition to binding cholesterol. In the future, these possibilities can be addressed by performing cross-linking studies using different photoaffinity probes in intact cells, and/or by using various biochemical procedures that allow investigators to study binding of NPC1 and various ligands in liposomes, or in detergent-solubilized conditions in vitro.
Supplementary Material
Acknowledgments
We thank Drs. Shigeki Sugii, Patrick C. Reid, and Yoshio Yamauchi for helpful discussions; Dr. Peter Lobel for the anti-NPC2 antibodies; and Dr. Yiannis A. Ioannou (Mount Sinai School of Medicine, New York) for the NPC2-deficient human fibroblast. We also thank Helina Morgan for careful editing of the manuscript. This work was supported by National Institutes of Health Grant HL 36709 (to T.-Y.C.), and a grant from the Ara Parseghian Medical Research Foundation (to M.P.S.). M.P.S. is an Investigator of the Howard Hughes Medical Institute. N.O. has been supported by American Heart Association Postdoctoral Fellowship 0225593T. D.C.K. has been supported by a Medical Scientist Training Grant.
Abbreviations: AC, 7,7-azocholestanol; FP, fluorescent protein; IP, immunoprecipitation; NPC, Niemann–Pick type C; SSD, sterol-sensing domain; CHO, Chinese hamster ovary; CD, methyl-β-cyclodextrin.
Note Added in Proof. Radhakrishnan et al. (49) very recently reported that SCAP, another polytopic membrane protein that contains the SSD, binds cholesterol with high affinity in vitro.
References
- 1.Patterson, M. C., Vanier, M. T., Suzuki, K., Morris, J. A., Carstea, E., Neufeld, E. B., Blanchette-Mackie, J. E. & Pentchev, P. G. (2001) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C. R., Beaudet, A. L., Sly, W. S. and Valle, D. (McGraw–Hill, New York), Vol. III, pp. 3611–3633. [Google Scholar]
- 2.Karten, B., Vance, D. E., Campenot, R. B. & Vance, J. E. (2003) J. Biol. Chem. 278, 4168–4175. [DOI] [PubMed] [Google Scholar]
- 3.Treiber-Held, S., Distl, R., Meske, V., Albert, F. & Ohm, T. G. (2003) J. Pathol. 200, 95–103. [DOI] [PubMed] [Google Scholar]
- 4.Reid, P. C., Sakashita, N., Sugii, S., Ohno-Iwashita, Y., Shimada, Y., Hickey, W. F. & Chang, T. Y. (2004) J. Lipid Res. 45, 582–591. [DOI] [PubMed] [Google Scholar]
- 5.Cadigan, K. M., Spillane, D. M. & Chang, T. Y. (1990) J. Cell Biol. 110, 295–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cruz, J. C., Sugii, S., Yu, C. & Chang, T. Y. (2000) J. Biol. Chem. 275, 4013–4021. [DOI] [PubMed] [Google Scholar]
- 7.Dahl, N. K., Reed, K. L., Daunais, M. A., Faust, J. R. & Liscum, L. (1992) J. Biol. Chem. 267, 4889–4896. [PubMed] [Google Scholar]
- 8.Millard, E. E., Srivastava, K., Traub, L. M., Schaffer, J. E. & Ory, D. S. (2000) J. Biol. Chem. 275, 38445–38451. [DOI] [PubMed] [Google Scholar]
- 9.Higaki, K., Ninomiya, H., Sugimoto, Y., Suzuki, T., Taniguchi, M., Niwa, H., Pentchev, P. G., Vanier, M. T. & Ohno, K. (2001) J. Biochem. (Tokyo) 129, 875–880. [DOI] [PubMed] [Google Scholar]
- 10.Carstea, E. D., Morris, J. A., Coleman, K. G., Loftus, S. K., Zhang, D., Cummings, C., Gu, J., Rosenfeld, M. A., Pavan, W. J., Krizman, D. B., et al. (1997) Science 277, 228–231.9211849 [Google Scholar]
- 11.Naureckiene, S., Sleat, D. E., Lackland, H., Fensom, A., Vanier, M. T., Wattiaux, R., Jadot, M. & Lobel, P. (2000) Science 290, 2298–2301. [DOI] [PubMed] [Google Scholar]
- 12.Ko, D. C., Binkley, J., Sidow, A. & Scott, M. P. (2003) Proc. Natl. Acad. Sci. USA 100, 2518–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friedland, N., Liu, H. L., Lobel, P. & Stock, A. M. (2003) Proc. Natl. Acad. Sci. USA 100, 2512–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watari, H., Blanchette-Mackie, E. J., Dwyer, N. K., Watari, M., Neufeld, E. B., Patel, S., Pentchev, P. G. & Strauss, J. F., III (1999) J. Biol. Chem. 274, 21861–21866. [DOI] [PubMed] [Google Scholar]
- 15.Ko, D. C., Gordon, M. D., Jin, J. Y. & Scott, M. P. (2001) Mol. Biol. Cell 12, 601–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sever, N., Yang, T., Brown, M. S., Goldstein, J. L. & DeBose-Boyd, R. A. (2003) Mol. Cell 11, 25–33. [DOI] [PubMed] [Google Scholar]
- 17.Xu, L. & Simoni, R. D. (2003) Arch. Biochem. Biophys. 414, 232–243. [DOI] [PubMed] [Google Scholar]
- 18.Hua, X., Nohturfft, A., Goldstein, J. L. & Brown, M. S. (1996) Cell 87, 415–426. [DOI] [PubMed] [Google Scholar]
- 19.Ingham, P. & McMahon, A. (2001) Genes Dev. 15, 3059–3087. [DOI] [PubMed] [Google Scholar]
- 20.Altmann, S. W., Davis, H. R. J., Zhu, L. J., Yao, X., Hoos, L. M., Tetzloff, G., Iyer, S. P., Maguire, M., Golovko, A., Zeng, M., et al. (2004) Science 303, 1201–1204. [DOI] [PubMed] [Google Scholar]
- 21.Osborne, T. F. & Rosenfeld, J. M. (1998) Curr. Opin. Lipidol. 9, 137–140. [DOI] [PubMed] [Google Scholar]
- 22.Kuwabara, P. E. & Labouesse, M. (2002) Trends Genet. 18, 193–201. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, M., Dwyer, N. K., Love, D. C., Cooney, A., Comly, M., Neufeld, E., Pentchev, P. G., Blanchette-Mackie, E. J. & Hanover, J. A. (2001) Proc. Natl. Acad. Sci. USA 98, 4466–4471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgins, M. E., Davies, J. P., Chen, F. W. & Ioannou, Y. A. (1999) Mol. Genet. Metab. 68, 1–13. [DOI] [PubMed] [Google Scholar]
- 25.Neufeld, E. B., Wastney, M., Patel, S., Suresh, S., Cooney, A. M., Dwyer, N. K., Roff, C. F., Ohno, K., Morris, J. A., Carstea, E. D., et al. (1999) J. Biol. Chem. 274, 9627–9635. [DOI] [PubMed] [Google Scholar]
- 26.Wojtanik, K. M. & Liscum, L. (2003) J. Biol. Chem. 278, 14850–14856. [DOI] [PubMed] [Google Scholar]
- 27.Sugii, S., Reid, P. C., Ohgami, N., Du, H. & Chang, T. Y. (2003) J. Biol. Chem. 278, 27180–27189. [DOI] [PubMed] [Google Scholar]
- 28.Lange, Y., Ye, J. & Steck, T. L. (1998) J. Biol. Chem. 273, 18915–18922. [DOI] [PubMed] [Google Scholar]
- 29.Cruz, J. C. & Chang, T. Y. (2000) J. Biol. Chem. 275, 41309–41316. [DOI] [PubMed] [Google Scholar]
- 30.Reid, P. C., Sugii, S. & Chang, T. Y. (2003) J. Lipid Res. 44, 1010–1019. [DOI] [PubMed] [Google Scholar]
- 31.Zervas, M., Somers, K. L., Thrall, M. A. & Walkley, S. U. (2001) Curr. Biol. 11, 1283–1287. [DOI] [PubMed] [Google Scholar]
- 32.Krishnamurthy, M., Higaki, K., Tinkelenberg, A. H., Balderes, D. A., Almanzar-Paramio, D., Wilcox, L., Erdeniz, N., Redican, F., Padamsee, M., Liu, Y., et al. (2004) J. Cell Biol. 164, 547–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies, J. P., Chen, F. W. & Ioannou, Y. A. (2000) Science 290, 2295–2298. [DOI] [PubMed] [Google Scholar]
- 34.Brunner, J. (1996) Trends Cell Biol. 6, 154–157. [DOI] [PubMed] [Google Scholar]
- 35.Thiele, C., Hannah, M. J., Fahrenholz, F. & Huttner, W. B. (2000) Nat. Cell Biol. 2, 42–49. [DOI] [PubMed] [Google Scholar]
- 36.Cruz, J., Thomas, M., Wong, E., Ohgami, N., Chang, C. C. Y., Curphey, T. & Chang, T. Y. (2002) J. Lipid Res. 43, 1341–1347. [PubMed] [Google Scholar]
- 37.Walter, M., Davies, J. P. & Ioannou, Y. A. (2003) J. Lipid Res. 44, 243–253. [DOI] [PubMed] [Google Scholar]
- 38.Kuniyasu, A., Itagaki, K., Shibano, T., IIino, M., Kraft, G., Schwartz, A. & Nakayama, H. (1998) J. Biol. Chem. 273, 4635–4641. [DOI] [PubMed] [Google Scholar]
- 39.Advani, R. J., Yang, B., Prekeris, R., Lee, K. C., Klumperman, J. & Scheller, R. H. (1999) J. Cell Biol. 146, 765–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strauss, J. F., III, Liu, P., Christenson, L. K. & Watari, H. (2002) Steroids 67, 947–951. [DOI] [PubMed] [Google Scholar]
- 41.Liscum, L. & Faust, J. R. (1989) J. Biol. Chem. 264, 11796–11806. [PubMed] [Google Scholar]
- 42.Yoshimori, T., Yamamoto, A., Moriyama, Y., Futai, M. & Tashiro, Y. (1991) J. Biol. Chem. 266, 17707–17712. [PubMed] [Google Scholar]
- 43.Goldstein, J. L. & Brown, M. S. (1990) Nature 343, 425–430. [DOI] [PubMed] [Google Scholar]
- 44.Horton, J. D., Goldstein, J. L. & Brown, M. S. (2002) J. Clin. Invest. 109, 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanier, M. T. & Suzuki, K. (1998) Brain Pathol. 8, 163–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanier, M. T. (1999) Neurochem. Res. 24, 481–489. [DOI] [PubMed] [Google Scholar]
- 47.Yano, T., Taniguchi, M., Akaboshi, S., Vanier, M. T., Tai, T., Sakuraba, H. & Ohno, K. (1996) Proc. Jpn. Acad. 72B, 214–219. [Google Scholar]
- 48.Zhang, M., Dwyer, N. K., Neufeld, E. B., Love, D. C., Cooney, A., Comly, M., Patel, S., Watari, H., Strauss, J. F., III, Pentchev, P. G., et al. (2001) J. Biol. Chem. 276, 3417–3425. [DOI] [PubMed] [Google Scholar]
- 49.Radhakrishnan, A., Sun, L. P., Kwon, H. J., Brown, M. S. & Goldstein, J. L. (2004) Mol. Cell 15, 259–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.