Abstract
Prostate cancer is the most common solid tumor in men, and it shares with all cancers the hallmark of elevated, nonhomeostatic cell proliferation. Here we have tested the hypothesis that the SONIC HEDGEHOG (SHH)–GLI signaling pathway is implicated in prostate cancer. We report expression of SHH–GLI pathway components in adult human prostate cancer, often with enhanced levels in tumors versus normal prostatic epithelia. Blocking the pathway with cyclopamine or anti-SHH antibodies inhibits the proliferation of GLI1+/PSA+ primary prostate tumor cultures. Inversely, SHH can potentiate tumor cell proliferation, suggesting that autocrine signaling may often sustain tumor growth. In addition, pathway blockade in three metastatic prostate cancer cell lines with cyclopamine or through GLI1 RNA interference leads to inhibition of cell proliferation, suggesting cell-autonomous pathway activation at different levels and showing an essential role for GLI1 in human cells. Our data demonstrate the dependence of prostate cancer on SHH–GLI function and suggest a novel therapeutic approach.
SONIC HEDGEHOG (SHH) signaling has been implicated in different aspects of animal development, acting through several components, including the transmembrane proteins PATCHED1 (PTCH1) and SMOOTHENED (SMOH), to activate the GLI zinc-finger transcription factors (1, 2). In addition, we and others have shown that SHH signaling is implicated in a number of tumors (reviewed in refs. 2 and 3), such as basal cell carcinomas (4–6), medulloblastomas (7, 8), gliomas (7), sarcomas (9, 10), tumors of the digestive tract (11), small cell lung cancers (12,) and pancreatic carcinomas (13). To date there is no direct evidence linking SHH signaling to prostate cancer, the most common solid cancer in men (14), although we have found that sporadic prostate tumors express GLI1 (7), a reliable marker of SHH signaling (15, 16). This observation allowed us to propose the hypothesis that the SHH–GLI pathway participates in prostate cancer (7). Consistently, Shh signaling has been found to be essential for prostate patterning and development (17–22), and genetic mapping data has revealed that at least two key components of the SHH–GLI pathway [SMOH and SUPPRESSOR OF FUSED (SUFUH)] are located in chromosomal regions implicated in familial human prostate cancer (23, 24). Here we have tested the involvement of SHH–GLI signaling in prostate cancer.
Methods
Cell Lines and Primary Cultures. The PC3, LNCaP, and DU145 cell lines (25–27) were purchased from American Type Culture Collection and grown as specified. All primary prostate tumors were obtained following approved protocols. Tumors in PBS were chopped with a razor blade and incubated with Papain for 1 h at 37°C, they were then dissociated by passing them through a fire-polished pipette and washed several times in serum containing media. All dissociated primary tumors were plated in polyornithin- and laminin-treated p16 plates in DMEM-F12 with 10% FBS at ≈30,000 cells per p16 well. Primary cultures were used 2–4 days after plating, when the cells reached 60–70% confluence.
In Situ Hybridization and Immunocytochemistry. Immunocytochemistry was performed with anti-BrdUrd (Beckton Dickinson), anti-SHH (Santa Cruz Biotechnology), and anti-Ki-67 (DAKO), using FITC- or horseradish peroxidase (HRP)-conjugated secondary antibodies (Boehringer Mannheim) as described (7). For tissue arrays, slides were baked and deparaffinized before blocking of endogenous peroxides. They were then developed with HRP-conjugated secondary antibodies and diaminobenzidine (DAB). In situ hybridizations on frozen sections with digoxygenin-labeled antisense RNA probes for GLI1, PTCH1, and SHH and a sense control GLI1 were as described (7).
Prostate Tissue Microarrays and Microdissection. After institutional review board approval, tissue microarrays (28) were prepared from archived paraffin blocks from 288 radical prostatectomy cases from the Medical College of Wisconsin. For each case, 0.6-mm cores of tumor were isolated and placed in the array blocks, and 5-μm slides were prepared for immunohistochemistry. Slides were reviewed by a trained urologic pathologist (M.W.D.) and scored for the presence of benign prostate glands, high-grade prostatic intraepithelial neoplasia, or invasive tumor. The presence of tumor or high-grade prostatic intraepithelial neoplasia was confirmed by immunohistochemical staining for high molecular mass cytokeratin (CK903 Ab, DAKO). Individual cores were examined as duplicates, and staining was correlated to a set of anonymous deidentified pathologic and outcomes data with χ2 and Fisher's exact or two-tailed ANOVA analyses.
Normal and tumor tissue from the same patients for real-time PCR analyses were microdissected from sections with a laser capture microscope after pathological assessment.
SHH, Anti-SHH Antibody, Cyclopamine, and Tomatidine Treatments. Commercial N-SHH (R & D Systems) was used at 100 nM because we have found that this commercial protein is ≈20 times less active than the octyl-modified SHH-N we had previously used from Curis in the C3H10T1/2 induction assay (data not shown). 5E1 anti-SHH blocking antibody (29) was purchased from the Hybridoma Bank at the University of Iowa and was used at 8 μg/ml. Cyclopamine (Toronto Research Chemicals) and Tomatidine (Sigma) were used at 10 μM unless otherwise noted; for cells in culture, they were dissolved in ethanol, and ethanol alone was used as control. Treated cells were in 2.5% serum for 48 h instead of the usual 10% routinely used for standard growth.
Proliferation Assays. BrdUrd (Sigma) was given at 4 μg/ml before fixation. The time of the BrdUrd pulse depended on the growth rate of the cells tested. Cell lines were given a 2-h pulse, whereas primary tumor cultures, which grow less rapidly, were given 16-h pulses. Proliferation in tissue arrays was measured by the level of Ki-67 antigen expression.
PCRs. For RT-PCRs, the following primers were used (all 5′ to 3′). GLI1s, GGGATGATCCCACATCCTCAGTC, and GLI1a, CTGGAGCAGCCCCCCCAGT at 60°C; PSAs, CTTGTAGCCTCTCGTGGCAG, and PSAa, GACCTTCATAGCATCCGTGAG at 56°C. Primers for PTCH1 and GAPDH were as described (7, 30).
For real-time PCR, total RNA was DNase treated (Invitrogen) and reverse transcribed with TaqMan (Applied Biosystems) using oligo(dT) primers as described by the manufacturer. Reactions were run by using SYBR Green (Applied Biosystems) on an ABI Prism 7700 machine. Each sample was run minimally at three concentrations in triplicate. All primer sets amplified 75- to 300-bp fragments. Sequences are available upon request. The raw data are available upon request from S.D.
RNA Interference. Double-stranded small interference RNAs (siRNAs, 21 nt long) were purchased from Dahrmacon, purified, and desalted. The sequences for the GLI1 siRNAs used was: AACUCCACAGGCAUACAGGAU; control siRNA was: AACGUACGCGGAAUACAACGA. This siRNA was also used FITC-tagged. siRNA transfections (0.2 μM) were with Oligofectamine (Invitrogen) as described by the manufacturer. Cells were treated for 60 h before fixation.
Results
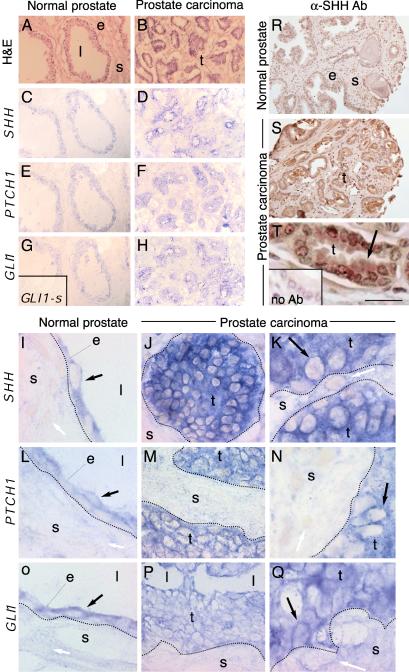
To begin to analyze the role of SHH–GLI signaling in prostate cancer, we first tested for the expression of SHH–GLI pathway components in prostate cancer resections and normal tissue from the same patients. In situ hybridization showed that GLI1, PTCH1, and SHH are normally coexpressed in epithelial cells and not in the surrounding stroma (Fig. 1 A, C, E, G, I, L, and O). Prostate tumors were uniformly SHH+/GLI1+/PTCH1+ (Fig. 1 B, D, F, H, J, K, M, N, P, and Q), although variable levels of expression were detected visually in the tumors. Coexpression of these markers in tumor cells is consistent with their derivation from the normal prostatic epithelium.
Fig. 1.
Expression of SHH–GLI pathway components in normal prostate tissue and prostate tumors. Sections of normal prostate tissue (A, C, E, G, I, L, and O) and prostate tumors (B, D, F, H, J, K, M, N, P, and Q) show hematoxylin and eosin (H&E) staining (A and B) or the expression of SHH (C, D, and I–K), PTCH1 (E, F, and L–N), and GLI1 (G, H, and O–Q). (G Inset) Sense GLI1 probe control showing no background. Prostate tumors have many small epithelial glandular structures. Black arrows point to expressing cells. White arrows point to nonexpressing cells. (R–T) Sections from the tissue microarrays of normal prostate tissue (R) and prostate tumors (S and T) showing expression of SHH protein with an anti-SHH antibody (αSHH Ab) (R–T) and a no-primary antibody control (T Inset). All sections were counterstained with hematoxylin to visualize nuclei and tissue structure. Arrow in T points to localization of SHH protein in the cytoplasm of epithelial cells. e, epithelium; l, lumen; s, stroma; t, tumor. (Scale bar in T is 150 μmin A–H, R, and S, 20 μmin J, M, P, and T, and 10 μmin I–L, N, O, and Q.)
More sensitive real-time PCR analyses of six of the same microdissected matched pairs showed up-regulation of the expression of SHH, PTCH1, GLI1, GLI2, and GLI3 (between 1.5- and ≈300-fold) in many tumor cases compared to normal tissue after normalization to the ubiquitous similar expression of β-actin (Table 1). Levels of expression within tumors were variable. Such differences could be related to the known heterogeneity of prostate cancer, because this is a general diagnosis that encompasses a broad range of histological phenotypes (31–33). Whereas varying levels have also been observed in other tumors (reviewed in refs. 2 and 3), the meaning of such differences is not known, although they have been proposed to correlate in a direct or inverse manner with tumor type or grade (34–36). What is important is that the loyal markers of an active SHH–GLI pathway, GLI1 and PTCH1 (refs. 15, 16, and 37), are consistently transcribed in the examined tumor cells, showing the presence of an active pathway.
Table 1. SHH, GLI1, GLI2, GLI3, and PTCH1 expression in human prostate cancer.
|
SHH
|
PTCH1
|
GLI1
|
GLI2
|
GLI3
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | Fold increase | Range | Fold increase | Range | Fold increase | Range | Fold increase | Range | Fold increase | Range |
| 829 | 0 | 0-0.01 | 1.5 | 1.1-2.1 | 26.1 | 20.8-32.7 | 0.02 | 0.02-0.02 | 72 | 53-99 |
| 887 | 0.2 | 0.05-0.9 | 8.5 | 7.6-9.5 | 0.09 | 0.07-0.13 | 0.43 | 0.37-0.51 | 1.1 | 0.8-1.6 |
| 921 | 2.9 | 1.3-6.3 | 50 | 30-84 | 2 | 1.1-3.4 | 3.8 | 2.4-6.1 | 12.5 | 7.7-20.4 |
| 945 | 9.8 | 6.2-15.7 | 7.8 | 5.7-10.7 | 22.7 | 21.6-23.9 | 0.7 | 0.5-1.0 | 2.2 | 2.1-2.4 |
| 1854 | 4.7 | 1.8-11.7 | 213 | 164-278 | 5.1 | 3.8-6.9 | 19.5 | 10.9-35.1 | 5.7 | 4.4-7.5 |
| 1866 | 4.6 | 4.1-5.2 | 3.4 | 3.1-3.7 | 299 | 260-342 | 0.03 | 0.02-0.03 | 0.18 | 0.15-0.2 |
Fold increase in gene expression in tumors versus matched normal tissue determined by real-time RT-PCR analyses as calculated by the ΔCT method. Range indicates ± 1 SD. Gene expression levels were normalized to β-actin. Increases of 2-fold or more are shown in bold.
To extend these findings, we performed immunohistochemistry for SHH, as a secreted and potentially useful systemic marker for prostate cancer, on tissue microarrays representing 239 prostate carcinomas, 15 precancerous lesion high-grade prostatic intraepithelial neoplasia (HGPIN), and 135 benign prostate tissues from 297 patients. SHH expression was increased in tumors and was present as a secreted protein in the glandular lumens made by tumor cells (Fig. 1 R–T), likely reflecting the origin of tumors from the SHH+ prostatic epithelia. Higher SHH levels, determined visually, were found in 33% of tumors compared to <1% of cases of normal adjacent tissue, indicating a significant correlation between high SHH levels and tumor presence. High SHH levels were also correlated with higher Ki-67+ cell proliferation (Table 2). The level of SHH expression was not correlated with Gleason score or other clinical parameters (Table 2). This finding may indicate that inappropriately maintained or elevated SHH expression is an early and general event in prostate cancer, reflecting the origin of tumors from the SHH+ prostatic epithelia.
Table 2. Correlation of elevated SHH expression with tumorigenesis and clinical features of prostate cancer.
| SHH
|
||||
|---|---|---|---|---|
| Expression low | Expression high | χ2 or Fisher's exact test | ||
| Histology | Tumor | 141 | 70 | |
| Normal | 126 | 1 | P < 0.00005 | |
| HGPIN | 13 | 1 | ||
| Normal | 126 | 1 | P = 0.0563 | |
| Clinical stage | cT2 | 16 | 6 | |
| cT3/4 | 2 | 1 | NS | |
| Tumor grade | Gleason 6 | 30 | 1 | |
| Gleason 7,8,9 | 57 | 7 | NS | |
| Pathologic stage | pT1-pT2 | 50 | 4 | |
| pT3 | 37 | 4 | NS | |
| Nodal status | pN0 | 27 | 12 | |
| pN1 | 1 | 0 | NS | |
| Outcomes | PSA Recurrence | 8 | 1 | |
| No PSA recurrence | 22 | 12 | NS | |
| Vital status | Alive | 42 | 18 | |
| Dead | 4 | 3 | NS | |
| Ki-67 expression | Sample no. | 275 | 69 | |
| Mean % Ki-67+ nuclei | 5.1 | 7.6 | P = 0.0141* | |
Significance was only found between SHH expression and tumorigenesis and SHH expression and higher proliferative levels as measured by Ki-67 staining. Tumor grade is presented as Gleason score. Pathologic staging uses the American Joint Comission on Cancer 2002 tumor staging criteria. HGPIN, high-grade prostatic intraepithelial neoplasia.
Two-tailed ANOVA.
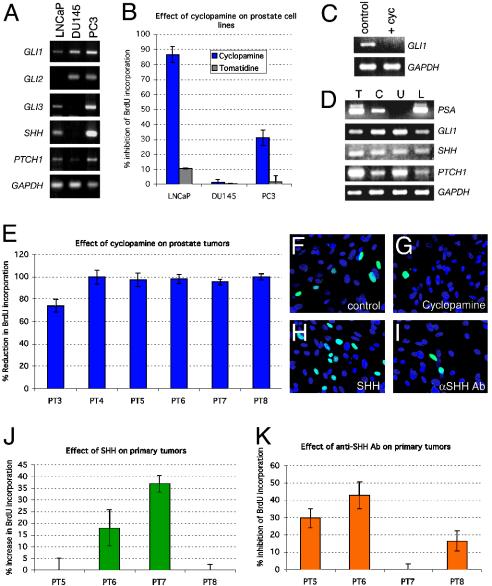
The difficulty of growing human prostate cancer cells in vitro translates into a dearth of available cancer cells to test. Here we have chosen the three most widely used prostate cancer cell lines, LNCaP, an androgen sensitive cell line derived from a prostate cancer lymph node metastasis; and PC3 and DU145, androgen insensitive cell lines derived from prostate cancer bone metastases, to assay for the expression of SHH–GLI pathway components. All of the cells expressed GLI1 and PTCH1 (Fig. 2A), consistent with our expression studies and indicating that they harbor an active pathway. Of these cell lines, only DU145 and PC3 cells expressed GLI2, and only LNCaP and PC3 cells expressed GLI3 and SHH at detectable levels (Fig. 2A). GLI1 is thus the only GLI gene consistently expressed at detectable levels in all of these cells, and thus, we have focused on GLI1.
Fig. 2.
Response of prostate tumor cell lines to alterations in the SHH–GLI pathway. (A) PCR analyses for the expression of SHH–GLI pathway components in three cell lines as indicated. In this and all other PCR assays, the expression of the ubiquitous gene GAPDH is measured as quantitative control. (B) Inhibition of prostate cell line proliferation as measured by BrdUrd incorporation in the three prostate cell lines used with cyclopamine. Tomatidine is used as control. (C and D) PCR analyses of the suppression of GLI1 expression in LNCaP cells by cyclopamine treatment at 36 h (C) or of the expression of prostate specific antigen (PSA), GLI1, SHH, and PTCH1 expression in whole prostate tumor tissue (T), primary culture (C), the glioblastoma cell line U87 (U), and LNCaP (L) cells (D). PSA is expressed in prostate but not in brain cells. All samples express GLI1 and SHH. The whole tissue and primary culture correspond to PT6. (E) Histogram of the inhibition of BrdUrd incorporation in primary cultures of prostate tumor (PT3-PT8) by cyclopamine treatment. (F–I) Immunocytochemistry for BrdUrd incorporation with secondary FITC-antibodies showing BrdUrd+ nuclei (green) in a field of primary prostate cells (PT6) in control cells (treated with ethanol as the carrier for cyclopamine, F), cyclopamine (G), SHH protein (H), or anti-SHH antibody (αSHH Ab, I). All nuclei are stained with 4′,6-diamidino-2-phenylindole (blue). (J and K) Histograms of the increase in (J) or inhibition of (K) BrdUrd incorporation of primary prostate tumors after treatment with SHH (J) or anti-SHH antibody (αSHH Ab, K) for 48 h. Histogram error bars represent SEM in all panels.
To interfere with SHH–GLI signaling, we first used cyclopamine, a selective inhibitor of SMOH (38). Effects of cyclopamine treatment after 48 h were tested by BrdUrd incorporation as a sensitive measure of cell proliferation. Such treatment led to a large (>80%) decrease in BrdUrd incorporation in LNCaP cells, and a significant decrease (≈30%) in PC3 cells but had no effect in DU145 cells (Fig. 2B). Treatment with tomatidine (38) served as control and had little or no effect on BrdUrd incorporation (Fig. 2B). The lack of effects of cyclopamine on DU145 cells shows that this drug is not non-specific. Because we used short-term assays to focus on early, direct effects on cell proliferation, the changes in total cell number were consequently relatively conservative. For instance, cyclopamine reduced total 4′,6-diamidino-2-phenylindole-positive LNCaP cell number by 22.1 ± 1.1% (P = 0.0001) after 48 h. No cytotoxic effects or significant cell death were observed during these experiments. Cyclopamine treatment also led to a decrease in GLI1 expression, consistent with the expected down-regulation of the SHH–GLI pathway (Fig. 2C).
Analyses of primary prostate tumors is complicated by the difficulty of growing primary human prostate cancer cultures (39). Nevertheless, we were able to dissociate and plate six of eight primary prostate tumors, although stable cultures were not obtained. Primary cells that remained attached after 2 days had a uniform cuboidal morphology, formed small clusters and expressed prostate-specific antigen (PSA), as well as SHH, PTCH1, and GLI1 (Fig. 2D), proving their prostatic epithelial origin. Cyclopamine treatment led to a major (>70%) decrease in BrdUrd incorporation in all primary cultures as compared with carrier-treated samples (Fig. 2 E–G), mimicking the results obtained in LNCaP cells. Here again, the insensitivity of DU145 to cyclopamine provides a control for the action of the drug. Indeed, although we have not tested the response of normal human prostate cells to cyclopamine, we expect that it would also inhibit the proliferation of normal SHH+/PTCH1+/GLI1+ prostate epithelial cells (Fig. 1). As with the cell lines, the total number of 4′,6-diamidino-2-phenylindole-positive primary tumor cells was similarly reduced by cyclopamine treatment [e.g., 26.7 ± 1.1% decrease in primary tumor 6 (PT6), P = 0.001] after 48 h. Although stromal cells are likely to be present in our primary cultures, their numbers appear to be small because >90% of the cells examined microscopically had a similar cuboidal morphology. Moreover, the high inhibition levels by cyclopamine would be inconsistent with effects only in contaminating stromal cells, which do not appreciably express PTCH1 or GLI1 (Fig. 1).
We then tested for the ability of exogenous SHH to stimulate prostate cancer cell proliferation and for the possible existence of autocrine signaling. Addition of recombinant SHH protein led to an increase in BrdUrd incorporation in two of four primary cultures after 48 h (Fig. 2 F, H, and J). In contrast, addition of the standard blocking antibody against SHH (5E1; ref. 29) resulted in a inhibition of BrdUrd incorporation by 15–40% for three of four tumors (Fig. 2 F, I, and K), suggesting that several tumors display autocrine signaling. Interestingly, the only primary culture that was insensitive to Shh Ab blockade, PT7, being sensitive to cyclopamine [which targets SMOH (38), Fig. 2E], was also the more sensitive to the addition of exogenous Shh. This might indicate that although the pathway is activated downstream of the site of ligand action in PT7, possibly affecting PTCH1 or SMOH, exogenous Shh can still increase the levels of signaling. Taken together, the functional heterogeneity that we detect parallels that found for GLI and SHH expression described above and may reflect independent activating events as well as the well known heterogeneity of prostate cancers.
Treatment of LNCaP, PC3 or DU145 cells with either blocking antibody or recombinant Shh protein did not result in significant changes in BrdUrd incorporation (data not shown). LNCaP and PC3 cells could thus display an activated pathway at the membrane level (being sensitive to cyclopamine inhibition) that has lost responsiveness to ligand. Cyclopamine-insensitive DU145 cells may have an activated pathway downstream of SMOH (or at the level of SMOH affecting its inhibition by cyclopamine), having lost also the ability to respond to SHH. It remains possible that the different behavior of primary cultures versus established cell lines also reflects unrelated transformation or immortalization events.
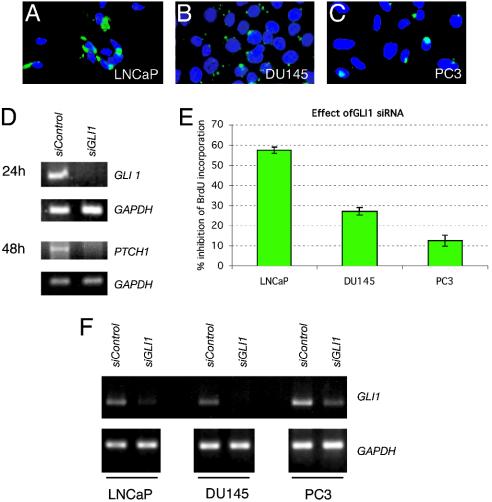
The GLI zinc-finger transcription factors have been suggested to be essential for the mediation of HH signals (reviewed in refs. 1, 2, and 40). However, Gli1 is apparently redundant in mouse development and tumorigenesis (41, 42), and there is to date no data on the requirement for GLI1 in human cells. Here, we tested the function of GLI1, the only GLI gene consistently expressed in all primary tumors and cell lines, by RNA interference to knockdown its function with a specific 21-nt-long small RNA. (This siRNA inhibits the effect of SHH on multipotent C3H10T1/2 cells; P.S. and A.R.A., unpublished data). Lipofection of primary cultures resulted in a negligible number of transfected cells, making it impractical to use siRNAs in such cultures. In contrast, lipofection of FITC–siRNA proved efficient (≈50–80%) in the LNCaP, PC3, and DU145 cell lines (Fig. 3 A–C). It is important to note that, because transfection efficiencies are <100%, the results of cell pool assays necessarily underestimate the effects of RNA interference. Transfection of a control siRNA at the same concentration served as control in all tests.
Fig. 3.
Response of prostate cell lines to GLI1 RNA interference. (A–C) Immunocytochemisty of the three prostate cell lines indicated showing the efficiency of lipofection of an FITC-tagged control siRNA (green). Note the lower efficiency in PC3 cells. (D) Effect of GLI1 siRNA on gene expression. RNA interference reduces GLI1 and PTCH1 mRNA levels as seen at 24 and 48 h, respectively (E) Histogram of the inhibition of BrdUrd incorporation in prostate tumor cell lines by GLI1 siRNA. (F) Specificity of the effects of GLI1 siRNA on GLI1 mRNA levels in the three prostate cell lines, compared with those of a control unrelated siRNA, 8 h after transfection. The levels of GAPDH are shown below as controls.
The specificity of the GLI1 siRNA was further tested in LNCaP cells. Reduction of GLI1 mRNA levels by the GLI1 siRNA was detected as early as 3 h after transfection and at 8 and 24 h, but not at 48 h (Fig. 3 D and F and data not shown), suggesting up-regulation of GLI1 after its inhibition, possibly because of the action of a rapid positive feedback loop (7, 43). GLI1 siRNA also robustly repressed PTCH1, a result most clearly seen at 48 h, but not the housekeeping gene GAPDH (Fig. 3D and data not shown). Because PTCH1 is a SHH target (37), and in particular of GLI1 (44), this result indicates that interference with GLI1 function by RNAi is selective and effective in prostate cancer cells. GLI1 siRNA also decreased GLI1 mRNA levels in DU145 and PC3 cells after 8 h (Fig. 3F).
Inhibition of GLI1 by RNA interference led to a variable reduction in BrdUrd incorporation in all three cell lines, with strongest effects (≈60%) in LNCaP cells (Fig. 3E). These cells are thus very sensitive to inhibition by cyclopamine and GLI1 interference, suggesting the presence of a fully active canonical pathway activated at the level of SMOH or upstream, but downstream of SHH, because treatment with the blocking anti-SHH Ab had no effect. DU145 cells are not sensitive to cyclopamine, but are sensitive to GLI1 interference, suggesting activation downstream of SMOH and upstream or at the level of GLI1 function. In contrast, PC3 cells are sensitive to cyclopamine and less so to GLI1 interference, perhaps suggesting compensation by the other GLI proteins because PC3 cells express GLI2 [and this GLI gene mediates SHH signals (45) and can behave like Gli1 in mice (46)] or the presence of alternate pathways for tumor cell proliferation. We note, however, that lipofection efficiencies in PC3 cells (Fig. 3C) are the lowest (≈50%) of the three cells tested, indicating that the real effects of GLI1 interference may be higher. Taken together, our results show the requirement of GLI1 in human prostate tumor cells.
Discussion
Here we demonstrate the dependence of prostate cancer cell proliferation on SHH–GLI pathway activity. The data suggest activation of the pathway at different levels in primary prostate tumors and cell lines derived from metastatic lesions. These findings, together with the involvement of this pathway in normal prostate development and growth (17–22), indicate that the normal patterning role of SHH–GLI signaling is deregulated in cancer. This idea is consistent with the proposed events in other tissues, including brain, lung, stomach, muscle, pancreas, and skin, in which the SHH–GLI pathway regulates patterned growth and when deregulated can give rise to SHH–GLI-dependent tumors (reviewed in refs. 2 and 3). Thus, there is a surprising and unexpected parallel in the requirement of SHH–GLI signaling of prostate tumors with those in organs of very different origin, function, and location.
The deduction that prostate tumors display activation at different levels is consistent with findings in brain (ref. 7 and P.S. and A.R.A., unpublished data) and pancreatic (14) tumors, even though the entire set of activating events or mutations have not been described in any case. Indeed, our data suggest that the regulation of the SHH–GLI pathway in the normal prostatic epithelium is altered away from homeostasis in the tumors by epigenetic events or mutations in components such as PTCH1, SMOH, or SUFUH, similar to those already found in other tumors (e.g., refs. 47–53). However, the finding that the pathway is active as assessed by the expression of GLI1 and PTCH1 [as in the case of basal cell carcinomas (6), medulloblastomas (7), and gliomas (7)] allows us to bypass the identification of the likely myriad of activating events to discern that tumor cells harbor an active pathway. Indeed, the finding that SHH expression levels are not correlated with Gleason score, but that all prostate tumor samples tested require continued pathway activity for proliferation, allows us to propose that this pathway is a critical and essential component of prostate cancer.
Specifically, we show the requirement for SHH, SMOH, and/or GLI1 for the proliferation of prostate cancer cells. The fact that all primary tumors tested are sensitive to cyclopamine indicates that SMOH, or upstream elements from it, are common targets leading to the activation of downstream mediators. Several primary cultures are also sensitive to inhibition by blocking anti-SHH Ab, suggesting that, like in stomach tumors (11), autocrine signaling is a frequent cause of pathway activation in prostate cancer. The consistent expression of GLI1 in tumor cell lines and in primary tumors together with the effects of RNA interference indicate that this GLI gene plays a central and general role in prostate tumor cell proliferation, and demonstrate its requirement in human tumorigenesis. In contrast, GLI2 and GLI3 do not appear to be consistently expressed in prostate cancer cells. When expressed, they could have complementary or compensatory roles in some cases, although their roles remains to be determined.
Prostate cancer is thought to develop from a lesion in the epithelial layer to become an invasive tumor that spreads within the prostate and subsequently acquires the potential to metastasize to distant sites, most often the lymph nodes and bone (54). Inhibition of testosterone-dependent tumor growth is the common treatment for advanced disease, but subsequent hormone-independent cell proliferation and metastasis often leads to patient death (55). Our data on the behavior of the three prostate cancer cell lines derived from metastatic lesions suggest that such tumors could harbor additional changes that may make them ligand-independent, albeit still being SHH–GLI pathway dependent, and explain their differential behavior in comparison with the primary cultures. Perhaps the gain of intracellular, cell-autonomous activation of the SHH–GLI pathway represents an advantage for metastatic cells, allowing efficient proliferation far from the prostatic epithelium, where SHH appears to be continually and abundantly produced.
The high inhibition of proliferation by SHH–GLI pathway blockade of the presumed androgen-sensitive primary tumors used in this study, which derive from patients that did not receive hormone treatments, and of the androgen-sensitive LNCaP cell line might be related to the proposed requirement of Shh signaling for normal androgen function, because defects derived from loss of Shh signaling in mice can be rescued by exogenous androgens (22). Prostate cancer could therefore initiate through inappropriate maintenance or enhanced activity of SHH–GLI signaling, and more aggressive (androgen insensitive) states may require additional alterations. Nevertheless, the inhibition of the androgen-insensitive DU145 cell line by RNA interference suggests that even highly aggressive tumors may be sensitive, albeit to different degrees, to GLI1 inhibition.
Prostate stem cells may play a critical role in the epithelial development and homeostasis (56, 57). Because cancer may be a disease of stem cell lineages (discussed in refs. 2, 3, 40, and 58) and SHH–GLI signaling controls the behavior of precursors and of cells with stem cell properties in the mammalian brain (e.g., refs. 30, 59, and 60 and V. Palma, D. Lim, N. Dahmane, N., P.S., Y. Gitton, A. Alvarez-Buylla, A., and A.R.A., unpublished data) and in other tissues and species (61, 62), prostate cancer might derive from inappropriate expansion of prostatic epithelial stem cell lineages caused by abnormal SHH–GLI function.
Finally, our data suggest that SHH and GLI1 may not only be useful markers for prostate cancer but also good targets for anticancer therapies, with emphasis on GLI function as the last and essential step of the pathway, the inhibition of which will likely block signaling by upstream events at any level. SHH–GLI pathway blocking agents should thus provide attractive therapeutic strategies to combat prostate cancer of any grade.
Acknowledgments
We thank Van Nguyen, Verónica Palma, Nadia Dahmane, Virginie Clement, Didier Trono, and Stylianos Antonarakis for discussion and comments on the manuscript. S.D. thanks Robert Chapkin for access to real-time PCR equipment. P.S. was a recipient of an American Brain Tumor Association postdoctoral fellowship. This work was supported by National Institutes of Health/National Cancer Institute grants and the Breast Cancer Showhouse Foundation (to M.W.D.), the Texas A&M University Vice President of Research (to S.D.), and the National Institutes of Health/National Institute of Neurological Disorders and Stroke, National Institutes of Health/National Cancer Institute, the Hirschl Foundation, and the Jeantet Foundation (to A.R.A.).
Abbreviations: SHH, SONIC HEDGEHOG; PTCH1, PATCHED1; SMOH, SMOOTHENED; siRNA, small interfering RNA.
References
- 1.Ingham, P. & McMahon, A. (2001) Genes Dev. 15, 3059–3087. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz i Altaba, A., Sanchez, P. & Dahmane, N. (2002) Nat. Rev. Cancer 2, 361–372. [DOI] [PubMed] [Google Scholar]
- 3.Pasca di Magliano, M. & Hebrok, M. (2003) Nat. Rev. Cancer 3, 903–911. [DOI] [PubMed] [Google Scholar]
- 4.Hahn, H., Wicking, C., Zaphiropoulous, P. G., Gailani, M. R., Shanley, S., Chidambaram, A., Vorechovsky, I., Holmberg, E., Unden, A. B., Gillies, S., et al. (1996) Cell 85, 841–851. [DOI] [PubMed] [Google Scholar]
- 5.Johnson, R. L., Rothman, A. L., Xie, J., Goodrich, L. V., Bare, J. W., Bonifas, J. M., Quinn, A. G., Myers, R. M., Cox, D. R., Epstein, E. H., Jr., & Scott, M. P. (1996) Science 272, 1668–1671. [DOI] [PubMed] [Google Scholar]
- 6.Dahmane, N., Lee, J., Robins, P., Heller, P. & Ruiz i Altaba, A. (1997) Nature 389, 876–881. [DOI] [PubMed] [Google Scholar]
- 7.Dahmane, N., Sanchez, P., Gitton, Y., Palma, V., Sun, T., Beyna, M., Weiner, H. & Ruiz i Altaba, A. (2001) Development (Cambridge, U.K.) 128, 5201–5212. [DOI] [PubMed] [Google Scholar]
- 8.Berman, D. M., Karhadkar, S. S., Hallahan, A. R., Pritchard, J. I., Eberhart, C. G., Watkins, D. N., Chen, J. K., Cooper, M. K., Taipale, J., Olson, J. M. & Beachy, P. A. (2002) Science 297, 1559–1561. [DOI] [PubMed] [Google Scholar]
- 9.Hahn, H., Wojnowski, L., Zimmer, A. M., Hall, J., Miller, G. & Zimmer, A. (1998) Nat. Med. 4, 619–622. [DOI] [PubMed] [Google Scholar]
- 10.Stein, U., Eder, C., Karsten, U., Haensch, W., Walther, W. & Schlag, P. M. (1999) Cancer Res. 59, 1890–1895. [PubMed] [Google Scholar]
- 11.Berman, D. M., Karhadkar, S. S., Maitra, A., Montes De Oca, R., Gerstenblith, M. R., Briggs, K., Parker, A. R., Shimada, Y., Eshleman, J. R., Watkins, D. N. & Beachy, P. A. (2003) Nature 425, 846–851. [DOI] [PubMed] [Google Scholar]
- 12.Watkins, D. N., Berman, D. M., Burkholder, S. G., Wang, B., Beachy, P. A. & Baylin, S. B. (2003) Nature 422, 313–317. [DOI] [PubMed] [Google Scholar]
- 13.Thayer, S. P., Pasca di Magliano, M. P., Heiser, P. W., Nielsen, C. M., Roberts, D. J., Lauwers, G. Y., Qi, Y. P., Gysin, S., Fernandez-del-Castillo, C., Yajnik, V., et al. (2003) Nature 425, 851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson, W. G., De Marzo, A. M. & Isaacs, W. B. (2003) N. Engl. J. Med. 349, 366–381. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J., Platt, K. A., Censullo, P. & Ruiz i Altaba, A. (1997) Development (Cambridge, U.K.) 124, 2537–2552. [DOI] [PubMed] [Google Scholar]
- 16.Hynes, M., Stone, D. M., Dowd, M., Pitts-Meek, S., Goddard, A., Gurney, A. & Rosenthal. A. (1997) Neuron 19, 15–26. [DOI] [PubMed] [Google Scholar]
- 17.Podlaseck, C. A., Barnett, H. Y., Wu, X. R., Laciak, R., Shapiro, E. & Bushman, W. (1999) Dev. Biol. 209, 28–39. [DOI] [PubMed] [Google Scholar]
- 18.Barnett, D. H., Huang, H. Y., Wu, X. R., Laciak, R., Shapiro, E. & Bushman, W. (2002) J. Urol. 168, 2206–2210. [DOI] [PubMed] [Google Scholar]
- 19.Lamm, M. L., Catbagan, W. S., Laciak, R. J., Barnett, D. H., Hebner, C. M., Gaffield, W., Walterhouse, D., Iannaccone, P. & Bushman, W. (2002) Dev. Biol. 249, 349–366. [DOI] [PubMed] [Google Scholar]
- 20.Wang, B. E., Shou, J., Ross, S., Kowppen, H., De Sauvage, F. J. & Gao, W. Q. (2003) J. Biol. Chem. 278, 18506–18513. [DOI] [PubMed] [Google Scholar]
- 21.Freestone, S. H., Marker, P., Grace, O. C., Tomlinson, D. C., Cunha, G. R., Harnden, P. & Thomson, A. A. (2003) Dev. Biol. 264, 352–362. [DOI] [PubMed] [Google Scholar]
- 22.Berman, D. M., Desai, N., Wang, X., Karhadkar, S. S., Reynon, M., Abate-Shen, C., Beachy, P. A. & Shen, M. M. (2004) Dev. Biol. 267, 387–398. [DOI] [PubMed] [Google Scholar]
- 23.Easton, D. F., Schaid, D. J., Whittemore, A. S. & Isaacs, W. J. (2003) Prostate 57, 261–269. [DOI] [PubMed] [Google Scholar]
- 24.Xu, J., Gillanders, E. M., Isaacs, S. D., Chang, B. L., Wiley, K. E., Zheng, S. L., Jones, M., Gildea, D., Riedesel, E., Albertus, J., et al. (2003) Prostate 57, 320–325. [DOI] [PubMed] [Google Scholar]
- 25.Stone, K. R., Mickey, D. D., Wunderli, H., Mickey, G. H. & Paulson, D. F. (1978) Int. J. Cancer 21, 274–281. [DOI] [PubMed] [Google Scholar]
- 26.Kaighn, M. E., Lechner, J. F., Narayan, K. S. & Jones, L. W. (1978) Natl. Cancer Inst. Monogr. 49, 17–21. [PubMed] [Google Scholar]
- 27.Horoszewicz, J. S., Leong, S. S., Chu, T. M., Wajsman, Z. L., Friedman, M., Papsidero, L., Kim, U., Chai, L. S., Kakati, S., Arya, S. K. & Sandberg, A. A. (1980) Prog. Clin. Biol. Res. 37, 115–132. [PubMed] [Google Scholar]
- 28.Matysiak, B. E., Brodzeller, T., Buck, S., French, A., Counts, C., Boorsma, B., Datta, M. W. & Kajdacsy-Balla, A. A. (2003) Appl. Immunohistochem. Mol. Morphol. 11, 269–273. [DOI] [PubMed] [Google Scholar]
- 29.Ericson, J., Morton, S., Kawakami, A., Roelink, H. & Jessell, T. M. (1996) Cell 87, 661–673. [DOI] [PubMed] [Google Scholar]
- 30.Palma, V. & Ruiz i Altaba, A. (2004) Development (Cambridge, U.K.) 131, 337–345. [DOI] [PubMed] [Google Scholar]
- 31.Bostwick, D. G., Shan, A., Qian, J., Darson, M., Maihle, N. J., Jenkins, R. B. & Cheng, L. (1998) Cancer 83, 1995–2002. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan-Lefko, P. J., Chen, T. M., Ittmann, M. M., Barrios, R. J., Ayala, G. E., Huss, W. J., Maddison, L. A., Foster, B. A. & Greenberg, N. M. (2003) Prostate 55, 219–237. [DOI] [PubMed] [Google Scholar]
- 33.DeMarzo, A. M., Nelson, W. G., Isaacs, W. B. & Epstein, J. I. (2003) Lancet 361, 955–964. [DOI] [PubMed] [Google Scholar]
- 34.Pomeroy, S. L., Tamayo, P., Gaasenbeek, M., Sturla, L. M., Angelo, M., McLaughlin, M. E., Kim, J. Y., Goumnerova, L. C., Black, P. M., Lau, C., et al. (2002) Nature 415, 436–442. [DOI] [PubMed] [Google Scholar]
- 35.Grachtchouk, V., Grachtchouk, M., Lowe, L., Johnson, T., Wei, L., Wang, A., de Sauvage, F. & Dlugosz, A. A. (2003) EMBO J. 22, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katayam, M., Yoshida, K., Ishimori, H., Katayama, M., Kawase, T., Motoyama, J. & Kamiguchi, H. (2002) J. Neurooncol. 59, 107–115. [DOI] [PubMed] [Google Scholar]
- 37.Goodrich, L. V., Johnson, R. L., Milenkovic, L., McMahon, J. A. & Scott, M. P. (1996) Genes Dev. 10, 301–312. [DOI] [PubMed] [Google Scholar]
- 38.Chen, J. K., Taipale, J., Cooper, M. K. & Beachy, P. A. (2002) Genes Dev. 16, 2743–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhim, J. S. (2000) Prostate Cancer Prostatic Dis. 3, 229–235. [DOI] [PubMed] [Google Scholar]
- 40.Ruiz i Altaba, A., Stecca, B. & Sanchez, P. (2004) Cancer Lett. 204, 145–157. [DOI] [PubMed] [Google Scholar]
- 41.Park, H. L., Bai, C., Platt, K. A., Matise, M. P., Beeghly, A., Hui, C. C., Nakashima, M. & Joyner, A. L. (2000) Development (Cambridge, U.K.) 127, 1593–1605. [DOI] [PubMed] [Google Scholar]
- 42.Weiner, H. L., Bakst, R., Hurlbert, M. S., Ruggiero, J., Ahn, E., Lee, W. S., Stephen, D., Zagzag, D., Joyner, A. L. & Turnbull, D. H. (2002) Cancer Res. 62, 6385–6389. [PubMed] [Google Scholar]
- 43.Regl, G., Neill, G. W., Eichberger, T., Kasper, M., Ikram, M. S., Koller, J., Hintner, H., Quuinn, A. G., Frischauf, A. M. & Aberger, F. (2002) Oncogene 21, 5529–5539. [DOI] [PubMed] [Google Scholar]
- 44.Ågren, M., Kogerman, P., Kleman, M. I., Wessling, M. & Toftgård, R. (2004) Gene 330, 101–114. [DOI] [PubMed] [Google Scholar]
- 45.Roessler, E., Du, Y. Z., Mullor, J. L., Casas, E., Allen, W. P., Gillessen-Kaesbach, G., Roeder, E. R., Ming, J. E., Ruiz i Altaba, A. & Muenke, M. (2003) Proc. Natl. Acad. Sci. USA 100, 13424–13429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bai, C. B. & Joyner, A. L. (2001) Development (Cambridge, U.K.) 128, 5161–5172. [DOI] [PubMed] [Google Scholar]
- 47.Pietsch, T., Waha, A., Koch, A., Kraus, J., Albrecht, S., Tonn, J., Sorensen, N., Berthold, F., Henk, B., Schmandt, N., et al. (1997) Cancer Res. 57, 2085–2088. [PubMed] [Google Scholar]
- 48.Raffel, R., Jenkins, B., Frederick, L., Hebrink, D., Alderete, B., Fults, D. W. & James, C. D. (1997) Cancer Res. 57, 842–845. [PubMed] [Google Scholar]
- 49.Wolter, M., Reifenberger, J., Sommer, C., Ruzicka, T. & Reifenberger, G. (1997) Cancer Res. 57, 2581–2585. [PubMed] [Google Scholar]
- 50.Reifenberger, J., Wolter, M., Weber, R. G., Megahed, M., Ruzicka, T., Lichter, P. & Reifenberger, G. (1998) Cancer Res. 58, 1798–1803. [PubMed] [Google Scholar]
- 51.Dong, J., Gailani, M. R., Pomeroy, S. L., Reardon, D. & Bale, A. E. (2000) Hum. Mutat. 16, 89–90. [DOI] [PubMed] [Google Scholar]
- 52.Zurawel, R. H., Allen, C., Chiappa, S., Cato, W., Biegel, J., Cogen, P., de Sauvage, F. & Raffel, C. (2000) Genes Chromosomes Cancer 27, 44–51. [DOI] [PubMed] [Google Scholar]
- 53.Taylor, M. D., Liu, L., Raffel, C., Hui, C. C., Mainprize, V. G., Zhang, X., Agatep, R., Chiappa, S., Gao, L., Lowrance, A., et al. (2002) Nat. Genet. 31, 306–310. [DOI] [PubMed] [Google Scholar]
- 54.Abate-Shen, C. & Shen, M. (2000) Genes Dev. 14, 2410–2434. [DOI] [PubMed] [Google Scholar]
- 55.Martel, C. L., Gumerlock, P. H., Meyers, F. J. & Lara, P. N. (2003) Cancer Treat. Rev. 29, 171–187. [DOI] [PubMed] [Google Scholar]
- 56.De Marzo, A. M., Nelson, W. G., Meeker, A. K. & Coffey, D. S. (1998) J. Urol. 160, 2381–2392. [DOI] [PubMed] [Google Scholar]
- 57.Bonkhoff, H. (1996) Eur. Urol. 30, 201–205. [DOI] [PubMed] [Google Scholar]
- 58.Reya, T., Morrison, S. J., Clarke, M. F. & Weissman, I. L. (2001) Nature 414, 105–111. [DOI] [PubMed] [Google Scholar]
- 59.Lai, K., Kaspar, B. K., Gage, F. H. & Schaffer, D. V. (2003) Nat. Neurosci. 6, 21–27. [DOI] [PubMed] [Google Scholar]
- 60.Machold, R., Hayashi, S., Rutlin, M., Muzumdar, M. D., Nery, S., Corbin, J. G., Gritli-Linde, A., Dellovade, T., Porter, J. A., Rubin, L. L., et al. (2003) Neuron 39, 937–950. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, Y. & Kalderon, D. (2001) Nature 410, 599–604. [DOI] [PubMed] [Google Scholar]
- 62.Park, Y., Rangel, C., Reynolds, M. M., Caldwell, M. C., Johns, M., Nayak, M., Welsh, C. J., McDermott, S. & Datta, S. (2003) Dev. Biol. 253, 247–257. [DOI] [PubMed] [Google Scholar]