Abstract
Coronaviruses are important pathogens that cause acute respiratory diseases in humans. Replication of the ≈30-kb positive-strand RNA genome of coronaviruses and discontinuous synthesis of an extensive set of subgenome-length RNAs (transcription) are mediated by the replicase-transcriptase, a barely characterized protein complex that comprises several cellular proteins and up to 16 viral subunits. The coronavirus replicase-transcriptase was recently predicted to contain RNA-processing enzymes that are extremely rare or absent in other RNA viruses. Here, we established and characterized the activity of one of these enzymes, replicative nidoviral uridylate-specific endoribonuclease (NendoU). It is considered a major genetic marker that discriminates nidoviruses (Coronaviridae, Arteriviridae, and Roniviridae) from all other RNA virus families. Bacterially expressed forms of NendoU of severe acute respiratory syndrome coronavirus and human coronavirus 229E were revealed to cleave single-stranded and double-stranded RNA in a Mn2+-dependent manner. Single-stranded RNA was cleaved less specifically and effectively, suggesting that double-stranded RNA is the biologically relevant NendoU substrate. Double-stranded RNA substrates were cleaved upstream and downstream of uridylates at GUU or GU sequences to produce molecules with 2′-3′ cyclic phosphate ends. 2′-O-ribose-methylated RNA substrates proved to be resistant to cleavage by NendoU, indicating a functional link with the 2′-O-ribose methyltransferase located adjacent to NendoU in the coronavirus replicative polyprotein. A mutagenesis study verified potential active-site residues and allowed us to inactivate NendoU in the full-length human coronavirus 229E clone. Substitution of D6408 by Ala was shown to abolish viral RNA synthesis, demonstrating that NendoU has critical functions in viral replication and transcription.
Human coronavirus 229E (HCoV-229E), a group 1 coronavirus, is one of the major viral pathogens causing upper respiratory tract illness in humans (1), whereas severe acute respiratory syndrome coronavirus (SARS-CoV), a group 2 coronavirus, has been identified as the causative agent of SARS, a life-threatening form of pneumonia that caused >8,000 fatalities in a worldwide epidemic in 2003 (2, 3). The extremely large positive-strand RNA genomes of HCoV-229E (27.3 kb) and SARS-CoV (29.7 kb) contain 8 and 14 ORFs, respectively (4, 5). The two most 5′-terminal ORFs, 1a and 1b, encode the major subunits of the replicative machinery, whereas the more downstream ORFs encode structural and virus-specific accessory proteins.
Coronavirus gene expression involves a series of complex transcriptional, translational, and posttranslational regulatory mechanisms (6, 7). After receptor-mediated entry, two large replicative polyproteins, pp1a (>450 kDa) and pp1ab (>750 kDa), are translated from the genome RNA. The polyproteins are encoded by the replicase gene (>20,000 bases) that comprise ORFs 1a and 1b (Fig. 1A) (8). Expression of pp1ab involves ribosomal frameshifting into the -1 frame just upstream of the ORF 1a translation termination codon (9). Human coronavirus polyproteins are autoproteolytically processed by two (SARS-CoV) or three (HCoV-229E) viral proteases called papain-like proteases and 3C-like main protease (5, 10-12). In total, at least 16 processing end-products called nonstructural proteins (nsp) 1-16 are derived from the pp1a and pp1ab polyproteins (8). Together with a number of processing intermediates and cellular proteins, nsp1-nsp16 are thought to form a huge membrane-anchored protein complex, called replicase-transcriptase, that is poorly characterized but generally accepted to mediate all of the functions required for genome replication and synthesis of a diverse set of subgenomic mRNAs (transcription) (13). Coronavirus subgenomic mRNAs have a common 5′-terminal leader sequence that is identical to the 5′ end of the genome, which are produced through a unique discontinuous transcription mechanism (14, 15).
Fig. 1.
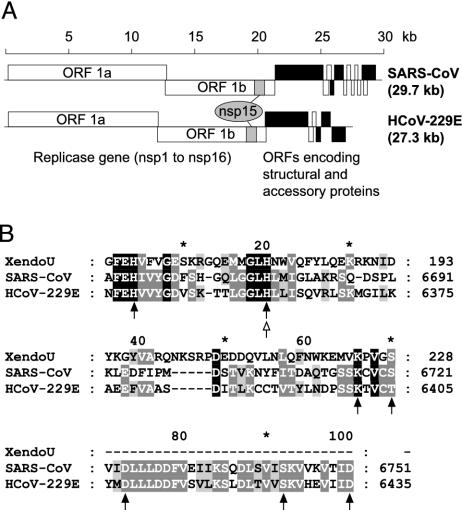
Human coronaviruses replicase genes encode a putative endoribonuclease. (A) Functional ORFs in the genomes of SARS-CoV and HCoV-229E are expressed from both genomic RNA and a set of subgenomic mRNAs. ORFs encoding the four structural proteins S, E, M, and N, are indicated in black. The ORF 1a- and ORF 1b-encoded proteins, pp1a and pp1ab, are cleaved by viral proteases to yield 16 processing end-products: nsp1-nsp16. The C-terminal part of nsp15 was predicted to harbor an endoribonuclease domain (20). (B) Partial sequence alignment of SARS-CoV and HCoV-229E NendoU domains with Xenopus laevis XendoU, a poly(U)-specific endoribonuclease. Conserved residues targeted in this study by site-directed mutagenesis are indicated by filled (HCoV-229E) and open (SARS-CoV) arrows.
Besides a putative RNA-dependent RNA polymerase activity (16) and the previously characterized helicase and protease activities (7, 17-19), which are all common to positive-strand RNA viruses, the coronavirus replicase-transcriptase also contains a set of putative enzymes that are rare or even unique among other RNA viruses. Thus, nsp3, nsp14, nsp15, and nsp16 were recently predicted to harbor ADP-ribose 1″-phosphatase, 3′-to-5′ exoribonuclease, endoribonuclease, and 2′-O-ribose methyltransferase domains (20). It seems logical to link the conservation of these additional and, in part, unique activities with the enormous size of the coronavirus RNA genome and the unusual RNA synthesis mechanisms used by coronaviruses (20). To a varying degree, the putative coronavirus RNA-processing activities are also conserved in distant relatives from the Nidovirales order that use similar RNA synthesis mechanisms and/or have comparably large genomes (20).
We initiated a broad analysis of these newly predicted enzymes by using HCoV-229E and SARS-CoV. In this paper, we describe the characterization of nsp15, which has been predicted to contain in its C-terminal region a nidovirus-wide conserved endoribonuclease domain called nidoviral uridylate-specific endoribonuclease (NendoU) (20). We show by site-directed mutagenesis of the nsp15 coding sequence in the infectious HCoV-229E clone that the domain is critically involved in coronavirus replication and transcription. Recombinant forms of both HCoV-229E and SARS-CoV nsp15 were revealed to possess Mn2+-dependent endoribonuclease activities that produce molecules with 2′-3′ cyclic phosphate ends. The data suggest a preference of NendoU for cleavage of double-stranded (ds)RNA substrates upstream and downstream of uridylates in GU or GUU sequences. RNA 2′-O-ribose methylation blocked these cleavages, indicating a functional link between the coronavirus NendoU and putative 2′-O-ribose methyltransferase activities. The coordinated expression of the NendoU and 2′-O-ribose methyltransferase activities, which reside adjacent to each other in the viral polyprotein in nsp15 and nsp16, further supports a functional interplay between these activities. Finally, the characterization of nsp15 mutants with substitutions of conserved residues allowed insights into the active site of this endoribonuclease family.
Materials and Methods
Protein Expression and Purification. Nucleotides 19,551-20,588 of SARS-CoV (strain Frankfurt 1) and 18,622-19,665 of HCoV-229E, respectively, followed by a translation stop codon, were inserted in the unique XmnI site of pMal-c2 plasmid DNA (New England Biolabs). Mutations in the nsp15 coding sequence were introduced by PCR-based methods. For protein expression, pMal-SARS-CoV-nsp15 and pMal-HCoV-229E-nsp15 plasmid DNA, respectively, or mutant derivatives were used to transform Escherichia coli TB1 cells (New England Biolabs). Maltose-binding protein (MBP) fusion proteins were expressed and purified by amylose affinity chromatography as described in ref. 10.
RNA and DNA Substrates and Size Markers. RNA oligonucleotides were purchased from Eurogentec (Seraing, Belgium): RNA2 (5′-CGCAGUGAGCUCCUAAUCGCCC-3′), RNA3 (5′-CGCAGUUAGCUCCUAAUCGCCC-3′), RNA8 (5′-GGGCGAUUAGGAGCUAACUGCG-3′), and RNA9 (2′-O-methyl RNA3) and marker RNAs m1 (5′-CGCAGUUA-3′), m2 (5′-CGCAGUU-3′), m3 (5′-CGCAGU-3′), m4 (5′-CGCAG-3′), and m5 (5′-CGCA-3′). To produce dsRNA forms of RNAs 2, 3, and 9, the oligonucleotides were annealed with a 1.5-fold excess of RNA8. Longer RNA and DNA substrates of 1 kbp were produced by in vitro transcription and PCR by using the protocols described in ref. 18. We performed 5′-labeling of single-stranded (ss)DNAs and ssRNAs by using T4 polynucleotide kinase (PNK) and [γ-32P]ATP.
Nuclease Assay. Typical reactions contained 300 nM of wild-type or mutant MBP-nsp15 and 50 nM (labeled) or 500 nM (unlabeled) (ribo)oligonucleotide. Longer DNA and RNA molecules of 1 kbp were used at 10 nM. Reactions were performed in 25 mM Hepes·KOH (pH 7.4)/50 mM NaCl/5 mM MnCl2/1 mM DTT. Following incubation at 37°C for up to 60 min, the reactions were stopped by adding an equal volume of loading buffer (formamide containing 10 mM EDTA) and the products were analyzed in 8% or 20% polyacrylamide gels (acrylamide/bisacrylamide ratio, 19:1) buffered with 0.5× Tris-borate-EDTA containing 0.1% SDS or 7.5 M urea.
Analysis of the 3′ Terminus. Gel-purified 5′-terminal cleavage product of RNA2 was incubated with (i) buffer P alone (70 mM Tris·HCl, pH 7.6/10 mM MgCl2/5 mM DTT), (ii) buffer P in the presence of 5 units of PNK or (iii) buffer C (50 mM Tris·HCl, pH 7.9/100 mM NaCl/10 mM MgCl2/1 mM DTT) in the presence of 5 units of calf intestine phosphatase (CIP). After incubation at 37°C for 30 min, the reaction mixtures were incubated at 94°C for 5 min. Thereafter, the RNAs were ethanol-precipitated and redissolved in 50 mM Tris·HCl, pH 7.8/10 mM MgCl2/10 DTT/10% dimethyl sulfoxide/5 μM ATP/20 μg/ml BSA. The mixtures were then incubated for 3.5 h at 4°C with 8 units of T4 RNA ligase and 1 μM [5′-32P]pCp [3,000 Ci/mmol (1 Ci = 37 GBq)] and analyzed by denaturing PAGE and autoradiography.
Primer Extension. Unlabeled dsRNAs 2 and 3, respectively, were cleaved with nsp15. The resulting products were annealed with 5′-[γ-32P]-labeled DNA14 and DNA15, respectively, in first-strand Superscript III buffer (Invitrogen) supplemented with 5 mM DTT and 1 mM dNTPs. After incubation with 200 units of Superscript III RNase H- reverse transcriptase at 40°C for 30 min, the reactions were stopped by adding equal volumes of loading buffer and the reaction products were analyzed by denaturing PAGE.
Mutagenesis of the HCoV-229E Full-Length cDNA Clone. The recombinant vaccinia virus vHCoV-inf-1 containing the full-length cDNA of HCoV-229E (21) was used to construct vHCoV-1ab-D6408A, a mutant derivative in which the HCoV-229E nucleotides 19513GAT (encoding pp1ab residue Asp-6408) were substituted with 19513GCT (Ala-6408). Mutagenesis of vHCoV-inf-1 involved several steps of vaccinia virus-mediated recombination by using the E. coli guanine phosphoribosyltransferase (gpt) gene as a marker for positive or negative selection (see Supporting Materials and Methods, which is published as supporting information on the PNAS web site). Genome-length RNA was prepared by in vitro transcription by using purified genomic DNA from vHCoV-inf-1 and vHCoV-1ab-D6408A, respectively. HCoV-inf-1 RNA and HCoV-1ab-D6408A RNA (15 μg), respectively, were used for electroporation of 4 × 106 BHK-21 cells expressing the HCoV-229E nucleocapsid protein [BHK-21 (N) cells] (V.T., unpublished data). At 72 h after transfection, viral RNA synthesis was analyzed by Northern blotting by using a 32P-labeled probe corresponding to HCoV-229E nucleotides 26,297-27,273.
Results
Conservation of nsp15-Associated Mn2+ Ion-Dependent Ribonuclease Activities in Human Coronaviruses. To verify the predicted endonuclease activity of NendoU, the coding sequences of nsp15 of two human coronaviruses, SARS-CoV and HCoV-229E, were expressed as N-terminal fusions with E. coli MBP. As presumptive negative controls, two nsp15 mutants with Ala replacements of the conserved His residues, His-6678 (for SARS-CoV) and His-6360 (for HCoV-229E) (Fig. 1B), were generated. The affinity-purified proteins were incubated with different types of nucleic acid substrates in the presence of Mn2+, a known cofactor for the endonucleolytic activity of the Xenopus laevis homolog poly(U)-specific endoribonuclease (XendoU) (22). Both wild-type proteins, but not their mutant derivatives, were revealed to cleave ssRNA and, even more effectively, dsRNA, whereas ssDNA and dsDNA remained uncleaved, demonstrating the predicted ribonuclease activity of nsp15 (Fig. 2). The NendoU activities strictly depended on divalent metal ions, with Mn2+ being strongly preferred over Mg2+ and Ca2+ (Fig. 7, which is published as supporting information on the PNAS web site).
Fig. 2.
RNA-specific endonuclease activity of SARS-CoV and HCoV-229E nsp15. ss and ds versions of 5′-[32P] RNA and DNA substrates, respectively, of 1 kb(p) were incubated for 20 min at 37°C without protein (lanes 1, 6, 11, and 16), with HCoV-229E MBP-nsp15_H6360A (HCoV nsp15_mut; lanes 2, 7, 12, and 17), with HCoV-229E MBP-nsp15 (HCoV nsp15; lanes 3, 8, 13, and 18), with SARS-CoV MBP-nsp15_H6678A (SCoV nsp15_mut; lanes 4, 9, 14, and 19) or with SARS-CoV MBP-nsp15 (SCoV nsp15; lanes 5, 10, 15, and 20). Reaction products and RNA size markers were separated in an 8% polyacrylamide gel containing 0.1% SDS.
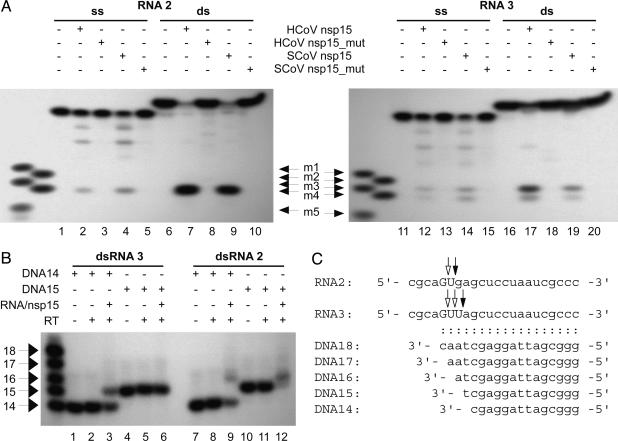
Uridylate Specificity of Coronavirus NendoU. To gain insights into possible biological substrates of coronavirus NendoU, we sought to determine its substrate specificity in more detail. By using a series of in vitro-transcribed and synthetic RNAs, evidence was obtained to suggest that NendoU may be specific for uridylates (data not shown). However, in contrast to data published on the poly(U)-specific cellular homolog, XendoU (22), we did not observe a strict requirement for poly(U) or oligo(U) sequences. Thus, for example, 5′-[32P]-labeled ssRNA2, which contained no UU dinucleotides in its sequence (Fig. 3C), was cleaved by coronavirus nsp15 to yield three major products and one minor product whose sizes (as determined by denaturing PAGE) were compatible with cleavages at the four monouridylates present in the sequence (Fig. 3A Left). Cleavage of RNA3, which differed from RNA2 by a G→U replacement at the seventh position, yielded one additional product (Fig. 3A Right), supporting the specificity of both SARS-CoV and HCoV-229E nsp15 for uridylate. dsRNA substrates produced by annealing RNAs 2 and 3 with complementary RNAs were cleaved more efficiently than their nonannealed ssRNA counterparts in repeated experiments by using different batches of substrates (illustrated in Fig. 3A).
Fig. 3.
Substrate specificity of coronavirus endoribonucleases. (A) ss and ds versions of 5′-[32P]-labeled RNAs 2 and 3 were cleaved with nsp15, and the sizes of the 5′-terminal reaction products were determined by denaturing PAGE with 5′-[32P]-labeled RNAs, m1-m5, as markers. Lanes 1, 6, 11, and 16, reactions without protein; lanes 2, 7, 12, and 17, reactions with HCoV-229E MBP-nsp15; lanes 3, 8, 13, and 18, reactions with HCoV-229E MBP-nsp15_H6360A; lanes 4, 9, 14, and 19, reactions with SARS-CoV (SCoV) MBP-nsp15; lanes 5, 10, 15 and 20, reactions with SARS-CoV MBP-nsp15_H6678A. (B) Primer extension analysis suggests cleavages also downstream of the most 3′ uridylate in GU and GUU sequences. dsRNAs 3 and 2, respectively, were cleaved with nsp15, and primer extension reactions were done by using the indicated 5′-[32P]-labeled DNA primers. Reaction products were analyzed by denaturing PAGE with 5′-[32P]-labeled DNAs 14-18 as size markers. Lanes 1, 4, 7, and 10, reverse transcription primers DNA14 and DNA15, respectively; lanes 2, 5, 8, and 11, reactions in the absence of RNA template; lanes 3, 6, 9, and 12, reactions with the indicated RT primer and the indicated nsp15-cleaved dsRNA molecule as template. (C) Sequences of synthetic oligonucleotides used in this experiment. ds forms of RNAs 2 and 3 were produced by annealing with complementary RNAs. Cleavages downstream of uridylates as determined by primer extension (B) are indicated by black arrows, and cleavages upstream of uridylates (A) are indicated by open arrows.
In the above experiment, the cleavage sites in dsRNA substrates were determined by comparison of the 5′-[32P]-labeled products with a nested set of 5′-[32P]-labeled marker RNAs (m1-m5) that were run in parallel in denaturing gels. The 5′-terminal cleavage product of dsRNA2 comigrated with RNA m4 (5 nucleotides), indicating efficient cleavage upstream of U in the 5GUGA8 sequence (Fig. 3 A Left and C, open arrows). By using dsRNA3 (5GUGA8→5GUUA8), we observed 5′-terminal cleavage products comigrating with RNAs m3 (six nucleotides) and m4, suggesting cleavages upstream of each of the uridylates present in the 5GUUA8 sequence (Fig. 3 A Right and C, open arrows).
The NendoU cleavage specificity was further analyzed by primer extension experiments. Surprisingly, determination of the 5′-terminal nucleotide of the 3′-terminal cleavage product(s) revealed that cleavage also occurred downstream of uridylate. Thus, by using nsp15-cleaved RNA3 as a template for reverse transcription, the 14-nt primer (DNA14) was extended by 1 nt, whereas the 15-nt primer (DNA15) was not extended, indicating complete cleavage downstream of the most 3′-terminal uridylate in the 5GUUA8 sequence. Likewise, analysis of the DNA14- and DNA15-primed extension products by using nsp15-cleaved RNA2 as a template showed that the 3′-terminal cleavage product of RNA2 has G7 at its 5′ terminus, suggesting that efficient cleavage in the 5GUGA8 sequence also occurred downstream of uridylate.
Despite complete cleavage downstream of the 3′-terminal uridylate of a given cleavage site, we failed to observe a corresponding 5′-terminal (intermediate) product of the expected size in the experiment shown in Fig. 3A (also at shorter reaction times). The pronounced stability of the larger of the two 5′-terminal cleavage products shown in Fig. 3A Right at extended reaction times (data not shown) further argued against sequential cleavages (”trimming”) of 3′ uridylates in 3′-to-5′ direction. Taken together, the data suggest that, after cleavage, which primarily occurs upstream of uridylates in GU(U) sequences of dsRNA substrates, remaining uridylate at the 5′ end of the 3′-terminal cleavage product is efficiently removed. The details of this activity remain to be investigated.
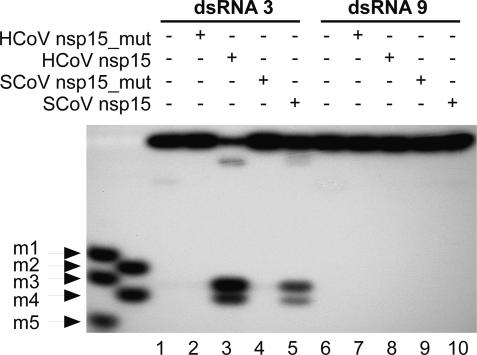
RNA 2′-O-Ribose Methylation Blocks Cleavage by Coronavirus NendoU. We next investigated whether 2′-O-ribose methyl groups present in the substrate RNA affect nsp15-mediated endonucleolytic cleavage. To this end, RNA3, which in previous experiments had proven to be an excellent substrate for NendoU, was synthesized as a 2′-O-ribose methylated derivative (RNA9). Fig. 4 shows that, in clear contrast to RNA3, the 2′-O-ribose methylated RNA9 was completely resistant to cleavage, demonstrating the critical role of the 2′ hydroxyl group for NendoU-mediated cleavage.
Fig. 4.
RNA 2′-O-ribose methylation blocks clearage by coronavirus nsp15. dsRNA substrates were produced by annealing RNA8 to the complementary 5′-[32P]-labeled RNA3 or its 2′-O-ribose-methylated derivative, RNA9. Substrates were incubated with SARS-CoV (SCoV) and HCoV-229E (HCoV) MBP-nsp15, respectively, or inactive control proteins. Reaction products were analyzed by denaturing PAGE and autoradiography. Lanes 1 and 6, reactions without protein; lanes 2 and 7, reactions with HCoV-229E MBP-nsp15_H6360A; lanes 3 and 8, reactions with HCoV-229E MBP-nsp15; lanes 4 and 9, reactions with SARS-CoV MBP-nsp15_H6678A; lanes 5 and 10, reactions with SARS-CoV MBP-nsp15.
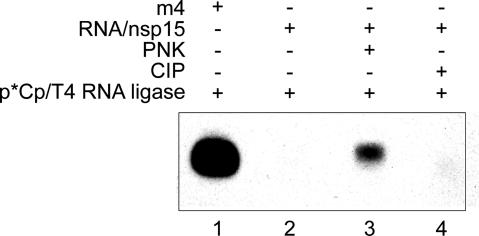
Products of NendoU-Mediated RNA Cleavage Contain 2′-3′ Cyclic Phosphate. Comparison with marker RNAs revealed a slight increase in the electrophoretic mobilities of 5′-terminal cleavage products (Figs. 3A and 4), indicating that the latter may have an additional negative charge because of the presence of 3′ phosphate or 2′-3′ cyclic phosphate ends. The latter structure has previously been identified in XendoU cleavage products (22). To test this hypothesis, the 5′-terminal dsRNA2 cleavage product was gel-purified and ligated with [5′-32P]pCp directly or after treatment with CIP or PNK. As expected, RNA m4, which had a 3′ hydroxyl group and was used as a positive control for the ligase reaction, could be ligated effectively with [5′-32P]pCp (Fig. 5, lane 1). In contrast, the nsp15 cleavage product required pretreatment with PNK, but not CIP, for effective ligation with [5′-32P]pCp (Fig. 5). Because PNK (in contrast to CIP) is capable of removing 2′-3′ cyclic phosphate (23), the data strongly suggest that products of NendoU-mediated cleavage reactions have 2′-3′ cyclic phosphate ends. This conclusion is consistent with the observed critical role of the substrate's 2′ hydroxyl group for cleavage.
Fig. 5.
Coronavirus NendoU activity produces molecules with 2′-3′ cyclic phosphate termini. Ligation of 5′-[32P]pCp with the 3′ terminus of coronavirus nsp15 cleavage products using T4 RNA ligase requires pretreatment with PNK that (in contrast to CIP) is capable of removing 2′-3′ cyclic phosphates. Reaction products were analyzed by denaturing PAGE and autoradiography. Lane 1, ligation reaction with control RNA m4; lanes 2-4, ligation reactions with purified 5′-terminal nsp15 cleavage product without further treatment (lane 2) and after treatment with PNK (lane 3) or CIP (lane 4).
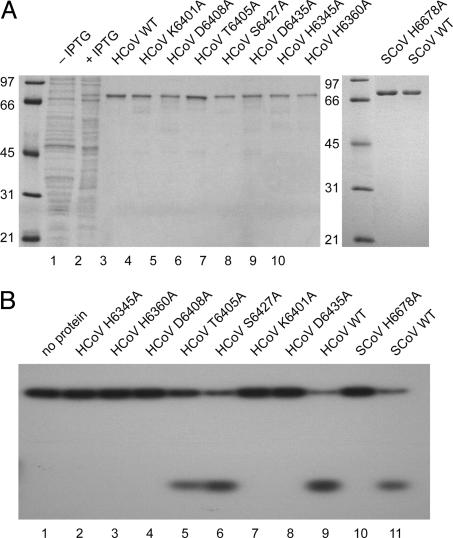
Mutagenesis Data Support the Alignment of Coronavirus NendoU with XendoU. To gain first insights in structural and functional features of enzymes of the XendoU/NendoU family, a set of mutant derivatives of HCoV-229E NendoU with Ala substitutions of conserved residues was characterized (Fig. 1B). The proteins were expressed and purified in an identical manner as the wild-type protein (Fig. 6A). Two His residues, 6345 and 6360, and Lys-6401, which are conserved across the entire XendoU/NendoU family, and the Asp-6408 and Asp-6435 residues conserved in NendoU proved to be indispensable for endonucleolytic activity with dsRNA2 (Fig. 6B) and dsRNA3 (data not shown) as substrates. By contrast, substitutions of two other less conserved residues, Ser-6427 and Thr-6405, were tolerated, albeit with slightly reduced activities. As mentioned above, substitution of the counterpart of HCoV-229E His-6360 in SARS-CoV nsp15 (SARS-CoV His-6678) also resulted in an inactive protein. Together, the data demonstrate that the endoribonucleolytic activities identified in this study are mediated by the recombinant coronavirus proteins rather than contaminating nucleases copurified from E. coli. Furthermore, the data support the critical role for enzymatic activity of three of the most conserved XendoU/NendoU residues (His/His/Lys) that we identified previously by sequence comparisons (20) and two Asp residues conserved in NendoU. The chemical nature of the five residues suggests their involvement in catalysis and/or metal ion coordination (24).
Fig. 6.
Activities of mutant forms of human coronavirus NendoU. (A) Coomassie brilliant blue-stained SDS-polyacrylamide gel showing the expression and purification of HCoV-229E (HCoV) and SARS-CoV (SCoV) MBP-nsp15 fusion proteins. Lanes 1 and 2, total lysates of E. coli cells transformed with pMal-HCoV-nsp15 plasmid DNA and grown in the absence (lane 1) or presence (lane 2) of isopropyl β-d-thiogalactoside (IPTG); lanes 3-12, purified MBP-nsp15 fusion proteins with nsp15 wild-type sequence (WT) or with the indicated substitutions of conserved residues. Numbering of residues is according to their positions in the replicase polyproteins of HCoV-229E and SARS-CoV, respectively. (B) HCoV-229E and SARS-CoV MBP-nsp15 fusion proteins or their mutant derivatives were incubated with dsRNA2, and reaction products were separated in a 20% polyacrylamide gel containing 7.5 M urea.
NendoU Is Critically Involved in Coronaviral RNA Synthesis. The nidovirus-wide conservation of NendoU suggests a key role for this protein in the life cycle of these viruses. To test this hypothesis, we introduced a single point mutation into the infectious HCoV-229E cDNA clone, leading to a replacement of the pp1ab residue Asp-6408 with Ala. As shown above (Fig. 6), this substitution completely inactivated the endonucleolytic activity of the MBP-nsp15 fusion protein in vitro. Analysis of viral RNA synthesis after transfection of mutant and wild-type RNA, respectively, into BHK-21 (N) cells revealed a complete block of viral RNA synthesis for the nsp15 mutant (Fig. 8, which is published as supporting information on the PNAS web site), demonstrating the critical role of nsp15 in coronavirus replication and transcription.
Discussion
We present here the results of the identification and characterization of an RNase of human coronaviruses designated NendoU. Besides Erns, a secreted and virion-associated RNase of pestiviruses (25), NendoU is just the second group of RNases identified in the >40 families of positive-strand RNA viruses. Erns was recently shown to be a pestivirus-specific cofactor of cell entry; it is not encoded in the related hepaciviruses and flavi-viruses. In contrast, NendoU belongs to the few replicative domains conserved in coronaviruses and all their distant nidovirus relatives (Arteriviridae and Roniviridae) (20, 26). In fact, it is one of only two unique genetic markers common to nidoviruses but absent in other RNA viruses (26, 27). Accordingly, NendoU is expected to mediate a nidovirus-specific step in RNA synthesis. Given the need to replicate positive-strand RNA genomes of extraordinary size, the presence of a replicative endoribonuclease in nidoviruses is a surprising finding that adds yet another dimension to the amazing plasticity of RNA viruses that is commonly associated with rapid evolution.
The data on two human coronavirus NendoU revealed striking parallels to a distant cellular homolog, XendoU, a poly(U)-specific endoribonuclease involved in small nucleolar RNA processing and utilization (22). The recently proposed relationship between XendoU and NendoU (20) is strongly supported by the biochemical data obtained in this study. Thus, like their cellular homolog, NendoU depended on Mn2+ ions and, rather untypical for Mn2+-dependent nucleases, generated molecules with 2′-3′ cyclic phosphate ends. Besides the similar chemistry of endonucleolytic cleavage, the cellular and viral homologs also share a similar specificity to uridylates in both ssRNAs and dsRNAs.
We found a strong preference of NendoU for cleavage at GU(U) sequences in dsRNA, whereas corresponding ssRNA molecules were cleaved (albeit less efficiently) also at uridylates preceded by cytidylate or adenylate residues. These observations suggest that the biologically relevant substrate of NendoU, which remains to be identified, may be dsRNA. Although more substrates must be tested to verify the sequence preference evident in this study, the complementarity of GUU to the strictly conserved core element, AAC, of nidovirus transcription-regulating sequences (TRSs) is noteworthy. TRSs are known to play a critical role in (discontinuous) minus-strand RNA synthesis in which they guide (by complementary basepairing) the transfer of the nascent minus strand to an upstream sequence (called leader TRS) on the template RNA (14, 15, 28). It is tempting to speculate that GUU, when part of dsRNA during minus-strand synthesis, is a candidate sequence to be cleaved by NendoU (before or after transfer of the nascent minus strand to the leader TRS). If confirmed, this cleavage may be used to remove potentially nonpaired nucleotides downstream of the minus-strand TRS complement upon its binding to the leader TRS.
Regardless of whether NendoU cleaves only GU(U) or also other short sequence(s), its activity must be sufficiently controlled to achieve an appropriate selectivity in the endonucleolytic processing of natural substrates. This control may include both protein and polynucleotide factors interacting with NendoU itself or NendoU substrates. In this respect, our finding that 2′-O-ribose methyl groups rendered RNA substrates insensitive to cleavage by NendoU is of particular interest as it suggests a cooperation between nsp15, which contains the NendoU domain, and nsp16, which is predicted to be a 2′-O-ribose methyltransferase. Should this (functional) cooperation be confirmed, it would provide a rationale for the coordinated release of the adjacent nsp15 and nsp16 subunits from the pp1ab polyprotein by 3C-like main protease (29).
Our data also provide support for parallels between ribosomal preRNA processing pathways (involving XendoU) and nidovirus RNA synthesis (involving NendoU) (20). In this context, it should be interesting to investigate whether NendoU has cellular substrates and is used to produce small regulatory RNAs (similar to the small nucleolar RNAs produced by XendoU) that may bind to specific (viral and/or cellular) target sequences by complementary base-pairing to trigger 2′-O-ribose methylation(s) by nsp16 (or cellular methylases).
To link the fundamental and applied aspects of the NendoU study, we characterized orthologs from two human coronaviruses and found very similar biochemical properties. These data together with the nidovirus-wide conservation of NendoU, the essential role in coronavirus RNA synthesis (demonstrated here for HCoV-229E), and the lack of close cellular homologs suggest that NendoU may be an attractive target for the development of selective antiviral drugs to combat respiratory infections caused by SARS-CoV and other known or emerging human nidoviruses.
Acknowledgments
We thank Eric Snijder and Clara Posthuma (Leiden University Medical Center) for sharing unpublished data. J.Z. thanks Stuart Siddell (University of Bristol) for the recombinant vaccinia virus vHCoV-inf-1 and for contributing to the development of the HCoV-229E reverse genetics system. The work was supported by Deutsche Forschungsgemeinschaft Grants SFB 479 and Zi 618/2-3 (to J.Z.).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ds, double-stranded; ss, single-stranded; HCoV-229E, human coronavirus 229E; MBP, Escherichia coli maltose-binding protein; NendoU, nidoviral uridylate-specific endoribonuclease; nsp, nonstructural protein; SARS-CoV, severe acute respiratory syndrome coronavirus; PNK, T4 polynucleotide kinase; CIP, calf intestine phosphatase; TRS, transcription-regulating sequence; XendoU, Xenopus laevis poly(U)-specific endoribonuclease.
References
- 1.Myint, S. H. (1995) in The Coronaviridae, ed. Siddell, S. G. (Plenum, New York), pp. 389-401.
- 2.Peiris, J. S., Yuen, K. Y., Osterhaus, A. D. & Stöhr, K. (2003) N. Engl. J. Med. 349, 2431-2441. [DOI] [PubMed] [Google Scholar]
- 3.Peiris, J. S., Lai, S. T., Poon, L. L., Guan, Y., Yam, L. Y., Lim, W., Nicholls, J., Yee, W. K., Yan, W. W., Cheung, M. T., et al. (2003) Lancet 361, 1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herold, J., Raabe, T., Schelle-Prinz, B. & Siddell, S. G. (1993) Virology 195, 680-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thiel, V., Ivanov, K. A., Putics, A., Hertzig, T., Schelle, B., Bayer, S., Weissbrich, B., Snijder, E. J., Rabenau, H., Doerr, H. W., Gorbalenya, A. E. & Ziebuhr, J. (2003) J. Gen. Virol. 84, 2305-2315. [DOI] [PubMed] [Google Scholar]
- 6.Lai, M. M. C. & Holmes, K. V. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), Vol. 1, pp. 1163-1185. [Google Scholar]
- 7.Ziebuhr, J., Snijder, E. J. & Gorbalenya, A. E. (2000) J. Gen. Virol. 81, 853-879. [DOI] [PubMed] [Google Scholar]
- 8.Ziebuhr, J. (2004) Curr. Top. Microbiol. Immunol. 287, in press. [DOI] [PMC free article] [PubMed]
- 9.Brierley, I., Digard, P. & Inglis, S. C. (1989) Cell 57, 537-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ziebuhr, J., Herold, J. & Siddell, S. G. (1995) J. Virol. 69, 4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziebuhr, J., Thiel, V. & Gorbalenya, A. E. (2001) J. Biol. Chem. 276, 33220-33232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herold, J., Gorbalenya, A. E., Thiel, V., Schelle, B. & Siddell, S. G. (1998) J. Virol. 72, 910-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thiel, V., Herold, J., Schelle, B. & Siddell, S. G. (2001) J. Virol. 75, 6676-6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawicki, S. G. & Sawicki, D. L. (1998) Adv. Exp. Med. Biol. 440, 215-219. [DOI] [PubMed] [Google Scholar]
- 15.Zúñiga, S., Sola, I., Alonso, S. & Enjuanes, L. (2004) J. Virol. 78, 980-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbalenya, A. E., Koonin, E. V., Donchenko, A. P. & Blinov, V. M. (1989) Nucleic Acids Res. 17, 4847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seybert, A., Hegyi, A., Siddell, S. G. & Ziebuhr, J. (2000) RNA 6, 1056-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov, K. A., Thiel, V., Dobbe, J. C., van der Meer, Y., Snijder, E. J. & Ziebuhr, J. (2004) J. Virol. 78, 5619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Anand, K., Ziebuhr, J., Wadhwani, P., Mesters, J. R. & Hilgenfeld, R. (2003) Science 300, 1763-1767. [DOI] [PubMed] [Google Scholar]
- 20.Snijder, E. J., Bredenbeek, P. J., Dobbe, J. C., Thiel, V., Ziebuhr, J., Poon, L. L., Guan, Y., Rozanov, M., Spaan, W. J. & Gorbalenya, A. E. (2003) J. Mol. Biol. 331, 991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thiel, V., Herold, J., Schelle, B. & Siddell, S. G. (2001) J. Gen. Virol. 82, 1273-1281. [DOI] [PubMed] [Google Scholar]
- 22.Laneve, P., Altieri, F., Fiori, M. E., Scaloni, A., Bozzoni, I. & Caffarelli, E. (2003) J. Biol. Chem. 278, 13026-13032. [DOI] [PubMed] [Google Scholar]
- 23.Pan, T. & Uhlenbeck, O. C. (1992) Biochemistry 31, 3887-3895. [DOI] [PubMed] [Google Scholar]
- 24.Bartlett, G. J., Porter, C. T., Borkakoti, N. & Thornton, J. M. (2002) J. Mol. Biol. 324, 105-121. [DOI] [PubMed] [Google Scholar]
- 25.Hausmann, Y., Roman-Sosa, G., Thiel, H. J. & Rümenapf, T. (2004) J. Virol. 78, 5507-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gorbalenya, A. E. (2001) Adv. Exp. Med. Biol. 494, 1-17. [PubMed] [Google Scholar]
- 27.Snijder, E. J., den Boon, J. A., Bredenbeek, P. J., Horzinek, M. C., Rijnbrand, R. & Spaan, W. J. (1990) Nucleic Acids Res. 18, 4535-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasternak, A. O., van den Born, E., Spaan, W. J. & Snijder, E. J. (2001) EMBO J. 20, 7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu, H. Y., Lim, K. P., Shen, S. & Liu, D. X. (2001) Virology 288, 212-222. [DOI] [PMC free article] [PubMed] [Google Scholar]