Abstract
The recent identification of multiple new genetic causes of neurological disorders highlights the need for model systems that give experimental access to the underlying biology. In particular, the ability to couple disease-causing mutations with human neuronal differentiation systems would be beneficial. Gene targeting is a well-known approach for dissecting gene function, but low rates of homologous recombination in somatic cells (including neuronal cells) have traditionally impeded the development of robust cellular models of neurological disorders. Recently, however, CRISPR/Cas9 gene editing technologies have expanded the number of systems within which gene targeting is possible. Here we adopt as a model system LUHMES cells, a commercially available diploid human female mesencephalic cell line that differentiates into homogeneous mature neurons in 1-2 weeks. We describe optimised methods for transfection and selection of neuronal progenitor cells carrying targeted genomic alterations using CRISPR/Cas9 technology. By targeting the endogenous X-linked MECP2 locus, we introduced four independent missense mutations that cause the autism spectrum disorder Rett syndrome and observed the desired genetic structure in 3-26% of selected clones, including gene targeting of the inactive X chromosome. Similar efficiencies were achieved by introducing neurodevelopmental disorder-causing mutations at the autosomal EEF1A2 locus on chromosome 20. Our results indicate that efficiency of genetic “knock-in” is determined by the location of the mutation within the donor DNA molecule. Furthermore, we successfully introduced an mCherry tag at the MECP2 locus to yield a fusion protein, demonstrating that larger insertions are also straightforward in this system. We suggest that our optimised methods for altering the genome of LUHMES cells make them an attractive model for the study of neurogenetic disorders.
Keywords: CRISPR, LUHMES, human, neurons, neurological, Cas9, MeCP2, EEF1A2
Introduction
The advent of technologies that introduce targeted mutations into the genome has dramatically changed the way in which genetic diseases can be modelled and studied. The most recent development in the genome editing field, the clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR associated 9 (Cas9) system, has proven to be extremely successful, due in part to its ease of use and efficient implementation in a variety of cell lines ( Cong et al., 2013; Jinek et al., 2013; Liang et al., 2015) and model organisms ( Friedland et al., 2013; Gratz et al., 2013; Hai et al., 2014; Hwang et al., 2013; Jiang et al., 2013; Li et al., 2013; Niu et al., 2014; Wang et al., 2013). The coupling of CRISPR gene editing technology with human induced pluripotent stem cells (iPSCs) has rapidly expanded the number of neurological disorders that can be modelled in a human neuronal background and is allowing researchers to probe the underlying molecular mechanisms in unprecedented detail ( Howden et al., 2016; Merkert & Martin, 2016; Mungenast et al., 2016). In particular, the ability to genetically modify a hiPSC line to create isogenic cell lines, which are genetically identical (bar the disease causing mutation), and differentiate these into neurons for phenotypic analysis is extremely powerful. However, despite advances in hiPSC culture and neuronal differentiation protocols, there are still some limitations to this strategy. One obstacle is the large variability of clonal iPS cell lines when they are derived, which can have negative downstream effects on CRISPR targeting efficiency, single cell cloning and particularly on phenotypic outcomes. Furthermore, there is still debate as to the robustness of the epigenome in iPSCs after reprogramming ( Kim et al., 2010; Ohi et al., 2011).
Alternative human neuronal progenitor cell lines are available including the SH-SY5Y line and neural stem cells derived from fetal human brain or human embryonic stem cells. Yet each of these models has drawbacks. SH-5YSY cells are a neuroblastoma cell line with multiple chromosomal duplications and deletions ( Krishna et al., 2014) and neural stem cells take a long time to mature during the differentiation process, expressing markers specific for neuronal progenitors for at least four weeks ( Shin & Vemuri, 2010; Sun et al., 2008; Tong et al., 2016). The LUHMES neuronal progenitor cell line is a recent alternative that is proving to be highly useful in the neuroscience field ( Lotharius et al., 2002). These female “pre-neuronal” cells are forced to proliferate in an immature state by expression of the retroviral element v-myc ( Hoshimaru et al., 1996). V-myc expression is under the control of tetracycline so by simple administration of the drug to cell culture medium, LUHMES cells undergo a rapid and robust differentiation into a homogeneous population of electrically active, post-mitotic, mature dopaminergic neurons in just 1 week ( Lotharius et al., 2002). The resulting neurons have thus far been used to model Parkinson’s disease ( Lotharius et al., 2005; Xiang et al., 2013), for cytotoxicity assays ( Tong et al., 2016) and for technology development ( Dinh et al., 2013; Hughes et al., 2014; Ilieva et al., 2013).
In order to make this cell line more widely applicable for the neuroscience field, it would be beneficial to routinely genetically modify LUHMES cells to create a variety of cell lines for disease modelling and drug-screening purposes. Historically genetic manipulation via homology-directed repair (HDR) of somatic cells has been difficult, with the most successful approaches involving rAAV-delivered homology arms to produce targeting efficiencies of ~1% ( Porteus & Baltimore, 2003; Porteus et al., 2003; Russel & Hirata, 1998). With the advent of CRISPR technologies, HDR targeting efficiencies in somatic cells has increased somewhat, although to different extents in different systems, for example 1.3% in primary neonatal fibroblasts ( Lin et al., 2014), 1.8% in vivo by AAV delivery to mouse lung tissue ( Platt et al., 2014) and 17% in T cells using Cas9 ribonucleoprotein complexes ( Schumann et al., 2015).
Here we describe a robust and reproducible method for the efficient transfection of LUHMES cells, and demonstrate various ways in which this cell line can be genetically manipulated using CRISPR engineering to create human models for the study of neurological disorders.
Results
Transfection of proliferating LUHMES cells using Nucleofection
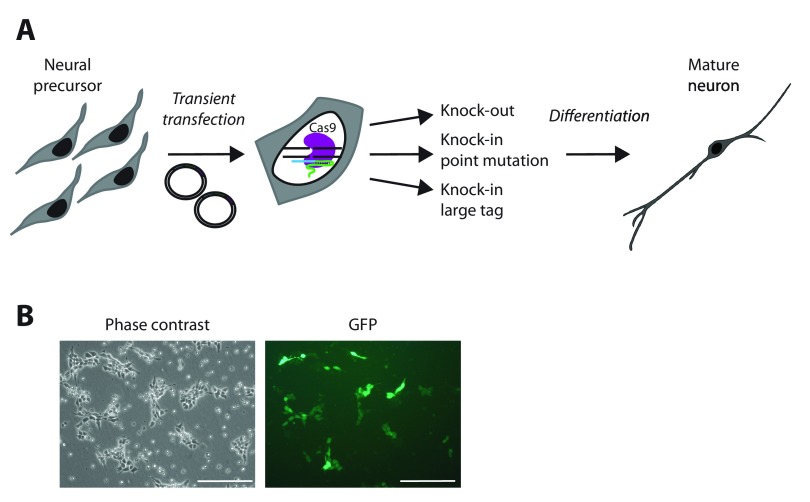
In this study we sought to edit the endogenous genome of the pre-neuronal somatic LUHMES cell line in three ways: i) by disrupting a target gene; ii) by introducing discrete mutations into the protein coding region; iii) by adding a relatively large protein tag to generate a fusion protein ( Figure 1A). Karyotyping confirmed that LUHMES cells have a normal diploid chromosome complement ( Supplementary Figure 1A) and RNA FISH demonstrated X inactivation to be established in the pre-neuronal cells prior to differentiation ( Supplementary Figure 1B). As a first step in editing the genome, a reproducible method of plasmid transfection needed to be established for these cells. LUHMES cells have proven to be difficult to transfect ( Schildknecht et al., 2013) and as a result previous studies relied on lentiviruses. In our hands transient transfection methods such as electroporation, Lipofectamine-2000, Neon transfection and JetPrime Polyplus all resulted in cell death or extremely low levels of transfection (Ruth R. Shah, Justine Cholewa-Waclaw, and Adrian Bird, unpublished report). Nucleofection has previously proven to be successful, but only after differentiating the cells for 2 days prior to trypsinisation and Nucleofection ( Schildknecht et al., 2013). For the generation of genetically modified cell lines, this protocol is undesirable as any transfected cells will be immediately differentiated into post-mitotic neurons, and no stock of proliferating, genetically modified cells remains. In order to optimise Nucleofection conditions in proliferating LUHMES neuronal progenitor cells, the Amaxa Basic Nucleofector Kit for primary neurons was used with 20 different Nucleofection programs to find the optimal balance between transfection efficiency and cell viability ( Supplementary Figure 1C). Program D33 reproducibly yielded transfection efficiencies of 25–30%, as judged by the number of GFP-positive cells in the population ( Figure 1B). Of note, transfection of plasmids that have been prepared in an endotoxin-free environment resulted in increased cell viability, but purification of plasmids by ethanol precipitation did not improve this ( Supplementary Figure 1E+F). In this way we achieved efficient and reproducible transfection of proliferating LUHMES cells using exogenous plasmids.
Figure 1. Nucleofection for efficient transfection of LUHMES cells.
( A) The aims of this study are to develop methods for genetic manipulation of the LUHMES pre-neuronal cell line, which in itself requires methods of transient transfection to be developed. After successful genetic modifications have been confirmed, LUHMES cells can then be differentiated into mature neurons for study. ( B) Representative images of plasmid transfection of LUHMES cells using program D33 with a Nucleofector device. Pictures were taken 47 hours post-Nucleofection. Scale bar is 200 μm.
Generation of a MeCP2 knock-out cell line
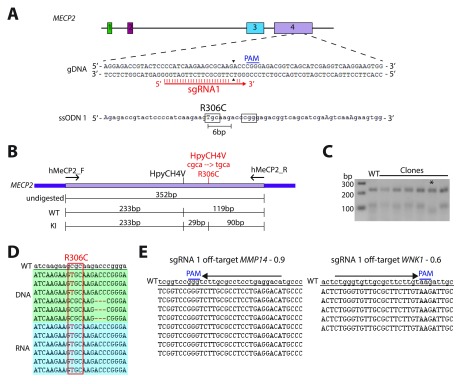
We next tested the ability of CRISPR/Cas9 to generate a knock-out LUHMES cell line. For this the MECP2 locus was chosen. The MeCP2 protein is highly expressed in neurons ( Shahbazian et al., 2002; Skene et al., 2010) and mutations within this protein lead to the autism-spectrum disorder Rett syndrome ( Amir et al., 1999). Multiple mouse models of Rett syndrome have been developed, including mice containing Rett syndrome-causing point mutations ( Brown et al., 2016) as well as knock-out alleles ( Chen et al., 2001; Guy et al., 2001). The MECP2 gene has four exons, with different isoforms being expressed from exons 1 and 2. As exon 3 is the first shared exon among all isoforms, this was chosen for targeting in order to ablate all MeCP2 protein isoforms. Two sgRNAs were designed within exon 3 ( Figure 2A) and were individually cloned into a plasmid that also encodes Cas9 and a puromycin resistance gene ( Figure 2B) ( Sanjana et al., 2014). LUHMES cells were Nucleofected ( Supplementary Figure 2A) and after selecting for positively transfected cells using puromycin resistance both sgRNAs were confirmed to be functional by the T7E1 assay ( Figure 2C) and single-cell colonies were established by serial dilution in 96-well plates. The genomic DNA from single cell colonies was extracted and sequenced in order to identify potential positive KO clones. The genomic DNA sequencing from two different cell lines is shown in Figure 2D, KO1 has a homozygous deletion of 9bp whereas KO2 has a heterozygous deletion of 14bp, with the second allele being unaltered. As MECP2 resides on the X chromosome and LUHMES cells are female cells with one X chromosome already in the inactive state ( Supplementary Figure 1B), the homozygous 9bp deletion in KO1 suggests that the inactive X chromosome can be edited by the CRISPR/Cas9 system. Overall, out of 13 colonies that were sequenced, 11 contained INDELs thus giving a targeting efficiency of 85%.
Figure 2. Generation of MeCP2 knock-out LUHMES cell lines.
( A) Schematic representation of the MECP2 locus with the targeting sequence of sgRNA A and sgRNA B labelled. Arrowheads indicate sites of double-strand break. ( B) Schematic representation of the plasmid used to deliver Cas9 and sgRNA to LUHMES cells. ( C) T7E1 assay of sgRNA A and sgRNA B. WT: non-transfected wild-type cells. NDC: wild-type cells Nucleofected without any DNA. Asterisks indicate non-specific bands. Arrows indicate bands specific to samples containing Cas9 and sgRNA. ( D) Sequencing of genomic DNA from two LUHMES single-cell clones. Wild-type genomic DNA is in lower case at the top. ( E) Sequencing of cDNA from two LUHMES single-cell clones. Wild-type cDNA is in lower case at the top. ( F) Sequencing of genomic DNA from a single LUHMES unedited clone that was transfected with Cas9 and sgRNA B. Wild-type gDNA is in lower case at the top. ( G) Western blot of MeCP2 protein and Histone H3 loading control from wild-type cells (WT), wild-type cells that went through the Nucleofection and cloning process (WTC), KO1 and KO2 cell lines. ( H) Sequencing of sgRNA off-target sites in KO1 and KO2 cell lines. Numbers next to each locus name indicate the off-target score as determined by crispr.mit.edu.
To determine the genotype of the actively expressed MECP2 mRNA in these cell lines, cDNA sequencing was performed ( Figure 2E). The 14bp deletion allele in KO2 appears to reside on the active X chromosome as all cDNA sequence reads from this cell line contained this out-of-frame deletion, highly indicative of a protein KO phenotype. Surprisingly the 9bp in-frame deletion in the middle of exon 3 of KO1 resulted in the whole of exon 3 being removed from the mature mRNA transcript, causing exons 2 and 4 to be spliced together in-frame. Western blot analysis confirmed the complete absence of any full length MeCP2 protein in both cell lines ( Figure 2G). In order to identify clones that might contain truncated protein, Western blot analysis was performed using two different antibodies, one against the N-terminus of MeCP2 and another against the C-terminus, and this revealed that KO1 has very low levels of a truncated protein ( Supplementary Figure 2B). Even though this cell line cannot technically be referred to as a protein KO cell line, the extremely low MeCP2 protein level that remains (and the removal of critical residues via deletion of exon 3) probably results in a cell line that is phenotypically null, as has been observed in mice ( Chen et al., 2001). Finally, the top off-target loci for each sgRNA were sequenced for off-target INDEL formation and as expected based on recent findings in hiPSCs ( Paquet et al., 2016), no off-target cutting was observed ( Figure 2H). These experiments confirm Cas9-induced INDEL formation to be successful, specific and highly efficient in LUHMES cells.
Insertion of Rett syndrome-causing point mutations into MECP2
We also explored the possibility of introducing specific point mutations into LUHMES cells, historically a more challenging procedure for somatic cells ( Hendrie & Russell, 2005). The MECP2 locus is an ideal candidate for use in optimising CRISPR knock-in (KI) conditions as there are a number of disease-causing point mutations throughout the locus ( Lyst & Bird, 2015). Furthermore the manipulation of this X-linked gene offers the opportunity to explore the ability of the CRISPR/Cas9 system to genetically manipulate genes on the inactive X chromosome.
In the previous experiment serial dilution was used to generate single cell colonies but we found that this method led to low efficiency of cloning and some colonies were derived from more than one genetically modified cell line, as several expressed alleles were detected in cDNA sequencing ( Supplementary Figure 2C). In order to improve clonal selection we used FACS sorting to cleanly isolate single cells into a 96-well plate. LUHMES cells were amenable to this manipulation, with approximately 50–60% of wells repopulating to produce single-cell colonies.
First, the ability to knock-in the Rett syndrome-causing missense mutation of arginine at position 306 to cysteine (R306C) was tested ( Figure 3A). The R306C mutation itself (CGC➔TGC) creates a novel target sequence for the restriction enzyme HpyCH4V ( Figure 3B). This allowed for easy screening of genomic DNA from single-cell clones using a restriction fragment length polymorphism assay (RFLP), with the positive clones from this assay being confirmed by sequencing. Initially, a plasmid targeting vector containing 2044bp of homology was used to deliver the R306C point mutation and a silent PAM-abolishing mutation to prevent re-cutting of a recombined allele. Out of 191 single cell clones that were screened using the HpyCH4V RFLP assay, 0 appeared to be positive (experiment 1 in Table 1). Next a 110bp single-stranded oligodeoxynucleotide (ssODN) was used in combination with a non-complementary sgRNA that cuts 6bp away from R306, and in this instance 1 cell line out of 69 was positive, giving a KI efficiency of 1.6% ( Figure 3A+C+D, experiment 2 in Table 1). Again analysis of this cell line demonstrated no off-target cutting ( Figure 3E). An alternative sgRNA:ssODN pair where sgRNA 2 cuts 31bp away from R306, but the sgRNA and ssODN were complementary to one another was also tested ( Figure 4A). This combination produced two positive R306C cell lines (KI efficiency of 2.9%, experiment 3 in Table 1), but both cell lines contained downstream deletions at the site of the double-strand break (DSB) ( Figure 4B). These results suggest that a large distance between the point mutation and the DSB could be more susceptible to error-prone recombination and therefore subsequent INDEL formation.
Figure 3. Generation of a human neuronal cell line containing a Rett syndrome-causing missense mutation in MECP2.
( A) Schematic representation of the MECP2 locus with the sgRNA 1 target sequence labelled and the ssODN 1 donor molecule with point mutation alterations indicated in upper case. The site of double strand break is labelled with two arrowheads and the distance between the point mutation of interest and the double-strand break site is indicated. ( B) Schematic representation of the RFLP screening assay used for identifying positive knock-in clones. Mutation of arginine at position 306 to cysteine results in the introduction of a novel target sequence for the restriction enzyme HpyCH4V. Primers used for PCR amplification are labelled. ( C) HpyCH4V digests of the PCR product ( Supplementary Figure 3A) to identify clones that have gained a novel HpyCH4V target sequence. A positive clone is identified with an asterisk. ( D) Sequencing of genomic DNA and cDNA from the RFLP-positive cell line confirms the cell line to be MECP2-R306C. ( E) Sequencing of the top two off-target sites for sgRNA 1 in the R306C cell line. Number next to the locus name is the off-target score as calculated by crispr.mit.edu.
Figure 4. Generation of human neuronal cell lines containing missense mutations that cause neurological disorders.
( A) Schematic representation of the MECP2 locus with the sgRNA 2 target sequence labelled. Site of double-strand break is indicated with arrowheads. ssODN molecule is shown with point mutation changes highlighted in upper case and the DSB to point mutation distance labelled. ( B) Genomic DNA sequencing of two R306C positive clones. Both clones have the correctly inserted R306C point mutation as well as downstream deletions induced by sgRNA 2. The site of DSB is indicated by an arrowhead. ( C) Schematic representation of a plasmid containing Cas9, sgRNA and EGFP expression constructs. ( D) Genomic DNA sequencing of two R111G positive cell lines. ( E) Genomic DNA sequencing of two R133C positive cell lines. ( F) Genomic DNA sequencing of two T158M positive cell lines. ( G) Schematic representation of part of the EEF1A2 locus, with sgRNA target sequences labelled and the ssODN donor molecule with point mutation alterations indicated in upper case. Sites of single-strand nicks are indicated with an arrowhead. Distances between each nick and the point mutation of interest are labelled. ( H) Genomic DNA sequencing of the EEF1A2-D252H positive cell line.
Table 1. Point mutation KI efficiencies in the MECP2 locus.
PM = point mutation. DSB = double-strand break.
| Point
mutation |
Selection,
plasmid amount |
Donor molecule
(ssODN is 10 μM) |
sgRNA:
ssODN |
PM ➔ DSB
distance |
Number of
upstream mutations |
Number of
KI clones |
KI efficiency | |
|---|---|---|---|---|---|---|---|---|
| 1 | R306C | Puro, 1.2 μg | 1.9 μg 2 kb plasmid | - | 31 bp | 1 | 0/191 | 0% |
| 2 | R306C | Puro, 2 μg | 4 μl 110 bp ssODN 1 | Not comp | 5 bp | 1 | 1/69 | 1.6% |
| 3 | R306C | GFP, 2.5 μg | 8 μl 110 bp ssODN 2 | Comp | 31 bp | 1 | 2/69 | 2.9% |
| 4 | R111G | GFP, 2 μg | 10 μl 100 bp ssODN 3 | Comp | 3 bp | 0 | 7/27 | 26% |
| 5 | R133C | GFP, 2 μg | 10 μl 100 bp ssODN 4 | Comp | 6 bp | 2 | 2/54 | 3.7% |
| 6 | T158M | GFP, 2 μg | 10 μl 100 bp ssODN 5 | Comp | 4 bp | 1 | 1/18 | 5.5% |
| 7 | T158M | GFP, 2 μg | 10 μl 100 bp ssODN 5 | Comp | 5 bp | 1 | 1/13 | 7.7% |
In an attempt to increase the efficiency of KI the CRISPR plasmid that encodes puromycin resistance was exchanged for a CRISPR plasmid that encodes for green fluorescent protein ( Ran et al., 2013b) ( Figure 4C). Thus, instead of subjecting the Nucleofected LUHMES cells to puromycin selection and FACS sorting, these two steps were combined into one by using the presence of GFP in cells two days after Nucleofection to identify positively transfected cells and to sort them into a 96-well plate. Using this new strategy three new Rett syndrome-causing point mutations, R111G, R133C and T158M, were targeted using ssODNs ( Supplementary Figure 3). Two of these three point mutations do not introduce a novel restriction enzyme target sequence and so one was engineered into the ssODN for ease of screening. Each 100bp ssODN contained point mutations to introduce the following motifs: the mutation of interest, a silent PAM abolishing mutation, and a silent mutation to insert a novel restriction enzyme target sequence ( Supplementary Figure 3). All three targeting experiments generated positive cell lines as judged by genomic DNA sequencing ( Figure 4D,E,F) and as can be seen in Table 1, the efficiency of KI for all three point mutations is significantly increased relative to the initial efficiency of 1.6%, reaching a maximum of 26% for KI of R111G.
As shown in Table 1, several factors could contribute to the large variability in targeting efficiency. Firstly, the distance of the sgRNA-induced DSB from the point mutation of interest varies, and secondly the number of mismatched residues in the ssODN that are upstream of the DSB also varies. This latter variable would be in line with evidence that mismatches in the non-sgRNA binding DNA strand upstream of the PAM are refractory to homology-directed repair (HDR) ( Richardson et al., 2016). Despite uncertainty regarding the exact constraints on efficient KI of point mutations using CRISPR technology, KI efficiencies in the somatic LUHMES neuronal progenitor cells are sufficient to allow the rapid generation of cell lines containing disease-causing point mutations with minimal clonal selection and screening.
Insertion of a neurodevelopmental disorder-causing point mutation into EEF1A2
To demonstrate the utility of LUHMES cells as a model system for other neurological disorders, and to confirm efficient KI at an autosomal locus, we targeted the D252H missense mutation in the EEF1A2 gene that causes severe neurodevelopmental delay and intellectual disability ( Nakajima et al., 2015). The approach was to use two sgRNAs that cut 51bp apart, combined with Cas9 nickase protein ( Ran et al., 2013a) ( Figure 4G) and a 200bp ssODN. Again the presence of GFP expression in LUHMES cells was used to identify positively transfected cells and to sort single cells into 96-well plates. In this experiment a KI efficiency of 14% was achieved and interestingly the D252H positive cell line has a KI on both alleles and is therefore a homozygous, true positive cell line ( Figure 4H). The successful knock-in of a point mutation into an autosomal locus and subsequent generation of an EEF1A2-D252H cell line demonstrates the ease of genetic manipulation of LUHMES cells and highlights its utility for modelling a variety of human neurogenetic disorders.
Knock-in of a large tag into the endogenous MECP2 locus
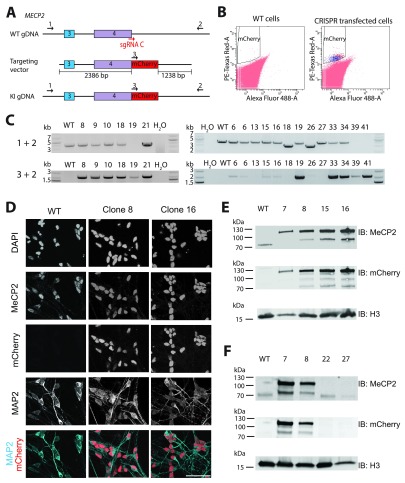
Finally, we assessed the ability of the CRISPR/Cas9 system to introduce a large tag into an endogenous locus in LUHMES cells. Again the MECP2 locus was targeted and a sgRNA that spans the stop codon was used, resulting in its targeting sequence being abolished once a successful KI has occurred ( Figure 5A). We chose to KI mCherry and use FACS analysis to provide an accurate estimate of KI efficiency, i.e. the percentage of mCherry positive cells in the whole population. Due to the large size of mCherry (711bp), a plasmid donor was used for targeting with 2.3kb and 1.2kb homology arms ( Figure 5A). FACS analysis determined the percentage of mCherry positive cells in the entire population to be 0.015% ( Figure 5B). Out of 29 single-cell clones assessed, 25 were MECP2-mCherry positive as judged by a PCR assay that used a forward primer in mCherry itself and a reverse primer in the MECP2 gene locus, outwith the targeting vector ( Figure 5C). Positive cell lines were confirmed by immunofluorescence and Western blot analysis ( Figure 5D+E; Supplementary Figure 4), and two negative cell lines (as determined by PCR analysis) were confirmed by Western blot ( Figure 5F, clones 22 + 27). These experiments demonstrate successful CRISPR-mediated KI of a large tag into LUHMES cells, thus highlighting the variety of genetic alterations that are feasible in this cell line.
Figure 5. Endogenous knock-in of an mCherry tag into the MECP2 locus in LUHMES cells.
( A) Schematic representation of the MECP2 locus with the sgRNA C target region labelled, the targeting vector, and the recombined genomic DNA allele. Positions of primers used for screening purposes in ( C) are indicated. ( B) Flow cytometry analysis of WT cells and cells that were transfected with Cas9, sgRNA and targeting plasmid. ( C) PCR screening of genomic DNA from single-cell clones that were identified as being mCherry positive by flow cytometry. ( D) Immunofluorescence imaging of WT cells and two single-cell clones using DAPI and antibodies probing for MeCP2 and MAP2. Images are slices through a z-stack. Scale bar represents 50 μm. ( E) Immunoblot analysis of WT cells and four MECP2-mCherry positive clones. ( F) Immunoblot analysis of WT cells, two MECP2-mCherry positive clones and two MECP2-mCherry negative clones. ( E + F) Top panel probe: MeCP2. Middle panel probe: mCherry. Bottom panel probe: Histone H3 as a loading control.
Discussion
The LUHMES cell line is an immortalised neuronal cell line derived from an 8-week old female foetus that is highly proliferative in a stem-cell like, yet neuronal-committed state, and can differentiate into mature dopaminergic neurons via addition of tetracycline to the cell culture medium ( Scholz et al., 2011). A key advantage of LUHMES cells compared to other neuronal differentiation systems is the near 100% homogeneity of differentiation into a population of mature, post-mitotic neurons, without the presence of astrocytes or other non-neuronal cell types. This homogeneity is extremely beneficial for “bulk population” experiments such as RNA-sequencing, Western blot analysis and Hi-C studies where mixed cell populations could result in skewed data and difficult-to-interpret results. Here we describe methods for the successful genetic manipulation of LUHMES cells in order to create targeted protein knock-out, disease-causing point mutation knock-in, and large tag knock-in cell lines.
The combination of targeted mutagenesis with rapid generation of mutant neuronal cells provides a potentially valuable tool for neuroscience. These manipulated cell lines may complement in vivo datasets as disease phenotypes obtained using mouse models can be coupled with electrophysiological and biochemical data from human neurons in order to bridge the gaps between disease causing mutations, neuronal malfunction and whole organism pathophysiology. The ease of high-throughput differentiation of this cell line in 96-well plates opens the door towards drug screening programs like those already being pursued using hiPSCs ( Cao et al., 2016; Lee et al., 2012). As a potential alternative, LUHMES cells simplify the differentiation procedure, speed up the differentiation time course, and ensure that a homogeneous population of mature neurons will be screened. Furthermore, although downstream applications of genetically modified LUHMES cells might be limited by their dopaminergic lineage, removal of supplements from the differentiation medium results in the production of tyrosine hydroxylase-negative cells, that differentiate into morphologically and immunocytologically mature neurons for study ( Scholz et al., 2011).
Bi-allelic X chromosome targeting
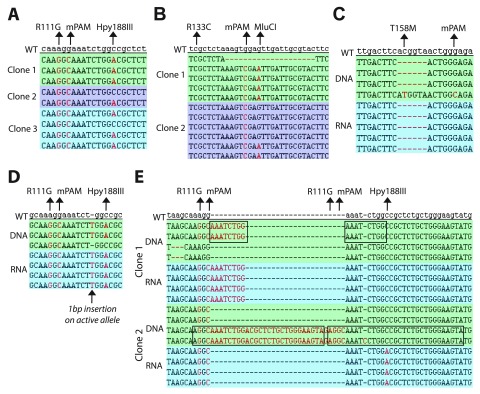
We observed that both alleles undergo HDR at a rather high frequency, regardless of whether the 2 nd allele is on an autosome or the inactive X chromosome. For the MECP2-R111G targeting experiment, out of the seven cell lines that contained a KI of R111G on the active X allele, six also contained a KI on the inactive X allele. Likewise for the MECP2-R133C targeting experiment, the two R133C positive cell lines had undergone HDR repair on both alleles. It is however important to note that not all HDR events result in a clean integration. Partial recombination within the short distance of a 100bp ssODN ( Figure 6A+B) and multiple integrations of the ssODN in tandem at a locus have been observed ( Figure 6E), as well as recombined alleles containing INDELs at the site of sgRNA DSB ( Figure 4B).
Figure 6. Analysis of range of mutations induced by CRISPR/Cas9 in LUHMES cells.
( A+ B) Partial recombination can occur with the ssODN molecule. ( A) Three single cell clones from the R111G targeting experiment, two of which contain a partially recombined allele. ( B) Two single cell clones from the R133C targeting experiment; clone 1 has an allele with 2/3 of the KI residues, clone 2 has an allele with only 1/3 of the KI residues. ( C) Genomic DNA sequencing of a single clone reveals one allele to contain two point mutations and the other allele to contain a 6bp deletion, yet cDNA sequencing identifies the HDR to have occurred with the allele on the inactive X chromosome. ( D) cDNA sequencing identifies the allele on the active X chromosome having undergone recombination, with all three point mutations integrated plus a single base pair insertion. ( E) Sequencing of single-cell clones reveals duplication events occurring as a result of multiple recombination events with the ssODN. ( A, B, C, D, E) mPAM - silent point mutation that abolishes the PAM site for each sgRNA. Rett-syndrome causing point mutations of interest and silent restriction enzyme target sequence point mutations are highlighted with arrows.
Surprisingly we found in more than one case that the MEPC2 allele on the inactive X chromosome underwent HDR, while the allele on the active X chromosome acquired an INDEL. It is expected that the active allele would be more open and accessible to recombination compared to the inactive X chromosome, however at least two cell lines were observed that have an active allele INDEL and an inactive allele KI ( Figure 6C+D). These data reflect the large variety of genomic alterations that can be induced by CRISPR/Cas9 and demonstrate the somewhat unpredictable nature of HDR-mediated gene targeting. Our experiments stress the need for a sgRNA-induced DSB as close as possible to the desired genetic alteration and highlight the importance of donor molecule design; in particular use of a sgRNA-complementary ssODN that has minimal mismatches upstream of the PAM seems to be most efficient.
Further optimisation of KI of large tags
In the mCherry KI experiment the plentiCRISPR plasmid ( Sanjana et al., 2014) was used to deliver Cas9 and sgRNA, and positively transfected cells were selected using the co-encoded puromycin resistance gene ( Figure 2B), while the mCherry targeting vector was delivered as a separate plasmid. It is possible that a proportion of transfected cells did not take up both plasmids ( Assur et al., 2012) and this could explain the low targeting efficiency of 0.015% observed in this experiment. As such, a double antibiotic selection method could increase HDR efficiencies when plasmid donors are necessary, for example by including an expression cassette for the bacterial blasticidin resistance gene (bsr) in the targeting plasmid and selecting with both puromycin and blasticidin.
Design of targeting vectors with alternative homology arm lengths could also improve the efficiency of KI. Indeed, others have reported comparable efficiencies with homology arms of 175 bp compared to 700–900 bp in human cell lines ( Natsume et al., 2016). Alternatively, the use of drugs to inhibit the NHEJ pathway could also boost HDR in LUHMES cells ( Maruyama et al., 2015; van Overbeek et al., 2016). Even without these enhancements, the power of FACS sorting allows efficient selection for the small number of positive cells within a large population and, as demonstrated here, this results in a stream-lined and efficient protocol for CRISPR-mediated tag KI in LUHMES cells.
Concluding remarks
In conclusion, we have demonstrated efficient genetic manipulation of the LUHMES female human neuronal cell line to create a number of lines harbouring neurological disease-causing point mutations ( Figure 7). The future phenotypic assessment of these cell lines will provide significant insight into the molecular mechanisms of these diseases. Using the methods described here LUHMES cells have the potential to be a valuable tool for exploration of the underlying biology of neurogenetic disorders and may pave the way for drug development and therapeutic strategies in the future.
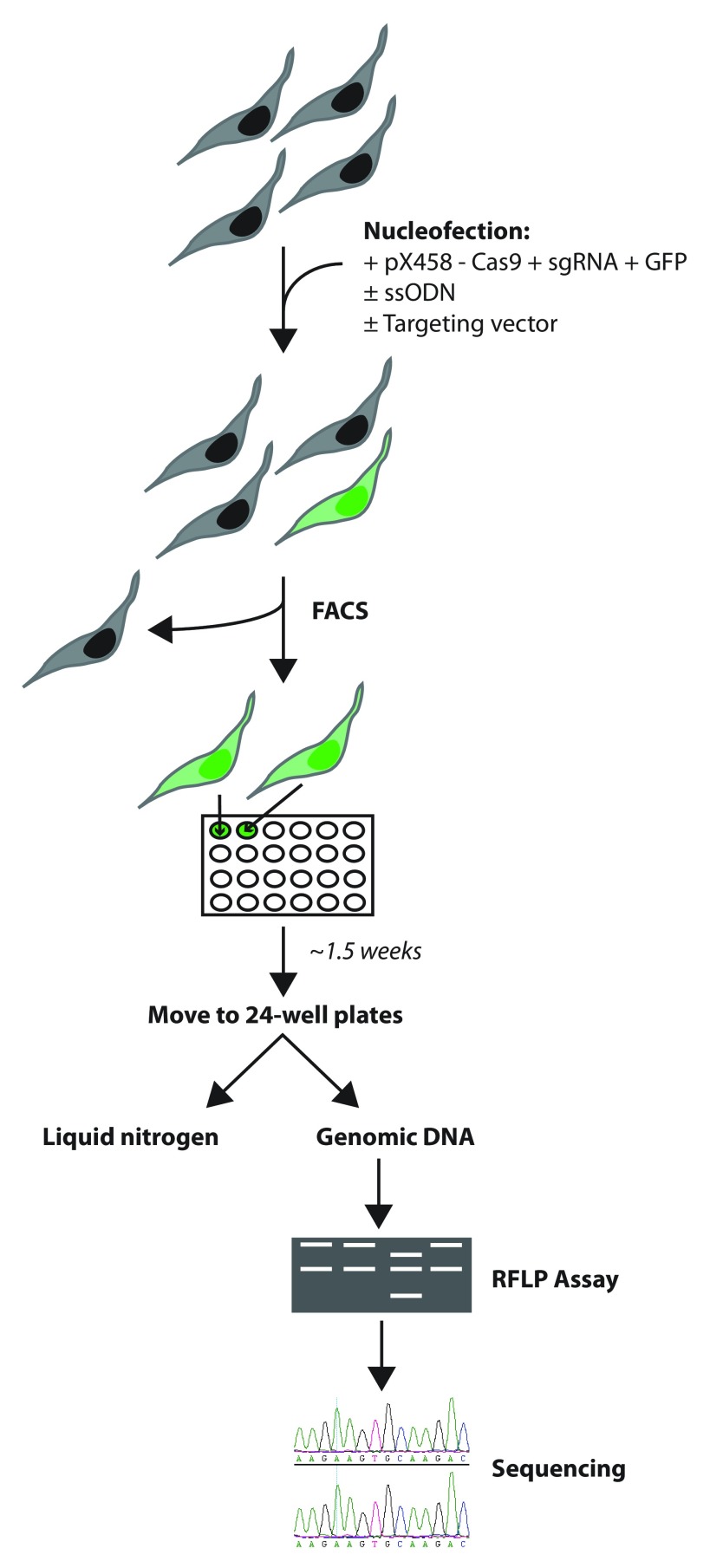
Figure 7. Simple targeting pipeline for generation of genetically modified LUHMES cell lines using CRISPR technology.
Cells are Nucleofected with the plasmids and ssODNs necessary for the specific targeting experiment and taken for FACS sorting two days later. After approximately 10 days single cell colonies will have expanded enough to require dissociation and transfer to 24-well plates. From 24-well plates cells can be frozen down for storage in liquid nitrogen and some cells harvested for genomic DNA. The first step of the screening strategy is to perform an RFLP assay to identify a subset of clones that will be taken for genomic DNA sequencing which will identify cell lines that are positive for a clean KI. For more detailed information see methods and materials.
Experimental procedures
Plasmids and cloning
All CRISPR plasmids were purchased from Addgene and originated from the Zhang lab; pX458 (48138), pX461 (48140) and plentiCRISPRv2 (52961). The crispr.mit.edu online webtool was used for sgRNA design ( Table 2). Cloning of sgRNAs into CRISPR plasmids was performed following the protocol from the Zhang lab, available online at genome-engineering.org and described in Ref ( Cong et al., 2013). All ssODNs were ordered as 100bp or 110bp (Sigma, desalted) or 200bp (IDT, PAGE-purified) oligonucleotides ( Table 3). Gateway cloning (Invitrogen) was used to create the mCherry targeting plasmid. The pmaxGFP plasmid for optimisation of Nucleofection techniques came in the Nucleofection kit provided by Lonza.
Table 2. Sequences of sgRNAs used in this study.
All sgRNAs are 20 nucleotides long. Those without a 5' G nucleotide had one added for efficient U6 promoter transcription to make a 21 nucleotide sequence.
| sgRNA name | Experiment | Sequence (5’ ➔ 3’) |
|---|---|---|
| sgRNA A | Knock-out of MeCP2 | AGAAGCTTCCGGCACAGCCG |
| sgRNA B | Knock-out of MeCP2 | CGCTCCATCATCCGTGACCG |
| sgRNA 1 | Knock-in of R306C-MECP2 | CCATCAAGAAGCGCAAGACC |
| sgRNA 2 | Knock-in of R306C-MECP2 | GACGGTCAGCATCGAGGTCA |
| sgRNA 3 | Knock-in of R111G-MECP2 | GGACACGGAAGCTTAAGCAA |
| sgRNA 4 | Knock-in of R133C-MECP2 | AAAAGCCTTTCGCTCTAAAG |
| sgRNA 5 | Knock-in of T158M-MECP2 | GATTTTGACTTCACGGTAAC |
| sgRNA 6 | Knock-in of T158M-MECP2 | ATTTTGACTTCACGGTAACT |
| sgRNA 1 | Knock-in of D252H-EEF1A2 | GCTTGTCCGTGGGGCGCGTG |
| sgRNA 2 | Knock-in of D252H-EEF1A2 | CAGGACGTGTACAAGATTGG |
| sgRNA C | Knock in of mCherry tag into MECP2 | TTAGCTGACTTTACACGGAG |
Table 3. Sequences of ssODNs used in this study.
Point mutation changes from the wild-type, endogenous sequence are highlighted in bold, with the point mutation of interest in bold red.
| ssODN | Experiment | Sequence (5’ ➔ 3’) |
|---|---|---|
| 1 | Knock-in of R306C-MECP2 | GAGTCTTCTAT
TCGATCTGTGCA
AGAGACCGTACTCCCCATCAAGAAG
TGCAAGACCCGGGAGACGGT
CAGCATCGA AGTCAA AGAAGTGGTGAAGCCCCTGCT AGTGTC |
| 2 | Knock-in of R306C-MECP2 | GACACCAGCAGGGGCTTCACCACTTCCTTGACCTCGATGCTGACCGTCTC
GCGGGTCTTGC
ACTTCT
TGATGGGGAGTACGGTCTCCTGCACAGATCG AATAGAAGACTC |
| 3 | Knock-in of R111G-MECP2 | CTTACTTACTTGATCAAATACACATCATACTTCCCAGCAGAGCG
TCCAGATTT
GC
CTTGCTTAAGCTTCCGT
GTCCAGCCTTCAGGCAGGGTGGGGTCAT |
| 4 | Knock-in of R133C-MECP2 | CAGGGATGTGTCGCCTACCTTTTCGAAGTACGCAATCAA
TTC
GACTTTAGAGC
AAAATGCTTTTCCCTGGG
GACTGTGGGGACAAACAGAAAGACACAAG |
| 5 | Knock-in of T158M-MECP2 | GGCTTCTTAGGTGGTTTCTGCTCTCGCCGGGAGGGGCTCCCTCT
GCCAGTTACC
ATGAAGTCAAAAT
CATTAGGGTCCAGGGATGTGTCGCCTACCTTTT |
| 6 | Knock-in of
D252H-EEF1A2 |
TAAGGAGGGCAACGCAAGCGGCGTGTCCCTGCTGGAGGCCCTGGACACCATCCTGCC
TCCCACG
CGCCCCACGGACAAGCCCCTGCGCCTGCCGCTGCAG CACGT TTA TAAGAT CGGCGGTGAGCAAGGGC GCTGTGCTGGAGCTCCTGCCTGGCCAGCTCTGCCTGCCCTAGACCAGGGGCCCCTACAAGGCATCTCAA |
The R306C targeting plasmid was created by firstly performing a PCR reaction using Phusion polymerase in GC buffer (NEB) with approximately 100–250 ng LUHMES genomic DNA in a 25 μl reaction volume ( Table 4). Subcloning was performed using 2 μl of this PCR mix with the Zero Blunt TOPO PCR cloning kit (Invitrogen) and 2 μl of the subcloning reaction was transformed into DH5α cells. The following day two colonies were picked into 3 ml cultures of LB/kanamycin and grown overnight by shaking at 37°C. The next day plasmids were extracted from 1.5 ml of culture using the Plasmid Miniprep kit (Qiagen) and sequenced using primers spanning the MECP2 locus. After sequencing confirmed error-free incorporation of the MECP2 fragment into the pTOPO vector, 50 ml cultures of LB/kanamycin were set up using the remaining 1.5 ml of bacterial culture and grown overnight by shaking at 37°C. The next day plasmids were extracted using the Plasmid Maxiprep kit (Qiagen). This plasmid was subjected to site-directed mutagenesis using the QuickChange II XL kit (Agilent) following the manufacturer’s instructions. Two rounds of mutagenesis PCRs were used to incorporate first the R306C point mutation, and then two PAM-abolishing point mutations for two separate sgRNAs to create the final targeting vector.
Table 4. Sequences of primers used in this study.
For Gateway cloning primers, sequence in red is the att tag, sequence in black is homologous to MECP2 or mCherry.
| Experiment | Primer name | Sequence (5’ ➔ 3’) |
|---|---|---|
| Sequence CRISPR
plasmids |
pLKO1_5 | GACTATCATATGCTTACCGT |
| Cloning of pR306C
targeting vector |
hMeCP2_6F | CGCTCTGCTGGGAAGTATGA |
| hMeCP2_5R | CCAACTACTCCCACCCTGAA | |
| Site-directed mutagenesis
of pR306C targeting vector |
R306C_F | TCCCCATCAAGAAGTGCAAGACCCGGGAG |
| R306C_R | CTCCCGGGTCTTGCACTTCTTGATGGGGA | |
| PAMs_F | ATCGAGGTCAAAGAAGTGGTGAAGCCCCTGCTAGTGTCCACCCTCG | |
| PAMs_R | CGAGGGTGGACACTAGCAGGGGCTTCACCACTTCTTTGACCTCGAT | |
| Gateway cloning of
mCherry targeting vector |
attB1_MeCP2_intron2F | GGGGACAAGTTTGTACAAAAAAGCAGGCTCCACAGCCCAAATTCCTAAA |
| attB4_MeCP2_stopR | GGGGACAACTTTGTATAGAAAAGTTGGGTG GCTAACTCTCTCGGTCACGG | |
| attB4r_mCherry_IF_Tm | GGGGACAACTTTTCTATACAAAGTTGTTATGGTGAGCAAGGGCGA | |
| attB3r_mCherry_IF_Tm | GGGGACAACTTTATTATACAAAGTTGTCTACTTGTACAGCTCGTCCATGC | |
| attB3_MeCP2_stopF | GGGGACAACTTTGTATAATAAAGTTGTGACTTTACACGGAGCGGAT | |
| attB2_MeCP2_8R | GGGGACCACTTTGTACAAGAAAGCTGGGTAGCAGAAATGGAAGGGGAGAA | |
| Phusion PCR for R306C
clones |
hMeCP2_F | AGCTCCTTGTCAAGATGCCT |
| hMeCP2_R | AGTCCTTTCCCGCTCTTCTC | |
| Phusion PCR for R111G
clones |
Intron2_2F | TCCCTTGAAGTGCGACTCAT |
| 1_R | CCTCTCCCAGTTACCGTGAA | |
| Phusion PCR for R133C
clones |
Intron3_2F | CAGACGAGTGAGTGGCTTTG |
| 2_R | AGTCCTTTCCCGCTCTTCTC | |
| Phusion PCR for T158M
clones |
Intron3_2F | CAGACGAGTGAGTGGCTTTG |
| 3_R | CAATCCGCTCCGTGTAAAGT | |
| Phusion PCR for
MECP2-
mCherry clones |
1 | TCCCTTGAAGTGCGACTCAT |
| 2 | GGACGGAGGAAGGGAAAGAA | |
| 3 | GGGGACAACTTTTCTATACAAAGTTGTTATGGTGAGCAAGGGCGA | |
| Phusion PCR for
EEF1A2-
D252H clones |
hD252H_1F | TTCCTCATCTCAAAGGGCACG |
| hD252H_2R | CAAGTTTAGCCTGAACAGCAGTA | |
| Sequencing of
EEF1A2-
D252H clones |
hD252H_2F | CCCACAGAAGTGTGTGGTAAG |
| hD252H_2R | TTGGAGACAGCCAGTCTTG |
Tissue culture
LUHMES cell (ATCC cat# CRL-2927, RRID CVCL_B057, kind gift from Dr Tanja Waldmann) tissue culture medium and methods were as described in Reference ( Scholz et al., 2011) with some minor alterations. All vessels were coated in poly-L-ornithine (PLO) and fibronectin overnight at 37°C. Proliferating LUHMES cells were seeded at 2x10 6 cells/T75 every 2 days. For differentiation, 2.5x10 6 cells were seeded in a T75 for the first 2 days of the protocol and on day 2 cells were seeded as follows: 8x10 6 cells/T75 and 0.15x10 6 cells/coverslip. During differentiation a half-media change was performed on day 6 and neurons were harvested for protein or fixed for immunostaining on day 9.
Transfection methods
LUHMES cells were transfected by Nucleofection (Lonza) using a Basic Nucleofector kit for primary neurons (VAPI-1003) and a Nucleofector II device. LUHMES cells were dissociated with 4ml of trypsin (Gibco), centrifuged at 13000 rpm for 5 minutes and resuspended in PBS for cell counting using a Scepter device (Millipore). Aliquots of 2x10 6 cells were pipetted into 15 ml falcons and these were centrifuged for 5 minutes at 13000 rpm. PBS was removed from all 15 ml falcons and the appropriate volume of each plasmid/ssODN was added to each tube (see Table 1). One by one cells and plasmids were then resuspended in 90 μl of the Nucleofection solution and immediately transferred into a cuvette (provided in the kit) for electroporation in the Nucleofector II device. After electroporation, RPMI medium (Sigma) was added using the pipette provided in the kit and cells were moved into 15 ml falcons and incubated at 37°C for 5 minutes before plating out into 6-well plates containing pre-warmed LUHMES proliferation media. Media was changed after a minimum of 4 hours.
FACS sorting
For flow cytometry sorting of single LUHMES cells into each well of a 96-well plate, Greiner plates were pre-coated overnight in PLO/fibronectin as described in Reference ( Scholz et al., 2011). Proliferation medium was supplemented with 1X B27 (Sigma), 100 U/ml penicillin (Gibco) and 100 μg/ml streptomycin (Gibco) and 100 μl was added to each well of the 96-well plates. LUHMES cells were trypsinised and centrifuged in a 15 ml falcon tube at 13000 rpm for 5 minutes and resuspended in 1 ml of Advanced DMEM/F12 supplemented with 10 μM HEPES. Cells were sorted using a 100 μm nozzle with a FACSaria machine at room temperature. Six days later 100 μl of proliferation medium was added to the 96-well plates.
Targeting pipeline in LUHMES cells
Day 1 – Thaw low passage number LUHMES
Day 3 – Passage once
Day 5 – Nucleofect and change media after 4 hours
Day 7 – Take for FACS sorting of single GFP-positive cells into a 96-well plate
Day 13 – Top up 96-well plate with 100 μl of media
Day 16 – Start moving clones from 96-well plates to 24-well plates
Day 17 onwards – Split individual clones in 24-well plates as they are confluent, freeze down half the well for liquid nitrogen, move the other half to a new well in a 24-well plate for genomic DNA.
Genomic DNA isolation
For extracting genomic DNA from tissue culture samples, Puregene Core Kit A (Qiagen) was used following the manufacturer’s instructions. Occasionally, when RNA and DNA were required from a single sample, the Allprep DNA/RNA Mini kit (Qiagen) was used.
For large scale genomic DNA extraction from LUHMES single cell clones in a 24-well plate an alternative approach was used. Individual clones were trypsinised and harvested when confluent and incubated in 400 μl lysis buffer (50 mM Tris pH 9.0, 20 mM EDTA pH 8.0, 40 mM NaCl, 1% SDS, 0.5 mg/ml proteinase K) overnight at 55°C. The next day 300 μl saturated NaCl was added to each sample, mixed by vigorous shaking for 1 minute, and centrifuged at 14000 rpm for 10 minutes at room temperature. The supernatant was transferred to an Eppendorf containing 500 μl isopropanol, mixed by inversion and centrifuged at 14000 rpm for 10 minutes at 4°C. The DNA pellet was washed with 750 μl 70% ethanol, mixed by inversion, and centrifuged at 14000 rpm for 10 minutes at 4°C. The DNA pellet was air dried, resuspended in 50 μl TE and allowed to dissolve at 55°C overnight.
RNA isolation and preparation of cDNA
RNA was extracted using an RNeasy Mini Kit (Qiagen) following manufacturer’s instructions. RNA concentration was measured using a Nanodrop 1000 (Thermo Scientific) and 10 μg RNA was taken for DNase1 treatment (Ambion) for 1 hour at 37°C. DNA-free RNA concentration was measured using a Nanodrop and 1 μg used for cDNA synthesis using qScript cDNA Supermix (Quanta).
T7 Endonuclease I assay
Phusion polymerase in GC buffer (NEB) was used to PCR amplify 100–250 ng genomic DNA in a 50 μl reaction volume ( Table 4) and 5 μl of the reaction was ran out on an agarose/TAE gel. To the remaining 45 μl PCR mix, 5 μl of Buffer 2 (NEB) was added and the reactions were heated at 95°C for 10 minutes in a PCR machine and cooled slowly to 25°C to produce heteroduplexes. Each reaction was split in half and to one half 1 μl of T7E1 (NEB) was added. All reactions were incubated at 37°C for 30 minutes, followed by addition of 10 μl of 6X loading dye and analysis on an agarose/TAE gel.
Restriction fragment length polymorphism assay
Phusion polymerase in GC buffer (NEB) was used to PCR amplify 100–250 ng genomic DNA in a 25 μl reaction volume ( Table 4) and 10 μl of each PCR was run out on an agarose/TAE gel. To the remaining PCR mix 29 μl of H 2O, 5 μl of CutSmart buffer (NEB) and 0.5 μl of the appropriate enzyme was added. Reactions were incubated at the necessary temperature and length of time for each enzyme and were analysed by electrophoresis in an agarose/TAE gel.
PCR and sequencing analysis
Phusion polymerase in GC buffer (NEB) was used to PCR amplify 100–250 ng genomic DNA in a 25 μl reaction volume ( Table 4) and 10 μl of the reaction was ran out on an agarose/TAE gel for confirmation of efficient PCR amplification. Each amplicon was subcloned using the Strataclone blunt PCR cloning kit (Agilent Technologies) following the manufacturer’s instructions and transformed into Strataclone Solopack competent bacteria (Agilent Technologies). The next day single colonies were picked and colony PCR was performed using either Phusion polymerase or DreamTaq polymerase (Thermo Fisher) in a 25 μl volume ( Table 4). For confirmation of PCR, 10 μl of each colony PCR reaction was analysed by agarose/TAE gel electrophoresis. Positive colony PCRs were treated with 0.25 μl Exonuclease I (NEB) and 0.25 μl FastAP alkaline phosphatase (Thermo) at 37°C for 15 minutes followed by inactivation at 85°C for 15 minutes. Sequencing was performed using BigDye Terminator v2.1 reaction mix with 3.5 μl of each sample and 3.2 pmol of primer.
Western blot analysis
Whole cell protein extracts were prepared by homogenising cell pellets in NE1 buffer (10 mM HEPES pH 7.9, 10 mM KCl, 1 mM MgCl 2, 0.5 mM DTT, 0.1% Triton-X 100, 20% glycerol and 1X protease inhibitor cocktail (Roche)). Homogenates were treated with benzonase for 15 minutes at room temperature and then measured for protein concentration using a Bradford assay (Protein Assay Dye Reagent concentrate, BioRad). Extracts were loaded onto pre-cast 4–20% Mini-PROTEAN TGX gels (BioRad) and ran at 200V for approximately 30–40 minutes. Gels were transferred onto nitrocelloluse membrane by transfer at 30V overnight at 4°C. Membranes were blocked in 5% milk, 0.1% Tween-20 in TBS for 30 minutes and probed with primary antibodies ( Table 5) in blocking solution for 1 hour at room temperature. IRDye 800CW α-mouse and IRDye 680LT α-rabbit secondary antibodies (Licor) were probed for 1 hour at room temperature in blocking buffer at a concentration of 1:10000 and scans were taken using a Licor Odyssey machine.
Table 5. Details of antibodies used in this study.
Antibodies used for Western blot analysis (WB) or immunofluorescence (IF). Cat number – catalogue number.
| Antibody | Company + Cat Number | Experiment | Dilution | Species | RRID |
|---|---|---|---|---|---|
| MeCP2 | Sigma 7443 | WB; IF | 1000X | Mouse monoclonal | AB_477235 |
| MeCP2 | Sigma 6818 | WB | 1000X | Mouse monoclonal | AB_262075 |
| GAPDH | Cell Signalling 5174 | WB | 5000X | Rabbit monoclonal | AB_10622025 |
| H3 | Abcam 1791 | WB | 5000X | Rabbit polyclonal | AB_302613 |
| mCherry | Abcam 167453 | WB, IF | 1000X | Rabbit polyclonal | AB_2571870 |
| Neurofilament | Covance SMI-311R-100 | IF | 500X | Mouse monoclonal | AB_509991 |
| MAP2 | Abcam 5392 | IF | 5000X | Chicken polyclonal | AB_2138153 |
Immunofluorescence imaging
Cells grown on coverslips were fixed in 4% formaldehyde for 10 minutes, permeabilised with 0.2% Triton-X/PBS for 10 minutes and blocked for 30 minutes in 10% fetal bovine serum in PBS (FBS/PBS). Coverslips were incubated in the presence of primary antibodies ( Table 5) in 1% FBS/0.1% Tween-20/PBS for one hour room temperature. Coverslips were washed with 0.1% Tween-20/PBS, and incubated for one hour at room temperature with Alexa Fluor secondary antibodies (1000X dilution, Invitrogen) in 1% FBS/0.1% Tween-20/PBS. Secondary antibodies: α-mouse 488, α-rabbit 555 and α-chicken 633. Coverslips were finally stained with DAPI (5000X dilution in PBS) for 10 minutes at RT and mounted onto microscope slides using Prolong Diamond solution (Thermo Fisher). Z-stack images were taken using a Leica SP5 confocal microscope and z-stacks were flattened and processed using ImageJ 1.47v software.
Other microscopy
Phase contrast images were taken on an Eclipse TS100 inverted microscope (Nikon) using QCapture Pro software version 5.1.1.14 (QImaging). UV fluorescence was used to take GFP pictures of cells. The IncuCyte microscope (Essen Biosciences) was used to take phase contrast and GFP fluorescent images and ZOOM software (Essen Biosciences) was used to extract and export the images.
RNA FISH
RNA FISH was performed as described in ( Chaumeil et al., 2008). BAC-based probes were used for detection of MECP2 and ATRX (RP11-119A22 and RP11-42M11 respectively). Probes for XIST RNA detection have been used previously, as described in ( Chaligne et al., 2015).
Data availability
Zenodo: Efficient and versatile CRISPR engineering of human neurons in culture to model neurological disorders, DOI 10.5281/zenodo.163342 ( Shah et al., 2016)
Acknowledgements
We are grateful to Dr Tanja Waldmann (University of Konstanz) for introducing us to the LUHMES system. We also thank Dr Martin Waterfall for FACS and Dr David Kelly for microscopy advice.
Funding Statement
This work was supported by the Wellcome Trust [099841], [091580], [107930], [92076]; Rett Syndrome Research Trust; and the Medical Research Council.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 2 approved]
Supplementary Material
Supplementary Figure 1. Optimisation of Nucleofection in LUHMES cells.
( A) Karyotyping of LUHMES pre-neuronal cells confirms a normal ploidy. ( B) RNA FISH of these female cells demonstrates that X inactivation is already established in the pre-neuronal cells. Scale bar is 5 μm. ( C) Trials of various electroporation programs using the Basic Nucleofector kit for primary neurons. ( D) Comparison of different plasmid amounts on cell viability. ( E) Comparison of phenol:chloroform:isoamylalcohol plasmid purification on cell viability. ( F) Comparison of cell viability when endotoxin-free plasmids are used, when the cells are incubated at 37°C in RPMI for 5 minutes (RPMI) and when they are centrifuged before plating (spin). All scale bars are 200 μm.
Supplementary Figure 2. Analysis of MeCP2 KO cell lines.
( A) Representative phase contrast and GFP fluorescence images of LUHMES cells 25 hours after Nucleofection. Scale bar is 200 μm. ( B) Immunoblot detection of MeCP2 protein in single cell clones derived from the plentiCRISPR targeting experiment using an N-terminal and a C-terminal antibody. GAPDH is probed for a loading control. Note: E10 = WTC, D10 = KO1 and H4 = KO2 in Figure 2. ( C) Sequencing of cDNA from single LUHMES clones identifies mixed cell populations by the presence of multiple cDNA alleles per clone. Each colour block denotes an individual clone.
Supplementary Figure 3. Optimisation of CRISPR-mediated point mutation knock-in of Rett syndrome causing missense mutations.
( A) hMeCP2_F and hMeCP2_R PCR of WT cells and clones that have been targeted with components shown in Figure 3A, in preparation for RFLP analysis. ( B, C, D) Schematic representation of R111G (B), R133C (C) and T158M (D) targeting experiments. Two sgRNAs are labelled in (D), each sgRNA was used in a separate targeting experiment. Sites of DSB and PAM sites are indicated for each sgRNA, with DSB to point mutation distances highlighted. Mutations that introduce a novel restriction enzyme target sequence are also labelled.
Supplementary Figure 4. MeCP2-mCherry positive cells lines as determined by immunofluorescence imaging.
Neurons were differentiated until day 9 and then fixed and stained for neurofilament (NF), MAP2, mCherry and DAPI. Images are flattened z-stacks. Scale bar is 50 μm.
References
- Amir RE, Van den Veyver IB, Wan M, et al. : Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–8. 10.1038/13810 [DOI] [PubMed] [Google Scholar]
- Assur Z, Hendrickson WA, Mancia F: Tools for coproducing multiple proteins in mammalian cells. Methods Mol Biol. 2012;801:173–187. 10.1007/978-1-61779-352-3_12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Selfridge J, Lagger S, et al. : The molecular basis of variable phenotypic severity among common missense mutations causing Rett syndrome. Hum Mol Genet. 2016;25(3):558–70. 10.1093/hmg/ddv496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L, McDonnell A, Nitzsche A, et al. : Pharmacological reversal of a pain phenotype in iPSC-derived sensory neurons and patients with inherited erythromelalgia. Sci Transl Med. 2016;8(335):335ra56. 10.1126/scitranslmed.aad7653 [DOI] [PubMed] [Google Scholar]
- Chaligné R, Popova T, Mendoza-Parra MA, et al. : The inactive X chromosome is epigenetically unstable and transcriptionally labile in breast cancer. Genome Res. 2015;25(4):488–503. 10.1101/gr.185926.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumeil J, Augui S, Chow JC, et al. : Chapter 18: Combined Immunofluorescence, RNA Fluorescent In Situ Hybridization, and DNA Fluorescent In Situ Hybridization to Study Chromatin Changes, Transcriptional Activity, Nuclear Organization, and X-Chromosome Inactivation. The Nucleus: Volume 1: Nuclei and Subnuclear Components2008. Reference Source [DOI] [PubMed] [Google Scholar]
- Chen RZ, Akbarian S, Tudor M, et al. : Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet. 2001;27(3):327–31. 10.1038/85906 [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, et al. : Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh ND, Chiang YY, Hardelauf H, et al. : Microfluidic construction of minimalistic neuronal co-cultures. Lab Chip. 2013;13(7):1402–1412. 10.1039/c3lc41224e [DOI] [PubMed] [Google Scholar]
- Friedland AE, Tzur YB, Esvelt KM, et al. : Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat Methods. 2013;10(8):741–743. 10.1038/nmeth.2532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, et al. : Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194(4):1029–35. 10.1534/genetics.113.152710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, et al. : A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27(3):322–326. 10.1038/85899 [DOI] [PubMed] [Google Scholar]
- Hai T, Teng F, Guo R, et al. : One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24(3):372–5. 10.1038/cr.2014.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrie PC, Russell DW: Gene targeting with viral vectors. Mol Ther. 2005;12(1):9–17. 10.1016/j.ymthe.2005.04.006 [DOI] [PubMed] [Google Scholar]
- Hoshimaru M, Ray J, Sah DW, et al. : Differentiation of the immortalized adult neuronal progenitor cell line HC2S2 into neurons by regulatable suppression of the v-myc oncogene. Proc Natl Acad Sci U S A. 1996;93(4):1518–1523. 10.1073/pnas.93.4.1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden SE, McColl B, Glaser A, et al. : A Cas9 Variant for Efficient Generation of Indel-Free Knockin or Gene-Corrected Human Pluripotent Stem Cells. Stem Cell Reports. 2016;7(3):508–17. 10.1016/j.stemcr.2016.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MA, Brennan PM, Bunting AS, et al. : Patterning human neuronal networks on photolithographically engineered silicon dioxide substrates functionalized with glial analogues. J Biomed Mater Res A. 2014;102(5):1350–1360. 10.1002/jbm.a.34813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, et al. : Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–229. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva M, Della Vedova P, Hansen O, et al. : Tracking neuronal marker expression inside living differentiating cells using molecular beacons. Front Cell Neurosci. 2013;7:266. 10.3389/fncel.2013.00266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Zhou H, Bi H, et al. : Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013;41(20):e188. 10.1093/nar/gkt780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, East A, Cheng A, et al. : RNA-programmed genome editing in human cells. eLife. 2013;2:e00471. 10.7554/eLife.00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Doi A, Wen B, et al. : Epigenetic memory in induced pluripotent stem cells. Nature. 2010;467(7313):285–290. 10.1038/nature09342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna A, Biryukov M, Trefois C, et al. : Systems genomics evaluation of the SH-SY5Y neuroblastoma cell line as a model for Parkinson’s disease. BMC Genomics. 2014;15(1):1154. 10.1186/1471-2164-15-1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee G, Ramirez CN, Kim H, et al. : Large-scale screening using familial dysautonomia induced pluripotent stem cells identifies compounds that rescue IKBKAP expression. Nat Biotechnol. 2012;30(12):1244–1248. 10.1038/nbt.2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Teng F, Li T, et al. : Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat Biotechnol. 2013;31(8):684–6. 10.1038/nbt.2652 [DOI] [PubMed] [Google Scholar]
- Liang P, Xu Y, Zhang X, et al. : CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–72. 10.1007/s13238-015-0153-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S, Staahl BT, Alla RK, et al. : Enhanced homology-directed human genome engineering by controlled timing of CRISPR/Cas9 delivery. eLife. 2014;3:e04766. 10.7554/eLife.04766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotharius J, Barg S, Wiekop P, et al. : Effect of mutant alpha-synuclein on dopamine homeostasis in a new human mesencephalic cell line. J Biol Chem. 2002;277(41):38884–38894. 10.1074/jbc.M205518200 [DOI] [PubMed] [Google Scholar]
- Lotharius J, Falsig J, van Beek J, et al. : Progressive degeneration of human mesencephalic neuron-derived cells triggered by dopamine-dependent oxidative stress is dependent on the mixed-lineage kinase pathway. J Neurosci. 2005;25(27):6329–6342. 10.1523/JNEUROSCI.1746-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyst MJ, Bird A: Rett syndrome: a complex disorder with simple roots. Nat Biotechnol. 2015;16(5):261–75. 10.1038/nrg3897 [DOI] [PubMed] [Google Scholar]
- Maruyama T, Dougan SK, Truttmann MC, et al. : Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33:538–542. 10.1038/nbt.3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkert S, Martin U: Site-Specific Genome Engineering in Human Pluripotent Stem Cells. Int J Mol Sci. 2016;17(7): pii: E1000. 10.3390/ijms17071000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mungenast AE, Siegert S, Tsai LH: Modeling Alzheimer's disease with human induced pluripotent stem (iPS) cells. Mol Cell Neurosci. 2016;73:13–31. 10.1016/j.mcn.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima J, Okamoto N, Tohyama J, et al. : De novo EEF1A2 mutations in patients with characteristic facial features, intellectual disability, autistic behaviors and epilepsy. Clin Genet. 2015;87(4):356–361. 10.1111/cge.12394 [DOI] [PubMed] [Google Scholar]
- Natsume T, Kiyomitsu T, Saga Y, et al. : Rapid Protein Depletion in Human Cells by Auxin-Inducible Degron Tagging with Short Homology Donors. Cell Rep. 2016;15:210–218. 10.1016/j.celrep.2016.03.001 [DOI] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, et al. : Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156(4):836–43. 10.1016/j.cell.2014.01.027 [DOI] [PubMed] [Google Scholar]
- Ohi Y, Qin H, Hong C, et al. : Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat Cell Biol. 2011;13(5):541–549. 10.1038/ncb2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, et al. : Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533(7601):125–129. 10.1038/nature17664 [DOI] [PubMed] [Google Scholar]
- Platt RJ, Chen S, Zhou Y, et al. : CRISPR-Cas9 knockin mice for genome editing and cancer modeling. Cell. 2014;159(2):440–455. 10.1016/j.cell.2014.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D: Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300(5620):763. 10.1126/science.1078395 [DOI] [PubMed] [Google Scholar]
- Porteus MH, Cathomen T, Weitzman MD, et al. : Efficient gene targeting mediated by adeno-associated virus and DNA double-strand breaks. Mol Cell Biol. 2003;23(10):3558–3565. 10.1128/MCB.23.10.3558-3565.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, et al. : Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013a;154(6):1380–1389. 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, et al. : Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013b;8(11):2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson CD, Ray GJ, DeWitt MA, et al. : Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34(3):339–344. 10.1038/nbt.3481 [DOI] [PubMed] [Google Scholar]
- Russel DW, Hirata RK: Human Gene Targeting by Viral Vectors. Nat Genet. 1998;18(4):325–30. 10.1038/ng0498-325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F: Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schildknecht S, Karreman C, Pöltl D, et al. : Generation of Genetically-Modified Human Differentiated Cells for Toxicological Tests and the Study of Neurodegenerative Diseases. ALTEX. 2013;30(4):427–44. 10.14573/altex.2013.4.427 [DOI] [PubMed] [Google Scholar]
- Scholz D, Poltl D, Genewsky A, et al. : Rapid, complete and large-scale generation of post-mitotic neurons from the human LUHMES cell line. J Neurochem. 2011;119(5):957–971. 10.1111/j.1471-4159.2011.07255.x [DOI] [PubMed] [Google Scholar]
- Schumann K, Lin S, Boyer E, et al. : Generation of knock-in primary human T cells using Cas9 ribonucleoproteins. Proc Natl Acad Sci U S A. 2015;112(33):10437–10442. 10.1073/pnas.1512503112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah RR, Cholewa-Waclaw J, Davies FCJ, et al. : Efficient and versatile CRISPR engineering of human neurons in culture to model neurological disorders. Zenodo. 2016. Data Source [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Antalffy B, Armstrong DL, et al. : Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet. 2002;11(2):115–24. 10.1093/hmg/11.2.115 [DOI] [PubMed] [Google Scholar]
- Shin S, Vemuri M: Culture and Differentiation of Human Neural Stem Cells. Protocols for Neural Cell Culture2010. 10.1007/978-1-60761-292-6_3 [DOI] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, et al. : Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state. Mol Cell. 2010;37(4):457–468. 10.1016/j.molcel.2010.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Pollard S, Conti L, et al. : Long-term tripotent differentiation capacity of human neural stem (NS) cells in adherent culture. Mol Cell Neurosci. 2008;38(2):245–258. 10.1016/j.mcn.2008.02.014 [DOI] [PubMed] [Google Scholar]
- Tong ZB, Hogberg H, Kuo D, et al. : Characterization of three human cell line models for high-throughput neuronal cytotoxicity screening. J Appl Toxicol. 2016. 10.1002/jat.3334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Overbeek M, Capurso D, Carter MM, et al. : DNA Repair Profiling Reveals Nonrandom Outcomes at Cas9-Mediated Breaks. Mol Cell. 2016;63(4):633–646. 10.1016/j.molcel.2016.06.037 [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, et al. : One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–918. 10.1016/j.cell.2013.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang W, Schlachetzki JC, Helling S, et al. : Oxidative stress-induced posttranslational modifications of alpha-synuclein: specific modification of alpha-synuclein by 4-hydroxy-2-nonenal increases dopaminergic toxicity. Mol Cell Neurosci. 2013;54:71–83. 10.1016/j.mcn.2013.01.004 [DOI] [PubMed] [Google Scholar]