Abstract
Receptors of the innate immune system detect conserved determinants of microbial and viral origin. Activation of these receptors initiates signaling events that culminate in an effective immune response. Recently, the view that innate immune signaling events rely on and operate within a complex cellular infrastructure has become an important framework for understanding the regulation of innate immunity. Compartmentalization within this infrastructure provides the cell with the ability to assign spatial information to microbial detection and regulate immune responses. Several cell biological processes play a role in the regulation of innate signaling responses; at the same time, innate signaling can engage cellular processes as a form of defense or to promote immunological memory. In this review, we highlight these aspects of cell biology in pattern-recognition receptor signaling by focusing on signals that originate from the cell surface, from endosomal compartments, and from within the cytosol.
Keywords: TLR, CLR, NLR, RLR, ALR
INTRODUCTION
Overview of Innate Immunity
The innate immune system detects the presence of microbes and initiates mechanisms to eliminate potentially infectious threats. Microbial detection is achieved through germline-encoded pattern-recognition receptors (PRRs) that survey both the extracellular and intracellular space for conserved microbial determinants that serve as indicators of infection (1). The model of microbial pattern recognition was proposed by Charles Janeway Jr. and describes two features of innate immunity: the ability to distinguish infectious nonself- molecules from self-molecules and the ability to activate adaptive immune responses to the former (2). Since Janeway made his prediction, investigators have shown that many microbial ligands, ranging from structural components of bacteria, fungi, and viruses to biosynthetic molecules such as nucleic acids, activate PRRs and induce the innate immune responses that protect us from infectious threats (3).
Most PRRs can be classified into one of five families based on protein domain homology. These five families consist of the Toll-like receptors (TLRs), C-type lectin receptors (CLRs), nucleotide-binding domain, leucine-rich repeat (LRR)-containing (or NOD-like) receptors (NLRs), RIG-I-like receptors (RLRs), and the AIM2-like receptors (ALRs) (Table 1) (1). These families can be separated into two main classes: membrane-bound receptors and unbound intracellular receptors. The former class consists of the TLRs and CLRs, which are found at the cell surface or on endocytic compartments. These receptors survey for the presence of microbial ligands in the extracellular space and within endosomes. The NLRs, RLRs, and ALRs form the latter group in that they are located in the cytoplasm, where they survey for the presence of intracellular pathogens.
Table 1.
Pattern-recognition receptor families
| Family | Members | Shared domains | Receptor locations |
|---|---|---|---|
| TLR | 1–10 in humans, 1–9 and 11–13 mice | LRR, TIR | Cell surface, endosomal compartments |
| CLR | Dectin-1, Dectin-2, … etc. (reviewed in Reference 4) | C-type lectin | Cell surface |
| NLR | NOD1 (NLRC1), NOD2 (NLRC2), NLRC3–5, NLRP1–9 and 11–14, NAIP1, -2, -5, -6 | Nucleotide binding, LRR | Cytoplasm, plasma, and endosomal membrane associated |
| RLR | RIG-I, MDA5, LGP2 | DExD/H helicase | Cytoplasm |
| ALR | AIM2, IFI16 | PYRIN, HIN-200 | Cytoplasm, nucleus (IFI16) |
Abbreviations: AIM, absent in melanoma; ALR, AIM2-like receptor; CARD, caspase recruitment domain; CLR, C-type lectin receptor; IFI, interferon, γ-inducible; LGP, laboratory of genetics and physiology; LRR, leucine-rich repeat; MDA, melanoma differentiation gene; NAIP, NLR family, apoptosis inhibitory protein; NLR, nucleotide-binding oligomerization domain receptor; NLRC, NLR family CARD domain containing; NLRP, NLR family PYD domain containing; NOD, nucleotide-binding oligomerization domain; RIG-I, retinoic acid–inducible gene I; RLR, RIG-I-like receptor; TIR, Toll/IL-1 receptor/resistance; TLR, Toll-like receptor.
A major component of a PRR-induced innate immune response is transcriptional, which leads to the production of proinflammatory cytokines and interferons (IFN); these chemical messages are critical for initiating innate and adaptive immune responses. PRR activation also initiates nontranscriptional responses such as the induction of phagocytosis, autophagy, cell death, and cytokine processing (5–7). These transcriptional and nontranscriptional innate immune responses are linked to PRR-mediated microbial detection by tightly controlled signal transduction pathways. The coordination of these signaling pathways orchestrates immune responses, which contain the spread of an initial infection and direct the appropriate adaptive response (8).
A common theme that has emerged in the study of innate immune receptor signaling is the requirement for adaptor proteins that link receptors to an enzymatic signal. Adaptors integrate a signal from more than one receptor and are therefore critical for the detection of numerous ligands. In this way, the adaptors perform a function that is more critical than the function of each receptor alone. Each adaptor or adaptor set contains domains that allow for protein-protein interactions with an upstream receptor as well as a downstream signaling protein. Although not every innate immune receptor requires the use of adaptor proteins for signaling, the cell biology of several adaptors is an area of current research interest. The TLRs utilize a set of sorting and signaling adaptors to engage the downstream enzymatic cascade. The TIR-containing adaptor protein (TIRAP) and the protein myeloid differentiation primary response 88 (MyD88) comprise one functional adaptor set for TLR signaling. A second set consists of the TIR domain–containing adaptor-inducing IFN-β (TRIF) and the TRIF-related adaptor molecule (TRAM). The adaptor protein for the antiviral RLR pathway was initially shown to localize to the outer membrane of mitochondria and is referred to as the mitochondrial antiviral signaling protein (MAVS). Lastly, the ASC [apoptosis-associated speck-like protein containing a CARD (caspase recruitment domain)] is an adaptor for receptors that activate inflammasomes (Table 2).
Table 2.
Adaptor proteins
| Adaptor or adaptor set | Receptor interaction | Signaling interaction | Localization |
|---|---|---|---|
| TIRAP/MyD88 | TIR domain | Death domain | Cell surface, endosomal compartments |
| TRAM/TRIF | TIR domain | TRAF binding, RHIM domain | Cell surface, endosomal compartments |
| MAVS | CARD domain | Proline-rich region, TRAF binding | Mitochondrial, peroxisomal, and mitochondria-associated membranes |
| ASC | PYRIN | CARD domain | Cytosol, mitochondria |
Abbreviations: ASC, apoptosis-associated speck-like protein containing a CARD; CARD, caspase recruitment domain; MAVS, mitochondrial antiviral signaling protein; RHIM, RIP homotypic interaction motif; TIR, Toll/IL-1 receptor/resistance; TIRAP, TIR-containing adaptor protein; TRAF, TNF receptor–associated factor; TRAM, TRIF-related adaptor molecule; TRIF, TIR domain–containing adaptor-inducing IFN-β.
In the 20 years since Janeway proposed his hypothesis, considerable progress has been made in identifying the molecular components involved in pattern recognition. In fact, so much has been identified that many innate immunity reviews limit their scope by focusing on a single PRR family. However, common among these families is an emerging theme that cellular processes and infrastructure can regulate and be regulated by receptor signaling. In this review, rather than focus on a single receptor family, we discuss the intersection of cell biology with innate immune signaling across receptor families. With this approach, we cannot be all-inclusive with the details from each receptor-signaling pathway, but these have been reviewed elsewhere (9–13).
The Importance of Cell Biology in Pattern Recognition
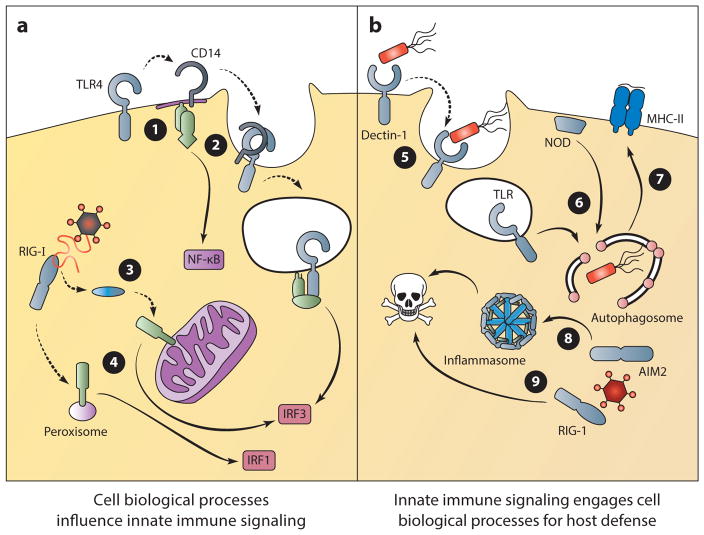
The idea that innate pattern recognition occurs in the context of the cellular infrastructure has two main implications. First, innate immune responses can be compartmentalized and regulated by both the structural and dynamic features of a cell (14). Second, these receptors can induce dynamic cellular processes that function as part of the immune response (Figure 1) (6, 7, 15). Receptor location can be viewed as a structural restriction that dictates the type of ligand that is accessible for detection. In addition, the location of a receptor may initiate a response with functional implications specific for that site. For instance, Dectin-1 (dendritic cell–associated C-type lectin 1) receptor activation at the cell surface can direct phagocytosis (5). This critical aspect of innate immunity aids in pathogen clearance and antigen acquisition for adaptive immune responses.
Figure 1.
Implications of cell biological processes on innate immune signaling. The trafficking of PRRs and the location of innate immune signaling can regulate signal transduction and dictate the outcome of microbial detection (a). ➊ TLR4 undergoes trafficking to specialized regions of the plasma membrane to activate MyD88-dependent signaling. ➋ CD14 mediates the endocytosis of TLR4, which results in TRIF-dependent signaling from the endosome. ➌ RIG-I undergoes trafficking to the site of MAVS signaling on mitochondria. ➍ MAVS signaling from mitochondria induces a type I and III IFN response, whereas signaling from peroxisomes induces the transcription of type III IFN only. Alternatively, innate immune signaling can engage cell biological processes to control infection (b). ➎ Dectin signaling can induce phagocytosis as an effective means of microbial clearance. ➏ NODs and TLRs recruit regulators of autophagy to the site of bacterial entry for pathogen elimination. ➐ Autophagosomes can deliver antigens of intracellular pathogens for MHC-II presentation to coordinate adaptive immune responses. ➑ Inflammasome activation induces a rapid form of cell death known as pyroptosis, which can limit the replication of intracellular pathogens. ➒ The RLR/MAVS signaling pathway can also induce cell death to limit viral replication. (Solid lines indicate signal transduction; dotted lines indicate trafficking events.) (Abbreviations: MAVS, mitochondrial antiviral signaling protein; MHC-II, major histocompatibility complex class II; NOD, nucleotide-binding oligomerization domain; RIG-I, retinoic acid–inducible gene I; TRIF, TIR domain–containing adaptor-inducing IFN-β.)
Differences in receptor localization are among the most commonly described cell biological features of innate immune signaling pathways. Correct receptor localization is one way to prevent inappropriate immune responses to self-molecules not associated with infection (16). Although most PRR ligands fit within Janeway’s nonself category, some molecules, such as nucleic acids, are inherent to the host as well. Detection of self-nucleic acids is a potentially dangerous situation for the host; however, receptor compartmentalization has emerged as one mechanism to prevent such responses. For example, the nucleic acid–sensing TLRs are compartmentalized within the intracellular space, which limits activation by self-nucleic acids that are abundantly present in the extracellular fluids of mammals (16, 17). Autoimmune disorders, such as systemic lupus erythematosus, have been linked to self–nucleic acid detection by PRRs and highlight the importance of regulatory mechanisms that restrict access of PRRs to self-encoded molecules (18). Therefore, the compartmentalization of PRRs helps to classify molecules as nonself and prevent autoactivation.
In addition to compartmentalized nucleic acid detection, receptor localization has implications for the type of response generated. Indeed, cytosolic PRRs activate responses that differ from cell surface receptors. For example, microbe-induced cell death is a common feature of cytosolic receptor detection but not generally a feature of cell surface signaling (7, 19). These differential responses may have to do with the type of microbe commonly encountered at these locations. Cell surface receptors survey for ligands in the extracellular space and therefore may encounter both pathogenic and nonpathogenic microbes. Cytosolic receptors, in contrast, are activated only when a microbe or virus accesses the host cytosol, a common feature of virulent pathogens. The induction of cell death can be an effective means of limiting the replication of viruses and other intracellular pathogens.
There are also implications for the localization of downstream adaptor proteins (Table 2) and ligand-induced receptor movement. As mentioned above, PRRs cannot function as signaling molecules alone, but rather require adaptor molecules to transmit a signal. The TLRs are the best-characterized PRR family and provide an example of adaptor localization and site-specific signaling. TLRs can activate different transcriptional responses depending on which adaptor set is utilized (Table 2) (20, 21). TLR4 is unique in that it utilizes both adaptor pairs in a sequential order. Following ligand binding, the receptor engages TIRAP/MyD88 to initiate signaling from the cell surface and then undergoes endocytosis and engages TRAM/TRIF-dependent signaling from endosomes (22, 23). This ligand-dependent movement of the receptor to a second signaling location is a mechanism that regulates innate immune responses. Indeed, many of the innate immune receptors (NLRs, RLRs, and ALRs) require ligand-dependent receptor movement to activate an adaptor molecule to initiate signal transduction.
In addition to regulating immune responses by requiring receptor movement to specific sub-cellular sites, the site of signaling itself dictates signaling outcomes. For example, TLR4-induced MyD88-dependent signaling originates from the cell surface, whereas TRIF-mediated signaling occurs from endosomes. The RLR adaptor MAVS provides another example of subcellular signaling specificity. MAVS is resident on mitochondria, peroxisomes, and mitochondria-associated membranes (MAMs) of the endoplasmic reticulum (24–26). However, the signaling events induced from mitochondria and peroxisomes differ, resulting in the induction of two distinct transcriptional profiles (25).
These examples highlight the importance of cell biology in innate immune responses and demonstrate that these receptors and signaling events do not take place within a void. In the following sections, we present our understanding of the cell biology of innate pattern recognition as it pertains to signaling from three cellular locations. First, we explore the signaling that originates at the cell surface. In a similar fashion to TLR4, we then translocate to the endosomal network to discuss the signals that originate there. Finally, we explore signals that originate from within the cytosol by discussing the biosynthetic sensing pathways that detect RNA, DNA, or nucleic acid derivatives within the cytosol.
SIGNALING FROM THE PLASMA MEMBRANE
Pathogens have a diverse set of replicative niches and life cycles that can range from the extracellular space to specific subcellular compartments. However, even intracellular pathogens commonly spend some time in the extracellular space, making this a critical location for pathogen detection by the innate immune system. PRRs on the plasma membrane are positioned facing outward to serve this purpose and detect structural components of foreign invaders.
TLRs are transmembrane glycoproteins characterized by an extracellular domain, a trans-membrane domain, and an intracellular TIR domain. The extracellular domain is responsible for ligand recognition and is characterized by a horseshoe-like structure with LRR modules (27). These LRRs are present in the inner concave surface of the extracellular domain, and their variation between different TLRs is thought to provide ligand specificity (28–30). The TLRs recruit and signal through different sets of adaptor proteins, and as mentioned above this can partially explain differences in transcriptional outcome. The receptors homo- or heterodimerize upon ligand binding and recruit adaptor proteins (Table 2) via TIR-TIR interactions. The TLR intracellular TIR domains are fundamental for signal transduction, and point mutations within these regions completely abolish TLR-dependent cellular responses. We begin by discussing signaling from the plasma membrane–associated TLRs—TLR1, -2, -4, -5, and -6—which recognize structural components present on the external surface of microorganisms.
TLR4, MD-2, CD14, and LBP: The Lipopolysaccharide-Multireceptor Complex
A prototype of the TLR family, TLR4 was the first member to be characterized functionally (31) and is the only TLR that uses all four adaptor proteins in signaling. Here we focus on TLR4’s ability to signal from the plasma membrane as part of the lipopolysaccharide (LPS)-multireceptor complex. Several exogenous and endogenous ligands can activate TLR4 and members of the LPS-multireceptor complex (Table 3). Depending on the ligand, coreceptors and accessory molecules may also be required for binding and signaling. The best-described ligand, LPS, was used to identify TLR4 mutations that rendered mice resistant to LPS-induced septic shock (32).
Table 3.
Ligands of the LPS-multireceptor complex
| Name | Origin | Components involveda | Reference(s) |
|---|---|---|---|
| LPS | Gram-negative bacteria | TLR4, LBP, CD14, MD-2 | See body of text |
| Taxol | Taxus brevifolia | TLR4, MD-2 | 33 |
| Fusion protein | Respiratory syncytial virus | TLR4, CD14 | 34 |
| Env protein | Murine Moloney leukemia virus | TLR4 | 35 |
| HSP60 | Chlamydia pneumoniae | TLR4, MD-2 | 36 |
| Cleaved fibrinogen | Aspergillus oryzae proteinase, thrombin | TLR4 | 37 |
| HSP60/70 | Endogenous | TLR4, MD-2 | 38, 39 |
| Type III repeat extra domain A | Fibronectin | TLR4, MD-2 | 40 |
| Polysaccharide | Hyaluronic acid, heparan sulfate proteoglycan | TLR4 | 41, 42 |
| Hyaluronan | Endogenous | TLR4 (TLR2) | 43 |
| Biglycan | Extracellular matrix | TLR4 (TLR2) | 44 |
| HMGB1 | Endogenous | TLR4 | 45, 46 |
| Beta-defensins | Endogenous | TLR4 | 47 |
| Minimally modified LDL | Endogenous | TLR4, MD-2, CD14 | 48 |
| Oxidized LDL | Endogenous | TLR4 (TLR6, CD36) | 49, 50 |
| Oxidized PAPC | Endogenous | TLR4, CD14, LBP | 51 |
| Surfactant protein A | Endogenous | TLR4 | 52 |
| Mrp8, Mrp14 | Endogenous | CD14, TLR4, MD-2 | 53 |
| Feutin A | Endogenous | TLR4 | 54 |
| Cold-inducible RNA-binding protein | Endogenous | TLR4, MD-2 | 55 |
The components in parentheses are not part of the LPS-multireceptor complex, but they can additionally detect the ligands listed.
Abbreviations: HMGB, high-mobility group protein B; HSP, heat shock protein; LBP, LPS-binding protein; LDL, low-density lipoprotein; LPS, lipopolysaccharide; PAPC, 1-palmitoyl-2-arachidonoyl-sn-glycero-3-phosphorylcholine; TLR, Toll-like receptor.
The TLR4 response to LPS requires a multireceptor complex consisting of LPS-binding protein (LBP), CD14, and MD-2, which act sequentially to extract LPS from bacteria and promote TLR4 signaling. LBP is a soluble molecule that can bind large LPS aggregates such as the bacterial outer membrane. With the aid of albumin, LBP somehow facilitates the transfer of LPS monomers to CD14 (56, 57). CD14 then transfers LPS to TLR4-bound MD-2, a process that dimerizes TLR4 and promotes signal transduction (58, 59). This sequential process of LPS transfer is likely important to ensure high sensitivity to the presence of bacteria. Indeed, it has been estimated that enough LPS monomers can be extracted from a single bacterium to activate TLR4 signaling on 1,000 macrophages (59). The mechanisms underlying the functions of MD-2 and CD14 have attracted much attention in recent years. MD-2 is a beta-cup folded protein with a hydrophobic pocket that can bind LPS with high affinity (30). MD-2 is required for TLR4 dimerization in response to LPS and is therefore essential for all TLR4-dependent responses to bacteria (60, 61).
CD14 was the first membrane protein identified as interacting with LPS, and it displays a dimerized structure that strongly resembles the TLR4 extracellular domain (62). This receptor can be released as a soluble factor or be anchored on the plasma membrane by glycosyl-phosphatidylinositol (GPI). The anchored pool of CD14 is localized to discrete regions enriched in cholesterol, called lipid rafts, which are important for protein trafficking and endocytosis and are believed to be sites of plasma membrane signaling (63). TLR4 is not found within these rafts prior to microbial encounters, but it does undergo LPS-induced movement to these sites. Thus, TLR4 movement to CD14-enriched lipid rafts is one example of subcellular receptor movement that promotes PRR signaling. The regulation of this movement is not well understood, however, and requires further investigation.
As a GPI-anchored protein, CD14 lacks an intracellular tail and was originally considered incapable of independently transducing an intracellular signal. However, recent studies have revealed two CD14-dependent signaling pathways that are activated by LPS, independent of TLR4 signaling. One pathway leads to calcium influx and gene regulation via nuclear factor of activated T cells (NFAT) activation (64–66), whereas the other controls endocytosis and moves TLR4 from the plasma membrane into endosomes (67–69). The second pathway controls LPS uptake and TLR4 internalization through a process dependent on immunoreceptor tyrosine-based activation motif (ITAM) adaptors, the tyrosine kinase Syk, PLCγ2, and the IP3 receptor (Figure 2) (67–70). This process of receptor endocytosis is critical for regulating TLR4-mediated type I IFN production (discussed in more detail in the following section), which specifically occurs from endosomes (23, 71), and it also explains previous reports that CD14 is required for LPS-mediated IFN production (72). In addition to CD14, other factors such as CD11b, Rab11a, and Arf6 can regulate TLR4 trafficking, in conjunction with or independently of CD14 (73–76). Functional studies have revealed that the primary function of CD14 in controlling TLR4 signaling from endosomes is to deliver the receptor to this compartment. This conclusion is supported by the finding that defects in TLR4 endocytosis associated with CD14 deficiency can be bypassed in dendritic cells (DCs) through the use of particulate LPS preparations, such as beads or intact bacteria (67). Under these conditions, TLR4 also signals via TRIF to induce IFN expression. CD14 can therefore be considered a bona fide microbe-inducible trafficking factor that regulates all cellular activities induced by TLR4 from the plasma membrane and endosomes. In conclusion, the movement of TLR4 to its second site of signaling within endosomes is a highly regulated cell biological process that provides a node of control for immune responses induced by this receptor.
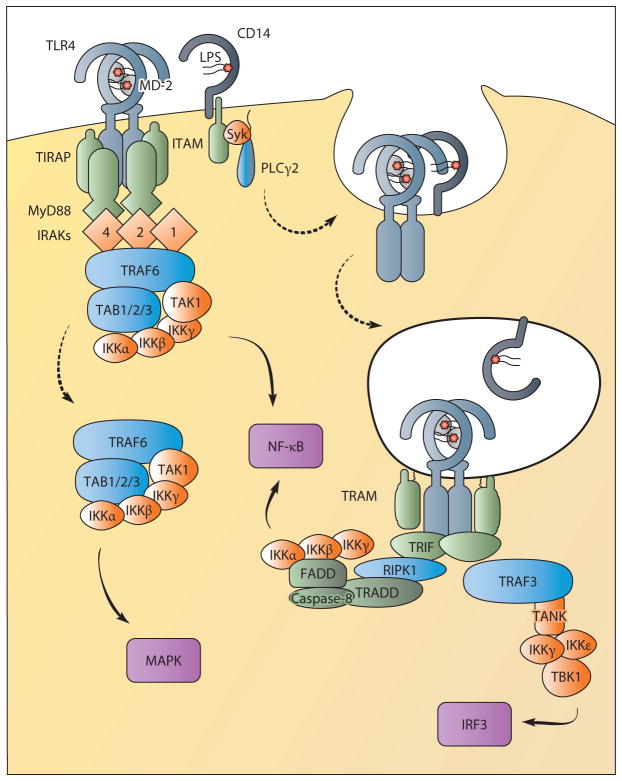
Figure 2.
TLR4 signals from the plasma membrane and endosomes. TLR4 requires translocation to lipid rafts enriched with TIRAP for signaling from the plasma membrane. This facilitates interactions with MyD88 upon ligand binding for the formation of the myddosome containing MyD88, TIRAP, and IRAKs. The IRAKs recruit the E3 ubiquitin ligase TRAF6, which interacts with a complex formed by TAB1, TAB2, TAB3, and TAK1. This complex regulates NF-κB activation via IKKs. TAK1 release into the cytoplasm also directs MAPK activation. CD14 controls the movement of TLR4 from the plasma membrane into endosomes through the activation of ITAM, Syk, and PLCγ2. From endosomes, TLR4 interacts with the sorting adaptor TRAM and the signaling adaptor TRIF to sustain NF-κB activation and to induce IRF3-mediated type I IFN production. TRIF-dependent NF-κB activation may proceed via the proteins RIPK1, TRADD, and the caspase-8 complex. IRF3 activation controls type I IFN production and requires TRAF3 recruitment to TRIF. TRAF3 then interacts with TANK (or TANK-related proteins) to recruit IKKγ, IKKε, and TBK1, which activate IRF3. (Solid lines indicate signal transduction; dotted lines indicate trafficking events.) (Abbreviations: FADD, Fas-associated protein with death domain; IKK, IκB kinase; IRAK, interleukin-1 receptor-associated kinase; IRF, IFN regulatory factor; ITAM, immunoreceptor tyrosine-based activation motif; MAPK, mitogen-activated protein kinase; NF-κB, nuclear factor-κB; PLCγ2, phospholipase Cγ2; RIPK1, receptor-interacting serine/threonine-protein kinase 1; TAB, TAK1-binding protein; TAK, TGF-β-activated kinase; TANK, TRAF family member–associated NF-κB activator; TBK, TANK-binding kinase; TIRAP, TIR-containing adaptor protein; TRADD, TNF receptor type 1–associated death domain; TRAF, TNF receptor–associated factor.)
TLR4 signal transduction itself requires adaptor proteins (Table 2) that can be subdivided into two groups: the sorting and the signaling adaptors. The sorting adaptors TIRAP and TRAM lack a signaling domain but can recruit the signaling adaptors MyD88 and TRIF. Together they initiate the signaling pathways that lead to the activation of NF-κB, adaptor protein (AP)-1, and IFN regulatory factor (IRF) 3, the major transcription factors regulating the TLR4-mediated gene expression response. TLR4 signaling and adaptor engagement are sequential, starting at the plasma membrane with CD14-dependent movement of LPS to TLR4 and the initiation of TIRAP/MyD88 signaling, followed by CD14-dependent receptor movement and TRAM/TRIF signaling from endosomes (67).
TLR4 Signal Transduction from the Plasma Membrane
TLR4-mediated MyD88-dependent signaling requires proper localization of the sorting adaptor TIRAP (22, 77–81), which is targeted to regions enriched with PI(4,5)P2 (PIP2), such as membrane ruffles and lipid rafts. TIRAP also contains a TIR domain that facilitates interaction with MyD88 and TLR4 to mediate signaling (77, 78). It is possible that TIRAP and MyD88 are prepositioned on lipid rafts poised for signal transduction and awaiting ligand-induced TLR4 movement to the region. In support of this, genetic targeting of TIRAP outside of PIP2-reach regions completely blocks TLR4 signaling (22, 81). Furthermore, ARF6, which regulates PI(4)P5K activity and PIP2 production (82), is required for proper TIRAP distribution and MyD88-dependent TLR4 signaling (22).
TLR4 signaling from the plasma membrane utilizes MyD88 and culminates in the activation of NF-κB and AP-1 through the spatial regulation of the ubiquitin-dependent kinase TAK1 (Figure 2). With the exception of TLR3, all TLRs use MyD88 for signal transduction, which highlights the importance of this adaptor. MyD88 interacts with TIRAP or with TLRs directly through a C-terminal TIR domain, whereas an N-terminal death domain is required for interaction with IL-1 receptor–associated kinase (IRAK) family members and signal transduction. Upon TLR4 activation, MyD88 oligomerizes to form a large signaling platform called a myddosome, which incorporates 6–8 MyD88 molecules, TIRAP, and members of the IRAK family (81, 83, 84). IRAKs contain two known functional domains: an N-terminal death domain for interaction with MyD88 and other IRAKs and a central domain with Ser/Thr kinase activity. IRAK4 is the first member of the family to be recruited to the myddosome, and its kinase activity is essential for signal transduction and activation of IRAK1 and IRAK2 (85, 86). Upon IRAK4 phosphorylation, IRAK1 and IRAK2 interact with MyD88 and TNF receptor–associated factor 6 (TRAF6) and regulate NF-κB activation (86–88). TRAF6 is an E3 ubiquitin ligase that functions with Uev1A:Ubc13 to generate K63-linked polyubiquitination chains (89). The linear ubiquitin assembly complex can bind these K63-linked chains and generate M1-linked chains to form K63/M1-linked hybrid chains (90). These hybrid chains recruit preassembled kinase complexes containing TAK1, TAB1, TAB2, and TAB3. Spatial regulation of this complex controls the activation of IκB kinase (IKK) and mitogen-activated protein kinase (MAPK) signaling to control the transcription factors NF-κB and AP-1, respectively (91). First, the IKK complex consisting of IKKα, IKKβ, and NEMO (NF-κB essential modulator, also known as IKKγ) is recruited to the TAK1-containing complex via the ubiquitin-binding domain of NEMO (92, 93). Once associated, TAK1 directly phosphorylates IKKβ, inducing IKK complex activity and releasing NF-κB from its inhibitor, IκBα (91). Second, the activation of MAPK signal transduction requires cIAP-mediated TRAF6 and TAK1 translocation to the cytoplasm to promote TAB2/TAB3-dependent TAK1 oligomerization (94, 95). From the cytosol, TAK1 acts as a MAPKKK and directs AP-1 activation. Thus, the translocation of TAK1 from the membrane to cytosol regulates gene expression and is another example of cell biological control of innate immune signaling by ligand-induced protein movement.
Plasma Membrane Signaling by Other Toll-Like Receptors
Because TLR4 is the best-characterized member of the TLR family, most of the signal transduction information from other plasma membrane TLRs is inferred from TLR4 biology. Nevertheless, some peculiarities and specific behaviors of TLR1, TLR2, TLR5, and TLR6 merit mention.
TLR2 mainly acts as a heterodimer in association with other TLRs (96), and its function depends on its TLR-binding partner, any coreceptor involved in the recognition process, and the type of bound ligand. TLR2 associates with TLR1 or TLR6 and utilizes CD14 and CD36 as major coreceptors (97, 98). This receptor complex flexibility allows TLR2 to recognize a diverse set of ligands (Table 4) (29, 99).
Table 4.
Ligands detected by TLR2 complexes
| TLR2 complex | Ligand | Reference(s) |
|---|---|---|
| TLR2/TLR1 | E. coli–based synthetic lipopeptides Pam3CSK4 | 100 |
| TLR2/TLR6 | E. coli–based synthetic lipopeptides Pam2CSK4 | 100 |
| TLR2/TLR6 | Mycoplasma fermentans–derived lipopeptide MALP-2 | 100 |
| TLR2/TLR6 | Mycoplasma salivarium–based lipopeptide FSL-1 | 100 |
| TLR2/TLR1 | Human (and mouse) cytomegalovirus | 101, 102 |
| TLR2/TLR1 | Herpes simplex virus | 103 |
| TLR2/TLR1/TLR6 | Herpes simplex virus, endogenous ligands | 104, reviewed in 105 |
At physiological ligand concentrations, TLR1, TLR2, and TLR6 require TIRAP for signaling via MyD88 (79, 80, 106). The formation of different heterocomplexes of TLRs is not believed to have major consequences on signal transduction, although several differences have been described between TLR2/TLR1 and TLR2/TLR6 signaling capacity (107, 108). Similar to TLR4, these TLR heterocomplexes signal from lipid rafts (109) and utilize coreceptors. CD36 is believed to be a major coreceptor for TLR2/TLR6 heterodimers (97), whereas CD14 appears to be involved mainly in TLR2/TLR1 signaling (98). Although the relevance of the signaling capacity of CD14 has never been directly associated with TLR2/TLR1 signaling, the capacity of CD36 to initiate a signaling pathway has been associated with cargo and receptor uptake (110, 111). CD36 is a trans-membrane protein that utilizes Fc receptor-γ (FcRγ) to induce internalization by Syk activation. Although TLR2/TLR1 and TLR2/TLR6 endocytosis is generally associated with downregulating receptor activity, TLR2 internalization has also been associated with type I IFN production (112, 113) in a cell type–dependent manner. Specifically, CD11b+Ly6C+ inflammatory monocytes induce TLR2-mediated type I IFN production in response to viral ligands. In contrast to TLR4, this process is MyD88-dependent and TRIF-independent.
TLR5 recognizes flagellin (114, 115), a major antigenic target of flagellated gram-negative and gram-positive bacteria. TLR5 is expressed on neutrophils, monocytes, DCs (116), and epithelial cells (117). Interestingly, intestinal epithelial cells regulate TLR5 signaling such that responses occur only when flagellin is detected from the basolateral cell surface (117).
Similar to other TLRs, TLR5 dimerizes in response to ligand binding and requires correct plasma membrane localization for its function (118). Localization of this receptor relies on the multispanning membrane protein Unc93B1, which also controls trafficking of endosomal TLRs (discussed in the following section) (119). Although TLR5 requires MyD88 for signaling (114), the role of other adaptors is not as clear; however, TIRAP is involved in proper TLR5 signaling in epithelial cells (120). In support of a requirement for TIRAP, TLR5 signaling is reduced in the absence of PTEN, a phosphoinositide-3-phosphatase that regulates PIP2 production. This suggests that proper distribution of the sorting adaptor is necessary for TLR5 signaling. Interestingly, some MyD88-independent activities have also been described for TLR5, such as flagellin uptake for MHC class II presentation and CD4+ T cell activation (121).
Dectins
Hundreds of CLRs form a heterogeneous group that recognizes a wide range of microorganisms. All CLRs share a characteristic C-type lectin-like domain (CTLD) and can be either soluble or membrane bound. The CTLD was originally identified as a double-loop domain able to bind calcium and carbohydrates. However, CTLD-bearing proteins have since been shown to bind other types of ligands, and today the CLR family is subdivided into 17 different groups (I-XVII) based on structure (4). Most of the CLRs act as opsonins without directly activating an NF-κB signaling cascade and are not discussed in this review. However, subgroups of these receptors initiate a proinflammatory response and promote skewing of T cell immune responses. Generally, this is achieved through a signaling cascade that relies on the function of a complex consisting of the CARD9/Bcl-10/MALT-1 proteins. Here we focus our attention on the PRR function of Dectin-1 and Dectin-2 because they are the best-characterized CLRs with PRR features.
Dectin-1 (Human CLEC7A; mouse Clec7a) is a receptor expressed by DCs, macrophages, neutrophils, and monocytes. Dectin-1 is a type II transmembrane protein containing an extracellular atypical CTLD that does not require calcium for ligand binding. The intracellular tail of Dectin-1 possesses a modified ITAM called hemITAM. The canonical ITAM domain contains a tandem repeat YxxL/I, whereas the intracellular tail of Dectin-1 possesses only one YxxL motif (122). Upon ligand binding, Dectin-1 promotes ligand uptake by phagocytosis and the initiation of a signaling cascade that regulates gene expression and cytokine production. The major ligands for Dectin-1 are fungal β-1,3-glucans, and both mice and human studies confirmed a crucial role for this receptor in antifungal defense (123, 124). Other than fungi, Dectin-1 also recognizes secretory IgA, mucins, and β-glucans from other microorganisms such as Listeria and Mycobacterium (125–127).
Similar to TLR4, this receptor also undergoes ligand-induced movement into lipid rafts where signaling occurs (128). Dectin-1 signaling requires receptor dimerization and initiates a Syk-dependent and a Syk-independent cascade (Figure 3). Syk-dependent signaling controls MAPK activation, canonical and noncanonical NF-κB activation, and NFAT activation. These pathways are initiated by recruitment of Syk to the intracellular hemITAM domains of Dectin-1 following its phosphorylation by Src family kinases (SFKs) (129). NFAT and canonical NF-κB activation require PLCγ2 activity (130), which hydrolyzes membrane PIP2 into inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG). IP3-induced calcium release from the endoplasmic reticulum regulates a calcium-release-activated calcium (CRAC)-mediated calcium influx for NFAT activation (131). Both IP3 and DAG act as second messengers able to regulate the canonical NF-κB pathway by activating protein kinase C δ (PKCδ) (132). PKCδ phosphorylates CARD9 to induce the formation of a complex with Bcl-10 and MALT-1. This recruits and activates a TRAF6-TAK1 complex for NF-κB activation (133). Additionally, the paracaspase activity of MALT-1 is particularly important for activation of the NF-κB subunit C-Rel (134). Syk also mediates the noncanonical activation of the NF-κB subunit p52-RelB through NF-κB-inducing kinase (NIK) (135). Dectin-1 also controls a Syk-independent pathway that represses the functions of the non-canonical NF-κB pathway (135). Upon receptor activation, Raf-1 induces the phosphorylation of the p65 subunit of NF-κB and the subsequent formation of a p65-RelB dimer that inhibits the formation of the p52-RelB complex.
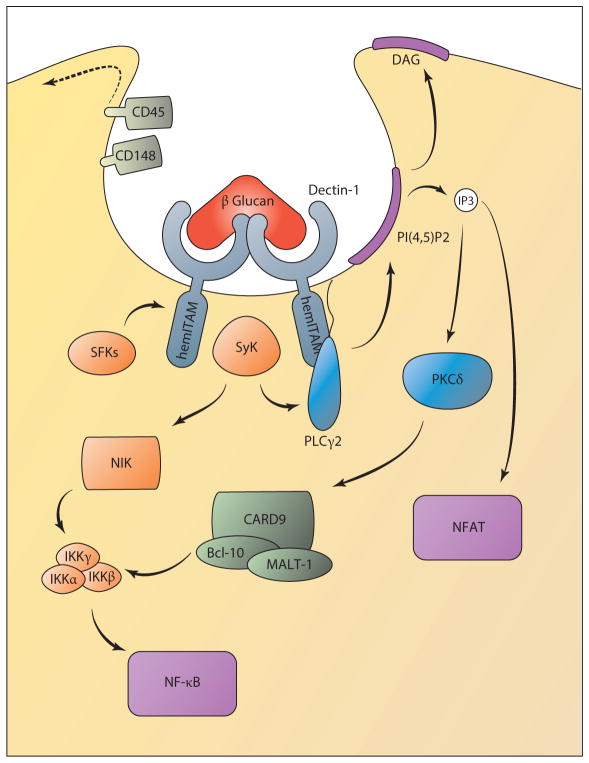
Figure 3.
Dectin-1 signaling controls NF-κB and NFAT activation. Upon ligand binding, the hemITAM domain of Dectin-1 is phosphorylated by Src family kinases (SFKs), which recruit the signaling kinase Syk. This requires trafficking of the phosphatases CD45 and CD148 away from the forming phagosome. Syk activation can control PLCγ2 activity to induce the formation of IP3 and DAG. IP3-mediated Ca2+ release from the endoplasmic reticulum induces a CRAC-dependent Ca2+ influx and consequent NFAT activation. Ca2+ and DAG also regulate PKCδactivation, which controls canonical NF-κB activation following induction of a CARD9/Bcl-10/MALT-1 complex. In parallel, Syk also controls noncanonical NF-κB activation through NIK. (Solid lines indicate signal transduction; dotted lines indicate trafficking events.) (Abbreviations: CARD, caspase recruitment domain; CRAC, calcium-release-activated calcium; DAG, diacylglycerol; IKK, IκB kinase; IP3, inositol 1,4,5-trisphosphate; ITAM, immunoreceptor tyrosine-based activation motif; NFAT, nuclear factor of activated T cells; NIK, NF-κB-inducing kinase; PKC, protein kinase C.)
The induction of phagocytosis by Dectin-1 signaling assists in microbial killing. However, although Dectin-1 can bind soluble ligands, only particulate ligands can induce Dectin-1 signaling. This feature may help to differentiate ligands directly associated with a microbe and those released from a distance (136). Signaling and phagocytosis require cell surface rearrangement of the phosphatases CD45 and CD148 away from Dectin-1 within the forming phagosome. Dectin-1-mediated internalization also requires tyrosine kinase activity and the Rho GTPases Cdc42 and Rac1 (137). Additionally, Dectin-1 activates PLCγ2, which is a regulator of phagocytosis and may contribute to the Dectin-1-mediated phagocytic process (130, 138, 139). This receptor also controls phagosome maturation and MHC class II loading through Syk-mediated release of reactive oxygen species (ROS), which induces LC3 association with phagosomes (140–142). Finally, FYCO1, a protein usually associated with autophagy, is required for the maturation of Dectin-1-Syk-ROS-induced LC3 phagosomes (143). This maturation step is an important point of regulation for the Dectin-1 response that limits signal duration and ROS production. Thus, an intricate link between phagocytosis and signaling exists for the function and regulation of Dectin-1 signaling.
The fine regulation of canonical and noncanonical NF-κB activation controls the release of cytokines that favor Th1 and Th17 responses (135, 144). Among these cytokines, IL-1β production merits discussion. Whereas the expression of pro-IL-1β can be triggered by the signaling and NF-κB activation described above, the processing and activation of this cytokine require the activity of caspase-8, which complexes with ASC, CARD9, Bcl-10, and MALT-1 following Dectin-1 activation (145). Interestingly, although ligand binding is necessary for Dectin-1-mediated caspase-8 activity, internalization is not required. The physiological relevance of this observation is less clear considering that Dectin-1 phagocytic activity is fundamental for its signaling.
Dectin-2 (human CLEC6A; mouse Clec4n) is the prototype receptor of the Dectin-2 family of CLRs that consists of Mincle, DCAR, BDCA-2, Dectin-3 (also called CLECSF8, MCL, or Clec4d), and DCIR. All the members of this CLR family, with the exception of DCIR, contain a short intracellular tail with no signal transduction capabilities. Therefore, Dectin-2 and most of the other receptors of this family associate with the ITAM-bearing molecule FcRγ for signal transduction to occur. Despite this distinction between Dectin-1 and -2 in terms of ITAM usage, the downstream signaling pathways induced by these receptors are quite similar. Dectin-2 binds α-mannans of fungal cell walls (146); however, it can also recognize other pathogens such as Schistosoma mansoni and Mycobacterium tuberculosis (127).
Similar to findings for Dectin-1, Dectin-2 receptor internalization and signaling require the action of SFKs, which target the ITAM residues of FcRγ for phosphorylation. However, Dectin-2 recognizes α-mannans on the surface of hyphal Candida albicans in association with Dectin-3 (147). Dectin-3 forms heterodimers with Dectin-2, conferring a higher sensitivity for fungal detection. The molecular mechanism of this increase in sensitivity is not known, but Dectin-3 is able to recruit both the adaptor FcRγ and another FcRγ-like adaptor protein, FcεRIγ, which may facilitate signal transduction.
SIGNALING FROM ENDOSOMES
As mentioned above, TLR4 signaling continues from a second location within the endocytic network. Professional phagocytes of the innate immune system routinely use the endocytic process to engulf and degrade foreign particles once they reach the hydrolytic lysosomal environment. In addition, many pathogenic microorganisms use the endocytic pathway for their own purposes; bacteria alter and reside within endosomes, whereas viruses may use the lowered pH as a signal to induce membrane fusion and gain access to the cytosol. For these reasons, the endocytic pathway is an ideal location for innate immune receptors to reside, as they are well positioned to detect ligands revealed during hydrolytic degradation or viral entry.
TLR4 Signal Transduction from Endosomes
The second site of TLR4 signaling from endosomes controls two important responses: a late wave of NF-κB activation and the production of type I IFN. The delayed wave of NF-κB activation can be demonstrated from cells deficient in MyD88 or TIRAP in response to LPS (79). Thus, TRAM and TRIF control the TLR4-driven late wave of NF-κB activation and type I IFN production (148, 149). The sorting adaptor TRAM contains a bipartate N-terminal localization signal containing a myristate group and a polybasic motif that control trafficking between the plasma membrane and endosomes (23, 150). The presence of TRAM in endosomes is absolutely required for TRAM/TRIF signaling, yet TRAM movement from the plasma membrane to endosomes is independent of CD14-mediated TLR4 movement to endosomes. It has been speculated that once it reaches endosomes, TIRAP is displaced from the TIR domain of TLR4 by TRAM, allowing for the TRIF-dependent pathway to be engaged (23, 151, 152).
The mechanism by which TRIF-dependent late-stage NF-κB activation occurs is unclear, but several studies have demonstrated biochemical or functional interactions between TRIF and RIPK1, TRADD, and caspase-8 that promote NF-κB activation (Figure 2).
TRIF also controls IRF3-mediated type I IFN production through the recruitment of TRAF3. TRAF3 interacts with TANK or TANK-related proteins (153) to recruit the kinases TBK1 and IKKε for IRF3 activation, thus regulating type I IFN production (154–156). The complete transcriptional response induced by TLR4 from endosomes requires late-stage activation of NF-κB and type I IFN production (157, 158).
Endosomal Signaling by Other Toll-Like Receptors
All TLRs induce NF-κB activation, but many can trigger alternative pathways that culminate in the activation of IRFs and the production of type I IFN. The triggering of IFN expression is accomplished in several ways. As described above, TLR4 initiates a TRIF-dependent pathway from endosomes through the sorting adaptor TRAM. TLR3 also activates TRIF-dependent signaling, but unlike TLR4, this process does not require TRAM (Figure 4) (159). Instead, the TIR domain of TLR3 has high affinity for the TIR domain of TRIF (160, 161) and is capable of direct binding. The reason for a differential requirement for TRAM is unclear, but it could be related to the need for TLR4 to engage different signaling adaptors in different compartments. Interestingly, a single point mutation in the TIR domain of TLR3 is sufficient to alter the specificity from TRIF to MyD88 (162).
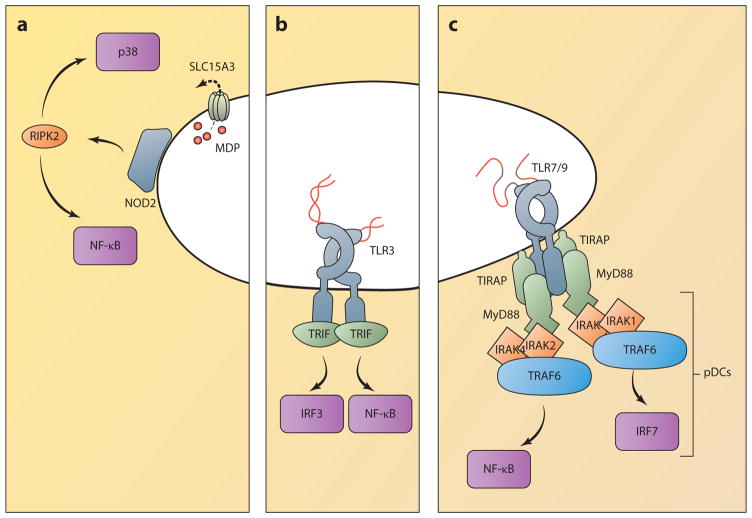
Figure 4.
Innate immune signaling from endosomes. (a) NOD receptors associate with endosomal membranes, where they are positioned to encounter PAMPs, as microbes escape from endosomes or as they are pumped out by transporters such as SLC15A3. NOD signaling activates RIPK2, which triggers proinflammatory cytokine production through NF-κB and MAPK activation. (b) TLR3 responds to double-stranded RNA and triggers an enzymatic signaling cascade through the adaptor protein TRIF. Unlike TLR4, TLR3 does not require the adaptor TRAM, but it can similarly activate IRF3 to produce type I IFN and ISGs, as well as NF-κB, to produce proinflammatory cytokines. (c) Other endosomal TLRs principally activate NF-κB through the adaptors MyD88 and TIRAP. In pDCs, TLR7 and -9 trigger IFN and ISG production through the activation of IRF7. (Abbreviations: IFN, interferon; IRAK, interleukin-1 receptor-associated kinase; IRF, IFN regulatory factor; ISG, IFN-stimulated gene; MAPK, mitogen-activated protein kinase; MDP, muramyl dipeptide; NOD, nucleotide-binding oligomerization domain; PAMP, pathogen-associated molecular pattern; pDC, plasmacytoid dendritic cell; RIPK2, RIP2 kinase; TIRAP, TIR-containing adaptor protein; TRAF, TNF receptor–associated factor; TRIF, TIR domain–containing adaptor-inducing IFN-β.)
In addition to producing NF-κB-mediated cytokines, endosomal TLRs also induce IFN expression in a cell type–specific manner. In particular, TLR7 and TLR9 trigger MyD88-dependent IFN production in plasmacytoid DCs (pDCs) (163, 164). Unlike TLR4-driven IFN production, pDCs do not use the TRAM/TRIF pathway. Instead, these cells initiate an alternative MyD88-dependent signaling pathway that activates the transcription factor IRF7 (Figure 4) (165). However, as in the TLR4 pathway, the production of IFN is spatially segregated from proinflammatory cytokines. Although both signaling pathways initiate from endosomes, pDCs from mice deficient for the adaptor complex AP-3 cannot produce IFN, but they do maintain their ability to produce proinflammatory cytokines (166). This suggests that TLR7 and -9 initiate distinct signaling pathways that originate from two distinct endosomal compartments, dubbed NF-κB endosomes and IRF7 endosomes because of their role in activating these distinct transcription factors (166). The bifurcation of signaling is also demonstrated by differential requirements for downstream signaling components. For example, IRF7 activation in pDCs requires IRAK1, whereas IRAK2 knockout cells have enhanced IRF7 activation (167). The reverse is true for NF-κB signaling; IRAK2 knockout cells cannot induce NF-κB activation, whereas IRAK1 knockouts have increased NF-κB signaling activity (168). These data suggest that distinct myddosomes activate distinct transcription factors and that these myddosomes may compete for upstream or downstream signaling resources.
The precise nature of the endosomes that permit TLR7/9 signaling is unknown, but several lines of evidence provide insight. The data from AP-3 knockout mice described above suggest two possibilities. First, signaling from the NF-κB endosome may occur early in the endocytic pathway, with AP-3 being required for the receptor to move to a later endosome for IRF7 activation. However, TLR9-mediated IRF7 activation occurs early in the endosomal pathway, and TLR9 signaling in macrophages proceeds normally when the sorting adaptor TIRAP is restricted to early endosomes (81, 169). Thus, a second possibility is that AP-3 delivers some component necessary for IRF7 signaling to a specific endosome, and without this component, only NF-κB activation is achieved. TRAF3 appears to be one such regulator controlled by AP-3, as artificially directing this ubiquitin ligase to membranes rich in 3′ phosphoinositides is sufficient to restore IFN production in AP-3 knockout cells (166). As the study of inter-endosome trafficking is technically complex, new technologies may need to be developed before a clear view of site-specific TLR signaling in endosomes can be achieved.
Regulation of Endosomal TLR Activity and Localization
Although TLR4 resides at the plasma membrane and migrates to endosomes upon ligand binding, many TLRs reside in and encounter their ligands from within endosomes. These TLRs predominantly recognize nucleic acid ligands, which are liberated from bacteria or viral capsids due to the hydrolytic activity of the endolysosomal compartment. Nucleic acids are a common feature of all pathogens and therefore ideal targets of PRRs. Yet recognition of nucleic acid is also problematic because there is little to distinguish self-RNA and -DNA from nonself. One solution to this apparent conundrum is to recognize particular chemical motifs that are not present on self-nucleic acids. For example, TLR9 predominantly recognizes unmethylated CpG DNA motifs, which are uncommon in the mammalian genome (170). Similarly, the ligand for TLR3 is double-stranded RNA, which is indicative of some viral genomes and replication intermediates but not often the product of mammalian cells (171).
As discussed above, an alternative (or supplement) to a strategy of chemical specificity is to restrict ligand recognition to particular subcellular locations in which self-nucleic acids are unlikely to be present. For this reason, nucleic acid recognition by TLRs is restricted to endolysosomal compartments through cell biological regulation. The clearest example of this regulation has been demonstrated for TLR3, -7, and -9. As already mentioned, TLR9 recognizes unmethylated CpG DNA motifs, whereas TLR7 is specific for single-stranded RNA and TLR3 for double-stranded RNA. However, these TLRs are synthesized in an inactive state and must be proteolytically cleaved to become active (172, 173). TLR cleavage is a multistep process, requiring multiple endosomal proteases, including asparagine endopeptidase and cathepsins (174, 175). This requirement may decrease the likelihood of detecting extracellular self-DNA by ensuring that endosomal TLRs function only within an endocytic compartment and not from the plasma membrane.
The transport of TLRs to endosomes is also tightly controlled and requires interaction with a multipass transmembrane chaperone called Unc93B1 to exit the endoplasmic reticulum (176, 177). Unc93B1 travels with TLRs to their final destination and interacts with numerous accessory molecules to mediate transport, but the route taken by each TLR is unique. For TLR9, this requires Unc93B1-mediated transport to the cell surface, where interaction with AP-2 mediates endocytosis and trafficking to endolysosomes (178). Once TLR9 enters the endocytic network, it is converted to the active form, as described above. Alternatively, Unc93B1-bound TLR7 associates with AP-4 in the Golgi complex, which delivers the receptor directly to endosomes without an intermediate cell surface step. It is unclear whether these differential trafficking patterns serve some regulatory purpose, but individual point mutants of Unc93B1 can uniquely abolish AP-2 binding or TLR7 binding (178). This indicates that trafficking for each TLR may be regulated through unique binding to Unc93B1.
NOD1/2 Signaling from Endosomes
NOD1 and NOD2 are the prototypical members of the NLR family of PRRs (179). Both are cytosolic proteins containing a series of C-terminal ligand-binding LRRs, a central NACHT domain, and a single (NOD1) or tandem (NOD2) N-terminal CARD domains. Both receptors detect components of bacterial outer membranes or cell walls, such as γD-glutamyl-meso-diaminopimelic acid (iE-DAP) or muramyl dipeptide (MDP) (180–182). Upon ligand binding, NODs associate with the kinase RIPK2 via CARD domain interactions, resulting in the formation of a signaling complex that mediates NF-κβ and MAPK activation (183, 184).
Though translated in the cytosol and lacking defined localization domains, NOD1 and NOD2 signaling occur in close association with endosomal membranes, as suggested by evidence (185–188) (Figure 4). The first indication of this came from the characterization of a Crohn’s disease–associated NOD2 variant, NOD2 3020insC, with impaired responsiveness to MDP (185). Whereas wild-type NOD2 displays colocalization with the plasma membrane in epithelial cells, a C-terminal truncation redistributes the hypomorphic variant to the cytosol. Furthermore, treatment of epithelial cells with bacterial outer membrane vesicles, a potent source of iE-DAP, caused recruitment of NOD1 and the signaling kinase RIPK2 to endosomes, and this recruitment correlated with cytokine production (186). A separate study showed that the endosomal transporter SLC15A3 was required to translocate the NOD2 ligand MDP into the cytoplasm in DCs (187). NOD2 also associates with SLC15A3 at phagosomal membranes containing MDP or Salmonella, and MDP-containing phagosomes associate with NOD1, NOD2, and RIPK2. Taken together, these data suggest that NOD1/2 are regulated by recruitment to endosomal membranes and that this is the site of signal transduction for these receptors.
Independent of their role in NF-κB activation, NOD1 and NOD2 can also direct autophagy to eliminate intracellular bacterial pathogens. Recruitment of the autophagy protein ATG16L1 to the site of bacterial entry at the plasma membrane requires functional NOD alleles (188). This incorporation of intracellular bacteria within autophagosomes is impaired in cells expressing the Crohn’s disease–associated NOD2 variant described above.
SIGNALING FROM THE BIOSYNTHETIC PATHWAY
As noted above, receptors at the plasma membrane and within endocytic compartments are poised to encounter both nonpathogenic and pathogenic microbes. However, two key features of the biosynthetic pathway help limit responses to true pathogens. First, these receptors are positioned within the cytosol (or nucleus), a location commonly accessed by pathogens to manipulate the host. Second, these receptors recognize foreign nucleic acids. Because active infections generate nucleic acids as a form of gene expression or replication, these molecules serve as markers of biosynthesis indicating an active infection. Several PRRs cooperate to survey this compartment and coordinate effective immune responses to viral, prokaryotic, and eukaryotic intracellular infection. Here we cover some of the cell biological features within the biosynthetic pathway.
RLR Signaling
The RLRs consist of three DExH/D box helicases [retinoic acid-inducible gene-I (RIG-I), melanoma differentiation gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2)], which detect the presence of foreign RNA within the cytosol (189, 190). RLRs are generally depicted as cytosolic proteins at steady state, but they may be poised at specific locations within the cytosol where viral entry and/or replication take place. To distinguish foreign RNA from endogenous RNA, these receptors detect features common to viral genomes and replication intermediates, including 5′ triphosphate RNA, long double-stranded RNA, and sequences specific to viral genomes such as the poly-uridine region of hepatitis C virus (191–196). These receptors are critical for host defense against RNA viral infection; however, they also function in defense against some DNA viruses and bacterial pathogens (197). The CARD domain–containing receptors RIG-I and MDA5 activate the adaptor MAVS through CARD-mediated interactions (24, 198–200). However, prior to MAVS interaction, RIG-I undergoes K63-linked ubiquitin modification by the E3 ubiquitin ligases tripartite motif-containing 25 (TRIM25) and RIPLET (also known as RNF135) (Figure 5) (201–203). Once activated, MAVS undergoes CARD-dependent self-polymerization, creating prion-like aggregates that are important for signal transmission (204). MAVS polymerization recruits a set of ubiquitin ligases (TRAF2, -5, and -6) required for the activation of TBK1 and the IKK complex (205). The kinases regulate the transcription factors NF-κB, IRF3, and IRF7, culminating in the expression of IFN, proinflammatory cytokines, and IFN-stimulated genes (ISGs).
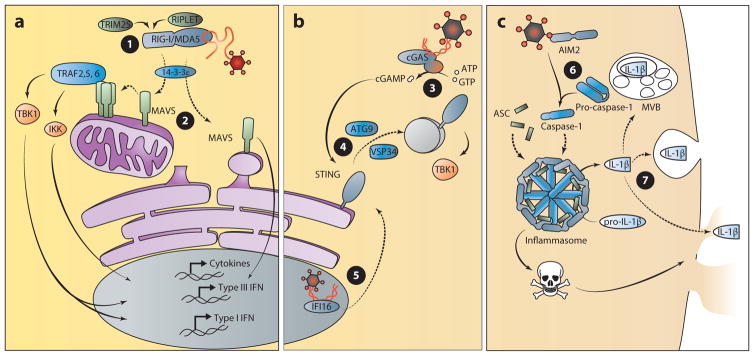
Figure 5.
The biosynthetic pathway: nucleic acid detection from within the cytosol. (a) The RLRs detect pathogen-derived RNA within the cytosol to induce the production of IFN and proinflammatory cytokines. The TBK1, IKK, and MAVS pathways lead to activation of the transcription factors for the induction of IFN and other cytokine genes. ➊ RIG-I signal transduction is regulated by TRIM25- and RIPLET-mediated ubiquitination and translocation to the site of signaling by 14-3-3ε. ➋ MAVS activity is regulated by polymerization, and signaling from mitochondria results in production of type I and III IFN, whereas peroxisomal signaling induces the production only of type III IFN. (b) cGAS and the ALRs detect pathogen-derived DNA from within the cytosol and nucleus to induce the production of IFN. ➌ In the presence of DNA, the enzyme cGAS converts ATP and GTP to the cyclic dinucleotide cGAMP. ➍ Production of cGAMP induces the activation and trafficking of STING to poorly defined sites of signaling in an ATG9- and VSP34-dependent manner. TBK1 is recruited to this site of signaling to induce the production of type I IFN. ➎ Viral DNA within the nucleus can be detected by IFI16 for the production of type I IFN in a STING-dependent manner. Therefore, the trafficking of this receptor or another factor from the nucleus to the cytosol may regulate IFI16-dependent signaling. (c) The inflammasome-mediated response to DNA within the cytosol. ➏ AIM2 detection of cytosolic DNA activates inflammasome formation, which induces cell death and the maturation of IL-1β. ➐ Secretion of this cytokine requires a noncanonical route that is independent of trafficking through the endoplasmic reticulum and Golgi apparatus. Several possibilities have been proposed for IL-1β release that include routes through lysosomal compartments, multivesicular bodies, or pores created during pyroptotic cell death. (Solid lines indicate signal transduction; dotted lines indicate trafficking events.) (Abbreviations: AIM, absent in melanoma; cGAMP, cyclic di-GMP/AMP; cGAS, cyclic GMP-AMP synthase; IFI, interferon, γ-inducible; IFN, interferon; IKK, IκB kinase; MAVS, mitochondrial antiviral signaling protein; MDA, melanoma differentiation gene; RIG-I, retinoic acid–inducible gene I; STING, stimulator of IFN gene; TBK, TANK-binding kinase; TRAF, TNF receptor–associated factor; TRIM25, tripartite motif-containing 25.)
Like the TLRs, RIG-I also undergoes ligand-induced translocation to specific sites of signaling. Following detection of viral RNA, RIG-I moves to sites of signaling on the limiting membranes of mitochondria, peroxisomes, and the MAM where the adaptor MAVS is located (24–26). The protein chaperone 14-3-3ε regulates this process of translocation by delivering RIG-I and TRIM25 to membrane-bound MAVS (Figure 5) (206). Correct localization to the aforementioned membranes is an absolute requirement for MAVS function and is dependent on its C-terminal transmembrane domain. Variants of MAVS lacking this domain are incapable of inducing an IFN signal, although they contain the necessary regions for interaction with downstream signaling proteins (24, 25). Taken together, these data suggest that MAVS signaling is regulated by receptor recruitment as well as by proper membrane localization.
The location from which MAVS signals dictates differences in antiviral gene expression. Signaling from the mitochondrial membrane induces the expression of type I and type III IFN genes, resulting in the subsequent expression of ISGs. In contrast, peroxisomal signaling induces the expression of ISGs independently of type I IFN, relying exclusively on type III IFN production (25, 207). An interesting feature of mitochondrial and peroxisomal biology is that these organelles interact at the MAM, a specialized subdomain of the endoplasmic reticulum (26, 208). Therefore, the MAM also participates in immune responses by providing a synaptic interface between mitochondria and peroxisomes.
The interaction of mitochondria and peroxisomes at the MAM demonstrates the dynamic nature of these organelles. Changes in the mitochondrial network have been observed following RLR activation, and mitochondrial dynamics appear to regulate or be regulated by RLR signaling (209). Mitochondrial fusion is one such event that plays a role in MAVS signaling, as demonstrated in cells deficient for Mitofusin 1 and 2 (MFN1, MFN2) (209–211). Given that the MFNs promote mitochondrial fusion, MFN-deficient cells have a fragmented mitochondrial network. However, IFN signaling was reduced in their absence during viral infection, indicating a link between the regulation of mitochondrial dynamics and RLR signaling outcome. One way in which fusion could regulate MAVS signaling is through its effects on inner mitochondrial membrane potential, a critical source of energy for ATP synthesis (212). Indeed, dissipation of membrane potential using the drug carbonyl cyanide m-chlorophenylhydrazone (CCCP) suppressed RLR-mediated IFN production (211). In contrast to its role in organelle fusion, MFN2 has been implicated as an inhibitor of MAVS signaling (213). However, this occurs through direct protein-protein interactions between MAVS and MFN2.
In addition to regulatory effects of mitochondria, regulation of peroxisome physiology can also have an effect on antiviral signaling, as observed in cells derived from patients with Zellweger syndrome, a peroxisome biogenesis disorder. Deficiencies in Pex19, which regulates peroxisome biogenesis and delivery of membrane proteins to the organelle, result in cells that lack distinct peroxisomes and in the development of Zellweger syndrome (214). Interestingly, these cells contain mitochondria with mistargeted peroxisomal membrane proteins and have a heightened antiviral response to reovirus infection (207). These data support the conclusion that changes in mitochondrial and peroxisomal organelle physiology can regulate MAVS signaling.
The catabolic process of autophagy, which is utilized for the maintenance of cellular health, degrades intracellular components at baseline levels but is also commonly induced in response to cellular stresses such as organelle dysfunction and viral infection. In the context of RLR signaling, autophagy is implicated as a negative regulator controlling IFN production (215). Several mediators of this process have been identified and include ATG5, ATG6, COX5B, NLRX1, and TUFM (216–218). Interfering with these regulators inhibits the formation of autophagosomes, resulting in higher levels of IFN production and viral replication. How autophagy functions to regulate RLR signaling requires further characterization, yet some proposed mechanisms include the removal of damaged mitochondria, the degradation of aggregated MAVS complexes, and the reduction in ROS.
Most research on RLR signaling has focused on IFN production, but the RLR pathway can also limit viral replication by initiating cell death (19, 219, 220). Control of cell death is MAVS dependent and requires correct localization of the adaptor. However, the induction of cell death does not require the CARD domain and is independent of MAVS-induced IFN signaling, which has been inferred from the characterization of N-terminal deletion mutants and a naturally encoded MAVS variant, miniMAVS (19, 221). Both the mutant and the variant lack the N-terminal CARD domain required for IFN production yet can still induce cell death. Although caspase activation is required, the specific form of cell death induced (apoptosis, necrosis, etc.) remains to be characterized (19, 222). Several proteins with known cell death function (TRADD, caspase-8, and VDAC1) interact with MAVS and regulate antiviral cell death (223–225). TRADD interacts with MAVS and recruits the proapoptotic proteins FADD and RIPK1 (198, 223). The detailed mechanism by which MAVS induces this process has yet to be determined; however, the induction of cell death independent of a CARD domain suggests that the process does not require polymerization of MAVS.
Interestingly, MAVS-dependent cell death can be induced in the absence of the RLRs. According to a recent study, MAVS-mediated cell death requires the kinases MKK7 and JNK2 but is independent of RIG-I and MDA5 (220). In addition, MAVS-dependent vesicular stomatitis virus–induced cell death does not require the CARD domain of MAVS (225). If MAVS-mediated cell death does not require its CARD domain or the upstream RLRs, this suggests that MAVS can detect viral infection directly or that other cell death–inducing receptors exist.
STING-Dependent Signaling
The search for cytosolic receptors that recognize DNA has implicated a bewildering number of genes with reported involvement in regulating the IFN response to this nucleic acid (226). Although further verification and characterization are needed for many of these genes, it is well established that STING (stimulator of IFN genes, also known as TMEM173, MPYS, MITA, and ERIS) plays an important role in this response (227–229). STING activates TBK1- and IRF3-mediated IFN production, contributing to the defense against viral and intracellular bacterial pathogens (230). Compared to other PRR signaling molecules, STING may function uniquely as both an adaptor and a receptor. As an adaptor, STING is required for antiviral responses emanating from proposed DNA receptors including IFI16 and DDX41 (231, 232), although the mechanisms by which these processes occur are unclear. The best understanding of STING functions derives from its ability to directly bind to cyclic dinucleotides (CDNs), which are second-messenger signaling molecules commonly produced by bacteria (233–238). Although CDN synthesis has been considered a unique feature of bacterial species, the recently identified mammalian enzyme cGAS can synthesize the CDN cyclic di-GMP/AMP (cGAMP) (239, 240). In the presence of DNA, cGAS utilizes ATP and GTP to produce cGAMP, which binds with high affinity to STING and activates IFN signaling more effectively than the bacterial-derived CDNs. Thus, it is likely that during infections with DNA viruses, cGAS produces cGAMP that directs STING to induce an antiviral cellular state.
STING is a multispanning transmembrane protein that is predominantly localized on the endoplasmic reticulum at steady state. Reports of STING on mitochondria and the MAM may be due to the dynamic interactions between these organelles as described above (228, 230). Following activation, STING undergoes trafficking to poorly defined vesicles or puncta via the Golgi apparatus (Figure 5) (230, 241). This trafficking event relies on ATG9a and VPS34, suggesting that autophagy may play a role in the regulation of STING (241, 242). Following STING movement, signaling occurs from these puncta through the activation of TBK1 and IRF3. Blocking movement of STING using Brefeldin A (BFA) or by siRNA-mediated knockdown of VPS34 inhibits the STING-dependent IFN response to cytosolic DNA and highlights the importance of this regulatory trafficking step (242).
Although the translocation of STING requires autophagy-related genes, there is a limited understanding of the role of autophagy in STING-dependent signaling. For example, the association between autophagy and STING activation has yet to be demonstrated with the formation of classic double membrane–bound autophagosomes. Additionally, ATG9a-deficient cells display a heightened response to DNA, indicating that autophagy is required to limit this signaling pathway (241). However, activation of STING by Mycobacterium tuberculosis DNA has been implicated in the resistance to infection through the induction of autophagy. Specifically, STING, TBK1, and ATG5 are important for limiting bacterial replication by targeting M. tuberculosis to autophagosomes (243). However, these experiments investigated bacterial infection rather than the response to purified ligand, so in addition to STING activation, other signals may contribute to autophagosome formation. Future studies are required to clearly define whether autophagy intersects with STING signaling as a form of regulation or as a defense mechanism against intracellular pathogens.
AIM2-Like Receptor Signaling
The ALRs participate in the detection of intracellular DNA. These receptors each have a PYHIN domain allowing for protein-protein interactions and a DNA-binding HIN-200 domain. The founding member of this family, AIM2, interacts with the adaptor ASC and promotes inflammasome formation following the detection of intracellular DNA (244–246). A more diverse set of functions has been attributed to a second member, IFI16, including the activation of STING-dependent IFN production and inflammasome formation (231, 247). Inflammasomes are multiprotein complexes that initiate an innate immune response characterized by the secretion of proinflammatory cytokines (IL-1β, IL-18) and a rapid form of cell death (pyroptosis) that contributes to inflammation (7). In addition to responding to intracellular DNA, inflammasomes are activated in response to several microbial patterns or danger signals. Discussing the numerous stimuli and regulators of inflammasome formation is beyond the scope of this review, but they have been reviewed extensively elsewhere (7, 248).
Generation of the proinflammatory cytokines IL-1β and IL-18 is a hallmark of inflammasome activation and a key component of the immune response to many viral and bacterial infections. However, the secretion of these cytokines is poorly defined. Protein secretion into the extracellular space usually depends on conventional transport through the endoplasmic reticulum and Golgi apparatus. However, IL-1β and IL-18 are part of a class of leaderless cytokines that do not contain the N-terminal signal sequence required for conventional protein trafficking. Instead, these cytokines are produced in an inactive form that must undergo processing by activated caspase-1 prior to secretion by nonconventional mechanisms (Figure 5) (249). Inflammasome formation provides a molecular scaffold for active caspase-1 to cleave the leaderless cytokines into their active form. It is not clear how these active cytokines are secreted, but there is some evidence that IL-1β is compartmentalized within vesicles and could be released through lysosome exocytosis or exosome release from multivesicular bodies (250–253). Another possibility is that these cytokines are released during inflammasome-mediated pyroptotic cell death (254). This form of cell death is hypothesized to result from osmotic pressure generated by membrane pore formation (255). Therefore, pore formation and/or cell death could also be involved in the release of these cytokines into the extracellular space. Understanding the cell biology regulating IL-1β/IL-18 release as well as pyroptosis will provide further insight into the immune response triggered by ALR-mediated inflammasome activation.
The second ALR to be identified, IFI16, has since been the focus of much attention in the study of antiviral defense (231). Several viruses, including herpes simplex virus (HSV), Kaposi’s sarcoma–associated herpesvirus (KSHV), cytomegalovirus, and HIV, are detected by IFI16 (231, 247, 256–258). Similar to other DNA receptors that activate STING or the inflammasome, IFI16 was originally classified as a cytoplasmic sensor. However, the subcellular localization of IFI16 has been found within the cytosol as well as the nucleus, depending on cell type (259). As several DNA viruses complete a portion of the replication cycle within the nucleus, it seems fitting that receptors identifying these viruses might also be found within the nucleus. Detection of KSHV and HSV1 by IFI16 occurs within the nucleus; however, signaling ultimately occurs in the cytosol through inflammasome or STING activation (Figure 5) (247, 260, 261). Thus, ligand-induced movement of IFI16 (or another regulator) to a cytosolic site of signaling is required to initiate antiviral defenses against herpesviruses. The identification of IFI16 acetylation sites that control its subcellular distribution provides some insight into the regulation of this receptor (260). This may allow IFI16 to function as both a nuclear and cytosolic sensor of foreign DNA. A major question that these findings highlight is that of how nuclear DNA sensors distinguish between self- and foreign DNA.
CONCLUDING REMARKS
This review was conceived with the goal of highlighting common cell biological properties that influence the activity of the innate immune system in mammals. The fact that numerous genetically distinct PRR families depend on microbe-induced protein trafficking pathways underscores the fundamental importance of these processes in signaling pathway design. As modern methods of scientific inquiry become more specialized, it is important for the field to continue to remember that signaling pathways within our cells did not evolve in a vacuum. Rather, all signaling pathways evolved to operate in the context of other cell biological processes that may be occurring simultaneously. As such, the fields of innate immune and protein/membrane trafficking have much to teach each other, and we hope that the ideas presented herein will help spur increased cross-disciplinary investigations.
FUTURE DIRECTIONS.
The ability of endosomal TLRs to induce IFN production in pDCs highlights cell type–specific differences in innate immune signal regulation. The extent of this signaling diversity and its implications for in vivo infection will be of future interest.
Do signals originating from a specific subcellular location require something inherent to that site, or does the site of signaling simply act as a scaffold that can be reconstituted in any location?
What are the cell biological aspects of inflammasome formation and function? It is understood that active inflammasomes are large multiprotein complexes, but much remains to be characterized in how these signaling complexes interact with the cellular infrastructure. It is becoming clear that the NLRP3 inflammasome can monitor mitochondrial dysfunction, and some reports implicate mitochondria as a site of NLRP3 inflammasome formation.
The cellular process of mRNA translation is emerging as a regulator of gene function. Recently, it has been reported that the MAVS transcript encodes two variants with distinct function. As the importance of translational control becomes more evident, it will be of interest to identify how this process controls other aspects of innate immunity during infection.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Contributor Information
Sky W. Brubaker, Email: skybrubaker@stanford.edu.
Kevin S. Bonham, Email: kbonham@fas.harvard.edu.
Ivan Zanoni, Email: ivan.zanoni@childrens.harvard.edu.
Jonathan C. Kagan, Email: jonathan.kagan@childrens.harvard.edu.
LITERATURE CITED
- 1.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30(1):16–34. doi: 10.3109/08830185.2010.529976. [DOI] [PubMed] [Google Scholar]
- 2.Janeway CA. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt. 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov R. Approaching the asymptote: 20 years later. Immunity. 2009;30(6):766–75. doi: 10.1016/j.immuni.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Zelensky AN, Gready JE. The C-type lectin-like domain superfamily. FEBS J. 2005;272(24):6179–217. doi: 10.1111/j.1742-4658.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 5.Drummond RA, Brown GD. The role of Dectin-1 in the host defence against fungal infections. Curr Opin Microbiol. 2011;14(4):392–99. doi: 10.1016/j.mib.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 6.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol. 2013;13(10):722–37. doi: 10.1038/nri3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamkanfi M, Dixit VM. Mechanisms and functions of inflammasomes. Cell. 2014;157(5):1013–22. doi: 10.1016/j.cell.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Palm NW, Medzhitov R. Pattern recognition receptors and control of adaptive immunity. Immunol Rev. 2009;227(1):221–33. doi: 10.1111/j.1600-065X.2008.00731.x. [DOI] [PubMed] [Google Scholar]
- 9.Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4(7):499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 10.Geijtenbeek TBH, Gringhuis SI. Signalling through C-type lectin receptors: shaping immune responses. Nat Rev Immunol. 2009;9(7):465–79. doi: 10.1038/nri2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, Shaw MH, Kim Y-G, Núñez G. NOD-like receptors: role in innate immunity and inflammatory disease. Annu Rev Pathol Mech Dis. 2009;4(1):365–98. doi: 10.1146/annurev.pathol.4.110807.092239. [DOI] [PubMed] [Google Scholar]
- 12.Loo Y-M, Gale M. Immune signaling by RIG-I-like receptors. Immunity. 2011;34(5):680–92. doi: 10.1016/j.immuni.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hornung V, Latz E. Intracellular DNA recognition. Nat Rev Immunol. 2010;10(2):123–30. doi: 10.1038/nri2690. [DOI] [PubMed] [Google Scholar]
- 14.Barton GM, Kagan JC. A cell biological view of Toll-like receptor function: regulation through compartmentalization. Nat Rev Immunol. 2009;9(8):535–42. doi: 10.1038/nri2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett H, McFadden G. Apoptosis: an innate immune response to virus infection. Trends Microbiol. 1999;7(4):160–65. doi: 10.1016/s0966-842x(99)01487-0. [DOI] [PubMed] [Google Scholar]
- 16.Mouchess ML, Arpaia N, Souza G, Barbalat R, Ewald SE, et al. Transmembrane mutations in Toll-like receptor 9 bypass the requirement for ectodomain proteolysis and induce fatal inflammation. Immunity. 2011;35(5):721–32. doi: 10.1016/j.immuni.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbalat R, Ewald SE, Mouchess ML, Barton GM. Nucleic acid recognition by the innate immune system. Annu Rev Immunol. 2011;29(1):185–214. doi: 10.1146/annurev-immunol-031210-101340. [DOI] [PubMed] [Google Scholar]
- 18.Lafyatis R, Marshak-Rothstein A. Toll-like receptors and innate immune responses in systemic lupus erythematosus. Arthritis Res Ther. 2007;9(6):222. doi: 10.1186/ar2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei Y, Moore CB, Liesman RM, O’Connor BP, Bergstralh DT, et al. MAVS-mediated apoptosis and its inhibition by viral proteins. PLOS ONE. 2009;4(5):e5466. doi: 10.1371/journal.pone.0005466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medzhitov R, Preston-Hurlburt P, Kopp E, Stadlen A, Chen C, et al. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol Cell. 1998;2(2):253–58. doi: 10.1016/s1097-2765(00)80136-7. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto M, Sato S, Mori K, Hoshino K, Takeuchi O, et al. Cutting edge: a novel Toll/IL-1 receptor domain-containing adapter that preferentially activates the IFN-β promoter in the Toll-like receptor signaling. J Immunol. 2002;169(12):6668–72. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 22.Kagan JC, Medzhitov R. Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell. 2006;125(5):943–55. doi: 10.1016/j.cell.2006.03.047. [DOI] [PubMed] [Google Scholar]
- 23.Kagan JC, Su T, Horng T, Chow A, Akira S, Medzhitov R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-β. Nat Immunol. 2008;9(4):361–68. doi: 10.1038/ni1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seth RB, Sun L, Ea C-K, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell. 2005;122(5):669–82. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 25.Dixit E, Boulant S, Zhang Y, Lee ASY, Odendall C, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141(4):668–81. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horner SM, Liu HM, Park HS, Briley J, Gale M. Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. PNAS. 2011;108(35):14590–95. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11(6):725–32. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 28.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, et al. Structural basis of Toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320(5874):379–81. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130(6):1071–82. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Park BS, Song DH, Kim HM, Choi B-S, Lee H, Lee J-O. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458(7242):1191–95. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 31.Medzhitov R, Preston-Hurlburt P, Janeway CA. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388(6640):394–97. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 32.Poltorak A, He X, Smirnova I, Liu MY, Van Huffel C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282(5396):2085–88. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 33.Kawasaki K, Akashi S, Shimazu R, Yoshida T, Miyake K, Nishijima M. Mouse Toll-like receptor 4·MD-2 complex mediates lipopolysaccharide-mimetic signal transduction by Taxol. J Biol Chem. 2000;275(4):2251–54. doi: 10.1074/jbc.275.4.2251. [DOI] [PubMed] [Google Scholar]
- 34.Kurt-Jones EA, Popova L, Kwinn L, Haynes LM, Jones LP, et al. Pattern recognition receptors TLR4 and CD14 mediate response to respiratory syncytial virus. Nat Immunol. 2000;1(5):398–401. doi: 10.1038/80833. [DOI] [PubMed] [Google Scholar]
- 35.Rassa JC, Meyers JL, Zhang Y, Kudaravalli R, Ross SR. Murine retroviruses activate B cells via interaction with Toll-like receptor 4. PNAS. 2002;99(4):2281–86. doi: 10.1073/pnas.042355399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bulut Y, Faure E, Thomas L, Karahashi H, Michelsen KS, et al. Chlamydial heat shock protein 60 activates macrophages and endothelial cells through Toll-like receptor 4 and MD2 in a MyD88-dependent pathway. J Immunol. 2002;168(3):1435–40. doi: 10.4049/jimmunol.168.3.1435. [DOI] [PubMed] [Google Scholar]
- 37.Millien VO, Lu W, Shaw J, Yuan X, Mak G, et al. Cleavage of fibrinogen by proteinases elicits allergic responses through Toll-like receptor 4. Science. 2013;341(6147):792–96. doi: 10.1126/science.1240342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohashi K, Burkart V, Flohé S, Kolb H. Cutting edge: heat shock protein 60 is a putative endogenous ligand of the Toll-like receptor-4 complex. J Immunol. 2000;164(2):558–61. doi: 10.4049/jimmunol.164.2.558. [DOI] [PubMed] [Google Scholar]
- 39.Vabulas RM, Ahmad-Nejad P, Ghose S, Kirschning CJ, Issels RD, Wagner H. HSP70 as endogenous stimulus of the Toll/interleukin-1 receptor signal pathway. J Biol Chem. 2002;277(17):15107–12. doi: 10.1074/jbc.M111204200. [DOI] [PubMed] [Google Scholar]
- 40.Okamura Y, Watari M, Jerud ES, Young DW, Ishizaka ST, et al. The extra domain A of fibronectin activates Toll-like receptor 4. J Biol Chem. 2001;276(13):10229–33. doi: 10.1074/jbc.M100099200. [DOI] [PubMed] [Google Scholar]
- 41.Termeer C, Benedix F, Sleeman J, Fieber C, Voith U, et al. Oligosaccharides of hyaluronan activate dendritic cells via Toll-like receptor 4. J Exp Med. 2002;195(1):99–111. doi: 10.1084/jem.20001858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Johnson GB, Brunn GJ, Kodaira Y, Platt JL. Receptor-mediated monitoring of tissue well-being via detection of soluble heparan sulfate by Toll-like receptor 4. J Immunol. 2002;168(10):5233–39. doi: 10.4049/jimmunol.168.10.5233. [DOI] [PubMed] [Google Scholar]
- 43.Jiang D, Liang J, Fan J, Yu S, Chen S, et al. Regulation of lung injury and repair by Toll-like receptors and hyaluronan. Nat Med. 2005;11(11):1173–79. doi: 10.1038/nm1315. [DOI] [PubMed] [Google Scholar]
- 44.Schaefer L, Babelova A, Kiss E, Hausser H-J, Baliova M, et al. The matrix component biglycan is proinflammatory and signals through Toll-like receptors 4 and 2 in macrophages. J Clin Investig. 2005;115(8):2223–33. doi: 10.1172/JCI23755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsung A, Sahai R, Tanaka H, Nakao A, Fink MP, et al. The nuclear factor HMGB1 mediates hepatic injury after murine liver ischemia-reperfusion. J Exp Med. 2005;201(7):1135–43. doi: 10.1084/jem.20042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bald T, Quast T, Landsberg J, Rogava M, Glodde N, et al. Ultraviolet-radiation-induced inflammation promotes angiotropism and metastasis in melanoma. Nature. 2014;507(7490):109–13. doi: 10.1038/nature13111. [DOI] [PubMed] [Google Scholar]
- 47.Biragyn A, Ruffini PA, Leifer CA, Klyushnenkova E, Shakhov A, et al. Toll-like receptor 4-dependent activation of dendritic cells by β-defensin 2. Science. 2002;298(5595):1025–29. doi: 10.1126/science.1075565. [DOI] [PubMed] [Google Scholar]
- 48.Miller YI, Viriyakosol S, Worrall DS, Boullier A, Butler S, Witztum JL. Toll-like receptor 4-dependent and -independent cytokine secretion induced by minimally oxidized low-density lipoprotein in macrophages. Arterioscler Thromb Vasc Biol. 2005;25(6):1213–19. doi: 10.1161/01.ATV.0000159891.73193.31. [DOI] [PubMed] [Google Scholar]
- 49.Stewart CR, Stuart LM, Wilkinson K, van Gils JM, Deng J, et al. CD36 ligands promote sterile inflammation through assembly of a Toll-like receptor 4 and 6 heterodimer. Nat Immunol. 2010;11(2):155–61. doi: 10.1038/ni.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheedy FJ, Grebe A, Rayner KJ, Kalantari P, Ramkhelawon B, et al. CD36 coordinates NLRP3 inflammasome activation by facilitating intracellular nucleation of soluble ligands into particulate ligands in sterile inflammation. Nat Immunol. 2013;14(8):812–20. doi: 10.1038/ni.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bochkov VN, Kadl A, Huber J, Gruber F, Binder BR, Leitinger N. Protective role of phospholipid oxidation products in endotoxin-induced tissue damage. Nature. 2002;419(6902):77–81. doi: 10.1038/nature01023. [DOI] [PubMed] [Google Scholar]
- 52.Guillot L, Balloy V, McCormack FX, Golenbock DT, Chignard M, Si-Tahar M. Cutting edge: the immunostimulatory activity of the lung surfactant protein-A involves Toll-like receptor 4. J Immunol. 2002;168(12):5989–92. doi: 10.4049/jimmunol.168.12.5989. [DOI] [PubMed] [Google Scholar]
- 53.Vogl T, Tenbrock K, Ludwig S, Leukert N, Ehrhardt C, et al. Mrp8 and Mrp14 are endogenous activators of Toll-like receptor 4, promoting lethal, endotoxin-induced shock. Nat Med. 2007;13(9):1042–49. doi: 10.1038/nm1638. [DOI] [PubMed] [Google Scholar]
- 54.Pal D, Dasgupta S, Kundu R, Maitra S, Das G, et al. Fetuin-A acts as an endogenous ligand of TLR4 to promote lipid-induced insulin resistance. Nat Med. 2012;18(8):1279–85. doi: 10.1038/nm.2851. [DOI] [PubMed] [Google Scholar]
- 55.Qiang X, Yang W-L, Wu R, Zhou M, Jacob A, et al. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat Med. 2013;19(11):1489–95. doi: 10.1038/nm.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schumann RR, Leong SR, Flaggs GW, Gray PW, Wright SD, et al. Structure and function of lipopolysaccharide binding protein. Science. 1990;249(4975):1429–31. doi: 10.1126/science.2402637. [DOI] [PubMed] [Google Scholar]
- 57.Eckert JK, Kim YJ, Kim JI, Gürtler K, Oh D-Y, et al. The crystal structure of lipopolysaccharide binding protein reveals the location of a frequent mutation that impairs innate immunity. Immunity. 2013;39(4):647–60. doi: 10.1016/j.immuni.2013.09.005. [DOI] [PubMed] [Google Scholar]
- 58.da Silva Correia J, Soldau K, Christen U, Tobias PS, Ulevitch RJ. Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex: transfer from CD14 to TLR4 and MD-2. J Biol Chem. 2001;276(24):21129–35. doi: 10.1074/jbc.M009164200. [DOI] [PubMed] [Google Scholar]
- 59.Gioannini TL, Weiss JP. Regulation of interactions of Gram-negative bacterial endotoxins with mammalian cells. Immunol Res. 2007;39(1–3):249–60. doi: 10.1007/s12026-007-0069-0. [DOI] [PubMed] [Google Scholar]
- 60.Nagai Y, Akashi S, Nagafuku M, Ogata M, Iwakura Y, et al. Essential role of MD-2 in LPS responsiveness and TLR4 distribution. Nat Immunol. 2002;3(7):667–72. doi: 10.1038/ni809. [DOI] [PubMed] [Google Scholar]
- 61.Meng J, Gong M, Björkbacka H, Golenbock DT. Genome-wide expression profiling and mutagenesis studies reveal that lipopolysaccharide responsiveness appears to be absolutely dependent on TLR4 and MD-2 expression and is dependent upon intermolecular ionic interactions. J Immunol. 2011;187(7):3683–93. doi: 10.4049/jimmunol.1101397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kim J-I, Lee CJ, Jin MS, Lee C-H, Paik S-G, et al. Crystal structure of CD14 and its implications for lipopolysaccharide signaling. J Biol Chem. 2005;280(12):11347–51. doi: 10.1074/jbc.M414607200. [DOI] [PubMed] [Google Scholar]
- 63.Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol. 2000;1(1):31–39. doi: 10.1038/35036052. [DOI] [PubMed] [Google Scholar]
- 64.Zanoni I, Ostuni R, Capuano G, Collini M, Caccia M, et al. CD14 regulates the dendritic cell life cycle after LPS exposure through NFAT activation. Nature. 2009;460(7252):264–68. doi: 10.1038/nature08118. [DOI] [PubMed] [Google Scholar]
- 65.Zanoni I, Ostuni R, Barresi S, Di Gioia M, Broggi A, et al. CD14 and NFAT mediate lipopolysaccharide-induced skin edema formation in mice. J Clin Investig. 2012;122(5):1747–57. doi: 10.1172/JCI60688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pugin J, Kravchenko VV, Lee JD, Kline L, Ulevitch RJ, Tobias PS. Cell activation mediated by glycosylphosphatidylinositol-anchored or transmembrane forms of CD14. Infect Immun. 1998;66(3):1174–80. doi: 10.1128/iai.66.3.1174-1180.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zanoni I, Ostuni R, Marek LR, Barresi S, Barbalat R, et al. CD14 controls the LPS-induced endocytosis of Toll-like receptor 4. Cell. 2011;147(4):868–80. doi: 10.1016/j.cell.2011.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lin Y-C, Huang D-Y, Chu C-L, Lin Y-L, Lin W-W. The tyrosine kinase Syk differentially regulates Toll-like receptor signaling downstream of the adaptor molecules TRAF6 and TRAF3. Sci Signal. 2013;6(289):ra71. doi: 10.1126/scisignal.2003973. [DOI] [PubMed] [Google Scholar]
- 69.Roy S, Karmakar M, Pearlman E. CD14 mediates Toll-like receptor 4 (TLR4) endocytosis and spleen tyrosine kinase (Syk) and interferon regulatory transcription factor 3 (IRF3) activation in epithelial cells and impairs neutrophil infiltration and Pseudomonas aeruginosa killing in vivo. J Biol Chem. 2014;289(2):1174–82. doi: 10.1074/jbc.M113.523167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chiang C-Y, Veckman V, Limmer K, David M. Phospholipase Cγ-2 and intracellular calcium are required for lipopolysaccharide-induced Toll-like receptor 4 (TLR4) endocytosis and interferon regulatory factor 3 (IRF3) activation. J Biol Chem. 2012;287(6):3704–9. doi: 10.1074/jbc.C111.328559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tanimura N, Saitoh S, Matsumoto F, Akashi-Takamura S, Miyake K. Roles for LPS-dependent interaction and relocation of TLR4 and TRAM in TRIF-signaling. Biochem Biophys Res Commun. 2008;368(1):94–99. doi: 10.1016/j.bbrc.2008.01.061. [DOI] [PubMed] [Google Scholar]
- 72.Jiang Z, Georgel P, Du X, Shamel L, Sovath S, et al. CD14 is required for MyD88-independent LPS signaling. Nat Immunol. 2005;6(6):565–70. doi: 10.1038/ni1207. [DOI] [PubMed] [Google Scholar]
- 73.Ling GS, Bennett J, Woollard KJ, Szajna M, Fossati-Jimack L, et al. Integrin CD11b positively regulates TLR4-induced signalling pathways in dendritic cells but not in macrophages. Nat Commun. 2014;5:3039. doi: 10.1038/ncomms4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Husebye H, Aune MH, Stenvik J, Samstad E, Skjeldal F, et al. The Rab11a GTPase controls Toll-like receptor 4-induced activation of interferon regulatory factor-3 on phagosomes. Immunity. 2010;33(4):583–96. doi: 10.1016/j.immuni.2010.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Acker T, Eyckerman S, Vande Walle L, Gerlo S, Goethals M, et al. The small GTPase Arf6 is essential for the Tram/Trif pathway in TLR4 signaling. J Biol Chem. 2014;289(3):1364–76. doi: 10.1074/jbc.M113.499194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aksoy E, Taboubi S, Torres D, Delbauve S, Hachani A, et al. The p110δ isoform of the kinase PI(3)K controls the subcellular compartmentalization of TLR4 signaling and protects from endotoxic shock. Nat Immunol. 2012;13(11):1045–54. doi: 10.1038/ni.2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Horng T, Barton GM, Medzhitov R. TIRAP: an adapter molecule in the Toll signaling pathway. Nat Immunol. 2001;2(9):835–41. doi: 10.1038/ni0901-835. [DOI] [PubMed] [Google Scholar]
- 78.Fitzgerald KA, Palsson-McDermott EM, Bowie AG, Jefferies CA, Mansell AS, et al. Mal (MyD88-adapter-like) is required for Toll-like receptor-4 signal transduction. Nature. 2001;413(6851):78–83. doi: 10.1038/35092578. [DOI] [PubMed] [Google Scholar]
- 79.Horng T, Barton GM, Flavell RA, Medzhitov R. The adaptor molecule TIRAP provides signalling specificity for Toll-like receptors. Nature. 2002;420(6913):329–33. doi: 10.1038/nature01180. [DOI] [PubMed] [Google Scholar]
- 80.Yamamoto M, Sato S, Hemmi H, Sanjo H, Uematsu S, et al. Essential role for TIRAP in activation of the signalling cascade shared by TLR2 and TLR4. Nature. 2002;420(6913):324–29. doi: 10.1038/nature01182. [DOI] [PubMed] [Google Scholar]
- 81.Bonham KS, Orzalli MH, Hayashi K, Wolf AI, Glanemann C, et al. A promiscuous lipid-binding protein diversifies the subcellular sites of Toll-like receptor signal transduction. Cell. 2014;156(4):705–16. doi: 10.1016/j.cell.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67(1):481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 83.Motshwene PG, Moncrieffe MC, Grossmann JG, Kao C, Ayaluru M, et al. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J Biol Chem. 2009;284(37):25404–11. doi: 10.1074/jbc.M109.022392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin S-C, Lo Y-C, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465(7300):885–90. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Suzuki N, Suzuki S, Duncan GS, Millar DG, Wada T, et al. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature. 2002;416(6882):750–56. doi: 10.1038/nature736. [DOI] [PubMed] [Google Scholar]
- 86.Kawagoe T, Sato S, Matsushita K, Kato H, Matsui K, et al. Sequential control of Toll-like receptor-dependent responses by IRAK1 and IRAK2. Nat Immunol. 2008;9(6):684–91. doi: 10.1038/ni.1606. [DOI] [PubMed] [Google Scholar]
- 87.Cao Z, Xiong J, Takeuchi M, Kurama T, Goeddel DV. TRAF6 is a signal transducer for interleukin-1. Nature. 1996;383(6599):443–46. doi: 10.1038/383443a0. [DOI] [PubMed] [Google Scholar]
- 88.Muzio M, Ni J, Feng P, Dixit VM. IRAK (Pelle) family member IRAK-2 and MyD88 as proximal mediators of IL-1 signaling. Science. 1997;278(5343):1612–15. doi: 10.1126/science.278.5343.1612. [DOI] [PubMed] [Google Scholar]
- 89.Deng L, Wang C, Spencer E, Yang L, Braun A, et al. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell. 2000;103(2):351–61. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 90.Emmerich CH, Ordureau A, Strickson S, Arthur JSC, Pedrioli PGA, et al. Activation of the canonical IKK complex by K63/M1-linked hybrid ubiquitin chains. PNAS. 2013;110(38):15247–52. doi: 10.1073/pnas.1314715110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature. 2001;412(6844):346–51. doi: 10.1038/35085597. [DOI] [PubMed] [Google Scholar]
- 92.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell. 2009;136(6):1098–109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 93.Laplantine E, Fontan E, Chiaravalli J, Lopez T, Lakisic G, et al. NEMO specifically recognizes K63-linked poly-ubiquitin chains through a new bipartite ubiquitin-binding domain. EMBO J. 2009;28(19):2885–95. doi: 10.1038/emboj.2009.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xia Z-P, Sun L, Chen X, Pineda G, Jiang X, et al. Direct activation of protein kinases by unanchored polyubiquitin chains. Nature. 2009;461(7260):114–19. doi: 10.1038/nature08247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tseng P-H, Matsuzawa A, Zhang W, Mino T, Vignali DAA, Karin M. Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nat Immunol. 2010;11(1):70–75. doi: 10.1038/ni.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ozinsky A, Underhill DM, Fontenot JD, Hajjar AM, Smith KD, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between Toll-like receptors. PNAS. 2000;97(25):13766–71. doi: 10.1073/pnas.250476497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoebe K, Georgel P, Rutschmann S, Du X, Mudd S, et al. CD36 is a sensor of diacylglycerides. Nature. 2005;433(7025):523–27. doi: 10.1038/nature03253. [DOI] [PubMed] [Google Scholar]
- 98.Manukyan M, Triantafilou K, Triantafilou M, Mackie A, Nilsen N, et al. Binding of lipopeptide to CD14 induces physical proximity of CD14, TLR2 and TLR1. Eur J Immunol. 2005;35(3):911–21. doi: 10.1002/eji.200425336. [DOI] [PubMed] [Google Scholar]
- 99.Kang JY, Nan X, Jin MS, Youn S-J, Ryu YH, et al. Recognition of lipopeptide patterns by Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31(6):873–84. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 100.Buwitt-Beckmann U, Heine H, Wiesmüller K-H, Jung G, Brock R, et al. TLR1- and TLR6- independent recognition of bacterial lipopeptides. J Biol Chem. 2006;281(14):9049–57. doi: 10.1074/jbc.M512525200. [DOI] [PubMed] [Google Scholar]
- 101.Boehme KW, Guerrero M, Compton T. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J Immunol. 2006;177(10):7094–102. doi: 10.4049/jimmunol.177.10.7094. [DOI] [PubMed] [Google Scholar]
- 102.Szomolanyi-Tsuda E, Liang X, Welsh RM, Kurt-Jones EA, Finberg RW. Role for TLR2 in NK cell-mediated control of murine cytomegalovirus in vivo. J Virol. 2006;80(9):4286–91. doi: 10.1128/JVI.80.9.4286-4291.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kurt-Jones EA, Chan M, Zhou S, Wang J, Reed G, et al. Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis. PNAS. 2004;101(5):1315–20. doi: 10.1073/pnas.0308057100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chang S, Dolganiuc A, Szabo G. Toll-like receptors 1 and 6 are involved in TLR2-mediated macrophage activation by hepatitis C virus core and NS3 proteins. J Leukoc Biol. 2007;82(3):479–87. doi: 10.1189/jlb.0207128. [DOI] [PubMed] [Google Scholar]
- 105.Erridge C. Endogenous ligands of TLR2 and TLR4: agonists or assistants? J Leukoc Biol. 2010;87(6):989–99. doi: 10.1189/jlb.1209775. [DOI] [PubMed] [Google Scholar]
- 106.Kenny EF, Talbot S, Gong M, Golenbock DT, Bryant CE, O’Neill LAJ. MyD88 adaptor-like is not essential for TLR2 signaling and inhibits signaling by TLR3. J Immunol. 2009;183(6):3642–51. doi: 10.4049/jimmunol.0901140. [DOI] [PubMed] [Google Scholar]
- 107.Couture LA, Piao W, Ru LW, Vogel SN, Toshchakov VY. Targeting Toll-like receptor (TLR) signaling by Toll/interleukin-1 receptor (TIR) domain-containing adapter protein/MyD88 adapter-like (TIRAP/Mal)-derived decoy peptides. J Biol Chem. 2012;287(29):24641–48. doi: 10.1074/jbc.M112.360925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Santos-Sierra S, Deshmukh SD, Kalnitski J, Küenzi P, Wymann MP, et al. Mal connects TLR2 to PI3Kinase activation and phagocyte polarization. EMBO J. 2009;28(14):2018–27. doi: 10.1038/emboj.2009.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Triantafilou M, Morath S, Mackie A, Hartung T, Triantafilou K. Lateral diffusion of Toll-like receptors reveals that they are transiently confined within lipid rafts on the plasma membrane. J Cell Sci. 2004;117(Pt. 17):4007–14. doi: 10.1242/jcs.01270. [DOI] [PubMed] [Google Scholar]
- 110.Triantafilou M, Gamper FGJ, Haston RM, Mouratis MA, Morath S, et al. Membrane sorting of Toll-like receptor (TLR)-2/6 and TLR2/1 heterodimers at the cell surface determines heterotypic associations with CD36 and intracellular targeting. J Biol Chem. 2006;281(41):31002–11. doi: 10.1074/jbc.M602794200. [DOI] [PubMed] [Google Scholar]
- 111.Heit B, Kim H, Cosío G, Castaño D, Collins R, et al. Multimolecular signaling complexes enable Syk-mediated signaling of CD36 internalization. Dev Cell. 2013;24(4):372–83. doi: 10.1016/j.devcel.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. 2009;10(11):1200–7. doi: 10.1038/ni.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dietrich N, Lienenklaus S, Weiss S, Gekara NO. Murine Toll-like receptor 2 activation induces type I interferon responses from endolysosomal compartments. PLOS ONE. 2010;5(4):e10250. doi: 10.1371/journal.pone.0010250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410(6832):1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 115.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4(12):1247–53. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 116.Shibata T, Takemura N, Motoi Y, Goto Y, Karuppuchamy T, et al. PRAT4A-dependent expression of cell surface TLR5 on neutrophils, classical monocytes and dendritic cells. Int Immunol. 2012;24(10):613–23. doi: 10.1093/intimm/dxs068. [DOI] [PubMed] [Google Scholar]
- 117.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167(4):1882–85. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 118.Yoon S-I, Kurnasov O, Natarajan V, Hong M, Gudkov AV, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335(6070):859–64. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Huh J-W, Shibata T, Hwang M, Kwon E-H, Jang MS, et al. UNC93B1 is essential for the plasma membrane localization and signaling of Toll-like receptor 5. PNAS. 2014;111(19):7072–77. doi: 10.1073/pnas.1322838111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Choi YJ, Jung J, Chung HK, Im E, Rhee SH. PTEN regulates TLR5-induced intestinal inflammation by controlling Mal/TIRAP recruitment. FASEB J. 2013;27(1):243–54. doi: 10.1096/fj.12-217596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Letran SE, Lee S-J, Atif SM, Uematsu S, Akira S, McSorley SJ. TLR5 functions as an endocytic receptor to enhance flagellin-specific adaptive immunity. Eur J Immunol. 2011;41(1):29–38. doi: 10.1002/eji.201040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Rogers NC, Slack EC, Edwards AD, Nolte MA, Schulz O, et al. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22(4):507–17. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 123.Taylor PR, Tsoni SV, Willment JA, Dennehy KM, Rosas M, et al. Dectin-1 is required for β-glucan recognition and control of fungal infection. Nat Immunol. 2007;8(1):31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ferwerda B, Ferwerda G, Plantinga TS, Willment JA, van Spriel AB, et al. Human dectin-1 deficiency and mucocutaneous fungal infections. N Engl J Med. 2009;361(18):1760–67. doi: 10.1056/NEJMoa0901053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Rochereau N, Drocourt D, Perouzel E, Pavot V, Redelinghuys P, et al. Dectin-1 is essential for reverse transcytosis of glycosylated SIgA-antigen complexes by intestinal M cells. PLOS Biol. 2013;11(9):e1001658. doi: 10.1371/journal.pbio.1001658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shan M, Gentile M, Yeiser JR, Walland AC, Bornstein VU, et al. Mucus enhances gut homeostasis and oral tolerance by delivering immunoregulatory signals. Science. 2013;342(6157):447–53. doi: 10.1126/science.1237910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hoving JC, Wilson GJ, Brown GD. Signalling C-type lectin receptors, microbial recognition and immunity. Cell Microbiol. 2014;16(2):185–94. doi: 10.1111/cmi.12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu S, Huo J, Gunawan M, Su I-H, Lam K-P. Activated dectin-1 localizes to lipid raft microdomains for signaling and activation of phagocytosis and cytokine production in dendritic cells. J Biol Chem. 2009;284(33):22005–11. doi: 10.1074/jbc.M109.009076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fuller GLJ, Williams JAE, Tomlinson MG, Eble JA, Hanna SL, et al. The C-type lectin receptors CLEC-2 and Dectin-1, but not DC-SIGN, signal via a novel YXXL-dependent signaling cascade. J Biol Chem. 2007;282(17):12397–409. doi: 10.1074/jbc.M609558200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tassi I, Cella M, Castro I, Gilfillan S, Khan WN, Colonna M. Requirement of phospholipase C-γ2 (PLCγ2) for Dectin-1-induced antigen presentation and induction of TH1/TH17 polarization. Eur J Immunol. 2009;39(5):1369–78. doi: 10.1002/eji.200839313. [DOI] [PubMed] [Google Scholar]
- 131.Goodridge HS, Simmons RM, Underhill DM. Dectin-1 stimulation by Candida albicans yeast or zymosan triggers NFAT activation in macrophages and dendritic cells. J Immunol. 2007;178(5):3107–15. doi: 10.4049/jimmunol.178.5.3107. [DOI] [PubMed] [Google Scholar]
- 132.Strasser D, Neumann K, Bergmann H, Marakalala MJ, Guler R, et al. Syk kinase-coupled C-type lectin receptors engage protein kinase C-σ to elicit Card9 adaptor-mediated innate immunity. Immunity. 2012;36(1):32–42. doi: 10.1016/j.immuni.2011.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gorjestani S, Darnay BG, Lin X. Tumor necrosis factor receptor-associated factor 6 (TRAF6) and TGFβ-activated kinase 1 (TAK1) play essential roles in the C-type lectin receptor signaling in response to Candida albicans infection. J Biol Chem. 2012;287(53):44143–50. doi: 10.1074/jbc.M112.414276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gringhuis SI, Wevers BA, Kaptein TM, van Capel TMM, Theelen B, et al. Selective C-Rel activation via Malt1 controls anti-fungal TH-17 immunity by dectin-1 and dectin-2. PLOS Pathog. 2011;7(1):e1001259. doi: 10.1371/journal.ppat.1001259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gringhuis SI, den Dunnen J, Litjens M, van der Vlist M, Wevers B, et al. Dectin-1 directs T helper cell differentiation by controlling noncanonical NF-κB activation through Raf-1 and Syk. Nat Immunol. 2009;10(2):203–13. doi: 10.1038/ni.1692. [DOI] [PubMed] [Google Scholar]
- 136.Goodridge HS, Reyes CN, Becker CA, Katsumoto TR, Ma J, et al. Activation of the innate immune receptor Dectin-1 upon formation of a ‘phagocytic synapse. ’ Nature. 2011;472(7344):471–75. doi: 10.1038/nature10071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Herre J, Marshall ASJ, Caron E, Edwards AD, Williams DL, et al. Dectin-1 uses novel mechanisms for yeast phagocytosis in macrophages. Blood. 2004;104(13):4038–45. doi: 10.1182/blood-2004-03-1140. [DOI] [PubMed] [Google Scholar]
- 138.Botelho RJ, Teruel M, Dierckman R, Anderson R, Wells A, et al. Localized biphasic changes in phosphatidylinositol-4,5-bisphosphate at sites of phagocytosis. J Cell Biol. 2000;151(7):1353–68. doi: 10.1083/jcb.151.7.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Underhill DM, Ozinsky A. Phagocytosis of microbes: complexity in action. Annu Rev Immunol. 2002;20(1):825–52. doi: 10.1146/annurev.immunol.20.103001.114744. [DOI] [PubMed] [Google Scholar]
- 140.Underhill DM, Rossnagle E, Lowell CA, Simmons RM. Dectin-1 activates Syk tyrosine kinase in a dynamic subset of macrophages for reactive oxygen production. Blood. 2005;106(7):2543–50. doi: 10.1182/blood-2005-03-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ma J, Becker C, Lowell CA, Underhill DM. Dectin-1-triggered recruitment of light chain 3 protein to phagosomes facilitates major histocompatibility complex class II presentation of fungal-derived antigens. J Biol Chem. 2012;287(41):34149–56. doi: 10.1074/jbc.M112.382812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mansour MK, Tam JM, Khan NS, Seward M, Davids PJ, et al. Dectin-1 activation controls maturation of β-1,3-glucan-containing phagosomes. J Biol Chem. 2013;288(22):16043–54. doi: 10.1074/jbc.M113.473223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Ma J, Becker C, Reyes C, Underhill DM. Cutting edge: FYCO1 recruitment to dectin-1 phagosomes is accelerated by light chain 3 protein and regulates phagosome maturation and reactive oxygen production. J Immunol. 2014;192(4):1356–60. doi: 10.4049/jimmunol.1302835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.LeibundGut-Landmann S, Gross O, Robinson MJ, Osorio F, Slack EC, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8(6):630–38. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 145.Gringhuis SI, Kaptein TM, Wevers BA, Theelen B, van der Vlist M, et al. Dectin-1 is an extracellular pathogen sensor for the induction and processing of IL-1β via a noncanonical caspase-8 inflammasome. Nat Immunol. 2012;13(3):246–54. doi: 10.1038/ni.2222. [DOI] [PubMed] [Google Scholar]
- 146.Saijo S, Ikeda S, Yamabe K, Kakuta S, Ishigame H, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32(5):681–91. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 147.Zhu L-L, Zhao X-Q, Jiang C, You Y, Chen X-P, et al. C-type lectin receptors Dectin-3 and Dectin-2 form a heterodimeric pattern-recognition receptor for host defense against fungal infection. Immunity. 2013;39(2):324–34. doi: 10.1016/j.immuni.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 148.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, et al. Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science. 2003;301(5633):640–43. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 149.Yamamoto M, Sato S, Hemmi H, Uematsu S, Hoshino K, et al. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4(11):1144–50. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 150.Rowe DC, McGettrick AF, Latz E, Monks BG, Gay NJ, et al. The myristoylation of TRIF-related adaptor molecule is essential for Toll-like receptor 4 signal transduction. PNAS. 2006;103(16):6299–304. doi: 10.1073/pnas.0510041103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Enokizono Y, Kumeta H, Funami K, Horiuchi M, Sarmiento J, et al. Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling. PNAS. 2013;110(49):19908–13. doi: 10.1073/pnas.1222811110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Piao W, Ru LW, Piepenbrink KH, Sundberg EJ, Vogel SN, Toshchakov VY. Recruitment of TLR adapter TRIF to TLR4 signaling complex is mediated by the second helical region of TRIF TIR domain. PNAS. 2013;110(47):19036–41. doi: 10.1073/pnas.1313575110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Häcker H, Redecke V, Blagoev B, Kratchmarova I, Hsu L-C, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439(7073):204–7. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 154.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, et al. IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4(5):491–96. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 155.Hemmi H, Takeuchi O, Sato S, Yamamoto M, Kaisho T, et al. The roles of two IκB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199(12):1641–50. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. PNAS. 2004;101(1):233–38. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Thomas KE, Galligan CL, Newman RD, Fish EN, Vogel SN. Contribution of interferon-β to the murine macrophage response to the Toll-like receptor 4 agonist, lipopolysaccharide. J Biol Chem. 2006;281(41):31119–30. doi: 10.1074/jbc.M604958200. [DOI] [PubMed] [Google Scholar]
- 158.Zanoni I, Spreafico R, Bodio C, Di Gioia M, Cigni C, et al. IL-15 cis presentation is required for optimal NK cell activation in lipopolysaccharide-mediated inflammatory conditions. Cell Rep. 2013;4(6):1235–49. doi: 10.1016/j.celrep.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 159.Fitzgerald KA, Rowe DC, Barnes BJ, Caffrey DR, Visintin A, et al. LPS-TLR4 signaling to IRF-3/7 and NF-κB involves the Toll adapters TRAM and TRIF. J Exp Med. 2003;198(7):1043–55. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Brown V, Brown RA, Ozinsky A, Hesselberth JR, Fields S. Binding specificity of Toll-like receptor cytoplasmic domains. Eur J Immunol. 2006;36(3):742–53. doi: 10.1002/eji.200535158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Ulrichts P, Peelman F, Beyaert R, Tavernier J. MAPPIT analysis of TLR adaptor complexes. FEBS Lett. 2007;581(4):629–36. doi: 10.1016/j.febslet.2007.01.026. [DOI] [PubMed] [Google Scholar]
- 162.Verstak B, Arnot CJ, Gay NJ. An alanine-to-proline mutation in the BB-loop of TLR3 Toll/IL-1R domain switches signalling adaptor specificity from TRIF to MyD88. J Immunol. 2013;191(12):6101–9. doi: 10.4049/jimmunol.1300849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kerkmann M, Rothenfusser S, Hornung V, Towarowski A, Wagner M, et al. Activation with CpG-A and CpG-B oligonucleotides reveals two distinct regulatory pathways of type I IFN synthesis in human plasmacytoid dendritic cells. J Immunol. 2003;170(9):4465–74. doi: 10.4049/jimmunol.170.9.4465. [DOI] [PubMed] [Google Scholar]
- 164.Cao W, Manicassamy S, Tang H, Kasturi SP, Pirani A, et al. Toll-like receptor-mediated induction of type I interferon in plasmacytoid dendritic cells requires the rapamycin-sensitive PI(3)K-mTOR-p70S6K pathway. Nat Immunol. 2008;9(10):1157–64. doi: 10.1038/ni.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Honda K, Yanai H, Negishi H, Asagiri M, Sato M, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–77. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 166.Sasai M, Linehan MM, Iwasaki A. Bifurcation of Toll-like receptor 9 signaling by adaptor protein 3. Science. 2010;329(5998):1530–34. doi: 10.1126/science.1187029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Uematsu S, Sato S, Yamamoto M, Hirotani T, Kato H, et al. Interleukin-1 receptor-associated kinase-1 plays an essential role for Toll-like receptor (TLR)7- and TLR9-mediated interferon-α induction. J Exp Med. 2005;201(6):915–23. doi: 10.1084/jem.20042372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wan Y, Kim TW, Yu M, Zhou H, Yamashita M, et al. The dual functions of IL-1 receptor-associated kinase 2 in TLR9-mediated IFN and proinflammatory cytokine production. J Immunol. 2011;186(5):3006–14. doi: 10.4049/jimmunol.1003217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Honda K, Ohba Y, Yanai H, Negishi H, Mizutani T, et al. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434(7036):1035–40. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 170.Hemmi H, Takeuchi O, Kawai T, Kaisho T, Sato S, et al. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408(6813):740–45. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- 171.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature. 2001;413(6857):732–38. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 172.Ewald SE, Lee BL, Lau L, Wickliffe KE, Shi G-P, et al. The ectodomain of Toll-like receptor 9 is cleaved to generate a functional receptor. Nature. 2008;456(7222):658–62. doi: 10.1038/nature07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Park B, Brinkmann MM, Spooner E, Lee CC, Kim Y-M, Ploegh HL. Proteolytic cleavage in an endolysosomal compartment is required for activation of Toll-like receptor 9. Nat Immunol. 2008;9(12):1407–14. doi: 10.1038/ni.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ewald SE, Engel A, Lee J, Wang M, Bogyo M, Barton GM. Nucleic acid recognition by Toll-like receptors is coupled to stepwise processing by cathepsins and asparagine endopeptidase. J Exp Med. 2011;208(4):643–51. doi: 10.1084/jem.20100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sepulveda FE, Maschalidi S, Colisson R, Heslop L, Ghirelli C, et al. Critical role for asparagine endopeptidase in endocytic Toll-like receptor signaling in dendritic cells. Immunity. 2009;31(5):737–48. doi: 10.1016/j.immuni.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 176.Brinkmann MM, Spooner E, Hoebe K, Beutler B, Ploegh HL, Kim Y-M. The interaction between the ER membrane protein UNC93B and TLR3, 7, and 9 is crucial for TLR signaling. J Cell Biol. 2007;177(2):265–75. doi: 10.1083/jcb.200612056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Kim Y-M, Brinkmann MM, Paquet M-E, Ploegh HL. UNC93B1 delivers nucleotide-sensing Toll-like receptors to endolysosomes. Nature. 2008;452(7184):234–38. doi: 10.1038/nature06726. [DOI] [PubMed] [Google Scholar]
- 178.Lee BL, Moon JE, Shu JH, Yuan L, Newman ZR, et al. UNC93B1 mediates differential trafficking of endosomal TLRs. Elife. 2013;2(0):e00291–91. doi: 10.7554/eLife.00291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Saleh M. The machinery of Nod-like receptors: refining the paths to immunity and cell death. Immunol Rev. 2011;243(1):235–46. doi: 10.1111/j.1600-065X.2011.01045.x. [DOI] [PubMed] [Google Scholar]
- 180.Girardin SE, Boneca IG, Carneiro LAM, Antignac A, Jéhanno M, et al. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300(5625):1584–87. doi: 10.1126/science.1084677. [DOI] [PubMed] [Google Scholar]
- 181.Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4(7):702–7. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 182.Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278(11):8869–72. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 183.Inohara N, Koseki T, Lin J, del Peso L, Lucas PC, et al. An induced proximity model for NF-κB activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem. 2000;275(36):27823–31. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 184.Park J-H, Kim Y-G, McDonald C, Kanneganti T-D, Hasegawa M, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178(4):2380–86. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- 185.Barnich N, Aguirre JE, Reinecker H-C, Xavier R, Podolsky DK. Membrane recruitment of NOD2 in intestinal epithelial cells is essential for nuclear factor-κB activation in muramyl dipeptide recognition. J Cell Biol. 2005;170(1):21–26. doi: 10.1083/jcb.200502153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Irving AT, Mimuro H, Kufer TA, Lo C, Wheeler R, et al. The immune receptor NOD1 and kinase RIP2 interact with bacterial peptidoglycan on early endosomes to promote autophagy and inflammatory signaling. Cell Host Microbe. 2014;15(5):623–35. doi: 10.1016/j.chom.2014.04.001. [DOI] [PubMed] [Google Scholar]
- 187.Nakamura N, Lill JR, Phung Q, Jiang Z, Bakalarski C, et al. Endosomes are specialized platforms for bacterial sensing and NOD2 signalling. Nature. 2014;509(7499):240–44. doi: 10.1038/nature13133. [DOI] [PubMed] [Google Scholar]
- 188.Travassos LH, Carneiro LAM, Ramjeet M, Hussey S, Kim Y-G, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11(1):55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 189.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–37. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 190.Yoneyama M, Kikuchi M, Matsumoto K, Imaizumi T, Miyagishi M, et al. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J Immunol. 2005;175(5):2851–58. doi: 10.4049/jimmunol.175.5.2851. [DOI] [PubMed] [Google Scholar]
- 191.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, et al. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314(5801):994–97. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 192.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, et al. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 193.Kato H, Takeuchi O, Mikamo-Satoh E, Hirai R, Kawai T, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–10. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, et al. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol. 2009;83(20):10761–69. doi: 10.1128/JVI.00770-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Saito T, Owen DM, Jiang F, Marcotrigiano J, Gale M. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature. 2008;454(7203):523–27. doi: 10.1038/nature07106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Uzri D, Gehrke L. Nucleotide sequences and modifications that determine RIG-I/RNA binding and signaling activities. J Virol. 2009;83(9):4174–84. doi: 10.1128/JVI.02449-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chiu Y-H, Macmillan JB, Chen ZJ. RNA polymerase III detects cytosolic DNA and induces type I interferons through the RIG-I pathway. Cell. 2009;138(3):576–91. doi: 10.1016/j.cell.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, et al. IPS-1, an adaptor triggering RIG-I-and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–88. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- 199.Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, et al. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437(7062):1167–72. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- 200.Xu L-G, Wang Y-Y, Han K-J, Li L-Y, Zhai Z, Shu H-B. VISA is an adapter protein required for virus-triggered IFN-β signaling. Mol Cell. 2005;19(6):727–40. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 201.Gack MU, Shin YC, Joo C-H, Urano T, Liang C, et al. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446(7138):916–20. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 202.Oshiumi H, Matsumoto M, Hatakeyama S, Seya T. Riplet/RNF135, a RING finger protein, ubiquitinates RIG-I to promote interferon-β induction during the early phase of viral infection. J Biol Chem. 2009;284(2):807–17. doi: 10.1074/jbc.M804259200. [DOI] [PubMed] [Google Scholar]
- 203.Oshiumi H, Miyashita M, Inoue N, Okabe M, Matsumoto M, Seya T. The ubiquitin ligase Riplet is essential for RIG-I-dependent innate immune responses to RNA virus infection. Cell Host Microbe. 2010;8(6):496–509. doi: 10.1016/j.chom.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 204.Hou F, Sun L, Zheng H, Skaug B, Jiang Q-X, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146(3):448–61. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Liu S, Chen J, Cai X, Wu J, Chen X, et al. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife. 2013;2(0):e00785. doi: 10.7554/eLife.00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Liu HM, Loo Y-M, Horner SM, Zornetzer GA, Katze MG, Gale M. The mitochondrial targeting chaperone 14-3-3ε regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11(5):528–37. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, et al. Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol. 2014;5:717–26. doi: 10.1038/ni.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Rowland AA, Voeltz GK. Endoplasmic reticulum-mitochondria contacts: function of the junction. Nat Rev Mol Cell Biol. 2012;13(10):607–25. doi: 10.1038/nrm3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Castanier C, Garcin D, Vazquez A, Arnoult D. Mitochondrial dynamics regulate the RIG-I-like receptor antiviral pathway. EMBO Rep. 2010;11(2):133–38. doi: 10.1038/embor.2009.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Onoguchi K, Onomoto K, Takamatsu S, Jogi M, Takemura A, et al. Virus-infection or 5′ppp-RNA activates antiviral signal through redistribution of IPS-1 mediated by MFN1. PLOS Pathog. 2010;6(7):e1001012. doi: 10.1371/journal.ppat.1001012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Koshiba T, Yasukawa K, Yanagi Y, Kawabata S-I. Mitochondrial membrane potential is required for MAVS-mediated antiviral signaling. Sci Signal. 2011;4(158):ra7. doi: 10.1126/scisignal.2001147. [DOI] [PubMed] [Google Scholar]
- 212.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280(28):26185–92. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 213.Yasukawa K, Oshiumi H, Takeda M, Ishihara N, Yanagi Y, et al. Mitofusin 2 inhibits mitochondrial antiviral signaling. Sci Signal. 2009;2(84):ra47. doi: 10.1126/scisignal.2000287. [DOI] [PubMed] [Google Scholar]
- 214.Sacksteder KA, Jones JM, South ST, Li X, Liu Y, Gould SJ. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol. 2000;148(5):931–44. doi: 10.1083/jcb.148.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Xia M, Gonzalez P, Li C, Meng G, Jiang A, et al. Mitophagy enhances oncolytic measles virus replication by mitigating DDX58/RIG-I-like receptor signaling. J Virol. 2014;88(9):5152–64. doi: 10.1128/JVI.03851-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. PNAS. 2007;104(35):14050–55. doi: 10.1073/pnas.0704014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Zhao Y, Sun X, Nie X, Sun L, Tang T-S, et al. COX5B regulates MAVS-mediated antiviral signaling through interaction with ATG5 and repressing ROS production. PLOS Pathog. 2012;8(12):e1003086. doi: 10.1371/journal.ppat.1003086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lei Y, Wen H, Yu Y, Taxman DJ, Zhang L, et al. The mitochondrial proteins NLRX1 and TUFM form a complex that regulates type I interferon and autophagy. Immunity. 2012;36(6):933–46. doi: 10.1016/j.immuni.2012.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Holm GH, Zurney J, Tumilasci V, Leveille S, Danthi P, et al. Retinoic acid-inducible gene-I and interferon-β promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J Biol Chem. 2007;282(30):21953–61. doi: 10.1074/jbc.M702112200. [DOI] [PubMed] [Google Scholar]
- 220.Huang Y, Liu H, Li S, Tang Y, Wei B, et al. MAVS-MKK7-JNK2 defines a novel apoptotic signaling pathway during viral infection. PLOS Pathog. 2014;10(3):e1004020. doi: 10.1371/journal.ppat.1004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Brubaker SW, Gauthier AE, Mills EW, Ingolia NT, Kagan JC. A bicistronic MAVS transcript highlights a class of truncated variants in antiviral immunity. Cell. 2014;156(4):800–811. doi: 10.1016/j.cell.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Yu C-Y, Chiang R-L, Chang T-H, Liao C-L, Lin Y-L. The interferon stimulator mitochondrial antiviral signaling protein facilitates cell death by disrupting the mitochondrial membrane potential and by activating caspases. J Virol. 2010;84(5):2421–31. doi: 10.1128/JVI.02174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Michallet M-C, Meylan E, Ermolaeva MA, Vazquez J, Rebsamen M, et al. TRADD protein is an essential component of the RIG-like helicase antiviral pathway. Immunity. 2008;28(5):651–61. doi: 10.1016/j.immuni.2008.03.013. [DOI] [PubMed] [Google Scholar]
- 224.Maadidi El S, Faletti L, Berg B, Wenzl C, Wieland K, et al. A novel mitochondrial MAVS/Caspase-8 platform links RNA virus-induced innate antiviral signaling to Bax/Bak-independent apoptosis. J Immunol. 2014;192(3):1171–83. doi: 10.4049/jimmunol.1300842. [DOI] [PubMed] [Google Scholar]
- 225.Guan K, Zheng Z, Song T, He X, Xu C, et al. MAVS regulates apoptotic cell death by decreasing K48-linked ubiquitination of voltage-dependent anion channel 1. Mol Cell Biol. 2013;33(16):3137–49. doi: 10.1128/MCB.00030-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Unterholzner L. The interferon response to intracellular DNA: why so many receptors? Immunobiology. 2013;218(11):1312–21. doi: 10.1016/j.imbio.2013.07.007. [DOI] [PubMed] [Google Scholar]
- 227.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455(7213):674–78. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Zhong B, Yang Y, Li S, Wang Y-Y, Li Y, et al. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity. 2008;29(4):538–50. doi: 10.1016/j.immuni.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 229.Sun W, Li Y, Chen L, Chen H, You F, et al. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. PNAS. 2009;106(21):8653–58. doi: 10.1073/pnas.0900850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ishikawa H, Ma Z, Barber GN. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461(7265):788–92. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Unterholzner L, Keating SE, Baran M, Horan KA, Jensen SB, et al. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11(11):997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhang Z, Yuan B, Bao M, Lu N, Kim T, Liu Y-J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011;12(10):959–65. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, et al. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478(7370):515–18. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Yin Q, Tian Y, Kabaleeswaran V, Jiang X, Tu D, et al. Cyclic di-GMP sensing via the innate immune signaling protein STING. Mol Cell. 2012;46(6):735–45. doi: 10.1016/j.molcel.2012.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ouyang S, Song X, Wang Y, Ru H, Shaw N, et al. Structural analysis of the STING adaptor protein reveals a hydrophobic dimer interface and mode of cyclic di-GMP binding. Immunity. 2012;36(6):1073–86. doi: 10.1016/j.immuni.2012.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Huang Y-H, Liu X-Y, Du X-X, Jiang Z-F, Su X-D. The structural basis for the sensing and binding of cyclic di-GMP by STING. Nat Struct Mol Biol. 2012;19(7):728–30. doi: 10.1038/nsmb.2333. [DOI] [PubMed] [Google Scholar]
- 237.Shang G, Zhu D, Li N, Zhang J, Zhu C, et al. Crystal structures of STING protein reveal basis for recognition of cyclic di-GMP. Nat Struct Mol Biol. 2012;19(7):725–27. doi: 10.1038/nsmb.2332. [DOI] [PubMed] [Google Scholar]
- 238.Shu C, Yi G, Watts T, Kao CC, Li P. Structure of STING bound to cyclic di-GMP reveals the mechanism of cyclic dinucleotide recognition by the immune system. Nat Struct Mol Biol. 2012;19(7):722–24. doi: 10.1038/nsmb.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science. 2013;339(6121):786–91. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Wu J, Sun L, Chen X, Du F, Shi H, et al. Cyclic GMP-AMP is an endogenous second messenger in innate immune signaling by cytosolic DNA. Science. 2013;339(6121):826–30. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Saitoh T, Fujita N, Hayashi T, Takahara K, Satoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. PNAS. 2009;106(49):20842–46. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155(3):688–98. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Watson RO, Manzanillo PS, Cox JS. Extracellular M. tuberculosis DNA targets bacteria for autophagy by activating the host DNA-sensing pathway. Cell. 2012;150(4):803–15. doi: 10.1016/j.cell.2012.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Fernandes-Alnemri T, Yu J-W, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458(7237):509–13. doi: 10.1038/nature07710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458(7237):514–18. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, et al. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10(3):266–72. doi: 10.1038/ni.1702. [DOI] [PubMed] [Google Scholar]
- 247.Kerur N, Veettil MV, Sharma-Walia N, Bottero V, Sadagopan S, et al. IFI16 acts as a nuclear pathogen sensor to induce the inflammasome in response to Kaposi sarcoma-associated herpesvirus infection. Cell Host Microbe. 2011;9(5):363–75. doi: 10.1016/j.chom.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.von Moltke J, Ayres JS, Kofoed EM, Chavarría-Smith J, Vance RE. Recognition of bacteria by inflammasomes. Annu Rev Immunol. 2013;31(1):73–106. doi: 10.1146/annurev-immunol-032712-095944. [DOI] [PubMed] [Google Scholar]
- 249.Eder C. Mechanisms of interleukin-1β release. Immunobiology. 2009;214(7):543–53. doi: 10.1016/j.imbio.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 250.Rubartelli A, Cozzolino F, Talio M, Sitia R. A novel secretory pathway for interleukin-1β, a protein lacking a signal sequence. EMBO J. 1990;9(5):1503–10. doi: 10.1002/j.1460-2075.1990.tb08268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Andrei C, Dazzi C, Lotti L, Torrisi MR, Chimini G, Rubartelli A. The secretory route of the leaderless protein interleukin 1β involves exocytosis of endolysosome-related vesicles. Mol Biol Cell. 1999;10(5):1463–75. doi: 10.1091/mbc.10.5.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.MacKenzie A, Wilson HL, Kiss-Toth E, Dower SK, North RA, Surprenant A. Rapid secretion of interleukin-1β by microvesicle shedding. Immunity. 2001;15(5):825–35. doi: 10.1016/s1074-7613(01)00229-1. [DOI] [PubMed] [Google Scholar]
- 253.Qu Y, Franchi L, Núñez G, Dubyak GR. Nonclassical IL-1β secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J Immunol. 2007;179(3):1913–25. doi: 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]
- 254.Hogquist KA, Unanue ER, Chaplin DD. Release of IL-1 from mononuclear phagocytes. J Immunol. 1991;147(7):2181–86. [PubMed] [Google Scholar]
- 255.Fink SL, Cookson BT. Caspase-1-dependent pore formation during pyroptosis leads to osmotic lysis of infected host macrophages. Cell Microbiol. 2006;8(11):1812–25. doi: 10.1111/j.1462-5822.2006.00751.x. [DOI] [PubMed] [Google Scholar]
- 256.Gariano GR, Dell’Oste V, Bronzini M, Gatti D, Luganini A, et al. The intracellular DNA sensor IFI16 gene acts as restriction factor for human cytomegalovirus replication. PLOS Pathog. 2012;8(1):e1002498. doi: 10.1371/journal.ppat.1002498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Jakobsen MR, Bak RO, Andersen A, Berg RK, Jensen SB, et al. IFI16 senses DNA forms of the lentiviral replication cycle and controls HIV-1 replication. PNAS. 2013;110(48):E4571–80. doi: 10.1073/pnas.1311669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Monroe KM, Yang Z, Johnson JR, Geng X, Doitsh G, et al. IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science. 2014;343(6169):428–32. doi: 10.1126/science.1243640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Veeranki S, Choubey D. Interferon-inducible p200-family protein IFI16, an innate immune sensor for cytosolic and nuclear double-stranded DNA: regulation of subcellular localization. Mol Immunol. 2012;49(4):567–71. doi: 10.1016/j.molimm.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Li T, Diner BA, Chen J, Cristea IM. Acetylation modulates cellular distribution and DNA sensing ability of interferon-inducible protein IFI16. PNAS. 2012;109(26):10558–63. doi: 10.1073/pnas.1203447109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. PNAS. 2012;109(44):E3008–17. doi: 10.1073/pnas.1211302109. [DOI] [PMC free article] [PubMed] [Google Scholar]