Abstract
Aims
Genetics plays an important role in coronary heart disease (CHD) but the clinical utility of genomic risk scores (GRSs) relative to clinical risk scores, such as the Framingham Risk Score (FRS), is unclear. Our aim was to construct and externally validate a CHD GRS, in terms of lifetime CHD risk and relative to traditional clinical risk scores.
Methods and results
We generated a GRS of 49 310 SNPs based on a CARDIoGRAMplusC4D Consortium meta-analysis of CHD, then independently tested it using five prospective population cohorts (three FINRISK cohorts, combined n = 12 676, 757 incident CHD events; two Framingham Heart Study cohorts (FHS), combined n = 3406, 587 incident CHD events). The GRS was associated with incident CHD (FINRISK HR = 1.74, 95% confidence interval (CI) 1.61–1.86 per S.D. of GRS; Framingham HR = 1.28, 95% CI 1.18–1.38), and was largely unchanged by adjustment for known risk factors, including family history. Integration of the GRS with the FRS or ACC/AHA13 scores improved the 10 years risk prediction (meta-analysis C-index: +1.5–1.6%, P < 0.001), particularly for individuals ≥60 years old (meta-analysis C-index: +4.6–5.1%, P < 0.001). Importantly, the GRS captured substantially different trajectories of absolute risk, with men in the top 20% of attaining 10% cumulative CHD risk 12–18 y earlier than those in the bottom 20%. High genomic risk was partially compensated for by low systolic blood pressure, low cholesterol level, and non-smoking.
Conclusions
A GRS based on a large number of SNPs improves CHD risk prediction and encodes different trajectories of lifetime risk not captured by traditional clinical risk scores.
Keywords: Genomic risk score, Coronary heart disease, Myocardial infarction, Framingham risk score, Primary prevention
Introduction
Early and accurate identification of individuals with increased risk of coronary heart disease (CHD) is critical for effective implementation of preventative lifestyle modifications and medical interventions, such as statin treatment.1,2 To this end, risk scores such as the Framingham Risk Score (FRS)3 and the American College of Cardiology/American Heart Association 2013 risk score (ACC/AHA13),1 based on clinical factors and lipid measurements, have been developed and are widely used. Although the scores can identify individuals at very high risk, a large proportion of individuals developing CHD during the next 10 years remain unidentified. In particular, they do not provide sufficient discrimination at a younger age when implementation of preventative measures is likely to provide the greatest long-term benefit.
Genetic factors have long been recognized to make a substantial contribution to CHD risk.4 Although a positive family history is an independent risk factor for CHD, it may not completely and solely capture genetic risk. Recently, genome-wide association studies (GWAS) have identified 56 genetic loci associated with CHD at genome-wide significance.5–9 Studies of the predictive power of the top single nucleotide polymorphisms (SNPs) at some of these loci either individually or in combination have typically shown small improvements in CHD risk prediction,10–17 probably because together these variants only explain less than 20% of CHD heritability.8 As demonstrated recently for other traits such as height and BMI,18,19 the majority of unexplained heritability is likely hidden amongst the thousands of SNPs that did not reach genome-wide significance. Indeed, recent advances have shown that genomic prediction models that consider all available genetic variants can more efficiently stratify those at increased risk of complex disease.20–24 To leverage the maximum amount of information, we examined whether a genomic risk score (GRS) comprising a large number of SNPs, including those with less than genome-wide significance, could produce clinically relevant predictive power for CHD risk.
Methods
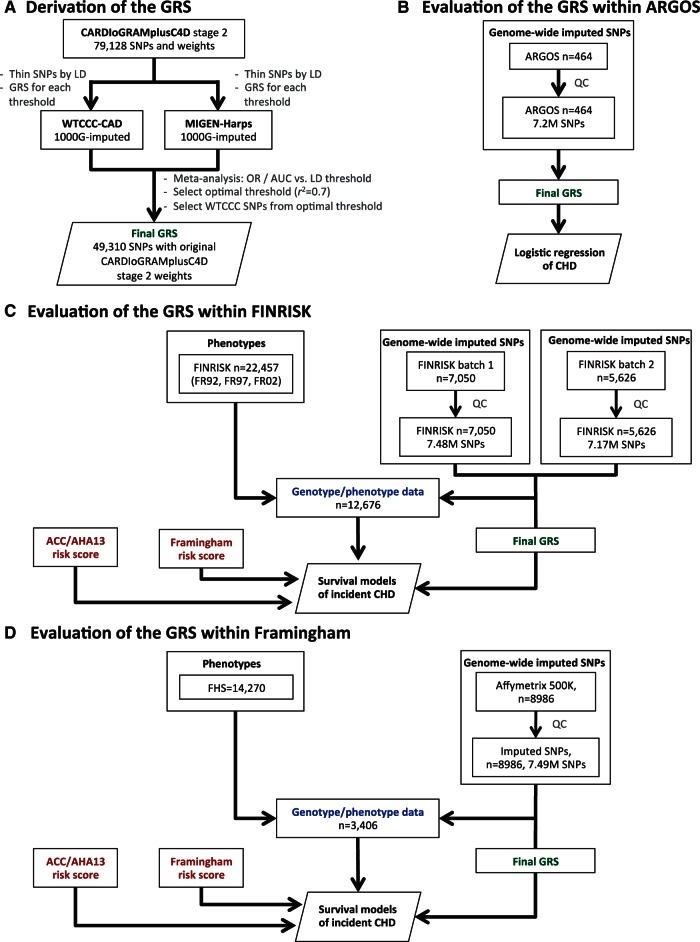
A summary of the key methods for the study is given here. The study design is given in Figure 1. Additional details are provided in the see Supplementary Data.
Figure 1.
Study workflow. (A) The procedure for deriving the GRS of incident CHD. The analysis workflow for evaluating the GRS within (B) ARGOS, (C) FINRISK, and (D) FHS.
Prospective study cohorts
We utilized two sets of prospective cohorts: (i) FINRISK, consisting of three prospective cohorts from Finland with 10–20 years of follow-up, from collections 1992, 1997, and 2002 (FR92, FR97, and FR02, respectively)25 and (ii) the Framingham Heart Study (FHS),26–28 with individuals of Western and Southern European ancestry taken from the Original and Offspring cohorts with 40–48 years of follow-up. In total, the FINRISK consisted of n = 12 676 individuals and the FHS of n = 3406 individuals, all of whom had the requisite data and were independent of the CARDIoGRAMplusC4D stage-2 meta-analysis utilized to generate the GRS (Table 1). The cohorts have been genome-wide SNP genotyped and further imputed to the 1000 Genomes reference panel (see Supplementary Data online, Supplementary Data). After genotype imputation and quality control, 69 044 autosomal SNPs of the 79 128 CARDIoGRAMplusC4D SNPs were available for subsequent analyses in the FINRISK, and 78 058 autosomal SNPs available in FHS.
Table 1.
Characteristics of the FINRISK and FHS cohorts
| Study | FINRISK |
Framingham Heart
Study |
|||||
|---|---|---|---|---|---|---|---|
| Cohort | FR92 (n=3547) | FR97 (n=4761) | FR02 (n=4368) | Total FINRISK (n=12,676) | FHS Original (n=950) | FHS Offspring (n=2456) | Total FHS (n=3406) |
| Men | 1578 (44%) | 2316 (49%) | 1919 (44%) | 5813 (46%) | 370 (39%) | 1179 (48%) | 1549 (45%) |
| Women | 1969 (56%) | 2445 (51%) | 2449 (56%) | 6863 (54%) | 580 (61%) | 1277 (52%) | 1857 (55%) |
| Baseline age, years | 43.59 (11.31) | 46.68 (13.15) | 47.12 (13.01) | 45.97 (12.7) | 53.7 (6.09) | 40.66 (7.47) | 44.3 (9.21) |
| Current smoker | 1027 (29%) | 1148 (24%) | 1162 (27%) | 3337 (26%) | 422 (44%) | 948 (39%) | 1370 (40%) |
| Blood pressure, systolic, mm Hg | 134.79 (19.13) | 135.02 (19.62) | 134.94 (20.24) | 134.93 (19.7) | 131.54 (19.35) | 122.64 (15.98) | 125.12 (17.45) |
| Cholesterol, total, mmol/L | 5.6 (1.12) | 5.54 (1.06) | 5.62 (1.14) | 5.58 (1.11) | 6.14 (1.08) | 5.21 (0.98) | 5.47 (1.09) |
| Cholesterol, HDL, mmol/L | 1.41 (0.35) | 1.42 (0.35) | 1.52 (0.43) | 1.45 (0.38) | 1.3 (0.37) | 1.33 (0.39) | 1.32 (0.39) |
| Prevalent type 2 diabetes | 119 (3%) | 299 (6%) | 278 (6%) | 696 (5%) | 19 (2%) | 39 (2%) | 58 (2%) |
| Lipid lowering treatment | 43 (1%) | 117 (2%) | 231 (5%) | 391 (3%) | – | – | – |
| Anti-hypertensive treatment | 302 (9%) | 569 (12%) | 582 (13%) | 1453 (11%) | 57 (6%) | 75 (3%) | 132 (4%) |
| Follow up, years | 18.49 (3.77) | 13.82 (2.88) | 9.47 (1.51) | 13.63 (4.53) | 29.91 (11.32) | 31.95 (8.44) | 31.38 (9.38) |
| Incident CHD event (before age 75) | 261 (7%) | 324 (7%) | 172 (4%) | 757 (6%) | 173 (18%) | 414 (17%) | 587 (17%) |
Categorical variables are shown as counts and percentages, continuous variables (age, follow-up time, cholesterol, and blood pressure) as means and standard deviations. Sample sizes are for participants with GWAS data after quality control and all other exclusions. Lipid lowering treatments were not assessed in FHS due to an insufficient number of exams with this information.
The outcome of interest in FINRISK was primary incident CHD event, defined as myocardial infarction (MI), a coronary revascularization procedure, or death from CHD, before age 75 years (see Supplementary Data online, Supplementary Methods). Individuals with prevalent cardiovascular disease (CVD) at baseline were excluded from the analysis. We censored events for individuals with an attained age of >75 years, as not all FINRISK cohorts had sufficient numbers of CHD events beyond that age. In FHS, we used the FHS definition of CHD, which included recognized/unrecognized MI or death from CHD as well as angina pectoris or coronary insufficiency (see Supplementary Data online, Supplementary Methods). FHS individuals with prevalent CHD or <30 years of age at baseline were excluded, and for consistency with the FINRISK analysis, a censoring age of 75 years was also applied to the FHS analyses.
Secondary external validation of the GRS was also performed in the ARGOS study, a Dutch case/control dataset where all individuals had familial hypercholesterolemia (248 young cases with early CHD, 216 elderly controls without CHD), imputed to 1000 Genomes reference panel (74 135 SNPs of the 79 128 CARDIoGRAMplusC4D SNPs were available; see Supplementary Data online, Supplementary Methods).
Statistical analysis
GRSs were generated via thinning the CARDIoGRAMplusC4D SNPs by linkage disequilibrium (LD) thresholds and evaluated using logistic regression and area under receiver-operating characteristic curve (AUC) for each threshold (see Supplementary Data online, Figure S1). To avoid overfitting we only used weights (log odds) from the CARDIoGRAMplusC4D stage-2 meta-analysis, which were not based on the WTCCC-CAD or MIGen studies (see Supplementary Data online, Supplementary Methods). We combined the estimates for WTCCC and MIGen-Harps using fixed-effects inverse-variance weighted meta-analysis.
Subsequent performance of the GRS was evaluated in external, independent validation data. For analysis of FINRISK, we used Cox proportional hazard models to evaluate the association of the GRS with time to incident CHD events, stratifying by sex and adjusting for geographic location and cohort, using age as the time scale. Secondary analyses adjusted for one of the clinical risk scores (FRS or ACC/AHA13), or individual baseline variables and known risk factors (cohort, geographical location, prevalent type-2 diabetes, log total cholesterol, log HDL, log systolic BP, smoking status, lipid treatment, and family history). Family history in FINRISK was self-reported and was defined as having a 1st-degree relative who had experienced MI before age 60. For FHS, we evaluated the association of the GRS with incident CHD using Cox proportional hazard models, stratifying by sex and adjusting for cohort (Original or Offspring), using age as the time scale. Family history was not available for both FHS cohorts and thus not considered in FHS analyses. Survival analyses allowing for competing risks were performed using the Aalen-Johansen estimator of survival and cause-specific Cox models (see Supplementary Data online, Supplementary Methods). Model discrimination of incident CHD event was evaluated in three groups of individuals: (i) all individuals (n = 12 676 in FINRISK, n = 3406 in FHS), (ii) individuals aged <60 years at baseline (n = 10 606 in FINRISK, n = 3218 in FHS), and (iii) individuals aged ≥60 years at baseline (n = 2070 in FINRISK, n = 188 in FHS).
Discrimination of incident CHD events within 10 years was assessed using Harrell’s C-index, and the difference in C-index between two models was assessed using the correlated jackknife test. Competing risk analyses were performed using the Aalen-Johansen empirical estimator of cumulative incidence and cause-specific Cox proportional hazard models. Risk reclassification was evaluated using continuous Net Reclassification Improvement (NRI), categorical NRI, and Integrated Discrimination Improvement. Meta-analysis of the discrimination statistics was performed using fixed-effect inverse-variance weighting. Additional details on the statistical methods are provided in the see Supplementary Data online, Supplementary Methods.
Results
To construct an optimized GRS using the WTCCC and MIGen-Harps datasets, we first generated a series of GRSs, starting with the 79 128 CARDIoGRAMplusC4D SNPs then progressively lowering the r2 threshold for LD to reduce the redundancy of predictive information and corresponding number of SNPs in the score (Methods and Figure 1). An r2 threshold of 0.7 provided optimal discrimination of CHD cases and controls (WTCCC and MIGen-Harps meta-analysis odds ratio(OR) = 1.70 per S.D. of GRS, 95% confidence interval (CI 1.61–1.80; meta-analysis AUC = 0.64, 95% CI 0.63–0.66), corresponding to 49 310 SNPs in WTCCC (see Supplementary Data online, Figure S1). Of these 49 310 SNPs, 85.9% (42 364 SNPs) and 95% (46 773 SNPs) were available in the FINRISK and FHS, respectively.
The 49K GRS showed similar odds ratios for incident CHD as a binary outcome in FINRISK (OR = 1.74, 95% CI 1.61–1.89, per S.D.), WTCCC (OR = 1.74, 95% CI 1.63–1.86, per S.D.), and MIGen-Harps (OR = 1.57, 95% CI 1.37–1.81, per S.D.) (Table 2). However in the FHS, the association was weaker, OR = 1.30 (95% CI 1.19–1.43, per S.D.) (Table 2). Density plots of the GRS in FINRISK and FHS for those with and without CHD <75 years are shown in see Supplementary Data online, Figure S2.
Table 2.
Association of the 49K GRS with incident CHD (binary outcome in logistic regression) in the five studies, per standard deviation of the GRS
| Dataset | # Incident CHD/Non-CHD | Odds Ratio (95% CI) |
|---|---|---|
| WTCCC-CAD1 | 1926/2938 | 1.74 (1.63–1.86) |
| MIGen-Harps | 488/531 | 1.57 (1.37–1.81) |
| ARGOS FH | 248/216 | 1.49 (1.21–1.84) |
| FINRISK | 757/11919 | 1.74 (1.61–1.89) |
| FHS | 587/2819 | 1.28 (1.17–1.41) |
WTCCC-CAD1: adjusted for sex and 5 PCs of the genotypes; MIGen-Harps: adjusted for sex and 5 PCs; ARGOS: adjusted for sex and 5 PCs; FINRISK: adjusted for sex, cohort, east/west, and 5 PCs; FHS: adjusted for sex, cohort, and 5 PCs.
Using survival analyses of time to incident CHD, within FINRISK the GRS had stronger association with CHD (HR = 1.74, 95% CI 1.61–1.86, per S.D.) than the 28 SNP score studied by Tikkanen et al.11 (HR = 1.21, 95% CI 1.13–1.30, per S.D.), the 27 SNP score used by Mega et al.29 (HR = 1.21, 95% CI 1.12–1.30 per S.D.), or the 153 SNPs found at FDR <0.05 by the CARDIoGRAMplusC4D consortium8 (HR = 1.25, 95% CI 1.16–1.39 per S.D.) (see Supplementary Data online, Supplementary Results). In FHS, the GRS showed weaker but statistically significant association with CHD (HR = 1.28 per S.D. of the GRS, 95% CI 1.18–1.38). The fixed-effect meta-analysis estimate for the GRS combining FINRISK and FHS was HR = 1.66 (95% CI 1.55–1.78), however, heterogeneity was high (I2 = 89.2%, Cochran’s Q P = 0.0023). The top vs. bottom quintiles of the GRS showed significantly different incident CHD risk overall (FINRISK HR = 4.51, 95% CI 3.47–5.85; FHS HR = 1.84 95% CI 1.43–2.37). For both FINRISK and FHS, the GRS showed improved prediction for incident CHD over the other risk scores composed of smaller numbers of SNPs (see Supplementary Data online, Supplementary Results and Table S3).
In both FINRISK and FHS, the hazard ratios for GRS were not substantially attenuated by adjusting for FRS or ACC/AHA13 clinical risk scores, lipid treatment at baseline, other established risk factors (including family history in FINRISK), or 5 principal components of the genotypes (see Supplementary Data online, Figures S3 and S4). The correlation between GRS and either FRS or ACC/AHA13 scores was close to zero with almost none of the variation in GRS explained by either clinical risk score (in both FINRISK and FHS, r2 < 0.004 between GRS and either FRS and ACC/AHA13; see Supplementary Data online, Figure S5). To further test that the CHD risk conferred by the GRS was largely independent of the effects of cholesterol, we further validated the GRS in the ARGOS familial hypercholesterolemia study, with comparable results to those obtained in WTCCC/MIGen (OR = 1.49, 95% CI 1.21–1.84 per S.D. of the GRS, adjusted for sex and five principal components) (see Supplementary Data online, Supplementary Methods).
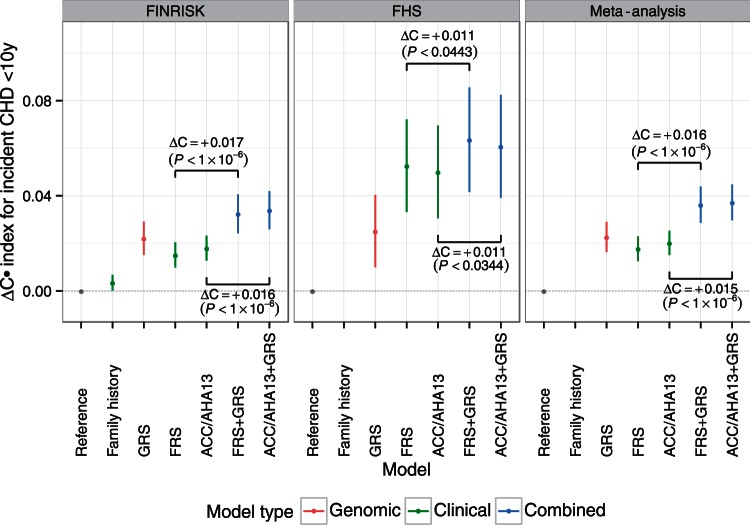
To assess the predictive power of the GRS, we compared its performance in discrimination of time to CHD event (C-index) with that of family history and the FRS and ACC/AHA13 clinical risk scores. We also assessed the incremental value of the GRS on top of the clinical risk scores. In both FINRISK and FHS, addition of GRS to either FRS or ACC/AHA13 scores provided statistically significant improvements in C-index, in FINRISK: +1.7% (P < 10 − 6) and +1.6% (P < 10 − 6) for FRS and ACC/AHA13, respectively; in FHS: +1.1% (P < 0.0443) and +1.1% (P < 0.0344) for FRS and ACC/AHA13, respectively (Figure 2). Overall, fixed-effects meta-analysis of the two studies showed that GRS improved the C-index by +1.6% (95% CI 0.01–0.02, P < 10 − 6; heterogeneity: I2 = 2.2%, Q = 1.02, P = 0.312) for FRS and GRS combined (FRS + GRS) over FRS alone and, similarly, +1.5% (95% CI 0.009–0.02, P < 10 − 6; heterogeneity: I2 = 0%, Q = 0.78, P = 0.378) for ACC/AHA13 + GRS over ACC/AHA13 alone (Figure 2). Larger increases in C-index were observed among older individuals, with the C-index of FRS + GRS compared with FRS alone increasing by 5.1% in individuals aged ≥60 years at baseline, while individuals aged <60 years at baseline showed C-index gains of 1.4% (see Supplementary Data online, Figure S6). Within FINRISK, the GRS had higher C-index than family history (+1.9%, P < 1.3 × 10 − 6).
Figure 2.
Difference in C-index (95% CI) for time to incident CHD event within 10 years, relative to the reference model in the FINRISK and FHS cohorts. Reference models used age as the time scale, stratified by sex (FINRISK: adjusted for cohort and geographic location; FHS: adjusted for cohort). Family history was not available for all of the FHS cohorts and thus not considered here. P-values are from the correlated jackknife test.
We assessed if the GRS improved the individual 10 years risk reclassification when added to clinical risk scores. Analyses within FINRISK and FHS are given in Table 3 for FRS and in Table 4 for ACC/AHA13. Overall, meta-analysis of the two datasets showed that the categorical Net Reclassification Improvement was 0.1 for both FRS + GRS and ACC/AHA13 + GRS, respectively (P < 0.0001; see Supplementary material online, Figure S7). Meta-analysis of continuous NRI was 0.344 (P < 0.001) and 0.334 (P < 0.001) for the FRS + GRS and ACC/AHA13 + GRS, respectively (see Supplementary Data online, Figure S8). Meta-analysis of IDI scores showed gains of 0.01 (P < 0.001) and 0.009 (P < 0.001) for FRS + GRS and ACC/AHA13 + GRS, respectively, however IDI scores showed high heterogeneity across FINRISK and FHS (I2 > 97%, Cochran’s Q P < 0.0001, see Supplementary Data online, Figure S9).
Table 3.
Reclassification of incident CHD event risk within 10 years for combined FRS + GRS compared with FRS only, in the FINRISK and FHS cohorts
| FINRISK |
FHS |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FRS+GRS |
FRS+GRS |
||||||||||||
| 0–7.5% | 7.5–10% | 10–20% | 20–100% | Total | Reclass % | 0–7.5% | 7.5–10% | 10–20% | 20–100% | Total | Reclass % | ||
| All
individuals | |||||||||||||
| FRS | 0–7.5% | 9566 | 218 | 138 | 6 | 9928 | 3.6 | 2482 | 88 | 4 | 0 | 2574 | 3.6 |
| 7.5–10% | 368 | 190 | 223 | 21 | 802 | 76.3 | 122 | 165 | 83 | 1 | 371 | 55.5 | |
| 10–20% | 299 | 290 | 767 | 298 | 1,654 | 53.6 | 11 | 74 | 339 | 19 | 443 | 23.5 | |
| 20–100% | 1 | 14 | 114 | 156 | 285 | 15.7 | 0 | 0 | 5 | 13 | 18 | 27.8 | |
| Total | 10,234 | 712 | 1,242 | 481 | 12669 | 15.7 | 2,615 | 327 | 431 | 33 | 3,406 | 11.9 | |
| Incident CHD present | |||||||||||||
| FRS | 0–7.5% | 110 | 21 | 19 | 2 | 152 | 27.6 | 67 | 6 | 0 | 0 | 73 | 8.2 |
| 7.5–10% | 22 | 12 | 28 | 4 | 66 | 81.8 | 5 | 11 | 6 | 0 | 22 | 50.0 | |
| 10–20% | 22 | 24 | 108 | 78 | 232 | 53.4 | 2 | 5 | 43 | 4 | 54 | 20.4 | |
| 20–100% | 0 | 2 | 17 | 48 | 67 | 28.4 | 0 | 0 | 0 | 1 | 1 | 0 | |
| Total | 154 | 59 | 172 | 132 | 517 | 46.2 | 74 | 22 | 49 | 5 | 150 | 18.7 | |
| Incident CHD absent | |||||||||||||
| FRS | 0–7.5% | 9456 | 197 | 119 | 4 | 9776 | 3.3 | 2415 | 82 | 4 | 0 | 2501 | 3.4 |
| 7.5–10% | 346 | 178 | 195 | 17 | 736 | 75.8 | 117 | 154 | 77 | 1 | 349 | 55.9 | |
| 10–20% | 277 | 266 | 659 | 220 | 1422 | 53.7 | 9 | 69 | 296 | 15 | 389 | 23.9 | |
| 20–100% | 1 | 12 | 97 | 108 | 218 | 50.5 | 0 | 0 | 5 | 12 | 17 | 29.4 | |
| Total | 10 080 | 653 | 1070 | 349 | 12 152 | 14.4 | 2541 | 305 | 382 | 28 | 3256 | 11.6 | |
|
All individuals | |||||||||||||
| FINRISK | FHS | ||||||||||||
| FRS+GRS | FRS+GRS | ||||||||||||
| NRI (categorical) [95% CI] |
|
|
|||||||||||
| NRI (continuous) [95% CI] |
|
|
|||||||||||
| IDI (continuous) [95% CI] | 0.028 [0.026–0.034]; P < 1 × 10−6 | 0.005 [0.002–0.008]; P < 0.00098 | |||||||||||
In FINRISK, 7 individuals of the 12 676 were excluded in this analysis due to missing clinical measurements.
Table 4.
Reclassification of incident CHD event risk within 10 years for combined ACC/AHA13 + GRS compared with ACC/AHA13 only, in the FINRISK and FHS cohorts
| FINRISK |
FHS |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ACC/AHA13+GRS |
ACC/AHA13+GRS |
||||||||||||
| 0–7.5% | 7.5–10% | 10–20% | 20–100% | Total | Reclass % | 0–7.5% | 7.5–10% | 10–20% | 20–100% | Total | Reclass % | ||
| All
individuals | |||||||||||||
| ACC/AHA13 | 0–7.5% | 9,588 | 211 | 144 | 7 | 9,950 | 3.6 | 2,513 | 78 | 7 | 0 | 2,598 | 3.3 |
| 7.5–10% | 381 | 176 | 199 | 14 | 770 | 77.1 | 112 | 159 | 66 | 1 | 338 | 53.0 | |
| 10–20% | 279 | 275 | 755 | 271 | 1,580 | 52.2 | 7 | 67 | 308 | 32 | 414 | 25.6 | |
| 20–100% | 2 | 10 | 127 | 230 | 369 | 37.7 | 0 | 0 | 16 | 40 | 56 | 28.6 | |
| Total | 10,250 | 672 | 1,225 | 522 | 12,699 | 15.2 | 2,632 | 304 | 397 | 73 | 3,406 | 11.3 | |
|
Incident CHD present | |||||||||||||
| ACC/AHA13 | 0–7.5% | 118 | 16 | 17 | 1 | 152 | 22.4 | 67 | 8 | 0 | 75 | 75 | 10.7 |
| 7.5–10% | 20 | 14 | 29 | 6 | 69 | 79.7 | 6 | 6 | 11 | 23 | 23 | 73.9 | |
| 10–20% | 15 | 29 | 104 | 60 | 208 | 50.0 | 1 | 6 | 34 | 46 | 46 | 26.1 | |
| 20–100% | 0 | 0 | 15 | 73 | 88 | 17.0 | 0 | 0 | 2 | 6 | 6 | 33.3 | |
| Total | 153 | 59 | 165 | 140 | 517 | 40.2 | 74 | 20 | 47 | 150 | 150 | 26.0 | |
|
Incident CHD absent | |||||||||||||
| ACC/AHA13 | 0–7.5% | 9,470 | 195 | 127 | 6 | 9,798 | 3.3 | 2,446 | 70 | 7 | 0 | 2,523 | 3.1 |
| 7.5–10% | 361 | 162 | 170 | 8 | 701 | 76.9 | 106 | 153 | 55 | 1 | 315 | 51.4 | |
| 10–20% | 264 | 246 | 651 | 211 | 1,372 | 62.6 | 6 | 61 | 274 | 27 | 368 | 25.5 | |
| 20–100% | 2 | 10 | 112 | 157 | 281 | 44.1 | 0 | 0 | 14 | 36 | 50 | 28.0 | |
| Total | 10,097 | 613 | 1,060 | 382 | 12,152 | 14.1 | 2,558 | 284 | 350 | 64 | 3,256 | 10.7 | |
|
All individuals | |||||||||||||
| FINRISK | FHS | ||||||||||||
| ACC/AHA13+GRS | ACC/AHA13+GRS | ||||||||||||
| NRI (categorical) [95% CI] |
|
|
|||||||||||
| NRI (continuous) [95% CI] |
|
|
|||||||||||
| IDI (continuous) [95% CI] | 0.028 [0.021–0.034]; P < 1 × 10−6 | 0.005 [0.002–0.008]; P = 0.00184 | |||||||||||
In FINRISK, 7 individuals of the 12,676 were excluded in this analysis due to missing clinical measurements.
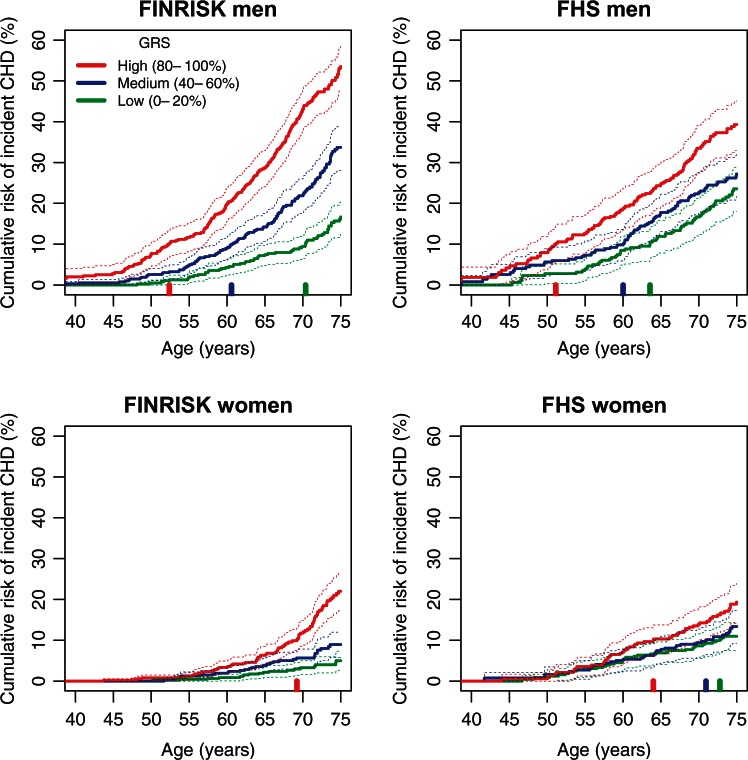
We next examined how variation in genomic risk translated into differences in cumulative lifetime risk of CHD, using Kaplan-Meier estimates stratified by GRS quintiles for men and women separately (Figure 3). As expected, cumulative risk increased with age for both sexes, with men displaying higher absolute risk than women. In both sexes there were substantial differences in cumulative risk between GRS groups with 1.7-fold (in FHS) to 3.2-fold (in FINRISK) higher cumulative risk by age 75 in those in the top quintile of GRS vs. bottom quintile. When considering clinically relevant levels of risk, FINRISK men in the top quintile of genomic risk achieved 10% cumulative risk 18 years earlier than those in the bottom quintile (ages 52 and 70, respectively), with a comparable difference of 12 years in FHS (ages 51 and 64). Women in the top quintile of genomic risk achieved 10% cumulative risk by age 69 (FINRISK) and 64 (FHS), whereas women in the bottom quintile did not achieve 10% risk by age 75 in FINRISK, or by age 73 in FHS. Estimated lifetime CHD risk in FINRISK showed no evidence of being affected by competing risks (incident CHD vs. non-CHD death) (see Supplementary Data online, Supplementary Methods and Supplementary Figure S10). Similarly, a cause-specific competing-risk Cox analysis of the GRS in FINRISK, adjusting for geographical location and cohort, resulted in a similar hazard ratio as standard Cox analysis (HR = 1.70, 95% CI 1.61–1.86).
Figure 3.
Kaplan-Meier cumulative risk of incident CHD event by genomic risk group for men and women in the FINRISK and FHS cohorts. Showing the cumulative risk in quintiles 0–20%, 40–60%, 80–100%. The vertical bars along the x-axis indicate the age at which each risk group attains a cumulative CHD risk of 10%. Dashed lines indicate 95% CI.
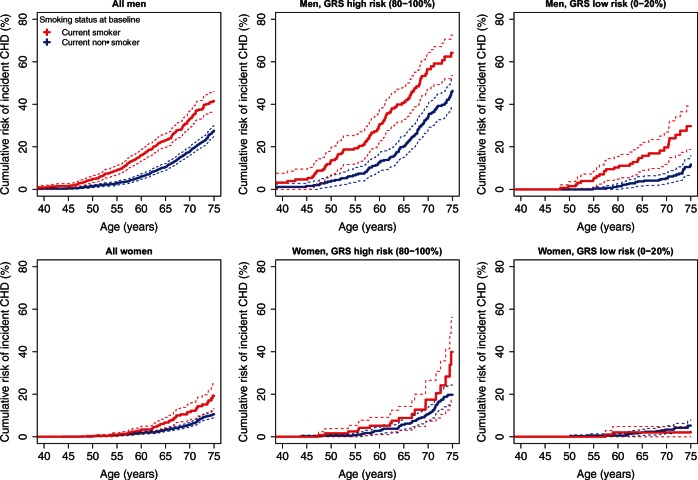
We next sought to investigate to what degree high genomic risk for CHD could be compensated for by low levels of clinical risk factors at baseline, and vice-versa. When considering baseline smoking status in both FINRISK and FHS, Kaplan-Meier analysis showed a substantial increase in cumulative risk of CHD in men who smoked and were also in the top quintile of genomic risk, relative to either non-smokers or smokers at low genomic risk (Figure 4 for FINRISK and see Supplementary Data online, Figure S11 for FHS). Similar but weaker trends were observed for women in the top vs. bottom quintiles of genomic risk. To test whether there was evidence for smoking affecting CHD hazard differently based on an individual’s genomic background, we used a Cox model allowing for an interaction term between the GRS and smoking; the interaction was not statistically significant in FINRISK (P = 0.91) and FHS (P = 0.49).
Figure 4.
Kaplan-Meier curves for incident CHD event risk stratified by GRS quintiles and smoking status at baseline, for men and women in the FINRISK cohorts.
We also examined the potential compensatory effects of baseline systolic blood pressure and total cholesterol, divided as tertiles of high, medium, and low levels (see Supplementary Data online, Figures S12 and S13). For both systolic blood pressure and total cholesterol, we observed the expected trends in CHD risk for high, medium and low levels. However, males with high vs. low levels of systolic blood pressure or total cholesterol showed greater absolute CHD risk if they were in the top vs. bottom quintiles of genomic risk.
Notably, in both FINRISK and FHS, women in the bottom quintile of genomic risk showed smaller differences in cumulative CHD risk when stratified by smoking. For tertiles of systolic blood pressure or total cholesterol, low genomic risk women in FINRISK showed similarly small differences in risk, but the effects in FHS for this subgroup were not consistent. Cox models allowing for interactions between the GRS and systolic blood pressure or total cholesterol did not show statistically significant interactions in either FINRISK or FHS (P > 0.2 for all).
Discussion
We have generated a GRS for CHD based on 49 310 SNPs and, using three prospective FINRISK and two FHS prospective cohorts, demonstrated that the GRS is associated with incident CHD events independently of established and widely-used clinical risk scores or individual CHD risk factors, including family history. Secondary validation in a familial hypercholesterolemia study (ARGOS) showed that GRS was also associated with CHD in this group of high-risk individuals. Subsequently, combining the GRS with established risk scores improved 10-year CHD risk prediction in FINRISK and FHS. We have also shown that the GRS can be leveraged to achieve meaningful lifetime CHD risk stratification, and that the impact of traditional CHD risk factors such as smoking, blood pressure, and cholesterol, vary substantially depending on the underlying genetic risk, thus offering the potential for both earlier and more targeted preventative efforts.
A distinctive feature of our analysis compared with several previous prospective studies11,29,30 examining the predictive utility of GRS for incident CHD is that the best predictive model was achieved here with SNPs that did not necessarily reach genome-wide or even statistical significance in previous GWA studies. The GRS outperformed other smaller SNP models, and shows greater promise in CHD prediction between top and bottom GRS quintiles than a recently published study testing a genetic risk score of 50 SNPs in Scandinavians30 (GRS50 HR = 1.92 vs. GRS49K HR = 4.51). Genome-wide SNP models have been applied successfully to other heritable human traits which seem to follow an “infinitesimal” genetic architecture, such as height.18 These results highlight the differing goals of GWAS and of genomic prediction: the stringent detection of causal genetic variants involved in the disease process vs. the construction of a model that robustly and maximally predicts future disease. While stringent procedures for minimizing the false positive rate of associated loci in GWAS are appropriate, these concerns are less relevant in construction of GRSs, especially when there are a large number of weakly correlated SNPs20 and when rigorous internal and external validation is performed.
While population stratification is a potential confounder of genomic prediction studies, our use of a large worldwide multi-ethnic meta-analysis to develop the GRS together with two fully independent prospective validation datasets and three independent case/control datasets minimizes this potential. Our GRS was constructed from the CARDIoGRAMplusC4D stage-2 meta-analysis and the FINRISK and FHS individuals are both independent of that study and of broadly European ancestry; thus it is unlikely that the GRS is substantially confounded by fine-scale population structure within these cohorts. Further, the LD-thinning threshold to maximize prediction was determined in the WTCCC and MIGen datasets prior to applying the GRS to ARGOS, FINRISK, or FHS. Nevertheless, for some measures, GRS gains were less pronounced in FHS than in FINRISK. This may partly be due to the different definitions of CHD in these studies, to differences in environmental exposures, or to differences in genetic effects.31 In addition, the FRS was developed in the FHS, leading to potential over-estimation of its association with CHD in the current analysis. Hence, there may be benefit from future development of population-specific GRSs, which may yield greater predictive power within each population.
The association of the GRS with incident CHD was not substantially attenuated by traditional risk factors or clinical risk scores derived from these risk factors. Furthermore, the GRS was strongly associated with CHD in a study consisting purely of individuals with familial hypercholesterolemia. These results suggest that genomic risk exerts its effect on CHD risk through molecular pathways that are largely independent of the effects of cholesterol, systolic blood pressure, and smoking. A hitherto unresolved question has been the extent to which a family history would capture any information that may be provided through genetic analysis. Here, we clearly demonstrate the superior performance of direct genetic information over self-reported family history of CHD, which is often incomplete and imprecise in practice and is influenced by family size and competing causes of death.
While we observed improvements in discrimination (C-index) resulting from adding the GRS to the clinical risk scores when considering adults of all ages, the improvements were substantially higher in older individuals (>60 years old). Rather than being driven by age-related differences in the effect of the GRS, these results are likely driven by differences in the clinical risk scores between the younger and older adults. Unlike the GRSs, the clinical risk scores showed substantial differences across ages, driven by temporal changes in the underlying risk factors as well as age itself. Beyond the aims of identifying older adults with high CHD risk, the invariance of genomic risk makes it particularly useful for CHD risk prediction earlier in life, in young adulthood or before, when traditional risk factors are typically not measured and less likely to be informative of risk later in life.
Our analyses focused on two clinical scores, the FRS and ACC/AHA13. While other scores exist, for example the SCORE system,32 we elected to use the FRS and ACC/AHA13 due to their widespread use and the fact that the FINRISK cohorts were a major contributor to the SCORE analysis, potentially biasing the analysis in FINRISK, in the same way that FRS seems to be biased towards the FHS, inflating its predictive power of the clinical risk scores there relative to the reference model.
Stratifying individual baseline smoking, systolic blood pressure, and total cholesterol levels measures into genomic risk groups revealed substantial differences in cumulative risk patterns. Importantly, this demonstrates that improved lifestyle may compensate for the innate increased CHD risk captured by the GRS. For men with high genomic risk, modifiable risk factors showed large effects on cumulative CHD risk. For women, the observed impacts of smoking, systolic blood pressure, and total cholesterol were low or not detectable in the low genomic risk group, particularly in FINRISK, however, we could not determine whether this was due to inadequate statistical power or other biological effects and further studies in larger cohorts of women are necessary to determine any clinical implications.
Our results, if validated in further studies and across different populations, suggest a potential paradigmatic shift in the current CHD screening strategy which has existed for over 40 years—namely determination of genomic risk at an early stage with screening later in life through traditional clinical risk scores to complement background genomic risk. Based on early genomic risk stratification, individuals at higher risk may benefit from earlier engagement with nutritionists, exercise regimes, smoking cessation programs or be initiated early on medical interventions such as statin therapy or blood-pressure lowering medications to minimize future CHD risk. In this context it is notable that Mega et al.29 recently demonstrated that the GRS of 27 CHD-associated SNPs better predicted which individuals would benefit most, both in relative and absolute terms, from statin treatment. In a study of type 2 diabetes, Florez et al.32 has shown that the effects of increased genetic susceptibility to disease can be ameliorated by lifestyle (diet and exercise) and therapeutic (metformin) interventions. Similar possibilities exist for CHD, whereby early targeted prevention strategies based on genomic CHD risk may be implemented well in advance of clinical risk scores attaining predictive capacity at later ages.33 Such early risk stratification will offer increased efficiency in allocating both therapeutic resources and lifestyle modifications with the potential for subsequent delay of onset of traditional risk factors and incident CHD risk.
While our study demonstrates both the independent and incremental predictive power provided by our GRS, it is important to note that even when combined with such scores, the overall positive predictive value still remains modest for an acceptable negative predictive value (see Supplementary Data online, Figure S14a). Furthermore, despite overall improved reclassification of 10 years risk, some individuals who went on to develop an incident event were reclassified at a lower risk by the addition of the GRS compared with their initial classification using a clinical score (Tables 3 and 4), emphasizing the limitations of the current GRS. The magnitude of the GRS effect was weaker in FHS than in the other datasets examined (FINRISK, WTCCC-CAD, MIGen-Harps, and ARGOS; Table 2). In addition to potential technical and clinical FHS differences discussed above, these results suggest that the benefit and clinical utility of the GRS may vary between populations; further evaluation in large prospective studies of varying ancestry will be required in order to assess these differences and how best to account for them in risk prediction. In this context, it should be noted that our GRS based on a starting list of 79 128 common SNPs tested by the CARDIOGRAMplusC4D consortium could be further improved. Future studies that construct GRSs using increased sample sizes and capturing the full spectrum of common and rare variants9,34 will likely provide additional gains in prediction and risk stratification.
In summary, this study has demonstrated the potential clinical utility of genome-scale GRS for CHD, both for early identification of individuals at increased CHD risk and for complementing existing clinical risk scores. Given recent advances and reduced cost of genotyping microarrays and sequencing-based technologies and their cost efficiency, determination of genome-wide SNP variants (including the 49 310 SNPs used here) is no longer beyond the realm of clinical application. In terms of technical feasibility, genome-wide genotyping of hundreds of thousands of SNPs is now both reliable and cost effective (<US$70 in bulk), and clinically certified genotyping services are now becoming available. Statistical SNP imputation will further expand the number of SNPs to an order of several million. Additionally, germline genotyping is a one-time cost for each individual. Further validation and cost-benefit analyses will be required in order to establish how this technology is deployed in clinical settings.
Supplementary Material
Acknowledgements
We thank Julie Simpson, Melbourne School of Population Health and Global Health (University of Melbourne), for advice regarding survival analyses.
Funding
National Health and Medical Research Council Early Career Fellowship (1090462 to G.A.); National Health and Medical Research Council and the National Heart Foundation of Australia (1061435 and 1062227 to M.I.); Finnish Foundation for Cardiovascular Research to V.S; British Heart Foundation and NIHR to N.J.S.; AP and SR are supported by the Academy of Finland (grant no. 251704, 286500, 293404 to AP, and 251217, 285380 to SR), Juselius Foundation, Finnish Foundation for Cardiovascular Research, NordForsk e-Science NIASC (grant no 62721) and Biocentrum Helsinki (to SR). The MI Genetics (MIGen) Consortium Study was funded by the National Heart, Lung, and Blood Institute of the United States National Institutes of Health (R01 HL087676). Genotyping was partially funded by The Broad Institute Center for Genotyping and Analysis, which was supported by grant U54 RR02027 from the National Center for Research Resources. This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475. The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement no. 261433 (Biobank Standardisation and Harmonisation for Research Excellence in the European Union—BioSHaRE-EU). We are grateful to the CARDIoGRAMplusC4D consortium for making their large-scale genetic data available. A list of members of the consortium and the contributing studies is available at www.cardiogramplusc4d.org.
Ethics statements
The FINRISK data and samples are part of the THL Biobank (https://www.thl.fi/en/web/thlfi-en/topics/information-packages/thl-biobank), which has been approved by the Coordinating Ethical Committee of The Helsinki and Uusimaa Hospital District (decision # 238/13/03/00/2014).
The FHS dataset was obtained from dbGaP (phs000007), approved by the University of Melbourne Health Sciences Human Ethics Sub-Committee (HREC 1442186).
The ARGOS study consisted of cases and controls recruited from a large study based on the Dutch nationwide screening program for familial hypercholesterolemia. All patients gave informed consent and the ethics committee of the Academic Medical Center of Amsterdam approved the protocol (MEC 00/41#00.17.628).
Conflict of interest: none declared.
References
- 1. Goff DC, Jr, Lloyd-Jones DM, Bennett G, Coady S, D'agostino RB, Gibbons R, Greenland P, Lackland DT, Levy D, O'donnell CJ, Robinson JG, Schwartz JS, Shero ST, Smith SC, Jr, Sorlie P, Stone NJ, Wilson PW, Jordan HS, Nevo L, Wnek J, Anderson JL, Halperin JL, Albert NM, Bozkurt B, Brindis RG, Curtis LH, DeMets D, Hochman JS, Kovacs RJ, Ohman EM, Pressler SJ, Sellke FW, Shen WK, Smith SC, Jr, Tomaselli GF. 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association task force on practice guidelines. Circulation 2014;129(25 Suppl 2):S49–S73. [DOI] [PubMed] [Google Scholar]
- 2. Stone NJ, Robinson JG, Lichtenstein AH, Bairey Merz CN, Blum CB, Eckel RH, Goldberg AC, Gordon D, Levy D, Lloyd-Jones DM, McBride P, Schwartz JS, Shero ST, Smith SC, Jr, Watson K, Wilson PW, Eddleman KM, Jarrett NM, LaBresh K, Nevo L, Wnek J, Anderson JL, Halperin JL, Albert NM, Bozkurt B, Brindis RG, Curtis LH, DeMets D, Hochman JS, Kovacs RJ, Ohman EM, Pressler SJ, Sellke FW, Shen WK, Smith SC, Jr, Tomaselli GF. 2013 ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascular risk in adults: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2014;129(25 Suppl 2):S1–45. [DOI] [PubMed] [Google Scholar]
- 3. Expert Panel on Detection, Evaluation, and Treatment of High Blood Cholesterol in Adults. Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA 2001;285:2486–2497. [DOI] [PubMed] [Google Scholar]
- 4. Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med 1994;330:1041–1046. [DOI] [PubMed] [Google Scholar]
- 5. The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 2007;447:661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H, Wtccc, the, Cardiogenics C. Genomewide association analysis of coronary artery disease. N Engl J Med 2007;357:443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Myocardial Infarction Genetics Consortium, Kathiresan S, Voight BF, Purcell S, Musunuru K, Ardissino D, Mannucci PM, Anand S, Engert JC, Samani NJ, Schunkert H, Erdmann J, Reilly MP, Rader DJ, Morgan T, Spertus JA, Stoll M, Girelli D, McKeown PP, Patterson CC, Siscovick DS, O'donnell CJ, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Melander O, Altshuler D, Ardissino D, Merlini PA, Berzuini C, Bernardinelli L, Peyvandi F, Tubaro M, Celli P, Ferrario M, Fetiveau R, Marziliano N, Casari G, Galli M, Ribichini F, Rossi M, Bernardi F, Zonzin P, Piazza A, Mannucci PM, Schwartz SM, Siscovick DS, Yee J, Friedlander Y, Elosua R, Marrugat J, Lucas G, Subirana I, Sala J, Ramos R, Kathiresan S, Meigs JB, Williams G, Nathan DM, MacRae CA, O'donnell CJ, Salomaa V, Havulinna AS, Peltonen L, Melander O, Berglund G, Voight BF, Kathiresan S, Hirschhorn JN, Asselta R, Duga S, Spreafico M, Musunuru K, Daly MJ, Purcell S, Voight BF, Purcell S, Nemesh J, Korn JM, McCarroll SA, Schwartz SM, Yee J, Kathiresan S, Lucas G, Subirana I, Elosua R, Surti A, Guiducci C, Gianniny L, Mirel D, Parkin M, Burtt N, Gabriel SB, Samani NJ, Thompson JR, Braund PS, Wright BJ, Balmforth AJ, Ball SG, Hall A, Wellcome Trust Case Control C, Schunkert H, Erdmann J, Linsel-Nitschke P, Lieb W, Ziegler A, Konig I, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Schunkert H, Samani NJ, Erdmann J, Ouwehand W, Hengstenberg C, Deloukas P, Scholz M, Cambien F, Reilly MP, Li M, Chen Z, Wilensky R, Matthai W, Qasim A, Hakonarson HH, Devaney J, Burnett MS, Pichard AD, Kent KM, Satler L, Lindsay JM, Waksman R, Knouff CW, Waterworth DM, Walker MC, Mooser V, Epstein SE, Rader DJ, Scheffold T, Berger K, Stoll M, Huge A, Girelli D, Martinelli N, Olivieri O, Corrocher R, Morgan T, Spertus JA, McKeown P, Patterson CC, Schunkert H, Erdmann E, Linsel-Nitschke P, Lieb W, Ziegler A, Konig IR, Hengstenberg C, Fischer M, Stark K, Grosshennig A, Preuss M, Wichmann HE, Schreiber S, Holm H, Thorleifsson G, Thorsteinsdottir U, Stefansson K, Engert JC, Do R, Xie C, Anand S, Kathiresan S, Ardissino D, Mannucci PM, Siscovick D, O'donnell CJ, Samani NJ, Melander O, Elosua R, Peltonen L, Salomaa V, Schwartz SM, Altshuler D.Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat Genet 2009;41:334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. CARDIoGRAMplusC4D Consortium, Deloukas P, Kanoni S, Willenborg C, Farrall M, Assimes TL, Thompson JR, Ingelsson E, Saleheen D, Erdmann J, Goldstein BA, Stirrups K, Konig IR, Cazier JB, Johansson A, Hall AS, Lee JY, Willer CJ, Chambers JC, Esko T, Folkersen L, Goel A, Grundberg E, Havulinna AS, Ho WK, Hopewell JC, Eriksson N, Kleber ME, Kristiansson K, Lundmark P, Lyytikainen LP, Rafelt S, Shungin D, Strawbridge RJ, Thorleifsson G, Tikkanen E, Van Zuydam N, Voight BF, Waite LL, Zhang W, Ziegler A, Absher D, Altshuler D, Balmforth AJ, Barroso I, Braund PS, Burgdorf C, Claudi-Boehm S, Cox D, Dimitriou M, Do R, Diagram Consortium Cardiogenics Consortium Doney AS, El Mokhtari N, Eriksson P, Fischer K, Fontanillas P, Franco-Cereceda A, Gigante B, Groop L, Gustafsson S, Hager J, Hallmans G, Han BG, Hunt SE, Kang HM, Illig T, Kessler T, Knowles JW, Kolovou G, Kuusisto J, Langenberg C, Langford C, Leander K, Lokki ML, Lundmark A, McCarthy MI, Meisinger C, Melander O, Mihailov E, Maouche S, Morris AD, Muller-Nurasyid M, MuTHeR Consortium Nikus K, Peden JF, Rayner NW, Rasheed A, Rosinger S, Rubin D, Rumpf MP, Schafer A, Sivananthan M, Song C, Stewart AF, Tan ST, Thorgeirsson G, van der Schoot CE, Wagner PJ, Wellcome Trust Case Control Consortium Wells GA, Wild PS, Yang TP, Amouyel P, Arveiler D, Basart H, Boehnke M, Boerwinkle E, Brambilla P, Cambien F, Cupples AL, de Faire U, Dehghan A, Diemert P, Epstein SE, Evans A, Ferrario MM, Ferrieres J, Gauguier D, Go AS, Goodall AH, Gudnason V, Hazen SL, Holm H, Iribarren C, Jang Y, Kahonen M, Kee F, Kim HS, Klopp N, Koenig W, Kratzer W, Kuulasmaa K, Laakso M, Laaksonen R, Lee JY, Lind L, Ouwehand WH, Parish S, Park JE, Pedersen NL, Peters A, Quertermous T, Rader DJ, Salomaa V, Schadt E, Shah SH, Sinisalo J, Stark K, Stefansson K, Tregouet DA, Virtamo J, Wallentin L, Wareham N, Zimmermann ME, Nieminen MS, Hengstenberg C, Sandhu MS, Pastinen T, Syvanen AC, Hovingh GK, Dedoussis G, Franks PW, Lehtimaki T, Metspalu A, Zalloua PA, Siegbahn A, Schreiber S, Ripatti S, Blankenberg SS, Perola M, Clarke R, Boehm BO, O'donnell C, Reilly MP, Marz W, Collins R, Kathiresan S, Hamsten A, Kooner JS, Thorsteinsdottir U, Danesh J, Palmer CN, Roberts R, Watkins H, Schunkert H, Samani NJ.Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 2013;45:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. CARDIoGRAMplusC4D Consortium. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet 2015;47:1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ripatti S, Tikkanen E, Orho-Melander M, Havulinna AS, Silander K, Sharma A, Guiducci C, Perola M, Jula A, Sinisalo J, Lokki ML, Nieminen MS, Melander O, Salomaa V, Peltonen L, Kathiresan S. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet 2010;376:1393–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tikkanen E, Havulinna AS, Palotie A, Salomaa V, Ripatti S. Genetic risk prediction and a 2-stage risk screening strategy for coronary heart disease. Arterioscler Thromb Vasc Biol 2013;33:2261–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thanassoulis G, Peloso GM, Pencina MJ, Hoffmann U, Fox CS, Cupples LA, Levy D, D'agostino RB, Hwang SJ, O'donnell CJ. A genetic risk score is associated with incident cardiovascular disease and coronary artery calcium: the Framingham Heart Study. Circ Cardiovasc Genet 2012;5:113–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ganna A, Magnusson PK, Pedersen NL, de Faire U, Reilly M, Arnlov J, Sundstrom J, Hamsten A, Ingelsson E. Multilocus genetic risk scores for coronary heart disease prediction. Arterioscler Thromb Vasc Biol 2013;33:2267–2272. [DOI] [PubMed] [Google Scholar]
- 14. Hughes MF, Saarela O, Stritzke J, Kee F, Silander K, Klopp N, Kontto J, Karvanen J, Willenborg C, Salomaa V, Virtamo J, Amouyel P, Arveiler D, Ferrieres J, Wiklund PG, Baumert J, Thorand B, Diemert P, Tregouet DA, Hengstenberg C, Peters A, Evans A, Koenig W, Erdmann J, Samani NJ, Kuulasmaa K, Schunkert H, Genetic markers enhance coronary risk prediction in men: the MORGAM prospective cohorts. PLoS One 2012;7:e40922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paynter NP, Chasman DI, Pare G, Buring JE, Cook NR, Miletich JP, Ridker PM. Association between a literature-based genetic risk score and cardiovascular events in women. JAMA 2010;303:631–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weijmans M, de Bakker PI, van der Graaf Y, Asselbergs FW, Algra A, Jan de Borst G, Spiering W, Visseren FL, Group SS. Incremental value of a genetic risk score for the prediction of new vascular events in patients with clinically manifest vascular disease. Atherosclerosis 2015;239:451–458. [DOI] [PubMed] [Google Scholar]
- 17. Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med 2008;358:1240–1249. [DOI] [PubMed] [Google Scholar]
- 18. Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 2010;42:565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Locke AE Kahali B Berndt SI Justice AE Pers TH Day FR Powell C Vedantam S Buchkovich ML Yang J Croteau-Chonka DC Esko T Fall T Ferreira T Gustafsson S Kutalik Z Luan J Magi R Randall JC Winkler TW Wood AR Workalemahu T Faul JD Smith JA Hua Zhao J Zhao W Chen J Fehrmann R Hedman AK Karjalainen J Schmidt EM Absher D Amin N Anderson D Beekman M Bolton JL Bragg-Gresham JL Buyske S Demirkan A Deng G Ehret GB Feenstra B Feitosa MF Fischer K Goel A Gong J Jackson AU Kanoni S Kleber ME Kristiansson K Lim U Lotay V Mangino M Mateo Leach I Medina-Gomez C Medland SE Nalls MA Palmer CD Pasko D Pechlivanis S Peters MJ Prokopenko I Shungin D Stancakova A Strawbridge RJ Ju Sung Y Tanaka T Teumer A Trompet S van der Laan SW van Setten J Van Vliet-Ostaptchouk JV Wang Z Yengo L Zhang W Isaacs A Albrecht E Arnlov J Arscott GM Attwood AP Bandinelli S Barrett A Bas IN Bellis C Bennett AJ Berne C Blagieva R Bluher M Bohringer S Bonnycastle LL Bottcher Y Boyd HA Bruinenberg M Caspersen IH Ida Chen YD Clarke R Daw EW de Craen AJ Delgado G Dimitriou M Doney AS Eklund N Estrada K Eury E Folkersen L Fraser RM Garcia ME Geller F Giedraitis V Gigante B Go AS Golay A Goodall AH Gordon SD Gorski M Grabe HJ Grallert H Grammer TB Grassler J Gronberg H Groves CJ Gusto G Haessler J Hall P Haller T Hallmans G Hartman CA Hassinen M Hayward C Heard-Costa NL Helmer Q Hengstenberg C Holmen O Hottenga JJ James AL Jeff JM Johansson A Jolley J Juliusdottir T Kinnunen L Koenig W Koskenvuo M Kratzer W Laitinen J Lamina C Leander K Lee NR Lichtner P Lind L Lindstrom J Sin Lo K Lobbens S Lorbeer R Lu Y Mach F Magnusson PK Mahajan A McArdle WL McLachlan S Menni C Merger S Mihailov E Milani L Moayyeri A Monda KL Morken MA Mulas A Muller G Muller-Nurasyid M Musk AW Nagaraja R Nothen MM Nolte IM Pilz S Rayner NW Renstrom F Rettig R Ried JS Ripke S Robertson NR Rose LM Sanna S Scharnagl H Scholtens S Schumacher FR Scott WR Seufferlein T Shi J Vernon Smith A Smolonska J Stanton AV Steinthorsdottir V Stirrups K Stringham HM Sundstrom J Swertz MA Swift AJ Syvanen AC Tan ST Tayo BO Thorand B Thorleifsson G Tyrer JP Uh HW Vandenput L Verhulst FC Vermeulen SH Verweij N Vonk JM Waite LL Warren HR Waterworth D Weedon MN Wilkens LR Willenborg C Wilsgaard T Wojczynski MK Wong A Wright AF Zhang Q LifeLines Cohort Study Brennan EP Choi M Dastani Z Drong AW Eriksson P Franco-Cereceda A Gadin JR Gharavi AG Goddard ME Handsaker RE Huang J Karpe F Kathiresan S Keildson S Kiryluk K Kubo M Lee JY Liang L Lifton RP Ma B McCarroll SA McKnight AJ Min JL Moffatt MF Montgomery GW Murabito JM Nicholson G Nyholt DR Okada Y Perry JR Dorajoo R Reinmaa E Salem RM Sandholm N Scott RA Stolk L Takahashi A Tanaka T Van't Hooft FM Vinkhuyzen AA Westra HJ Zheng W Zondervan KTADIPOGen Consortium, Agen-Bmi Working Group, CARDIOGRAMplusC4D Consortium, CKDGen Consortium, GLGC, ICBP, MAGIC Investigators, MuThER Consortium, MIGen Consortium, PAGE Consortium, ReproGen Consortium, GENIE Consortium, International Endogene Consortium, Heath AC Arveiler D Bakker SJ Beilby J Bergman RN Blangero J Bovet P Campbell H Caulfield MJ Cesana G Chakravarti A Chasman DI Chines PS Collins FS Crawford DC Cupples LA Cusi D Danesh J de Faire U den Ruijter HM Dominiczak AF Erbel R Erdmann J Eriksson JG Farrall M Felix SB Ferrannini E Ferrieres J Ford I Forouhi NG Forrester T Franco OH Gansevoort RT Gejman PV Gieger C Gottesman O Gudnason V Gyllensten U Hall AS Harris TB Hattersley AT Hicks AA Hindorff LA Hingorani AD Hofman A Homuth G Hovingh GK Humphries SE Hunt SC Hypponen E Illig T Jacobs KB Jarvelin MR Jockel KH Johansen B Jousilahti P Jukema JW Jula AM Kaprio J Kastelein JJ Keinanen-Kiukaanniemi SM Kiemeney LA Knekt P Kooner JS Kooperberg C Kovacs P Kraja AT Kumari M Kuusisto J Lakka TA Langenberg C Le Marchand L Lehtimaki T Lyssenko V Mannisto S Marette A Matise TC McKenzie CA McKnight B Moll FL Morris AD Morris AP Murray JC Nelis M Ohlsson C Oldehinkel AJ Ong KK Madden PA Pasterkamp G Peden JF Peters A Postma DS Pramstaller PP Price JF Qi L Raitakari OT Rankinen T Rao DC Rice TK Ridker PM Rioux JD Ritchie MD Rudan I Salomaa V Samani NJ Saramies J Sarzynski MA Schunkert H Schwarz PE Sever P Shuldiner AR Sinisalo J Stolk RP Strauch K Tonjes A Tregouet DA Tremblay A Tremoli E Virtamo J Vohl MC Volker U Waeber G Willemsen G Witteman JC Zillikens MC Adair LS Amouyel P Asselbergs FW Assimes TL Bochud M Boehm BO Boerwinkle E Bornstein SR Bottinger EP Bouchard C Cauchi S Chambers JC Chanock SJ Cooper RS de Bakker PI Dedoussis G Ferrucci L Franks PW Froguel P Groop LC Haiman CA Hamsten A Hui J Hunter DJ Hveem K Kaplan RC Kivimaki M Kuh D Laakso M Liu Y Martin NG Marz W Melbye M Metspalu A Moebus S Munroe PB Njolstad I Oostra BA Palmer CN Pedersen NL Perola M Perusse L Peters U Power C Quertermous T Rauramaa R Rivadeneira F Saaristo TE Saleheen D Sattar N Schadt EE Schlessinger D Slagboom PE Snieder H Spector TD Thorsteinsdottir U Stumvoll M Tuomilehto J Uitterlinden AG Uusitupa M van der Harst P Walker M Wallaschofski H Wareham NJ Watkins H Weir DR Wichmann HE Wilson JF Zanen P Borecki IB Deloukas P Fox CS Heid IM O'Connell JR Strachan DP Stefansson K van Duijn CM Abecasis GR Franke L Frayling TM McCarthy MI Visscher PM Scherag A Willer CJ Boehnke M Mohlke KL Lindgren CM Beckmann JS Barroso I North KE Ingelsson E Hirschhorn JN Loos RJ Speliotes EK.. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Abraham G, Kowalczyk A, Zobel J, Inouye M. Performance and robustness of penalized and unpenalized methods for genetic prediction of complex human disease. Genetic Epidemiol 2013;37:184–195. [DOI] [PubMed] [Google Scholar]
- 21. Abraham G, Tye-Din JA, Bhalala OG, Kowalczyk A, Zobel J, Inouye M. Accurate and robust genomic prediction of celiac disease using statistical learning. PLoS Genet 2014;10:e1004137.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simonson MA, Wills AG, Keller MC, McQueen MB. Recent methods for polygenic analysis of genome-wide data implicate an important effect of common variants on cardiovascular disease risk. BMC Med Genet 2011;12:146.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. International SC, Purcell SM, Wray NR, Stone JL, Visscher PM, O'donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 2009;460:748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goldstein BA, Yang L, Salfati E, Assimes TL. Contemporary considerations for constructing a genetic risk score: an empirical approach. Genet Epidemiol 2015;39:439–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vartiainen E, Laatikainen T, Peltonen M, Juolevi A, Mannisto S, Sundvall J, Jousilahti P, Salomaa V, Valsta L, Puska P. Thirty-five-year trends in cardiovascular risk factors in Finland. Int J Epidemiol 2010;39:504–518. [DOI] [PubMed] [Google Scholar]
- 26. Feinleib M, Kannel WB, Garrison RJ, McNamara PM, Castelli WP. The Framingham offspring study. Design and preliminary data. Prev Med 1975;4:518–525. [DOI] [PubMed] [Google Scholar]
- 27. Kannel WB, Dawber TR, Kagan A, Revotskie N, Stokes J., III Factors of risk in the development of coronary heart disease–six year follow-up experience. The Framingham Study. Ann Intern Med 1961;55:33–50. [DOI] [PubMed] [Google Scholar]
- 28. Dawber TR, Kannel WB, Lyell LP. An approach to longitudinal studies in a community: the Framingham Study. Ann N Y Acad Sci 1963;107:539–556. [DOI] [PubMed] [Google Scholar]
- 29. Mega JL, Stitziel NO, Smith JG, Chasman DI, Caulfield MJ, Devlin JJ, Nordio F, Hyde CL, Cannon CP, Sacks FM, Poulter NR, Sever PS, Ridker PM, Braunwald E, Melander O, Kathiresan S, Sabatine MS. Genetic risk, coronary heart disease events, and the clinical benefit of statin therapy: an analysis of primary and secondary prevention trials. Lancet 2015;385:2264–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tada H, Melander O, Louie JZ, Catanese JJ, Rowland CM, Devlin JJ, Kathiresan S, Shiffman D. Risk prediction by genetic risk scores for coronary heart disease is independent of self-reported family history. Eur Heart J 2016;37:561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dehghan A, Bis JC, White CC, Smith AV, Morrison AC, Cupples LA, Trompet S, Chasman DI, Lumley T, Volker U, Buckley BM, Ding J, Jensen MK, Folsom AR, Kritchevsky SB, Girman CJ, Ford I, Dorr M, Salomaa V, Uitterlinden AG, Eiriksdottir G, Vasan RS, Franceschini N, Carty CL, Virtamo J, Demissie S, Amouyel P, Arveiler D, Heckbert SR, Ferrieres J, Ducimetiere P, Smith NL, Wang YA, Siscovick DS, Rice KM, Wiklund PG, Taylor KD, Evans A, Kee F, Rotter JI, Karvanen J, Kuulasmaa K, Heiss G, Kraft P, Launer LJ, Hofman A, Markus MR, Rose LM, Silander K, Wagner P, Benjamin EJ, Lohman K, Stott DJ, Rivadeneira F, Harris TB, Levy D, Liu Y, Rimm EB, Jukema JW, Volzke H, Ridker PM, Blankenberg S, Franco OH, Gudnason V, Psaty BM, Boerwinkle E, O'donnell CJ. Genome-Wide Association Study for incident myocardial infarction and coronary heart disease in prospective cohort studies: the CHARGE consortium. PLoS One 2016;11:e0144997.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PI, Shuldiner AR, Knowler WC, Nathan DM, Altshuler D. Diabetes Prevention Program Research G. TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 2006;355:241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Berry JD, Dyer A, Cai X, Garside DB, Ning H, Thomas A, Greenland P, Van Horn L, Tracy RP, Lloyd-Jones DM. Lifetime risks of cardiovascular disease. N Engl J Med 2012;366:321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 2015;526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.