Abstract
In many species, under varying ecological conditions, social interactions among individuals result in the formation of dominance hierarchies. Despite general similarities, there are robust differences among dominance hierarchies across species, populations, environments, life stages, sexes, and individuals. Understanding the proximate mechanisms underlying the variation is an important step toward understanding the evolution of social behavior. However, physiological changes associated with dominance, such as gonadal maturation and somatic growth, often complicate efforts to identify the specific underlying mechanisms. Traditional gene expression analyses are useful for generating candidate gene lists, but are biased by choice of significance cut-offs and difficult to use for between-study comparisons. In contrast, complementary analysis tools allow one to both test a priori hypotheses and generate new hypotheses. Here we employ a meta-analysis of high-throughput expression profiling experiments to investigate the gene expression patterns that underlie mechanisms and evolution of behavioral social phenotypes. Specifically, we use a collection of datasets on social dominance in fish across social contexts, sex, and species. Using experimental manipulation to produce female dominance hierarchies in the cichlid Astatotilapia burtoni, heralded as a genomic model of social dominance, we generate gene lists, and assess molecular gene modules. In the dominant female gene expression profile, we demonstrate a strong pattern of up-regulation of genes previously identified as having male-biased expression and furthermore, compare expression biases between male and female dominance phenotypes. Using a threshold-free approach to identify correlation throughout ranked gene lists, we query previously published datasets associated with maternal behavior, alternative reproductive tactics, cooperative breeding, and sex-role reversal to describe correlations among these various neural gene expression profiles associated with different instances of social dominance. These complementary approaches capitalize on the high-throughput gene expression profiling from similar behavioral phenotypes in order to address the mechanisms associated with social dominance behavioral phenotypes.
Introduction
By comparing gene expression profiles across contexts and even across species, we can both generate and test biological hypotheses with high-throughput expression profiling. In many species, social interactions among individuals result in the formation of dominance hierarches that can sustain individual behavioral differences and reinforce social status (Sapolsky 2005) yet the degree to which similar mechanisms are employed is not known on a genome-wide basis. A meta-analysis approach to gene expression profiling has been highly profitable in biomedical research comparing human cases and mouse models of cancer (e.g., Sweet-Cordero et al. 2005) for investigating the evolution of the human brain (Oldham et al. 2006; Bauernfeind et al. 2015), and in studies of differing ecological situations (Aubin-Horth et al. 2009; Schumer et al. 2011; Leichty et al. 2012).
The majority of studies on aggression in the context of social dominance focus on males, despite clear evidence that aggressive dominant phenotypes have their own selective advantage in females. In both sexes, dominance and aggression varies among species, populations, environments, life stages, and individuals (Nelson 2006). Understanding the proximate mechanisms of social dominance is necessary in order to understand the evolution of social behavior and to explain mechanisms mitigating the observed tradeoffs among other fitness-linked traits (Knapp et al. 1999; McGlothlin and Ketterson 2008; Rosvall 2013). In accordance with the complexity of the dominance phenotype at the behavioral level, we observe substantial differences in brain gene expression patterns between social states (Renn et al. 2008; Toth et al. 2010; O'Connell and Hofmann 2012). However, confounding physiological changes associated with dominance (e.g., gonadal maturation, somatic growth) complicate efforts to identify the underlying mechanisms. Indeed, the physiological changes associated with aggressive behaviors and dominance hierarchies are often mediated through hormones (Wingfield 1994; Goymann et al. 2008; Goymann et al. 2015; Teles and Oliveira 2016) including, but not limited to, sex steroids that may cause profound changes in gene expression (O'Connell and Hofmann 2012). It is thus nearly impossible to know to what extent different aspects of neural gene expression pattern are related specifically to social behavior when only a single system is studied. In order to parse the context-specific from the invariant mechanisms related to the production of social dominance, one must study the molecular mechanisms in alternate contexts selected to control for confounding factors, such as age, sexual maturity, reproductive state, and social context.
In order to disentangle reproductive physiology from the neuroendocrine mechanism of social dominance, we take advantage of a highly social fish that has plastic behavioral phenotypes and readily forms dominance hierarchies. The African cichlid fish Astatotilapia burtoni has become a particularly powerful model system in social neuroscience (reviewed in Hofmann 2003; Fernald 2004) while at the same time its ecology and behavior have been well characterized (Fernald and Hirata 1977a, 1977b). At any specific time, only ∼ 30% of the males in a population is reproductively active and maintain a spawning territory through heightened aggressiveness, accompanied by bright body coloration and facial markings, a social phenotype referred to here as “dominant” (Hofmann et al. 1999). The remaining males are socially subordinate, directing resources toward growth and schooling with females until there is an opportunity to acquire a territory (Hofmann et al. 1999; Maruska and Fernald 2013). Female A. burtoni normally behave much like non-territorial males, schooling until spawning with dominant males. After spawning, females brood developing fry in their mouths for approximately 2 weeks and display maternal aggression and care upon release of the fry (Renn et al. 2009). Female A. burtoni provide an excellent opportunity to dissect the mechanisms of social status from the mechanisms regulating reproduction because female dominance hierarchies can be experimentally formed by removing males from the population in a manner independent of their reproductive state (Renn et al. 2012). As previously described (Renn et al. 2012), dominant and subordinate females produced in this way are reproductively competent and have equal growth rates (GRs), and modest change in body color. The dominant females exhibit increased aggressive behaviors and even male-like courtship behavior, without undergoing sex change (Renn et al. 2012; O'Connell et al. 2013) although steroid (both estradiol and testosterone) titers increase despite no difference in reproductive status. Gene expression differences associated with social dominance are well established for males of this species (Burmeister et al. 2007; Greenwood et al. 2008; Renn et al. 2008; O'Connell and Hofmann 2012; Maruska et al. 2013) providing a readily available appropriate dataset to compare to gene expression profiles in these experimental dominant females.
By minimizing the confounding influence of reproduction and GR, we can address the invariant molecular mechanisms of dominance in this species. Comparing results from independently collected profiling experiments is complicated by variables that may differ between experiments (Rittschof et al. 2014), even when working within a single species. In order to identify patterns beyond single gene level, it is necessary to use algorithms that are sensitive and robust to experimental variation, statistical noise, and cofounding factors (Plaisier et al. 2010; Pfenning et al. 2014). While most traditional methods of comparing gene expression datasets require setting arbitrary cut-offs based either on fold-differences or statistical significance, analysis based on rank-rank hypergeometric overlap (RRHO; Plaisier et al. 2010) allows the identification of statistically significant overlap (either negative or positive correlation) between gene signatures. Overlap between ranked gene lists is measured as the degree of statistical enrichment using a hypergeometric distribution while stepping across all possible thresholds through the two ranked lists. This approach complements threshold algorithms by providing more information about the strength and regions of overlap between two datasets without artificially truncating gene expression profiles. A meta-analysis assisted by RRHO analysis allows us to visualize the degree of coordinated regulation overlap between dominant phenotypes within and between both species and sexes (Aubin-Horth et al. 2007; Renn et al. 2008; Schumer et al. 2011) extending beyond cichlids (Stiver et al. 2015). Understanding the similarities and differences among these mechanisms will illuminate the evolution of these aggressive behaviors, uncover correlation in neural gene expression patterns associated with dominance regardless of the sex, species, or specific conditions, and provide valuable functional information and annotation for behaviorally relevant genes.
Methods
Housing and observation of fish
Laboratory stock fish derived from a wild population ∼40 years earlier (Fernald and Hirata 1977b) were kept under standard conditions (110 L aquaria at 28°C and pH 8.5 under 12 h light/12 h dark and simulated dawn and dusk). Fish were fed daily and provided with gravel and terracotta pot shards to simulate natural shelters. All experiments were conducted in accordance with Harvard University's Institutional Animal Care and Use Committee (IACUC protocol number 22-22).
The females considered in this current study are those described as “short-term paradigm” in our previous report of female dominance behavior (Renn et al. 2012). For dominant and subordinate samples, each 110-L tank was divided to house two populations of five females separated by a clear divider. Brooding, females were derived from two mixed-sex communities, each composed of five to six males and five to six females. Females were individually tagged for behavioral observations, completed over the course of 45 days as previously described using the ethogram adapted from A. burtoni male studies (Fernald and Hirata 1977a; Renn et al. 2008).
Tissue sample collection
Gonadosomatic index (GSI) was calculated as gonad mass/body mass. To calculate GR, the weekly relative change in standard length for the final 2 weeks was averaged. Each female was sacrificed by rapid decapitation between 11:00 and 13:00 following 4 weeks in a consistent behavioral phenotype. Whole brains were dissected within 10 min and stored in RNAlater (Ambion) at −20°C. Blood samples for hormone measures were not collected for these females.
Statistical analysis of female A. burtoni behavior and physiology
Statistical analyses of behavioral and physiological data for the 4 weeks preceding euthanasia were performed in R, using one-way analysis of variance (ANOVA) in a randomization test based on 10,000 replications to calculate P-values, followed by pairwise comparison of dominant, subordinate and brooding females. The probability of obtaining a difference at least as great as observed was calculated from the randomization, and those P-values were adjusted for multiple comparisons (Benjamini and Hochberg 1995; Table 1). While the 15 fish used for gene expression analysis derived from a total of five different communities, inspection of the behavioral data did not reveal any co-variation between animals derived from the same community.
Table 1.
Behavioral and physiological measures averaged over the final 4 weeks for females used in gene expression studies (C = control; D = dominant; S = subordinate; n = 5 each). Bold indicates statistically significant contrasts.
| Behavior | Mean (+/− S.E.M) |
One-way ANOVA |
P-value (BH corrected randomization) |
|||||
|---|---|---|---|---|---|---|---|---|
| Control | Subordinate | Dominant | F2,12 | P | C vs. S | C vs. D | S vs. D | |
| Chase (events/5 min) | 1.057 | 0.301 | 9.595 | 43.82 | <0.0001 | 0.8067 | 0.0006 | 0.0003 |
| (0.308) | (0.116) | (1.309) | ||||||
| Border fight | 0 | 0 | 0.457 | 9.12 | <0.0001 | 0.9700 | 0.0003 | 0.0003 |
| (0) | (0) | (0.151) | ||||||
| Threat (events/5 min) | 0.025 | 0.109 | 1.898 | 14.06 | <0.0001 | 0.9098 | 0.0009 | 0.0012 |
| (0.025) | (0.109) | (0.476) | ||||||
| Flee (events/5 min) | 5.046 | 5.303 | 0.252 | 22.04 | 0.0004 | 0.8821 | 0.0025 | 0.0025 |
| (0.638) | (0.829) | (0.081) | ||||||
| School % time | 88.821 | 62.011 | 6.605 | 25.75 | <0.0001 | 0.2835 | <0.0001 | 0.0344 |
| (2.647) | (13.951) | (1.79) | ||||||
| GSI | 0.009 | 0.052 | 0.094 | 10.23 | 0.0099 | 0.1413 | 0.0006 | 0.1413 |
| (0.002) | (0.017) | 0.011 | ||||||
| Growth % change/week | 0.006 | 0.009 | 0.018 | 3.41 | 0.0585 | 0.5986 | 0.0744 | 0.1632 |
| (0.0005) | (0.005) | (0.003) | ||||||
| DI | −3.964 | −4.893 | 11.186 | 71.34 | <0.0001 | 0.860 | 0.0006 | 0.0003 |
| (0.917) | (0.869) | (1.353) | ||||||
Bold indicates statistically significant contrasts.
Microarray analysis of neural gene expression
RNA was extracted from homogenized brains (Tissue Tearor, Biospec products) according to the standard Trizol protocol (Invitrogen: Thermo Scientific, Waltham MA) and inspected for integrity on the Bioanalyzer (Agilent: Santa Clara CA) prior to indirect RNA labeling. Briefly, each RNA sample (2 ug each) was labeled twice, once with Cy3 and once with Cy5 through a balanced loop design (gene expression omnibus [GEO] GEO accession: GSE84470).
Samples were competitively hybridized to the brain-specific cDNA array from A. burtoni (Renn et al. 2004; GEO PLATFORM GLP928) at 65°C for 12–16 hours. Arrays were scanned on an Axon 4000B arrays scanner (Genepix 4.0; Molecular Devices: Sunnyvale CA). The features of this array were annotated by BLASTing the EST sequences to the set of NCBI (National Center for Biotechnology Information) predicted genes from the Tilapia genome sequence (Brawand et al. 2014). The Tilapia genome was selected over A. burtoni due to its superior assembly and annotation.
After flagging for bad feature morphology and hybridization artifacts, raw data were imported into R (R Development Core Team, 2010) for quality filtering and loess normalization using the Linear Models for Microarray Data package (LIMMA v 1.6.6; Smyth et al. 2004). Hybridization ratios were averaged across features that represent the same predicted Tilapia gene prior to fitting a linear model accounting for biological and technical replicates to extract the pairwise contrasts of interest (Smyth et al. 2004). For statistical analysis, LIMMA uses an empirical Bayes method to moderate the standard errors of the estimated log fold-differences (Smyth 2005). Venn Diagrams scaled to relative group numbers were drawn according to (Micallef and Rodgers 2014).
Meta-analyses
For meta-analyses of gene expression profiles associated with social dominance, we applied the same analysis pipeline described above to re-analyze additional datasets (Table 5) that have been produced with the A. burtoni spotted cDNA microarray. We then used the gene expression profile for social dominance in females as a “signature query” to mine other whole brain gene expression studies related to social dominance (see Supplementary Table 2 for analysis results). By focusing our meta-analysis on studies that have employed the A. burtoni cDNA microarray we avoid the need to determine orthology across species.
Table 5.
Datasets queried in meta-analyses
| Species | Contrasts analyzed | No. of arrays | Citation | Dataset |
|---|---|---|---|---|
| A. burtoni | territorial male vs. non-territorial male | 30 | Renn et al. 2008 | GSE10624 |
| A. burtoni | male vs. female | 30 in duplicate | Renn et al. 2008 | GSE10624 |
| A. burtoni | Maternal female vs. brooding female | 24 | Unpublished (Renn et al. 2016) | |
| J. marlieri | Dominant female vs. subordinate male | 10 | Schumer et al. 2011 | GSE23094 |
| N. brichardi | Brooding female vs. helper female | 31 | Aubin-Horth et al. 2007 | |
| S. ocellatus | Nest male vs. sneaker male | 9 (used) | Stiver et al. 2015 | Fungal gene database ID no. 164 |
RRHO analysis
Most existing methods of comparing gene expression datasets require setting arbitrary cut-offs based either on fold-differences or on statistical significance. In order to compare the underlying mechanisms of social dominance in a less biased approach, we used the nonparametric RRHO (Plaisier et al. 2010). This method allows the identification of statistically significant overlap (correlation either negative or positive) between gene signatures across the different species and conditions. The genes sets were first ranked according to LIMMA estimated coefficient (fold difference) with rank 1 being assigned to the gene with greatest expression bias toward the phenotype hypothesized to be most dominant-like. Genes were then plotted according to rank using x-axis and y-axis for the two experiments. Overlap between ranked gene lists is measured as the degree of statistical enrichment using a hypergeometric distribution while stepping across all possible thresholds through the two ranked lists generated in separate experiments. The gray scale values represent the Benjamini–Yekutieli (BY) corrected false discovery rate (FDR) (Benjamini and Yekutieli 2001) log transformed hypergeometric P-value for the likelihood of observing the given number of overlapping genes between the two rank thresholds at each step along the plot. Step size was set at 50 genes for all comparisons except the smaller wrasse dataset for which a step size of 20 was used. Analyses were conducted with the RRHO package, (Rosenblatt and Stein 2014) in R (R Development Core Team 2010) and plots were created with the filled.contour function in R (Cleveland 1993).
This approach complements threshold algorithms by providing more information about the strength and regions of overlap between two datasets without truncating gene expression profiles. Spearman rank correlation P-values were also calculated between the pair-wise compared gene signatures for RRHO plots, revealing weak but significant correlations in several comparisons. RRHO plots appeared very similar when genes were ranked according to the signed rank P-values obtained from the LIMMA analyses (data not shown).
Results and discussion
Female phenotypes resemble those of males
While not intended to accurately mimic ecologically relevant conditions, male removal reproducibly promotes a dominant female phenotype in A. burtoni (Renn et al. 2012; O'Connell et al. 2013). A subset of the females display overt aggressive behaviors, allowing individuals to be classified as dominant or subordinate according to standard measures of dominance (Fernald and Hirata 1977a). Dominant females display increased chasing behavior, border conflicts, and threat displays, and reduced fleeing behavior and shoaling. Subordinate females do not differ significantly from control brooding females housed in standard mixed sex tanks (Table 1; Supplementary Figure 1). These behavioral differences resulted in the expected relative dominance indices (DI; DOM: 11.2 ± 1.4; SUB: −4.9 ± +0.9 CON: −3.9 ± 0.9). The three female phenotypes did not differ significantly with regard to GR. As previously reported (Renn et al. 2012; O'Connell et al. 2013), despite a slight trend toward a greater gonosomatic index (GSI) for dominant females relative to subordinate females, the GSI for these phenotypes did not differ significantly. However, dominant females did have a significantly greater GSI than control brooding females that do not feed nor ripen eggs. In summary, social phenotypes of dominance and subordinance in female quantitatively parallel the well-studied male phenotypes associated with access to mates and reproductive success. In females, these phenotypes can be produced independent of reproductive state and other confounding physiological variables.
Gene expression profile of female dominance
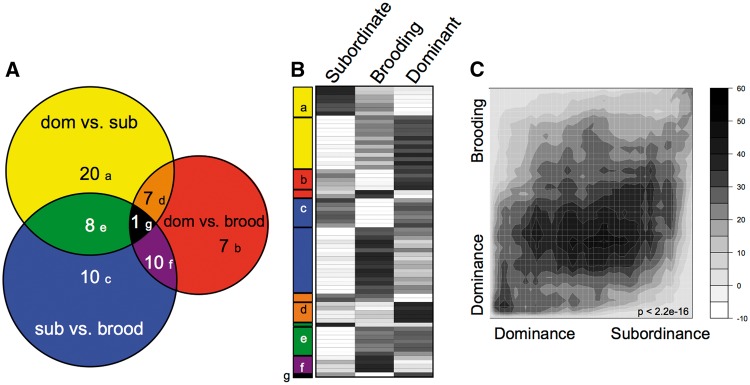
To identify the gene expression profile associated with social dominance in females, we competitively hybridized whole brain samples from dominant, subordinate, and brooding females to the A. burtoni spotted cDNA microarray (Renn et al. 2004). It should be noted that the more comprehensive expression profiles enabled by the use of whole brains comes at the cost of specificity with regard to localized gene regulation in different brain regions. The A. burtoni spotted cDNA microarray contains features representing 1981 different annotated cichlid genes, 1753 of which genes passed quality filters and were analyzed. A total of 70 genes showed differential expression among pairwise comparisons at an alpha value of 0.05 FDR (Fig. 1A). Dominant and subordinate phenotypes showed significant differences in gene expression level for a total of 36 genes with a bias toward increased expression in the dominant phenotype (26 genes) and fewer genes with increased expression in the subordinate phenotype (10 genes; Fig. 1B; Supplementary Table 1). The comparison of dominant and brooding females showed a similar number of differentially expressed genes with a total of 25 genes. Again, there was a bias toward increased gene expression level in the dominant phenotype (17 genes) and fewer genes showing increased expression in the subordinate phenotype (8 genes). When the two gene sets are compared, they share 8 genes, which is a small but statistically significant number (hypergeometric test: P < 0.00001). Interestingly, when the statistical threshold for the second list is relaxed (P < 0.05 no FDR), we find 44% of the dominance-related genes to show the same expression bias, i.e., regulated in a concordant manner in subordinate and control females relative to dominant females (hypergeometric test: P < 0.00001).
Fig. 1.
Socially regulated and reproductively regulated neural gene expression in A. burtoni females. (A) Scaled Venn diagram depicts overlap of differentially expressed genes (P < 0.05 FDR) for each pair-wise comparison in this study (P < 0.05 FDR; Supplementary Table 1). (B) Heatmap indicates the relative expression level (black: high; white: low) in each phenotype for these 70 differentially expressed genes, grouped according to the Venn diagram modules and further subdivided by direction of regulation within the module. (C) Rank-rank hypergeometric map demonstrates an overall correlation between the gene expression profile for dominance compared with subordinance and to the brooding reproductive phenotype. Scale bar depicts the relative range of BY-corrected, −log10 transformed hypergeometric P-values for the RRHO analysis. P-value indicates Spearman rank correlation of the fold-change data.
This apparent correlation between the two gene lists becomes very clear in the RRHO analysis (Fig. 1C). RRHO analyses (Plaisier et al. 2010) identify statistically significant overlap (correlation either negative or positive) between gene signatures in the absence of an arbitrary statistical cut-off or classification of genes. The RRHO analysis (see methods) reveals a clear overlap (positive correlation) between the full ranked gene lists for dominance relative to subordinance and dominance relative to brooding phenotype across a range of possible thresholds of these two ranked lists.
Genes associated with social status in females
A traditional approach to analyzing gene sets from ecologically relevant transcriptome studies involves comparing the gene annotations (derived largely from BLAST similarity) to the literature to derive novel hypotheses for the current study. For functional annotation of the gene sets, we rely on the Tilapia genome annotations rather than those for A. burtoni due to the superior assembly and annotation of the former (Brawand et al. 2014). Among the gene lists, we find repeated functional themes, expected candidates, and novel new hypotheses.
As predicted, several hormonal and neuropeptide genes showed increased expression in dominant females. Both prolactin paralogs were strongly up-regulated in dominant females compared with subordinate females. Elevated prolactin levels are associated with territorial aggression in stickleback males (Sanogo et al. 2012) and hostility and aggression in women (Barry et al. 2015). Arginine vasotocin was also up-regulated in dominant females, which is consistent with findings in other fishes and in A. burtoni males (Greenwood et al. 2008; Silva et al. 2013; Yaeger et al. 2014). The glycoprotein alpha polypeptide subunit (CGA), a necessary precursor step in the production of active gonadotropin-releasing hormone (GnRH), luteinizing hormone, follicle-stimulating hormone, and thyrotropin, was up-regulated in dominant females. Notably, differential GnRH levels in the preoptic area have been associated with the transition from subordinate to dominant phenotype in male A. burtoni (Maruska and Fernald 2010) and independent of reproductive context in the pituitaries of male Cichlasoma dimerus (Alonso et al. 2012). CGA is also up-regulated in territorially challenged stickleback males (Sanogo et al. 2012) and seasonally up-regulated in aggressive song sparrow males (Mukai et al. 2009). Interestingly, neither form of GnRH, nor the GnRH receptor, showed differential expression according to female social status as is seen in males. However, GnRH was significantly down-regulated in the control brooding females, which are not allocating energy to egg maturation. This result would suggest that among females regulation of GnRH is not associated with the aggressive dominance; however, soma size has been correlated with female dominance in a similar iso-sex aggressive female paradigm with A. nigrofasciatus, a bi-parental cichlid species (Nesjan et al. 2014), which might suggest that the neuropeptide levels are regulated elsewhere in the pathway, but not at the level of gene expression. Interestingly, somatolactin, a pituitary protein hormone that, in coordination with GnRH (Canepa et al. 2008), is associated with both reproductive status and dominance status in fishes (Trainor and Hofmann 2006), was up-regulated in dominant females. Also concurring with previous studies in male A. burtoni (Renn et al. 2008; Huffman et al. 2013; Goppert et al. 2016), brain aromatase, a key enzyme catalyzing the local biosynthesis of estrogens from testosterone, was up-regulated in dominant females. There are generally positive, but species- and receptor-type-specific effects of brain estrogen and aromatase levels on aggressive behavior in rodents (reviewed in Nelson and Trainor 2007; Wu et al. 2009), birds (reviewed in Trainor et al. 2006), and fish (e.g., peacock blenny males: Goncalves et al. 2008; midshipmen: Forlano et al. 2015).
In many vertebrates, social status is associated with the production and differentiation of neurons, particularly in brain regions associated with behavior and learning. In rats, dominance is associated with increased survival of new neurons. Among the list of genes associated with dominance status in our female A. burtoni, we find several genes that are implicated in neuroplasticity. Four of these genes—FK506-binding protein 1, cell cycle associated protein 1, neuromodulin, and dynamin-1—were up-regulated in dominant females, while one neuroplasticity gene, voltage-dependent N-type calcium channel subunit alpha-1B, was expressed at higher levels in subordinate females. Neuroplasticity gene networks have also been found to be differentially regulated in response to territorial intrusion and season in song sparrow males (Mukai et al. 2009), and in response to both social status and aggressive interactions in zebrafish (Teles et al. 2016). In addition to the relationship between neuroplasticity and aggression, up-regulation of neuroplasticity genes has been associated with dominant phenotypes in fish (Sorensen et al. 2013) and mammals (Kozorovitskiy and Gould 2004; Hoshaw et al. 2006).
Transcriptional modules of territorial dominance in A. burtoni
Another traditional approach to analyze gene expression results is to consider coordinately regulated gene sets as “transcriptional modules” (Segal et al. 2004) and to look for interesting intersection with other predefined categories (often based on protein structure, or Gene Ontology assignment). Alternatively, modules identified beforehand in previously published datasets that were linked to a phenotype of interest can be used as a hypothesis to be tested in a new context (Landry and Aubin-Horth 2007). To test the hypothesis that there are “core” modules of neural gene expression associated specifically with social dominance, we examined the concordance between the gene set produced in the current study and that produced from a parallel analysis of whole brain gene regulation in dominant and subordinate males of A. burtoni (Renn et al. 2008). Importantly, the dominant and subordinate female phenotypes examined in the current study are not confounded by differences in GR or reproductive activity; by comparing the gene sets identified in the two studies, we aim to identify gene modules that underlie shared aspects of dominance under the two contexts.
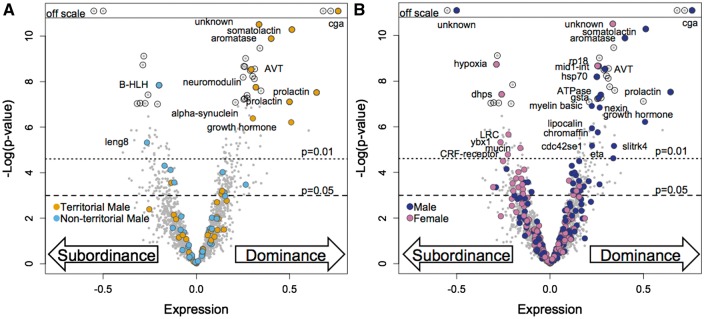
Rather than directly comparing the lists of male dominance-related genes to the lists of female dominance-related genes generated by a similar P-value threshold, we visualize gene expression bias by querying the behavior of the previously defined gene set in the current study by mapping those genes onto a volcano plot (Fig. 2). This approach is less constrained than applying a statistical threshold to both datasets, which may have different statistical power (Townsend 2004). With this approach, it is readily apparent that a subset of the most up-regulated female dominance-related genes derives from the gene set previously identified as male dominance-related (Fig. 2A). It is also evident that this bias is strongest for genes of high statistical significance in the current study. The previous study also identified genes associated with sex. Those genes previously identified as male-biased appear to be dominance-related in females even at weaker P-values (Fig. 2B). At a P-value threshold of 0.05 without FDR 70 sex-biased dominance-related genes followed this pattern (39 male-biased genes up-regulated in dominant females and 31 female-biased genes down-regulated in dominant females; Tables 2 and 3) and only 26 genes showed the opposite pattern (nine male-biased genes are up-regulated in dominant females, and 17 female-biased genes are down-regulated in dominant females; Table 4). This apparent “masculinization” of dominant females may not be surprising given the lack of consistent sex-bias in neural gene expression patterns that has been observed in other organisms (Yang et al. 2006; Baker et al. 2007; Mank and Ellegren 2009), including fish (Manousaki et al. 2014) as demonstrated by microarray (Machado et al. 2009) and RNAseq (Boehne et al. 2014) studies in cichlids. Many sex-biased genes are highly regulated according to social phenotype. This corroborates studies at the single gene level that show that sex-specific patterns expression for important neuropeptides are often species-specific (Kelly and Goodson 2014; Almeida and Oliveira 2015).
Fig. 2.
Volcano plots representing gene expression bias associated with dominance phenotypes. Genes (gray circles) are plotted according to expression level in the comparison of dominant versus subordinate A. burtoni females (x-axis) and the −log10 P-value for differential expression (y-axis). P-value thresholds are indicated (dotted line = 0.01, dashed line = 0.05) and genes meeting the FDR correction at P < 0.05 are highlighted with black circles. In each plot, the genes are highlighted with color if they were also found to be differentially expressed in the comparison of (A) territorial and non-territorial males (P < 0.01) (B) male and female (P < 0.01) Abbreviated gene names are provided for the overlapping genes with significant gene expression bias in both comparisons.
Table 2.
In total, 39 genes show consistent expression biases between female dominance and sex (P < 0.05)
| Male-biased genes associated with female dominance | |
|---|---|
| GenBank accession number | Abbreviated gene annotation |
| NM_001279786.1 | prolactin |
| NM_001279792.1 | prolactin |
| XM_003439275.3 | carbonic anhydrase 1-like |
| XM_013274638.1 | tubulin beta chain |
| XM_003441601.3 | somatolactin |
| XM_003442542.3 | growth hormone |
| XM_003443505.3 | lipocalin |
| XM_003445209.3 | mid1-interacting protein 1 |
| XM_003445709.3 | ATP synthase F(0) |
| XM_003446089.3 | vasotocin-neurophysin |
| XM_003448890.3 | hsp70 |
| XM_003452055.3 | pleiotrophin |
| XM_003454957.3 | alpha-synuclein |
| XM_003455030.3 | tubulin, beta 4B |
| XM_005450809.2 | brain aromatase |
| XM_003456457.3 | RING finger protein 150 |
| XM_005468883.2 | uncharacterized |
| XM_005473790.2 | glutathione S-transferase |
| XM_005450664.1 | myelin basic protein |
| XM_005476705.2 | sorting nexin |
| XM_005455478.2 | ubiquitin specific peptidase 5 |
| XM_005460357.2 | cat eye syndrome chromosome region |
| XM_005461967.2 | CDC42 small effector 1 (cdc42se1) |
| XM_005467407.1 | ATPase, Na+/K+ transp. beta 4 polypeptide |
| XM_005450176.2 | CXXC-type zinc finger protein |
| XM_005448790.2 | basic helix-loop-helix family (bhlhe23) |
| XM_003455305.3 | phosphoserine aminotransferase 1 (psat1) |
| XM_005476734.2 | ATP-sensitive inward rectifier potassium channel |
| XM_005477075.2 | glycoprotein hormones, alpha polypeptide (cga) |
| XM_013265264.1 | arf-GAP w/GTPase, ANK repeat & PH domain |
| XM_013269879.1 | corticotropin-releasing factor receptor |
| XM_013270440.1 | myelin basic protein |
| XM_013271142.1 | Y box binding protein 1 (ybx1) |
| XM_013271984.1 | heat shock protein 90kDa alpha (cytosolic) |
| XM_013272174.1 | receptor-type tyrosine-protein phosphatase |
| XM_003439383.3 | coactosin-like F-actin binding protein 1 (cotl1) |
| XM_013275731.1 | serine/threonine-protein phosphatase 2A |
| XM_013276263.1 | kinectin 1 (kinesin receptor) |
| XR_269170.2 | uncharacterized |
Table 3.
In total, 31 genes that show consistent expression biases between female subordinance and sex (P < 0.05)
| Female-biased genes associated with female subordinance | |
|---|---|
| GenBank accession number | Abbreviated gene annotation |
| XM_003442007.3 | LIM and SH3 protein 1 (lasp1) |
| XR_268729.2 | uncharacterized |
| XM_005475529.1 | von Willebrand factor A domain-containing protein |
| XM_005461142.2 | hypoxia-inducible factor 1-alpha |
| XR_001223047.1 | uncharacterized |
| XM_003445583.3 | ans-golgi network protein 2 (tgoln2) |
| XM_013274594.1 | leukocyte receptor cluster (LRC) member 8 (leng8) |
| XM_003452809.3 | deoxyhypusine synthase (dhps) |
| XM_013275471.1 | cytokinesis protein 3 |
| XR_001224604.1 | acyl-coenzyme A thioesterase |
| XM_003448942.2 | cytochrome c oxidase subunit 7A2 |
| XM_013266972.1 | pleckstrin homology domain (plekha6) |
| XM_005469874.2 | mucin-5AC |
| XM_005473424.2 | proprotein convertase subtilisin |
| XM_013277052.1 | human C3orf58 (clg9h3orf58) |
| XM_013264267.1 | chromaffin granule amine transporter |
| XM_003438067.3 | neuronal differentiation 6 (neurod6) |
| XM_005458596.1 | uncharacterized |
| XM_013273869.1 | inositol polyphosphate phosphatase-like 1 (inppl1) |
| XM_003445682.3 | transmembrane protein 127 |
| XM_013269342.1 | clathrin heavy chain |
| XM_003456230.3 | nicolin 1 (nicn1) |
| XM_005454863.2 | nuclear GTPase SLIP-GC |
| XM_005461516.2 | SLIT and NTRK (slitrk4) |
| XM_005462511.1 | olfactory receptor 2K2 |
| XM_005458825.2 | shisa family member 4 (shisa4) |
| XM_003443990.3 | dehydrogenase E1 & transketolase (dhtkd1) |
| XM_005450930.2 | coiled-coil domain - 136 |
| XM_013271758.1 | acyl-CoA synthetase long-chain (acsl4) |
| XM_003455175.3 | early growth response 1 (egr1) |
| XM_003458435.3 | MAM domain-containing glycosylphosphatidylinositol anchor |
Table 4.
In total, 26 genes that show contradictory expression biases between dominance and sex (P < 0.05)
| Female-biased genes associated with female dominance | |
|---|---|
| GenBank accession number | Abbreviated gene annotation |
| XM_003438153.3 | schwannomin-interacting protein |
| XM_003446848.2 | ADP/ATP translocase |
| XM_005453621.2 | protein-glutamine gamma-glutamyltransferase |
| XM_003456372.3 | niloticus coagulation factor X |
| XM_003442872.3 | visinin |
| XM_005478262.2 | N-terminal EF-hand calcium-binding |
| XM_005475970.2 | glucagon family neuropeptides |
| XR_001224304.1 | uncharacterized |
| XM_003452424.2 | niloticus ribosomal protein S5 (rps5) |
| XR_134814.3 | uncharacterized |
| XM_005452080.2 | transmembrane emp24 |
| XM_013269458.1 | ADP-ribosylation factor 1 (arf1) |
| XM_013277498.1 | DnaJ (Hsp40) homolog (dnajc13) |
| XM_003444320.3 | uncharacterized |
| XM_003450625.2 | EPH receptor B6 (ephb6) |
| XM_003451557.2 | ribosomal protein L8 (rpl8) |
| XM_013274785.1 | uncharacterized |
| Male-biased genes associated with female subordinance | |
| XM_003437687.2 | synaptosomal-associated protein 25 |
| XM_003438309.3 | uncharacterized |
| XM_003448308.3 | glutamate receptor |
| XM_003459428.3 | zinc finger and BTB (zbtb14) |
| XM_005454355.1 | late endosomal/lysosomal adaptor (lamtor1) |
| XM_005469157.2 | syntaxin 1B (stx1b) |
| XM_005476752.2 | myelin protein zero (mpz) |
| XM_013272974.1 | nuclear factor 1 X |
| XM_005450398.2 | v-ral simian leukemia viral oncogene homolog B (ralb) |
By name, we see that some of these male-biased, female dominance-related genes are the same genes associated with male dominance (e.g., aromatase, AVT, prolactin, cga, and somatolactin) previously referred to as a “super male module” being up-regulated in males relative to females and further up-regulated in dominant males relative to subordinate males (Renn et al. 2008). We also see many male-biased genes that were not associated with the male dominant phenotype (e.g., Hsp70, nexin, eta, lipocalin). While it is tempting to speculate that genes identified as a module in one study and not in another study represent mechanisms that are specific to the unique aspects on the phenotype, such hypotheses must be evaluated cautiously because lack of statistical difference does not imply equivalence (Eijgelaar et al. 2010; Qiu and Cui 2010). Also, genome-scale data are often inherently noisy, and the functional threshold of equivalence will vary greatly across genes. Nonetheless, it is interesting to ask about consistent trends or expression biases that fail to meet our statistical thresholds.
Meta-analysis
Transcriptome analyses facilitate genome-wide investigation of mechanisms that influence higher-order phenotypes. One approach to capitalize on such large datasets and genome-wide studies is to move beyond the discussion of single genes or even discrete identified modules and examine the patterns of gene expression free from the constraints imposed by setting arbitrary cutoffs, or relying on oversimplified classification systems (Landry and Aubin-Horth 2007). Applied to transcriptomics in wild populations (Aubin-Horth et al. 2005; Aubin-Horth et al. 2009; Whitehead 2012; Ledon-Rettig et al. 2013; Alvarez et al. 2015), these tools can extract a wealth of untapped “functional” information for thousands of genes. The RRHO approach (Plaisier et al. 2010) can be applied across experiments and even across species to identify correlation between gene signatures by testing for significant overlap in two ranked gene lists using a moving threshold across the full range. Here, we show how this approach can complement the interrogation of gene modules described above.
We collected available expression data for other whole brain gene expression studies of social dominance phenotypes and applied the same pipeline for reanalysis to generate ranked gene lists according to fold-difference without applying a statistical threshold (Table 5). We used our own expression data, ranked for female social dominance, to query these other gene expression studies for similarity. In doing so, we reveal interesting correlations between dominance signature in females and dominance signatures in other contexts and other species. While largely championed in the biomedical literature, this approach lends itself well to studies of the evolution of gene expression associated with complex phenotypes. Here, we avoid issues of orthology by focusing on studies that employed the A. burtoni cDNA microarray.
Overlap with sex and social dominance in A. burtoni
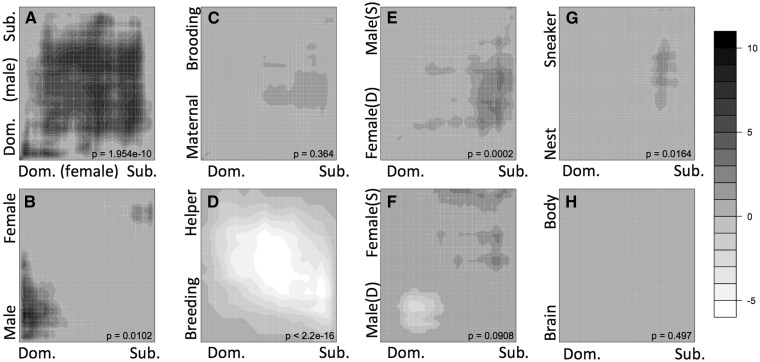
To complement the fruitful gene set level comparison of female and male dominance-related transcriptomes, we used RRHO analysis to determine the extent of correlation as a further test of the hypothesis that the molecular mechanisms for male and female dominance are shared (in part). A weak correlation would refute this hypothesis, instead suggesting that selection on females has resulted in female-specific mechanisms, either in reaction to maladaptive sexual conflict or in response to female-specific adaptive pressures. Interestingly, while the highest ranked genes in both lists produce a region of strong overlap (bottom left corner of the plot), there is also detectable significant overlap in lower ranges of the ranked gene lists (dark vertical and horizontal bands; Fig. 3A). This correlation is not as strong as was seen comparing two lists for dominant females (Fig. 1C), highlighting the substantial differences between expression profiles for male and female dominance.
Fig. 3.
RRHO plots demonstrate that expression patterns for different forms of dominance show patterns of overlap. These representations are based on the full set of genes analyzed in each experiment pair and do not apply any statistical threshold cut-offs. For both lists, the genes were ranked according to the degree of expression bias (fold-difference). In all datasets, genes are ranked from 1 being the greatest bias toward dominant-like phenotype (shown in the lower left corner of each plot) to greater rank numbers for expression bias toward subordinate-like phenotypes. The rank list of female social dominance (x-axis) is compared to eight gene lists derived from (A) male social dominance in male A. burtoni (Renn et al. 2008), (B) sex bias in male versus female A. burtoni (Renn et al. 2008), (C) maternal aggression post release of fry versus mouth brooding stage in A. burtoni (Renn unpublished), (D) female social status of breeder versus helper females in the cooperatively breeding cichlid N. brichardi (Aubin-Horth et al. 2007), (E) sex-role dominance of aggressive females versus submissive males in the sex-role reversed cichlid J. marlieri (Schumer et al. 2011), (F) sex-role dominance of aggressive males versus submissive females in the sex-role conventional cichlid J. transcriptus (Schumer et al. 2011), (G) male breeding tactic of nest males versus sneaker males in the wrasse S. ocellatus (Stiver et al. 2015), (H) non-social regulation of gene expression between brain and body in A. burtoni (Renn et al. 2004) showed no overlap. Overlap was evaluated using a step size of 50 genes for all comparisons except G where step = 20. Heat map gray scale intensity indicates the significance of overlap between the set of genes from each list that are ranked above that sliding rank threshold position (i.e., between that point and the lower left corner). Scale bar indicates BY-corrected, negative natural log transformed hypergeometric P-values for each comparison except that D is plotted as −log10. By convention of the RRHO analysis, negative values indicate under enrichment and positive values indicate over-enrichment such that D represents an overall negative correlation. P-values provided are Spearman rank correlation of the fold change data.
As applied to the “masculinization” in dominant females seen in the threshold gene module analysis above, RRHO analysis (Fig. 3B) revealed correlation only between genes at the tops of the gene lists (bottom left corner) and not elsewhere throughout the ranked lists. This pattern demonstrates that the genes most over-expressed in males relative to females are the primary genes that are coordinately regulated in the female comparison. In this case, the traditional thresholds used to identify modules and gene sets did present a fairly comprehensive picture for the relationship between the two forms of dominance.
Overlap with maternal aggression in A. burtoni
While female dominance in the current study was induced by manipulating the social environment, females that engage in maternal care naturally become highly aggressive toward intruders and dominant toward other females while interacting socially with their fry (Renn et al. 2009). RRHO analysis allows us to determine the extent to which our induced female dominance outside of the reproductive context recapitulates the rich social phenotype of maternal care (Carleton 2009, unpublished; Fig. 3C). In that study, the brooding females showed low levels of aggression, while post-release, maternal females showed high levels of aggression, similar to our socially dominant females. Perhaps surprisingly, the correlation in these datasets is modest, and restricted to a narrow range of gene ranks suggesting that maternal aggression involves dramatically different sets of genes to produce the full maternal phenotype including care as well as dominance. These results suggest that RRHO analysis will be a useful tool with which to quantify similarity across stages of the female reproductive cycle as behavioral phenotype varies.
Overlap with breeding status in Neolamprologus brichardi
In the cooperatively breeding cichlid species N. brichardi, there is a dominance hierarchy established between the dominant breeding pair and the subordinate helpers (both male and female; Balshine-Earn et al. 1998; Taborsky and Grantner 1998; Buchner et al. 2004; Desjardins et al. 2008). Clustering analysis and Principal Component Analysis of microarray results demonstrated that whole brain gene expression profiles are driven by phenotype rather than by sex (Aubin-Horth et al. 2007). Our RRHO analysis reveals an overall negative correlation between transcription profiles of status in N. brichardi females and social dominance in A. burtoni females (Fig. 3D), a pervasive characteristic of genes throughout the mid-rank ranges (moderate fold-differences). The complex social phenotypes in N. brichardi revolve around parental care and nest defense in a breeding system that is very different from that of A. burtoni, which might have predicted a lack of correlation, but the observed negative correlation is quite surprising. The same negative correlation occurs when we query the transcription profile for male status in N. brichardi (data not shown). A negative correlation could be caused by a need for dominant breeding pairs to rein in aggression in order to provide for parental care. At the level of hormones, it is known that the high hormone titers for testosterone that are associated with aggression compromise parental care (Trainor and Marler 2001; Lynn 2008).
Overlap with sex-bias phenotypes in Julidochromis
The evolvability of mating systems in cichlids (Koblmuller et al. 2005) is evident in the genus Julidochromis, comprised of biparental substrate brooding species (Koning 1998) that show either conventional or reversed sex-biased behaviors, with sex roles enforced through aggression from the dominant territorial individual toward the subordinate individual that provides egg care. Julidochromis transcriptus exhibits the conventional sex-biased behavior, with males primarily performing the territory defense and females the egg care; the sister species J. marlieri exhibits reversed sex-biased behavior, where females tend to be larger, behave aggressively, and engage in the territory defense (Yamagishi and Kohda 1996; Barlow and Lee 2005; Wood et al. 2014). Therefore, the ranked gene list for J. marlieri female versus male (Schumer et al. 2011) represents an expression profile of female dominance that, according to inspection of the RRHO analysis, shares a weak but significant correlation with the expression profile of female social dominance in A. burtoni (Fig. 3E). This correlation is strongest for genes that are up-regulated in subordinate A. burtoni females (right side of the plot) and distributed through a range of the J. marlieri ranked list. Similarly, regions of significant overlap are detected when the behaviorally conventional sex-biased J. transcriptus male versus female gene list is compared with the A. burtoni female social dominance expression profile (Fig. 3F). Here, ranges of both over-representation (black) and under-representation (white) can be seen, revealing a complex relationship between these two instances of social dominance.
Overlap with alternative reproductive tactics in wrasse
A recent study in the ocellated wrasse Symphodus ocellatus allows comparison to dominance relationships associated with alternative male reproductive tactics (ARTs; Stiver et al. 2015). In this species, males exhibit one of three tactics (nesting, satellite, and sneaker) throughout a reproductive season, but may switch tactics between years (Alonzo et al. 2000). Brain transcriptomes of satellites and females were most similar to each other, and intermediate to nesting and sneaker males, which show both the greatest difference in gene expression profile and the greatest difference in dominance-related behaviors (Stiver et al. 2015). These social phenotypes vary in a number of traits, including aggression, territoriality, and cooperation, and provide an additional comparison for the current dataset. Here, the RRHO heatmap reveals only a very weak and restricted range of overlap (Fig. 3G). While this lack of correlation may reflect the dramatic behavioral differences between dominance mechanisms that underlie wrasse ARTs and female social dominance in A. burtoni, these results may be partially obscured by phylogenetic distance and complications of heterologous hybridization (Buckley 2007; Machado et al. 2009; Renn et al. 2010).
Brain versus body tissue comparison
As a negative control, we selected a dataset comparison largely free from the influence of social phenotype. This comparison of brain and body tissues (mixed skin, muscle and blood vessel; Renn et al. 2004) was previously used to asses heterologous hybridization across a phylogenetic range. As expected, the RRHO plot reveals an absence of any detectable correlation (Fig. 3H). While this negative control does not thoroughly validate the significance of the other detected correlations among social phenotype signatures, it is encouraging to see that gene expression differences between tissues do not correlate with those studied within a tissue, namely the brain.
Summary
We demonstrate a meta-analysis of high-throughput expression profiling experiments to investigate the gene expression patterns that underlie social dominance across contexts, sex, and species using by analyses that rely on significance thresholds to differing degrees. We were able to control for GR, sex, and reproduction by artificially inducing a social hierarchy among females. This experiment provided an opportunity to address the proximate mechanisms of social dominance while controlling for confounding variables. We demonstrate a traditional approach comparing our gene set its male counterpart to reveal a core module of genes associated with social dominance, and an interesting up-regulation of genes previously identified as male-biased. This finding supports many recent studies that suggest sex-bias in gene expression pattern is highly evolvable and highly plastic undermining the traditional “masculine” and “feminine” designations for neural gene expression profiles. Further experiments are needed to uncover the discrete behaviors shared or altered between sexes and contexts that are associated with these gene expression patterns. This module approach is complemented by threshold free approaches that allow us to both test a priori hypotheses and generate new hypotheses. Beyond modules of highly expressed genes, RRHO analyses show robust correlation throughout the gene expression profile for male and female dominance. Such analyses also reveal the degree to which the maternal phenotype differs from simple female dominance within A. burtoni. In comparison to dominance phenotypes in other cichlid species female dominance in A. burtoni is actually negatively correlated with female dominance mechanisms in a cooperatively breeding species and shows complex relationships with sex-biased patterns in conventional and sex-role reversed cichlid species. These results suggest that RRHO analysis will be a useful tool with which to compare the mechanisms underlying outwardly similar behavioral phenotypes across contexts and species.
Supplementary Material
Acknowledgments
We wish to thank Josiah Altshuler for fish husbandry, Albyn Jones for statistical consulting, and Rose Driscoll, Caitlin Friesen, Rebecca Young-Brim, and two anonymous reviewers for editorial suggestions on the manuscript. This research was supported by the Harvard College Research Program to E.J.F.; an FQRNT postdoctoral fellowship, an NSERC postdoctoral fellowship and a Natural Sciences and Engineering Research Council of Canada thorough the Discovery grant program to NAH; NIH grant NIGMS-GM068763, Harvard’s Bauer Center for Genomics Research, NSF grants IOS-1354942 & IOS-1501704, and by the NSF BEACON Center for Science and Technology to HAH; and an NIH-NRSA post-doctoral fellowship, NSF grant IOS-0818957 and Murdock Life Sciences Grant to SCPR.
Funding
This research was supported by the Harvard College Research Program to E.J.F.; an FQRNT postdoctoral fellowship, an NSERC postdoctoral fellowship and a Natural Sciences and Engineering Research Council of Canada thorough the Discovery grant program to NAH; NIH grant NIGMS-GM068763, Harvard's Bauer Center for Genomics Research, NSF grants IOS-1354942 & IOS-1501704, and by the NSF BEACON Center for Science and Technology to HAH; and an NIH-NRSA post-doctoral fellowship, NSF grant IOS-0818957 and Murdock Life Sciences Grant to SCPR.
Supplementary data
Supplementary data available at ICB online.
References
- Almeida O, Oliveira RF. 2015. Social status and arginine vasotocin neuronal phenotypes in a cichlid fish. Bra Behav Evol 85:203–13. [DOI] [PubMed] [Google Scholar]
- Alonso F, Honji RM, Moreira RG, Pandolfi M. 2012. Dominance hierarchies and social status ascent opportunity: anticipatory behavioral and physiological adjustments in a Neotropical cichlid fish. Physiol Behav 106:612–8. [DOI] [PubMed] [Google Scholar]
- Alonzo SH, Taborsky M, Wirtz P. 2000. Male alternative reproductive behaviours in a Mediterranean wrasse, Symphodus ocellatus: evidence from otoliths for multiple life-history pathways. Evol Ecol Res 2:997–1007. [Google Scholar]
- Alvarez M, Schrey AW, Richards CL. 2015. Ten years of transcriptomics in wild populations: what have we learned about their ecology and evolution? Mol Ecol 24:710–25. [DOI] [PubMed] [Google Scholar]
- Aubin-Horth N, Desjardins JK, Martei YM, Balshine S, Hofmann HA. 2007. Masculinized dominant females in a cooperatively breeding species. Mol Ecol 16:1349–58. [DOI] [PubMed] [Google Scholar]
- Aubin-Horth N, Landry CR, Letcher BH, Hofmann HA. 2005. Alternative life histories shape brain gene expression profiles in males of the same population. Proc Roy Soc B Biol Sci 272:1655–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Horth N, Letcher BH, Hofmann HA. 2009. Gene-expression signatures of Atlantic salmon’s plastic life cycle. Gen Comp Endocrinol 163:278–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DA, Meadows LA, Wang J, Dow JA, Russell S. 2007. Variable sexually dimorphic gene expression in laboratory strains of Drosophila melanogaster. BMC Genomics 8:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balshine-Earn S, Neat FC, Reid H, Taborsky M. 1998. Paying to stay or paying to breed? Field evidence for direct benefits of helping behavior in a cooperatively breeding fish. Behav Ecol 9:432–8. [Google Scholar]
- Barlow GW, Lee JSF. 2005. Sex-reversed dominance and aggression in the cichlid fish Julidochromis marlieri. Ann Zool Fenn 42:477–83. [Google Scholar]
- Barry JA, Moran E, Thomas M, Hardiman PJ. 2015. Prolactin and hostility in hospitalised patients and healthy women: A systematic review and meta-analysis. J Obstet Gynaecol 35:499–507. [DOI] [PubMed] [Google Scholar]
- Bauernfeind AL, Reyzer ML, Caprioli RM, Ely JJ, Babbitt CC, Wray GA, For PR, Sherwood CC. 2015. High spatial resolution proteomic comparison of the brain in humans and chimpanzees. J Comp Neurol 523:2043–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc, Series B. 57:289–300. [Google Scholar]
- Benjamini Y, Yekutieli D. 2001. The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–88. [Google Scholar]
- Boehne A, Sengstag T, Salzburger W. 2014. Comparative transcriptomics in East African cichlids reveals sex- and species-specific expression and new candidates for sex differentiation in fishes. Gen Biol Evol 6:2567–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, Wagner CE, Li YI, Malinsky M, Keller I, Fan S, Simakov O, Ng AY, Lim ZW, Bezault E. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513:375–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner AS, Sloman KA, Balshine S. 2004. The physiological effects of social status in the cooperatively breeding cichlid Neolamprologus pulcher . J Fish Biol 65:1080–95. [Google Scholar]
- Buckley BA. 2007. Comparative environmental genomics in non-model species: using heterologous hybridization to DNA-based microarrays. J Exp Biol 210:1602–6. [DOI] [PubMed] [Google Scholar]
- Burmeister SS, Kailasanath V, Fernald RD. 2007. Social dominance regulates androgen and estrogen receptor gene expression. Horm Behav 51:164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canepa M, Pozzi A, Astola A, Maggese MC, Vissio P. 2008. Effect of salmon melanin-concentrating hormone and mammalian gonadotrophin-releasing hormone on somatolactin release in pituitary culture of Cichlasoma dimerus. Cell Tiss Res 333:49–59. [DOI] [PubMed] [Google Scholar]
- Carleton JB. 2009. One bad mother: functional genomics of maternal aggression in Astatotilapia burtoni [Thesis]. Reed College Portland OR USA. [Google Scholar]
- Cleveland WS. 1993. Visualizing Data. Hobart Press, Summit, New Jersey, USA. [Google Scholar]
- Desjardins JK, Stiver KA, Fitzpatrick JL, Milligan N, Van Der Kraak GJ, Balshine S. 2008. Sex and status in a cooperative breeding fish: behavior and androgens. Behav Ecol Sociobiol 62:785–94. [Google Scholar]
- Eijgelaar WJ, Horrevoets AJG, Bijnens A-PJJ, Daemen MJAP, Verhaegh WFJ. 2010. Equivalence testing in microarray analysis: similarities in the transcriptome of human atherosclerotic and nonatherosclerotic macrophages. Physiol Gen 41:212–23. [DOI] [PubMed] [Google Scholar]
- Fernald RD. 2004. Social influences on the brain. Horm Behav 46:129–30. [Google Scholar]
- Fernald RD, Hirata NR. 1977a. Field study of Haplochromis burtoni: quantitative behavioral observations. Anim Behav 25:964–75. [Google Scholar]
- Fernald RD, Hirata NR. 1977b. Field study of Haplochromis burtoni: habitats and co-habitants. Environ Biol Fish 2:299–308. [Google Scholar]
- Forlano PM, Sisneros JA, Rohmann KN, Bass AH. 2015. Neuroendocrine control of seasonal plasticity in the auditory and vocal systems of fish. Front Neuroendocrinol 37:129–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goncalves D, Teles M, Alpedrinha J, Oliveira RF. 2008. Brain and gonadal aromatase activity and steroid hormone levels in female and polymorphic males of the peacock blenny Salaria pavo . Horm Behav 54:717–25. [DOI] [PubMed] [Google Scholar]
- Goppert C, Harris RM, Geis A, Hofmann HA, Salzburger W, Bohne A. 2016. Inhibition of aromatase induces partial sex change in a cichlid fish: Distinct functions for sex steroids in brains and gonads. Sex Devel 10:97–110. [DOI] [PubMed] [Google Scholar]
- Goymann W, Villavicencio CP, Apfelbeck B. 2015. Does a short-term increase in testosterone affect the intensity or persistence of territorial aggression? - An approach using an individual’s hormonal reactive scope to study hormonal effects on behavior. Physiol Behav 149:310–6. [DOI] [PubMed] [Google Scholar]
- Goymann W, Wittenzellner A, Schwabl I, Makomba M. 2008. Progesterone modulates aggression in sex-role reversed female African black coucals. Proc Roy Soc B Biol Sci 275:1053–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood AK, Wark AR, Fernald RD, Hofmann HA. 2008. Expression of arginine vasotocin in distinct preoptic regions is associated with dominant and subordinate behaviour in an African cichlid fish. Proc Roy Soc B Biol Sci 275:2393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann HA. 2003. Functional Genomics of Neural and Behavioral Plasticity. J Neurobiol 54:272–282. [DOI] [PubMed] [Google Scholar]
- Hofmann HA, Benson ME, Fernald RD. 1999. Social status regulates growth rate: Consequences for life-history strategies. Proc Nat Acad Sci USA 96:14171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshaw BA, Evans JC, Mueller B, Valentino RJ, Lucki I. 2006. Social competition in rats: Cell proliferation and behavior. Behav Bra Res 175:343–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman LS, O'Connell LA, Hofmann HA. 2013. Aromatase regulates aggression in the African cichlid fish Astatotilapia burtoni . Physiol Behav 112:77–83. [DOI] [PubMed] [Google Scholar]
- Kelly AM, Goodson JL. 2014. Social functions of individual vasopressin-oxytocin cell groups in vertebrates: What do we really know? Front Neuroendocrinol 35:512–29. [DOI] [PubMed] [Google Scholar]
- Knapp R, Wingfield JC, Bass AH. 1999. Steroid hormones and paternal care in the plainfin midshipman fish (Porichthys notatus). Horm Behav 35:81–9. [DOI] [PubMed] [Google Scholar]
- Koblmuller S, Duftner N, Katongo C, Phiri H, Sturmbauer C. 2005. Ancient divergence in bathypelagic Lake Tanganyika deepwater cichlids: Mitochondrial phylogeny of the tribe Bathybatini. J Mol Evol 60:297–314. [DOI] [PubMed] [Google Scholar]
- Koning A. 1998. Tanganyika Cichlids in their natural habitat. El Paso (TX): Cichlid Press. [Google Scholar]
- Kozorovitskiy Y, Gould E. 2004. Dominance hierarchy influences adult neurogenesis in the dentate gyrus. J Neurosci 24:6755–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry CR, Aubin-Horth N. 2007. Ecological annotation of genes and genomes through ecological genomics. Mol Ecol 16:4419–21. [DOI] [PubMed] [Google Scholar]
- Ledon-Rettig CC, Richards CL, Martin LB. 2013. Epigenetics for behavioral ecologists. Behav Ecol 24:311–24. [Google Scholar]
- Leichty AR, Pfennig DW, Jones CD, Pfennig KS. 2012. Relaxed genetic constraint is ancestral to the evolution of phenotypic plasticity. Int Comp Biol 52:16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn SE. 2008. Behavioral insensitivity to testosterone: why and how does testosterone alter paternal and aggressive behavior in some avian species but not others? Gen Comp Endocrinol 157:233–40. [DOI] [PubMed] [Google Scholar]
- Machado HE, Pollen AA, Hofmann HA, Renn SCP. 2009. Interspecific profiling of gene expression informed by comparative genomic hybridization: A review and a novel approach in African cichlid fishes. Int Comp Biol 49:644–59. [DOI] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. 2009. Are sex-biased genes more dispensable? Biol Lett 5:409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manousaki T, Tsakogiannis A, Lagnel J, Sarropoulou E, Xiang JZ, Papandroulakis N, Mylonas CC, Tsigenopoulos CS. 2014. The sex-specific transcriptome of the hermaphrodite sparid sharpsnout seabream (Diplodus puntazzo). BMC Genomics 15:655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruska KP, Fernald RD. 2010. Reproductive status regulates expression of sex steroid and GnRH receptors in the olfactory bulb. Behav Bra Res 213:208–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruska KP, Fernald RD. 2013. Social regulation of male reproductive plasticity in an African cichlid fish. Int Comp Biol 53:938–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruska KP, Zhang A, Neboori A, Fernald RD. 2013. Social opportunity causes rapid rranscriptional changes in the social behaviour network of the brain in an African cichlid fish. J Neuroendocrinol 25:145–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlothlin JW, Ketterson ED. 2008. Hormone-mediated suites as adaptations and evolutionary constraints. Phil Tran Roy Soc B Biol Sci 363:1611–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micallef L, Rodgers P. 2014. eulerAPE: Drawing area-proportional 3-Venn diagrams using ellipses. Plos One 9:e101717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukai M, Replogle K, Drnevich J, Wang G, Wacker D, Band M, Clayton DF, Wignfield JC. 2009. Seasonal differences of gene expression profiles in song sparrow (Melospiza melodia) hypothalamus in relation to territorial aggression. Plos One 4:e8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ. 2006. The Biology of Aggression. New York (NY: ): Oxford University Press. [Google Scholar]
- Nelson RJ, Trainor BC. 2007. Neural mechanisms of aggression. Nat Rev Neurosci 8:536–46. [DOI] [PubMed] [Google Scholar]
- Nesjan E, Gutierrez-Ibanez C, Cameron JR, Merrigan S, Wylie DR, Hurd PL. 2014. Social status and GnRH soma size in female convict cichlids (Amatitlania nigrofasciatus). Behav Bra Res 272:205–8. [DOI] [PubMed] [Google Scholar]
- O'Connell LA, Ding JH, Hofmann HA. 2013. Sex differences and similarities in the neuroendocrine regulation of social behavior in an African cichlid fish. Horm Behav 64:468–76. [DOI] [PubMed] [Google Scholar]
- O'Connell LA, Hofmann HA. 2012. Social status predicts how sex steroid receptors regulate complex behavior across levels of biological organization. Endocrinology 153:1341–51. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. 2006. Conservation and evolution of gene colexpression networks in human and chimpanzee brains. Proc Nat Acad Sci USA 103: 17973–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfenning AR, Hara E, Whitney O, Rivas MV, Wang R, Roulhac PL, Howard JT, Wirthlin M, Lovell PV, Ganapathy G. 2014. Convergent transcriptional specializations in the brains of humans and song-learning birds. Science 346:1256846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plaisier SB, Taschereau R, Wong JA, Graeber TG. 2010. Rank-rank hypergeometric overlap: identification of statistically significant overlap between gene-expression signatures. Nuc Aci Res 38:e169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J, Cui X. 2010. Evaluation of a statistical equivalence test applied to microarray data. J Biopharm Stat 20:240–66. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. 2010. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; (http://www.R–project.org). [Google Scholar]
- Renn SCP, Aubin-Horth N, Hofmann HA. 2004. Biologically meaningful expression profiling across species using heterologous hybridization to a cDNA microarray. BMC Genomics 5:42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SCP, Aubin-Horth N, Hofmann HA. 2008. Fish and chips: functional genomics of social plasticity in an African cichlid fish. J Exp Biol 211:3041–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SCP, Carleton JB, Magee H, Nguyen MLT, Tanner ACW. 2009. Maternal care and altered social phenotype in a recently collected stock of Astatotilapia burtoni cichlid fish. Int Comp Biol 49:660–73. [DOI] [PubMed] [Google Scholar]
- Renn SCP, Fraser EJ, Aubin-Horth N, Trainor BC, Hofmann HA. 2012. Females of an African cichlid fish display male-typical social dominance behavior and elevated androgens in the absence of males. Horm Behav 61:496–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renn SCP, Machado HE, Jones A, Soneji K, Kulathinal RJ, Hofmann HA. 2010. Using comparative genomic hybridization to survey genomic sequence divergence across species: a proof-of-concept from Drosophila. BMC Genomics 11:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rittschof CC, Bukhari SA, Sloofman LG, Troy JM, Caetano-Anolles D, Cash-Ahmed A, Kent M, Lu X, Sanogo YO, Weisner PA. 2014. Neuromolecular responses to social challenge: Common mechanisms across mouse, stickleback fish, and honey bee. Proc Nat Acad Sci USA 111:17929–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt JD, Stein JL. 2014. RRHO: Test overlap using the Rnak-Rank hypergeometric test. R package version 1.12.0.
- Rosvall KA. 2013. Proximate perspectives on the evolution of female aggression: good for the gander, good for the goose? Roy Soc Phil Tran Biol Sci 368:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanogo YO, Band M, Blatti C, Sinha S, Bell AM. 2012. Transcriptional regulation of brain gene expression in response to a territorial intrusion. Proc Roy Soc B Biol Sci 279:4929–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapolsky RM. 2005. The influence of social hierarchy on primate health. Science 308:648–52. [DOI] [PubMed] [Google Scholar]
- Schumer M, Krishnakant K, Renn SCP. 2011. Comparative gene expression profiles for highly similar aggressive phenotypes in male and female cichlid fishes (Julidochromis). J Exp Biol 214:3269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Friedman N, Koller D, Regev A. 2004. A module map showing conditional activity of expression modules in cancer. Nat Gen 36:1090–8. [DOI] [PubMed] [Google Scholar]
- Silva AC, Perrone R, Zubizarreta L, Batista G, Stoddard PK. 2013. Neuromodulation of the agonistic behavior in two species of weakly electric fish that display different types of aggression. J Exp Biol 216:2412–20. [DOI] [PubMed] [Google Scholar]
- Smyth GK. 2005. Limma: linear models for microarray data In:Bioinformatics and Computational Biology Solutions using R and Bioconductor, Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W. (eds.), Springer, New York, 397–420. [Google Scholar]
- Smyth GK, Thorne NP, Wettenhall J. 2004. LIMMA: Linear Models for Microarray Data Version 1.6.6, User’s Guide.
- Sorensen C, Johansen IB, Overli O. 2013. Neural plasticity and stress coping in teleost fishes. Gen Comp Endocrinol 181:25–34. [DOI] [PubMed] [Google Scholar]
- Stiver KA, Harris RM, Townsend JP, Hofmann HA, Alonzo SH. 2015. Neural gene expression profiles and androgen levels underlie alternative reproductive ractics in the ccellated wrasse, Symphodus ocellatus . Ethology 121:152–67. [Google Scholar]
- Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T. 2005. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Gen 37:48–55. [DOI] [PubMed] [Google Scholar]
- Taborsky M, Grantner A. 1998. Behavioural time-energy budgets of cooperatively breeding Neolamprologus pulcher (Pisces: Cichlidae). Ani Behav 56:1375–82. [DOI] [PubMed] [Google Scholar]
- Teles MC, Cardoso SD, Oliveira RF. 2016. Social plasticity relies on different neuroplasticity mechanisms across the brain social decision-making network in Zebrafish. Front Behav Neurosci 10:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teles MC, Oliveira RF. 2016. Androgen response to social competition in a shoaling fish. Horm Behav 78:8–12. [DOI] [PubMed] [Google Scholar]
- Toth AL, Varala K, Henshaw MT, Rodriguez-Zas SL, Hudson ME, Robinson GE. 2010. Brain transcriptomic analysis in paper wasps identifies genes associated with behaviour across social insect lineages. Proc Roy Soc B Biol Sci 277:2139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend JP. 2004. Resolution of large and small differences in gene expression using models for the Bayesian analysis of gene expression levels and spotted DNA microarrays. BMC Bioinform 5:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor BC, Hofmann HA. 2006. Somatostatin regulates aggressive behavior in an African cichlid fish. Endocrinology 147:5119–25. [DOI] [PubMed] [Google Scholar]
- Trainor BC, Kyomen HH, Marler CA. 2006. Estrogenic encounters: How interactions between aromatase and the environment modulate aggression. Front Neuroendocrinol 27:170–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor BC, Marler CA. 2001. Testosterone, paternal behavior, and aggression in the monogamous California mouse (Peromyscus californicus). Horm Behav 40:32–42. [DOI] [PubMed] [Google Scholar]
- Whitehead A. 2012. Comparative genomics in ecological physiology: toward a more nuanced understanding of acclimation and adaptation. J Exp Biol 215:884–91. [DOI] [PubMed] [Google Scholar]
- Wingfield JC. 1994. Regulation of territorial behavior in the sedentary song sparrow, Melospiza-Melodia Morphna . Horm Behav 28:1–15. [DOI] [PubMed] [Google Scholar]
- Wood KJ, Zero VH, Jones A, Renn SCP. 2014. Social reversal of sex-biased aggression and dominance in a biparental cichlid rish (Julidochromis marlieri). Ethology 120:540–50. [Google Scholar]
- Wu MV, Manoli DS, Fraser EJ, Coats JK, Tollkuhn J, Honda S, Harada N, Shah NM. 2009. Estrogen masculinizes neural pathways and sex-specific behaviors. Cell 139:61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeger C, Ros AM, Cross V, Deangelis RS, Stobaugh DJ, Rhodes JS. 2014. Blockade of argenine vasotocin signaling reduces aggressive behavior and c-fos expression in the preoptic area and periventricular nucleus of the posterior tuberculum in male. Neuroscience 267:205–18. [DOI] [PubMed] [Google Scholar]
- Yamagishi S, Kohda M. 1996. Is the cichlid fish Julidochromis marlieri polyandrous? Ichthyological Res 43:469–71. [Google Scholar]
- Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. 2006. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Gen Res 16:995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.