Zika virus, chikungunya virus, and dengue virus result in similar clinical presentations, and coinfections may be relatively common. Accurate, multiplex diagnostics are necessary to detect and differentiate these arboviruses for patient care and epidemiologic surveillance.
Keywords: dengue virus, chikungunya virus, Zika virus, coinfection, viremia
Abstract
Background. Zika virus (ZIKV), chikungunya virus (CHIKV), and dengue virus (DENV) cocirculate in Nicaragua. In this study, we sought to compare the quantified viremia and clinical presentation of patients infected with 1 or more of these viruses.
Methods. Acute-phase serum samples from 346 patients with a suspected arboviral illness were tested using a multiplex real-time reverse-transcription polymerase chain reaction for ZIKV, CHIKV, and DENV. Viremia was quantitated for each detected virus, and clinical information from request forms submitted with each sample was recorded.
Results. A total of 263 patients tested positive for 1 or more viruses: 192 patients tested positive for a single virus (monoinfections) and 71 patients tested positive for 2 or all 3 viruses (coinfections). Quantifiable viremia was lower in ZIKV infections compared with CHIKV or DENV (mean 4.70 vs 6.42 and 5.84 log10 copies/mL serum, respectively; P < .001 for both comparisons), and for each virus, mean viremia was significantly lower in coinfections than in monoinfections. Compared with patients with CHIKV or DENV, ZIKV patients were more likely to have a rash (P < .001) and less likely to be febrile (P < .05) or require hospitalization (P < .001). Among all patients, hospitalized cases had higher viremia than those who did not require hospitalization (7.1 vs 4.1 log10 copies/mL serum, respectively; P < .001).
Conclusions. ZIKV, CHIKV, and DENV result in similar clinical presentations, and coinfections may be relatively common. Our findings illustrate the need for accurate, multiplex diagnostics for patient care and epidemiologic surveillance.
Dengue has been endemic in Nicaragua since 1985, with all 4 dengue virus serotypes (DENV1–4) circulating, generally with 1 serotype dominant in each epidemic [1–4]. Chikungunya virus (CHIKV) was first detected in Nicaragua in July 2014, and the first autochthonous cases were confirmed in September 2014 [5]. Limited CHIKV transmission occurred in 2014–2015, followed by a larger epidemic in 2015–2016. The first autochthonous cases of Zika virus (ZIKV) in Nicaragua were reported in January 2016, and ZIKV and CHIKV now cocirculate with dengue virus (DENV) throughout the country [6]. This complicates the diagnosis of patients with an acute febrile illness, as the spectra of clinical manifestations that result from infection with these viruses overlap significantly [7–10]. Diagnosis is further complicated by cross-reactions observed in ZIKV-positive patients tested using immunoglobulin M or non-structural protein 1 assays for DENV and vice versa and by limited data on the duration of anti-CHIKV immunoglobulin M positivity following acute infection [7, 8].
Molecular diagnostics can be used to detect and differentiate ZIKV, CHIKV, and DENV in the acute phase, and real-time reverse-transcription polymerase chain reaction (rRT-PCR) can provide quantitative data in addition to qualitative detection [8, 11–14]. Quantitation of viremia has been widely reported in the DENV literature, where higher viremia at presentation has been associated with secondary DENV infections and disease severity [15, 16]. Fewer data are available regarding the level of viremia at presentation for patients with ZIKV and CHIKV [8, 9, 17–20]. As such, it is unknown how the level of viremia may correlate with clinical manifestations or clinical outcomes in these infections and whether the level of viremia varies in coinfections compared with monoinfections.
Our group previously described a single-reaction, multiplex rRT-PCR for the detection and differentiation of ZIKV, CHIKV, and DENV (referred to as the ZCD assay) [6]. The ZCD assay demonstrated similar sensitivity to a pan–DENV-CHIKV rRT-PCR and higher sensitivity than a published ZIKV rRT-PCR when evaluated using samples collected from Nicaraguan patients with suspected arboviral infections. The ZCD assay can also be performed as a quantitative test, though accurate quantitation of DENV viremia requires identification of the serotype [6, 21, 22].
Here, the ZCD assay and a companion serotype-specific DENV multiplex assay were used to study the level of viremia in patients infected with ZIKV, CHIKV, and/or DENV [21, 22]. This provides an extensive evaluation of the quantified viremia detected in patients presenting with ZIKV and CHIKV and how this compares to viremia in patients with DENV infections.
METHODS
Clinical Samples
Serum samples, collected at Ministry of Health facilities as part of routine care, were sent to the Centro Nacional de Diagnóstico y Referencia (CNDR) in Managua, Nicaragua, for reference ZIKV, CHIKV, and/or DENV molecular testing. Samples were obtained at the discretion of care providers from patients with suspected ZIKV, CHIKV, and/or DENV infections. Deidentified, acute-phase (collected within 7 days of symptom onset) serum samples collected between 1 September 2015 and 3 April 2016 were tested for this study. Serum was separated from whole blood at the collection site and then stored at −20°C and shipped to CNDR on ice. Serum was stored at CNDR at −20°C until thawed for nucleic acid extraction.
Clinical information was obtained from the epidemiologic records that are submitted along with each sample. These documents contain the following information: age, gender, pregnancy status, dates of symptom onset and sample collection, temperature, symptoms, clinical diagnosis, and date of hospitalization. Submission of an epidemiologic form is required for specimen processing, but completion of all data fields is voluntary. The Nicaraguan Ministry of Health and the Stanford University Institutional Review Board reviewed and approved the research protocol for this study.
Nucleic Acid Extraction and rRT-PCR Performance
RNA was extracted from 140 µL of serum using the QIAamp Viral RNA Mini kit (Qiagen) with a 60-µL elution volume. RNA extracts were stored at −80°C. Samples were tested for ZIKV, CHIKV, and/or DENV using the ZCD assay as previously described [6]. Viremia for ZIKV- and CHIKV-positive samples was quantitated in the ZCD assay, and DENV-positive samples were serotyped and viremia was quantitated using a serotype-specific DENV multiplex assay [21, 22]. For quantitation, the ZCD and DENV multiplex assays were performed with 4-point standard curves (8.0, 6.0, 4.0, and 2.0 log10 copies/µL of eluate). Standard curves were prepared using quantitated ssDNA (Integrated DNA Technologies) containing the target sequences for ZIKV, CHIKV, and the identified DENV serotypes. For quantitation, clinical samples and the standard curve were tested in duplicate on a single run. The mean cycle threshold (Ct) was used for all calculations. The concentration of RNA in the eluate (expressed as log10 copies/µL of eluate) was calculated from the linear regression equation for the standard curve. Viremia in log10 copies/mL of serum was then calculated from this value, accounting for volumes used in extraction. Data regarding the precision of ZIKV and CHIKV detection, linear range of the ZCD assay, and performance of external controls are provided in the Supplementary Data.
Mixing studies were performed to evaluate possible interference between channels in the ZCD assay. Quantitated ssDNA standards for ZIKV, CHIKV, and DENV-2 were diluted in nuclease-free water at concentrations equivalent to 8.0, 6.0, and/or 4.0 log10 copies/mL serum; standards were mixed in different combinations to create simulated coinfections, including triple infections (Supplementary Table 3).
Definitions
The term viremia is used generally to describe the presence of detectable viral RNA in serum using the ZCD assay. Quantifiable viremia is used to denote viremia that is within the linear range of the ZCD assay. The linear range for each target in the ZCD assay extends from 8.0–1.0 log10 copies/µL of eluate (Supplementary Figure 1). For serum samples tested in this study, this linear range corresponds to viremias of 10.6–3.6 log10 copies/mL. Low-positive viremia describes viremia that is detected but falls below the lower limit of quantitation for the ZCD assay (3.6 log10 copies/mL of serum) [6].
Statistics
Basic statistics were calculated using Excel software (Microsoft). Categorical variables were compared using Fisher exact tests, and continuous clinical variables were compared using t tests. Kruskal–Wallis tests were performed to compare viremia distributions that included low-positive viremia (below the limit of quantitation in the ZCD assay), and Welch t tests were used to compare quantifiable viremia. Fisher exact tests, t tests, and Kruskal–Wallis tests were performed with GraphPad software (GraphPad, San Diego, California). Pearson and Spearman correlation coefficients were calculated at socscistatistics.com. For the multivariable analysis of factors associated with hospitalization, R software was used to evaluate generalized linear models that included the following variables: viral etiology (CHIKV and/or DENV infection vs ZIKV infection), age, gender, viremia, temperature, and day post-onset of symptoms.
RESULTS
A total of 346 samples were tested using the ZCD assay. Among patients with information regarding day of symptom onset, 303/306 (99.0%) presented on days 1–6, where day 1 was defined as the day on which symptoms began. A total of 263 patients (76.0%) tested positive for 1 or more viruses (Table 1), and the mean day post-onset of symptoms at presentation was not significantly different for patients with positive samples and those with negative samples (Supplementary Table 4). RNA from a single virus (monoinfection) was detected in 192 patients (55.5% of all patients tested), and RNA from more than 1 virus (coinfections) was detected in 71 patients (20.5%; Table 1).
Table 1.
ZCD Assay Results for 346 Patients With Suspected Zika Virus, Chikungunya Virus, and/or Dengue Virus Infections
| ZCD Assay Result | Number, n (% of all Samples) |
|---|---|
| Positive | 263 (76.0) |
| Monoinfections | 192 (55.5) |
| ZIKV | 47 (13.6) |
| CHIKV | 91 (26.3) |
| DENVa | 54 (15.6) |
| Coinfections | 71 (20.5) |
| ZIKV-CHIKV | 16 (4.6) |
| ZIKV-DENVa | 6 (1.7) |
| CHIKV-DENVa | 43 (12.4) |
| ZIKV-CHIKV-DENVa | 6 (1.7) |
| Negative | 83 (24.0) |
Abbreviations: CHIKV, chikungunya virus; DENV, dengue virus; ZCD, multiplex real-time reverse-transcription polymerase chain reaction for the detection and differentiation of ZIKV, CHIKV, and DENV; ZIKV, Zika virus.
a Serotypes of 109 DENV-positive samples: DENV-2, 107; DENV-1, 1; DENV-4, 1.
Clinical Presentation
Clinical data were available for 163 positive (62.0% of all positives) and 41 negative patients (49.4% of all negatives). Patients with positive ZCD assay results were clinically similar to patients with negative results based on the variables analyzed (Supplementary Table 4). A comparison of clinical data for patients with ZIKV, CHIKV, and DENV is shown in Table 2. Patients with ZIKV were significantly older than patients with either CHIKV or DENV (aged 36.0 vs 20.4 years; P < .001). Rash, conjunctivitis, fever, and headache were the most common symptoms in patients with ZIKV, but only rash was significantly more common among ZIKV-positive patients (32/35, 91.4%) than among CHIKV-positive (49/87, 56.3) or DENV-positive patients (25/50, 50.0%; P < .001 for both comparisons). Although a similar proportion of patients with each virus had a history of fever, patients with ZIKV were significantly less likely to be febrile (≥38°C) at the time of presentation.
Table 2.
Comparison of the Clinical Presentation for all Patients With Zika Virus, Chikungunya Virus, and Dengue Virus Infections
| Patient Data | Zika Virus | Chikungunya Virus | Dengue Virus | P Valuea |
|---|---|---|---|---|
| Number, n | 37 | 103 | 66 | … |
| Gender, % female, % male | 72.2, 27.8 | 60.8, 39.2 | 63.6, 36.4 | NS |
| Age in years, mean (SD) | 36.0 (16.0) | 20.4 (15. 8) | 20.4 (15.2) | <.001, <.001 |
| Day post symptom onset, mean (SD) | 3.4 (1.1) | 3.2 (1.3) | 3.5 (1.3) | NS |
| Pregnant, n | 6 | 8 | 6 | … |
| Symptoms, positive/total (%)b | ||||
| Rash | 32/35 (91.4) | 49/87 (56.3) | 25/50 (50.0) | <.001, <.001 |
| Conjunctivitis | 18/22 (81.8) | 13/17 (76.5) | 8/11 (72.7) | NS |
| History of fever | 28/35 (80.0) | 81/90 (90.0) | 45/50 (90.0) | NS |
| Headache | 27/34 (79.4) | 54/85 (63.5) | 34/49 (69.4) | NS |
| Arthralgia | 21/30 (70.0) | 59/85 (69.4) | 32/48 (66.7) | NS |
| Myalgia | 20/31 (64.5) | 44/83 (53.0) | 28/48 (58.3) | NS |
| Retro-orbital pain | 13/27 (48.1) | 25/66 (37.9) | 18/42 (42.9) | NS |
| Nausea | 7/26 (26.9) | 24/58 (41.4) | 17/39 (43.6) | NS |
| Abdominal painc | 2/17 (11.8) | 10/57 (17.5) | 16/41 (38.1) | NS |
| Hemorrhage | 1/17 (5.9) | 2/56 (3.6) | 4/42 (9.5) | NS |
| Vomiting | 0/17 (0.0) | 6/58 (10.3) | 6/42 (14.3) | NS |
| Clinical Data | ||||
| Temperature, mean (SD) | 36.9 (0.6) | 37.4 (0.9) | 37.3 (0.8) | .005, .018 |
| Febriled, number/total (%)b | 2/27 (7.4) | 29/86 (33.7) | 16/56 (28.6) | .007, .044 |
| Clinical diagnosis, n | 35 | 97 | 63 | … |
| Chikungunya | 1 | 29 | 7 | … |
| Dengue | 0 | 41 | 40 | … |
| Zika | 27 | 15 | 14 | … |
| Multiple viruses Listede | 7 | 11 | 2 | … |
| Hospitalized, positive/total (%)b | 7/23 (30.4) | 55/70 (78.6) | 43/55 (78.2) | <.001, <.001 |
Abbreviations: NS, not significant; SD, standard deviation.
a P values are shown for comparisons of Zika virus (ZIKV) and chikungunya virus (CHIKV; first value) and ZIKV and dengue virus (DENV; second value). If results of both comparisons were not significant, only NS is shown.
b Reported as the number of positives over the total number with recorded information for each variable.
c P = .022 for comparison of CHIKV and DENV.
d Defined as a temperature ≥38°C.
e One CHIKV-positive patient had suspected leptospirosis.
A suspected clinical diagnosis was recorded for 153/163 (93.9%) positive samples (Table 2), and sensitivity of the clinical diagnosis varied by the virus identified. ZIKV infection was correctly diagnosed in a higher percentage of cases (32/35, 91.4%) than DENV infection (42/63, 66.7%; P = .007), and both ZIKV and DENV infections were correctly diagnosed in a higher percentage of cases than CHIKV (40/97, 41.2%; P < .001 and P = .002, respectively).
Quantitation of Viremia
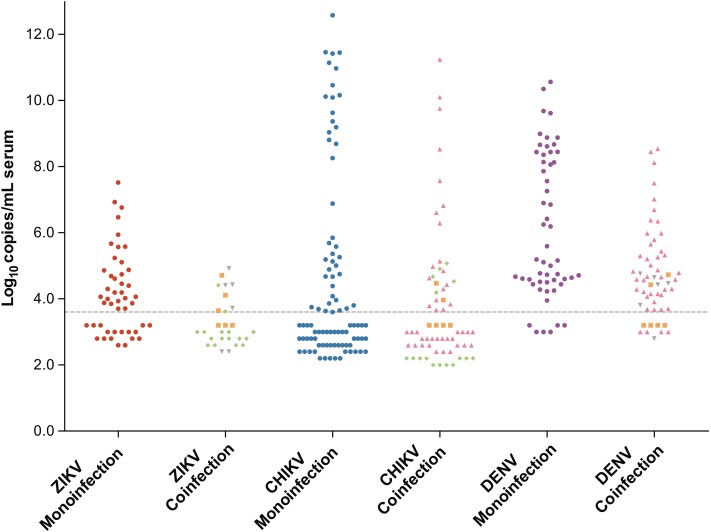
The distribution of viremia detected in ZIKV, CHIKV, and DENV monoinfections and coinfections is shown in Figure 1, and a comparison of quantifiable viremia in monoinfections and coinfections is shown in Table 3. Mean ZIKV viremia was significantly lower than mean viremia for either CHIKV or DENV (P < .001 for both comparisons). Six pregnant women tested positive for ZIKV, and mean ZIKV viremia in those women (5.05 log10 copies/mL serum; standard deviation [SD], 0.91) was higher than viremia detected in patients who were not pregnant (3.73 log10 copies/mL serum; SD, 1.02; P = .006). The highest viremias identified were in CHIKV-positive samples. In particular, neonates diagnosed with CHIKV in the first month of life had the highest levels of CHIKV detected (6/6 patients with >11.0 log10 copies/mL serum). Despite these findings, the mean quantifiable viremia for CHIKV and DENV were not significantly different (Table 3, P = .1).
Figure 1.
Levels of viremia in Zika virus (ZIKV), chikungunya virus (CHIKV), and dengue virus (DENV) monoinfections and coinfections. Monoinfections are represented by filled circles (•): ZIKV, red; CHIKV, blue; and DENV, purple. Coinfections are represented by the following: ZIKV-CHIKV (♦), ZIKV-DENV (▾), CHIKV-DENV (▴), and ZIKV-CHIKV-DENV (▪). Viremia for each virus detected in a coinfection is displayed. The limit of quantitation for the multiplex real-time reverse-transcription polymerase chain reaction for the detection and differentiation of ZIKV, CHIKV, and DENV assay is displayed as a dashed gray line (3.6 log10 copies/mL serum). Samples with viral RNA that was detectable but below the limit of quantitation (low positives) are shown below this line; marker positions for these samples do not represent estimated viremia.
Table 3.
Quantifiable Viremia for Monoinfections and Coinfections With Zika Virus, Chikungunya Virus, and Dengue Virus
| ZCD Assay Result | Number of Samples, n (%)a | Quantifiable Viremia, n (%)b | Viremia, mean (standard deviation) | P Value |
|---|---|---|---|---|
| Zika virus | 75 | 39 (52.0) | 4.70 (0.97) | … |
| Monoinfections | 47 (62.7) | 30 (63.8) | 4.84 (1.04) | .018 |
| Coinfections | 28 (37.3) | 9 (32.1) | 4.22 (0.48) | |
| Chikungunya virus | 156 | 67 (42.9) | 6.42 (2.72) | … |
| Monoinfections | 91 (58.3) | 41 (45.1) | 6.92 (2.94) | .040 |
| Coinfections | 65 (41.7) | 26 (40.0) | 5.62 (2.15) | |
| Dengue virus | 109 | 94 (86.2) | 5.84 (1.82) | … |
| Mono-infections | 54 (49.5) | 48 (88.9) | 6.53 (2.01) | <.001 |
| Co-infections | 55 (50.5) | 46 (83.6) | 5.11 (1.25) |
Abbreviation: ZCD, multiplex real-time reverse-transcription polymerase chain reaction for the detection and differentiation of ZIKV, CHIKV, and DENV.
a % of samples positive for a given virus with mono- or coinfections.
b % of samples in each category with quantifiable viremia.
The distribution of DENV viremia differed significantly from distributions for ZIKV and CHIKV (P ≤ .001 for both comparisons, Kruskal–Wallis). Of all DENV-positive samples, only 15 (13.8%) were low positives. This was significantly lower than the proportion of ZIKV low-positive (36/75, 48.0%) or CHIKV low-positive samples (89/156, 57.1%; P < .001 for both comparisons).
The numbers of CHIKV samples with either quantifiable viremia or low-positive viremia were sufficient to investigate these categories further. The percentage of samples with quantifiable and low-positive CHIKV viremia was similar for monoinfections and coinfections (Table 3). Detection of quantifiable CHIKV viremia decreased from a high of 58.3% (7/12) of samples collected on the first day of symptoms to 19.0% (4/21) of samples collected on days 5 and 6 post-symptom onset (Supplementary Figure 2; Spearman, R = −0.943; P = .005). The documented clinical presentations of patients with quantifiable or low-positive CHIKV viremia were similar. However, among patients with low-positive viremia, chikungunya was the clinical diagnosis significantly less often and Zika was the clinical diagnosis significantly more often than among patients with quantifiable viremia (Supplementary Table 5).
Coinfections in the ZCD Assay
For all 3 viruses, mean quantifiable viremia in monoinfections was significantly higher than viremia for the same virus detected in coinfections (Table 3). There was no evidence of interference in mixing studies using samples with viremias similar to those observed in these clinical samples. Specifically, there was no interference observed among simulated triple infections with viruses mixed at equal concentrations of 8.0, 6.0, or 4.0 log10 copies/mL of serum (Supplementary Table 3). The clinical presentations of patients with coinfections and monoinfections were similar (data not shown), though there was a trend toward more frequent hospitalization in patients with coinfections (25/30, 83.3%) compared with those with monoinfections (55/86, 64.0%; P = .066).
Hospitalization
A total of 116 patients with positive ZCD test results had hospitalization status available: 80 hospitalized and 36 not hospitalized. Factors associated with hospitalization in univariate analysis are shown in Table 4. The best-fit multivariable model included temperature, viremia, and ZIKV monoinfection. Viremia and temperature were higher among hospitalized cases. Although there was a weak, positive correlation between viremia and recorded temperature (Pearson R = 0.207, P = .026), both variables remained significant in the final model. Patients with ZIKV monoinfections were significantly less likely to be hospitalized than patients with CHIKV and/or DENV infections, regardless of whether the latter group of patients had mono- or coinfections. Results from the multivariable analysis were similar when all patients without a documented hospitalization status were considered to be “not hospitalized.”
Table 4.
Univariate and Multivariable Analysis of Factors Associated With Hospitalization Among Patients With Zika Virus, Chikungunya Virus, and/or Dengue Virus Infections
| Patient Variables | Hospitalized | Not Hospitalized | Univariate, P Value | Multivariable Analysisa |
||
|---|---|---|---|---|---|---|
| Log Odds (Ln) | Standard Error | P Value | ||||
| Number, n | 80 | 36 | ||||
| Female gender, n (%) | 42 (52.5) | 26 (74.3) | .039 | … | … | … |
| Age in years, mean (SD) | 18.2 (15.5) | 28.2 (14.4) | .002 | … | … | … |
| Day post symptom onset, mean (SD) | 3.1 (1.5) | 3.6 (1.2) | .079 | … | … | … |
| Viremia, log10 copies/mL serum, mean (SD) | 7.1 (2.7) | 4.1 (1.5) | <.001 | 0.339 | 0.151 | .025 |
| Temperature, mean (SD) | 37.5 (0.9) | 36.8 (0.6) | <.001 | 0.978 | 0.391 | .012 |
| Zika virus monoinfection, n (%) | 4 (0.5) | 12 (33.3) | <.001 | −1.536 | 0.714 | .032 |
Abbreviation: SD, standard deviation.
a Variables indicated by a ellipse (…) were not included in the best-fit multivariable model.
DISCUSSION
Here, we present a comparative analysis of the level of viremia and clinical manifestations that resulted from infections with ZIKV, CHIKV, and/or DENV among patients in a single endemic country. The clinical presentations caused by these viruses were similar, and only the presence of a rash and a documented fever differed significantly between patients with ZIKV and both patients with CHIKV and patients with DENV. As a result, clinical suspicion was only correct in 41.2% of CHIKV infections and 66.7% of DENV infections. The apparent sensitivity of a clinical diagnosis of Zika (91.4%) may have resulted from heightened awareness of and concern for Zika during the study period, as 38/70 patients (54.3%) with suspected Zika tested negative for ZIKV RNA. However, a portion of these cases may have been detected by serology, which was unavailable for this study. Taken together, our findings highlight the difficulty of providing an accurate clinical diagnosis in a region where patients may be infected with any one of these pathogens and support the use of a testing protocol for ZIKV, CHIKV, and DENV in all suspected cases. This would be expected to improve case detection, inform management decisions, and allow for appropriate patient referral and follow-up.
Patients infected with ZIKV in the current study had lower quantifiable viremia, on average, than patients with CHIKV and/or DENV. Additionally, the maximum viremia detected among ZIKV-positive patients was 1000 to 100 000-fold lower than the maximum viremia detected for DENV and CHIKV, respectively. Studies to date have focused on the qualitative detection of ZIKV in serum or plasma, and fewer data have been published regarding the level of viremia in ZIKV-positive cases [8–10, 23]. In a study by Lanciotti et al, the mean estimated ZIKV viremia detected in 17 patients on Yap Island (4.4 log10 copies/mL of serum, SD 0.94) was very similar to the mean quantifiable viremia observed in our patients (4.7 log10 copies/mL of serum, SD 0.97) [8]. Such low-level viremia at presentation provides a likely explanation for the short window of ZIKV RNA detection in serum or plasma that has been reported [8, 9, 23, 24]. However, it remains possible, though unlikely, that peak viremia in ZIKV infections is similar to peaks observed in CHIKV and/or DENV infections but occurs earlier relative to symptom onset.
Patients with low-positive CHIKV viremia have been identified in other series, but the clinical presentation of and outcomes for such cases have not been evaluated [17–20, 25]. In our study, 57.1% of CHIKV-positive patients had viremia that was below the quantifiable range in the ZCD assay. The documented clinical presentation of patients with quantifiable and low-positive viremia was similar (Supplementary Table 5). Patients with low-positive viremia were more likely to present later in the course of illness, which is consistent with declining viremia in acute CHIKV infections over the first week of illness [17–19]. It is important to note that the lower limit of quantitation in the ZCD assay is dependent on characteristics of the test and the volumes of serum and elution buffer used during RNA extraction. This value bears no a priori biological significance.
Zika was suspected in a higher proportion of patients with low-positive CHIKV viremia (and chikungunya was suspected less often) compared with patients with quantifiable viremia. This may indicate that patients with low-positive CHIKV viremia had a milder clinical presentation despite similar reported symptoms. In addition, this finding is consistent with the associations between higher viremia (among all patients), fever at presentation, and hospitalization. These data, when considered along with a documented association between viremia and disease severity in the dengue literature [15, 16, 26, 27], suggest a more general correlation between viremia at the onset of clinical symptoms and illness severity in acute arboviral infections.
Despite transmission of ZIKV, CHIKV, and DENV in many regions, reports of coinfections between these viruses have been rare, particularly in comparison to the 20.5% of patients who had detectable RNA from 2 or all 3 viruses in our study. This figure is consistent with our earlier findings, which included a subset of patients from this analysis, as well as those from Guayaquil, Ecuador, where the ZCD assay has also been implemented [6, 28]. The apparent difference in rates of coinfection may then result from a number of factors, including a reliance on serological testing in many endemic areas; the performance of individual tests for each virus, which increases test cost and may decrease utilization; and a lack of signs or symptoms that clinically distinguish co-infections from monoinfections. This latter point was illustrated in findings from our patients and has also been observed in the few cases described in the literature [29, 30].
Given the nature of national surveillance sample collection in Nicaragua, only acute-phase specimens were available for testing. Therefore, serological testing on paired acute and convalescent serum samples could not be performed. An additional limitation to this study is the reliance on voluntarily completed epidemiologic forms for clinical information. Given that many forms were not fully completed, we focused the statistical analysis on clinical variables for which data were available for ≥50% of patients in at least 1 category (positive or negative ZCD result, viral etiology, low-positive or quantifiable CHIKV viremia). Finally, in simulated triple infections, high concentrations of CHIKV and DENV (8.0 log10 copies/mL of 1 virus plus 8.0 or 6.0 log10 copies/mL of the second) interfered with the detection of ZIKV at 4.0 log10 copies/mL (Supplementary Table 3). Although viremia in detected triple infections was far lower than these simulated infections, we cannot rule out that low-level ZIKV viremia was missed in CHIKV-DENV coinfections with high viremia.
In conclusion, the co-circulation of ZIKV, CHIKV, and DENV presents a number of challenges for clinical care and laboratory diagnosis in endemic areas. Patients infected with 1 or more of these viruses can present with similar clinical manifestations over a wide range of viremia. In addition, coinfections may be quite common, requiring that all patients be tested for each virus. This demonstrates the need for sensitive, accurate, multiplex diagnostics for clinical care, disease research, and epidemiologic surveillance of these arboviral diseases.
Supplementary Data
Supplementary materials are available at http://cid.oxfordjournals.org. Consisting of data provided by the author to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the author, so questions or comments should be addressed to the author.
Notes
Acknowledgments. We thank the staff of the National Virology Laboratory in the Centro Nacional de Diagnóstico y Referencia of the Nicaraguan Ministry of Health and the Sustainable Sciences Institute in Managua, Nicaragua, for their assistance with sample preparation and maintenance of the specimen collection used in this study. Additionally, we thank them for accommodating one of the authors (J. J. W.) throughout the course of this project.
Disclaimer. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Financial support. This work was supported by National Institute of Allergy and Infectious Diseases of the National Institutes of Health (NIH) (K08AI110528; J. J. W.). Studies in Nicaragua were supported by NIH (R01AI099631; A. B. and U54AI65359; A. B.).
Potential conflicts of interest. All authors: No potential conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Hammond SN, Balmaseda A, Perez L et al. Differences in dengue severity in infants, children, and adults in a 3-year hospital-based study in Nicaragua. Am J Trop Med Hyg 2005; 73:1063. [PubMed] [Google Scholar]
- 2.Harris E, Videa E, Perez L et al. Clinical, epidemiologic, and virologic features of dengue in the 1998 epidemic in Nicaragua. Am J Trop Med Hyg 2000; 63:5–11. [DOI] [PubMed] [Google Scholar]
- 3.OhAinle M, Balmaseda A, Macalalad AR et al. Dynamics of dengue disease severity determined by the interplay between viral genetics and serotype-specific immunity. Sci Transl Med 2011; 3:114ra28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouri G, Valdez M, Arguello L et al. Dengue epidemic in Nicaragua, 1985. Rev Inst Med Trop Sao Paulo 1991; 33:365–71. [PubMed] [Google Scholar]
- 5.Balmaseda A, Gordon A, Gresh L et al. Clinical attack rate of chikungunya in a cohort of Nicaraguan children. Am J Trop Med Hyg 2016; 94:397–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waggoner JJ, Gresh L, Mohamed-Hadley A et al. Single-reaction multiplex reverse transcription PCR for detection of Zika, chikungunya, and dengue viruses. Emerg Infect Dis 2016; 22:1295–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Waggoner JJ, Pinsky BA. Zika virus: diagnostics for an emerging pandemic threat. J Clin Microbiol 2016; 54:860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanciotti RS, Kosoy OL, Laven JJ et al. Genetic and serologic properties of Zika virus associated with an epidemic, Yap State, Micronesia, 2007. Emerg Infect Dis 2008; 14:1232–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Musso D, Roche C, Nhan TX, Robin E, Teissier A, Cao-Lormeau VM. Detection of Zika virus in saliva. J Clin Virol 2015; 68:53–5. [DOI] [PubMed] [Google Scholar]
- 10.Brasil P, Calvet GA, Siqueira AM et al. Zika virus outbreak in Rio de Janeiro, Brazil: clinical characterization, epidemiological and virological aspects. PLoS Negl Trop Dis 2016; 10:e0004636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faye O, Faye O, Dupressoir A, Weidmann M, Ndiaye M, Alpha Sall A. One-step RT-PCR for detection of Zika virus. J Clin Virol 2008; 43:96–101. [DOI] [PubMed] [Google Scholar]
- 12.Lanciotti RS, Kosoy OL, Laven JJ et al. Chikungunya virus in US travelers returning from India, 2006. Emerg Infect Dis 2007; 13:764–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santiago GA, Vergne E, Quiles Y et al. Analytical and clinical performance of the CDC real-time RT-PCR assay for detection and typing of dengue virus. PLoS Negl Trop Dis 2013; 7:e2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hue KDT, Tuan TV, Thi HTN et al. Validation of an internally controlled one-step real-time multiplex RT-PCR assay for the detection and quantitation of dengue virus RNA in plasma. J Virol Meth 2011; 177:168–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pozo-Aguilar JO, Monroy-Martinez V, Diaz D et al. Evaluation of host and viral factors associated with severe dengue based on the 2009 WHO classification. Parasit Vectors 2014; 7:590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tricou V, Minh NN, Farrar J, Tran HT, Simmons CP. Kinetics of viremia and NS1 antigenemia are shaped by immune status and virus serotype in adults with dengue. PLoS Negl Trop Dis 2011; 5:e1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chusri S, Siripaitoon P, Silpapojakul K et al. Kinetics of chikungunya infections during an outbreak in southern Thailand, 2008–2009. Am J Trop Med Hyg 2014; 90:410–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laurent P, Le Roux K, Grivard P et al. Development of a sensitive real-time reverse transcriptase PCR assay with an internal control to detect and quantify chikungunya virus. Clin Chem 2007; 53:1408–14. [DOI] [PubMed] [Google Scholar]
- 19.Panning M, Grywna K, van Esbroeck M, Emmerich P, Drosten C. Chikungunya fever in travelers returning to Europe from the Indian Ocean region, 2006. Emerg Infect Dis 2008; 14:416–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Riswari SF, Ma'roef CN, Djauhari H et al. Study of viremic profile in febrile specimens of chikungunya in Bandung, Indonesia. J Clin Virol 2016; 74:61–5. [DOI] [PubMed] [Google Scholar]
- 21.Waggoner JJ, Abeynayake J, Sahoo MK et al. Comparison of the FDA-approved CDC DENV-1–4 real-time reverse transcription-PCR with a laboratory-developed assay for dengue virus detection and serotyping. J Clin Microbiol 2013; 51:3418–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waggoner JJ, Abeynayake J, Sahoo MK et al. Single-reaction, multiplex, real-time rt-PCR for the detection, quantitation, and serotyping of dengue viruses. PLoS Negl Trop Dis 2013; 7:e2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gourinat AC, O'Connor O, Calvez E, Goarant C, Dupont-Rouzeyrol M. Detection of Zika virus in urine. Emerg Infect Dis 2015; 21:84–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fonseca K, Meatherall B, Zarra D et al. First case of Zika virus infection in a returning Canadian traveler. Am J Trop Med Hyg 2014; 91:1035–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simmons G, Bres V, Lu K et al. High incidence of chikungunya virus and frequency of viremic blood donations during epidemic, Puerto Rico, USA, 2014. Emerg Infect Dis 2016; 22:1221–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Libraty DH, Young PR, Pickering D et al. High circulating levels of the dengue virus nonstructural protein NS1 early in dengue illness correlate with the development of dengue hemorrhagic fever. J Infect Dis 2002; 186:1165–8. [DOI] [PubMed] [Google Scholar]
- 27.Vaughn DW, Green S, Kalayanarooj S et al. Dengue viremia titer, antibody response pattern, and virus serotype correlate with disease severity. J Infect Dis 2000; 181:2. [DOI] [PubMed] [Google Scholar]
- 28.Zambrano H, Waggoner JJ, Almeida C, Rivera L, Quintana Benjamin J, Pinsky BA. Zika virus and chikungunya virus co-infections: a series of three cases from a single-center in Ecuador. Am J Trop Med Hyg 2016; doi:10.4269/ajtmh.16-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupont-Rouzeyrol M, O'Connor O, Calvez E et al. Co-infection with Zika and dengue viruses in 2 patients, New Caledonia, 2014. Emerg Infect Dis 2015; 21:381–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Villamil-Gomez WE, Gonzalez-Camargo O, Rodriguez-Ayubi J, Zapata-Serpa D, Rodriguez-Morales AJ. Dengue, chikungunya and Zika co-infection in a patient from Colombia. J Infect Public Health 2016; 9:684–6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.