Abstract
Rickettsiales are important zoonotic pathogens, causing severe disease in humans globally. Although mosquitoes are an important vector for diverse pathogens, with the exception of members of the genus Wolbachia little is known about their role in the transmission of Rickettsiales. Herein, Rickettsiales were identified by PCR in five species of mosquitoes (Anopheles sinensis, Armigeres subalbatus, Aedes albopictus, Culex quinquefasciatus and Cu. tritaeniorhynchus) collected from three Chinese provinces during 2014–2015. Subsequent phylogenetic analyses of the rrs, groEL and gltA genes revealed the presence of Anaplasma, Ehrlichia, Candidatus Neoehrlichia, and Rickettsia bacteria in mosquitoes, comprising nine documented and five tentative species bacteria, as well as three symbionts/endosybionts. In addition, bacteria were identified in mosquito eggs, larvae, and pupae sampled from aquatic environments. Hence, these data suggest that Rickettsiales circulate widely in mosquitoes in nature. Also of note was that Ehrlichia and Rickettsia bacteria were detected in each life stage of laboratory cultured mosquitoes, suggesting that Rickettsiales may be maintained in mosquitoes through both transstadial and transovarial transmission. In sum, these data indicate that mosquitoes may have played an important role in the transmission and evolution of Rickettsiales in nature.
The order Rickettsiales (Alphaproteobacteria) comprises of a group of obligate intracellular bacteria that are common parasites of eukaryotes. The order contains three documented families (Anaplasmataceae, Holosporaceae, and Rickettsiaceae) as well as one tentative family (Candidatus Midichloriaceae)1,2. Rickettsiales are well known as zoonotic pathogens, causing such severe human diseases as anaplasmosis, ehrlichiosis, rickettsioses, and scrub typhus3, as well as being associated with extensive agricultural losses4,5. The global incidence and geographic range of rickettsial diseases is seemingly experiencing its second pronounced increase in the last 40 years6,7, and the incidence of human monocytotropic ehrlichiosis (HME) and human granulocytic anaplasmosis (HGA) have increased steadily since their discovery in the 1980s and 1990s, respectively8,11. Due to better diagnostic techniques and enhanced surveillance, the identification of new Rickettsiales and/or their associated diseases has increased markedly over the last 10 years7,12,13,14,15, and bacteria that had previously been considered nonpathogenic to humans are now associated with disease6,7,14,15,16. Clearly, Rickettsiales will present a considerable public health challenge for the forseeable future.
One of the most striking features of Rickettsiales is their diverse host range that includes protists, hydra, annelids, arthropods, vertebrates, and even plants15,17,18,19,20. However, only ticks have been found to act as the vectors for bacteria of the genera Anaplasma and Ehrlichia3,14,18. In nature, Rickettsia bacteria are spread by either transstadial and transovarial (i.e. vertical) transmission in their arthropod hosts, or by horizontal (co-feeding) transmission through an infected vertebrate15,17,21,22. To date, although Anaplasma and Ehrlichia bacteria are passed transstadially in ticks, definitive evidence for transovarial transmission is not yet available3,14, such that vertebrates (e.g. rodents and ruminants) are required as their main amplifying hosts.
Mosquitoes are members of the family Culicidae and comprise more than 3,500 species. Many species (although only females) feed on blood from various vertebrate hosts, including mammals and birds. Mosquitoes are the most important vector of disease-causing pathogens in humans and animals23, with, for example, more than one-half of the global population at risk for mosquito-borne infections such as dengue and malaria24,25. However, despite their significance in pathogen transmission, with the sole exception of the genus Wolbachia24,26, little is known about the circulation and transmission of Rickettsiales bacteria in mosquitoes. Indeed, there is only a single description of the 16 S rRNA (rrs) gene of Anaplasma bacteria in the midgut of Anopheles mosquitoes, although it could not be excluded that the bacteria were from ingested blood27, while a recent experimental study reported the transmission potential of R. felis by Anopheles gambiae mosquitoes28.
To determine whether mosquitoes are indeed hosts of Rickettsiales bacteria, we collected mosquitoes at different life stages – adults, eggs, larvae, and pupae – from a variety of locations in Hubei, Jiangxi, and Zhejiang provinces, China, and examined the prevalence of Anaplasma, Ehrlichia, Candidatus Neoehrlichia, and Rickettsia bacteria. In addition, we conducted a laboratory investigation of the potential mosquito transmission of Rickettsiales.
Results
Collection of mosquitoes and detection of Rickettsiales DNA
During 2014–2015, 971 adult mosquitoes were collected from three regions in China: (i) Wuhan city in Hubei province, (ii) Yudu county in Jiangxi province, and (iii) Cixi city in Zhejiang province (see Supplementary Fig. S1). The numbers, species, and geographic distributions of the adult mosquitoes collected are shown in Table 1. After morphological examination and sequence analysis of the 18S rRNA gene, Anopheles sinensis, Armigeres subalbatus, Aedes albopictus, Culex quinquefasciatus and Cu. tritaeniorhynchus mosquitoes were identified. In addition, approximately 950 eggs, 528 larvae, and 554 pupae of these mosquitoes were collected from aquatic environments in Wuhan city (Table 2). The eggs were pooled into groups of approximately 19 according to species.
Table 1. Prevalence of Rickettsiales bacteria in adult mosquitoes collected in Hubei, Jiangxi, and Zhejiang provinces, China, during 2014–2015.
| Species | Location |
Anaplasmaa |
Ehrlichia |
Rickettsia |
C.N.mik | Totalb | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C.A.bol | A.bov | A.mar | A.pha | A.pla | C.A.rod | E.cha | E.ehh | E.ns | C.R.as | R.bel | R.mon | R.nc | ||||
| Ae.a. | Hubei | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1/144 |
| Zhejiang | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4/6 | |
| An.s. | Hubei | 1 | 5 | 25 | 0 | 8 | 2 | 0 | 1 | 0 | 1 | 1 | 0 | 0 | 2 | 46/192 |
| Jiangxi | 0 | 2 | 4 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8/38 | |
| Zhejiang | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1/48 | |
| Ar.s. | Hubei | 0 | 5 | 4 | 4 | 3 | 1 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 7 | 27/103 |
| Jiangxi | 2 | 0 | 0 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 7/42 | |
| Zhejiang | 1 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4/14 | |
| Cu.q. | Hubei | 0 | 0 | 0 | 0 | 0 | 2 | 1 | 2 | 0 | 0 | 0 | 1 | 0 | 1 | 7/144 |
| Cu.t. | Hubei | 0 | 0 | 2 | 1 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 10/144 |
| Zhejiang | 0 | 0 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5/96 | |
| Total | 5 | 12 | 43 | 11 | 21 | 5 | 1 | 5 | 1 | 1 | 1 | 1 | 2 | 11 | 120/971 | |
aMosquito species are abbreviated as follows: Aedes albopictus, Ae.a.; Anopheles sinensis, An.s.; Armigeres subalbatus, Ar.s.; Culex quinquefasciatus, Cu.q.; Culex tritaeniorhynchus, Cu.t. Bacterial species are abbreviated as follows: Candidatus Anaplasma boleense, C. A.bol; Anaplasma bovis, A.bov; Anaplasma marginale, A.mar; Anaplasma phagocytophilum, A.pha; Anaplasma platys, A.pla; Candidatus Anaplasma rodmosense, C.A.rod; Ehrlichia chaffensis, E.cha; Ehrlichia sp. EHh317, E.ehh; Ehrlichia sp. NS101, E.ns; Candidatus Rickettsia sp. Anopheles sinensis, C. R.as; Rickettsia bellii, R.bel; Rickettsia monacensis, R.mon; Rickettsia symbiont of Nephotettix cincticeps, R.nc; Candidatus Neoehrlichia mikurensis, C.N.mik.
bPCR positive/mosquitoes collected.
Table 2. Prevalence of Rickettsiales bacteria in eggs, larvae and pupae collected from aquatic environments in Wuhan (Hubei province), China, during 2014–2015.
| Species |
Anaplasmaa |
Ehrlichia |
Rickettsia C.R.ct | C.N.mik | Total (mean ± 95% CI) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C.A.bol | A.bov | A.mar | A.pha | A.pla | C.A.rod | E.cha | E.ehh | E.ns | ||||
| Egg | ||||||||||||
| Ae.a | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/0 |
| An.s | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2/9 (10.5 ± 13.8) |
| Ar.s | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/0 |
| Cu.q | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/0 |
| Cu.t | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0/10 |
| Subtotal | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2/19 (10.5 ± 13.8) |
| Larvae | ||||||||||||
| Ae.a | 1 | 4 | 4 | 7 | 1 | 1 | 0 | 6 | 1 | 0 | 2 | 27/144 (18.8 ± 6.5) |
| An.s | 0 | 2 | 5 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 13/48 (27.1 ± 12.6) |
| Ar.s | 1 | 6 | 6 | 10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 25/48 (52.1 ± 14.4) |
| Cu.q | 1 | 0 | 2 | 3 | 0 | 2 | 1 | 0 | 0 | 0 | 2 | 11/144 (7.6 ± 4.4) |
| Cu.t | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 4/144 (2.8 ± 2.7) |
| Subtotal | 3 | 12 | 17 | 25 | 6 | 3 | 1 | 6 | 1 | 1 | 5 | 80/528 (15.1 ± 3.1) |
| Pupae | ||||||||||||
| Ae.a | 0 | 0 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 2 | 7/144 (2.8 ± 2.7) |
| An.s | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 4/88 (4.5 ± 4.4) |
| Ar.s | 1 | 2 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 6/48 (12.5 ± 9.5) |
| Cu.q | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 3/144 (2.1 ± 2.4) |
| Cu.t | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 2 | 3/130 (2.3 ± 2.6) |
| Subtotal | 2 | 3 | 4 | 0 | 3 | 1 | 1 | 3 | 0 | 0 | 6 | 23/554 (4.2 ± 1.7) |
aAbbreviations are the same as those given in Table 1. Candidatus Rickettsia sp. Culex tritaeniorhynchus, C. R.ct.
PCR products of the expected size for the rrs gene were successfully amplified from the adult and juvenile mosquitoes collected in the field and reared in the laboratory (Tables 1, 2, 3, , , , see Supplementary Table S1). Genetic analyses of these bacterial rrs gene sequences revealed that all the bacteria identified in mosquitoes belonged to the order Rickettsiales (see below) and exhibited high similarity in the rrs gene with those of the Anaplasma, Ehrlichia, Candidatus Neoehrlichia, and Rickettsia genera, with percentage identities greater than 95.6%, 97.6%, 99.0%, and 95.2%, respectively. Hence, these data clearly reveal that multiple generations of Rickettsiales naturally co-circulate in both adult and juvenile mosquitoes.
Table 3. Co-circulation of Rickettsiales in eggs, larvae, pupae, and adult mosquitoes reared in the laboratory.
| Species | |
Ehrlichia |
Rickettsia |
Total | |||
|---|---|---|---|---|---|---|---|
| Life stage | Ehrlichia sp. EHh317 | R. japonica | R. monacensis | R. sibirica | |||
| Ae. albopictus | Parent | Adult | 1 | 8 | 0 | 0 | 9/84 |
| Offspring | Egg | 0 | 0 | 0 | 0 | 0/4 | |
| Larvae | 2 | 10 | 0 | 0 | 12/57 | ||
| Pupae | 0 | 2 | 0 | 0 | 2/105 | ||
| Adult | 0 | 1 | 0 | 0 | 1/66 | ||
| An. sinensis | Parent | Adult | 0 | 4 | 0 | 0 | 4/120 |
| Offspring | Egg | 0 | 0 | 0 | 0 | 0/7 | |
| Larvae | 0 | 3 | 0 | 0 | 3/120 | ||
| Pupae | 0 | 4 | 0 | 0 | 4/114 | ||
| Adult | 0 | 1 | 0 | 0 | 1/120 | ||
| Ar. subalbatus | Parent | Adult | 0 | 4 | 0 | 0 | 4/120 |
| Offspring | Egg | 0 | 1 | 0 | 0 | 1/7 | |
| Larvae | 0 | 8 | 0 | 0 | 8/146 | ||
| Pupae | 0 | 7 | 0 | 0 | 7/116 | ||
| Adult | 0 | 1 | 0 | 0 | 1/118 | ||
| Cu. quinquefasciatus | Parent | Larvae | 0 | 5 | 0 | 0 | 5/120 |
| Adult | 0 | 6 | 0 | 0 | 6/120 | ||
| Offspring | Egg | 0 | 2 | 0 | 0 | 2/19 | |
| Larvae | 0 | 3 | 0 | 0 | 3/120 | ||
| Pupae | 0 | 6 | 0 | 0 | 6/106 | ||
| Adult | 0 | 1 | 0 | 0 | 1/138 | ||
| Cu. tritaeniorhynchus | Parent | Adult | 0 | 3 | 1 | 1 | 5/216 |
| Offspring | Egg | 0 | 0 | 1 | 1 | 2/5 | |
| Larvae | 0 | 2 | 2 | 2 | 6/120 | ||
| Pupae | 0 | 1 | 10 | 1 | 12/120 | ||
| Adult | 0 | 15 | 1 | 6 | 22/120 | ||
| Total | 3 | 98 | 15 | 11 | 127/2508 | ||
Phylogenetic analysis of Rickettsiales gene sequences recovered from mosquitoes
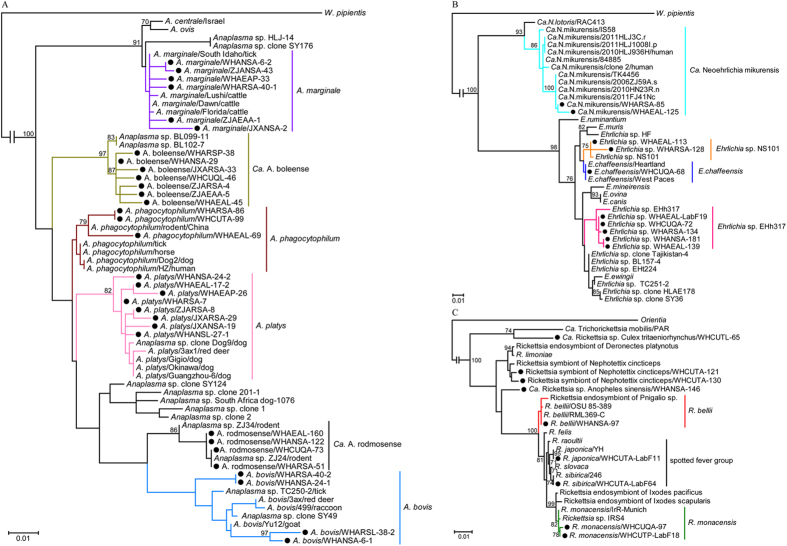
Phylogenetic analysis of the rrs gene sequences revealed that the Chinese mosquitoes collected in this study contained bacteria from the Anaplasma, Ehrlichia, or Candidatus Neoehrlichia genera of the family Anaplasmataceae, and from the genus Rickettsia of the family Rickettsiaceae (Fig. 1). Within the genus Anaplasma, the rrs gene sequences fell into six clusters on the phylogeny (Fig. 1A), corresponding to (i) four documented species: A. bovis, A. marginale, A. phagocytophilum, and A. platys, and (ii) two novel candidate species: Anaplasma sp. Bole (designed as Candidatus Anaplasma boleense) and Anaplasma sp. rodmos (designed as Candidatus Anaplasma rodmosense). Candidatus A. boleense bacteria, which were first identified in ticks sampled from Bole in the Xinjiang Uygur Autonomous Region, China18, formed two distinct lineages according to their vector origins. It is notable that the mosquito Candidatus A. rodmosense bacteria were closely related to Anaplasma sp. ZJ24 identified in Rattus losea rat from Zhejiang (FJ182047). Within the clusters of A. bovis, and A. platys bacteria, the mosquito Anaplasma bacteria were clearly distinct from those described previously (with 0.6–2.9% and 0.4–2.1% differences, respectively). A. bovis bacteria formed two lineages, one of which was most closely related to an A. bovis isolated from goats in China29. Particularly notable was that the A. platys bacteria from mosquitoes were diverse, showing 97.9–99.6% identity with known A. platys, and occupied the basal position in this cluster. A. phagocytophilum bacteria were also distinct but most closely related to a strain isolated from a R. norvegicus rat30. Finally, A. marginale bacteria were closely related to a cattle isolate sampled in Henan province31, which adjoins Hubei province, as well as those sampled from cattle in Australia and the USA.
Figure 1. Phylogenetic trees of Rickettsiales rrs gene sequences.
Phylogenetic trees for bacteria of the genera Anaplasma (A), Ehrlichia and Candidatus Neoehrlichia (B), and Rickettsia (C). Numbers at each node indicate bootstrap values. Trees (A–C) were mid-point rooted for clarity and the scale bar represents the number of nucleotide substitutions per site. Lineages shown in purple in panel A and taxa marked by circles in panels A–C depict sequences obtained in this study.
The Ehrlichia bacteria from mosquitoes formed three clusters, compatible with the existence of three species (Fig. 1B). One cluster comprised the mosquito rrs gene sequences WHARSA-128 and HHAEAL-113 and Ehrlichia sp. NS101 identified from deer in Japan. In contrast, the mosquito rrs gene sequence WHCUPA-68 was more closely related to those of human E. chaffeensis32. The remaining mosquito Ehrlichia bacteria formed a distinct cluster most closely related to the tick bacteria Ehrlichia sp. EHh317. Candidatus Neoehrlichia mikurensis bacteria were also identified in mosquitoes, showing a close relationship with those previously identified in rodents and humans33.
The Rickettsia bacteria sampled from the mosquitoes in this study were closely related to R. monacensis (sequence WHCUQA-97), R. bellii (WHANSA-97), R. japonica (WHCUTA-LabF11), R. sibirica (WHCUTA-Lab64), respectively (Fig. 1C). The sequences WHCUTA-121 and WHCUTA-130 exhibited a close evolutionary relationship with those of the Rickettsia symbiont of Nephotettix cincticeps34. Interestingly, the sequence of WHCUTL-65 was distinct, but most closely related to Candidatus Trichorickettsia mobilis isolated from Paramecium nephridiatum (97.0% identity)35, while the sequence WHANSA-146 was distinct from all known Rickettsial bacteria. Hence, our data suggest the presence of two novel Rickettsia species (or symbionts/endosybionts) in mosquitoes, designated as Candidatus Rickettsia sp. Anopheles sinensis and Candidatus Rickettsia sp. Culex tritaeniorhynchus to reflect their host species.
We also attempted to amplify other two genes – citrate synthase (gltA) and heat shock protein (groEL) – from all Rickettsiales DNA-positive mosquitoes (see Supplementary Table S1). However, compared to the rrs gene, these gene sequences were difficult to amplify from mosquito samples. Consequently, groEL gene sequences were only recovered from some samples positive for Anaplasma, Ehrlichia, and Rickettsia, while gltA gene sequences were obtained only from some samples positive for Anaplasma and Rickettsia.
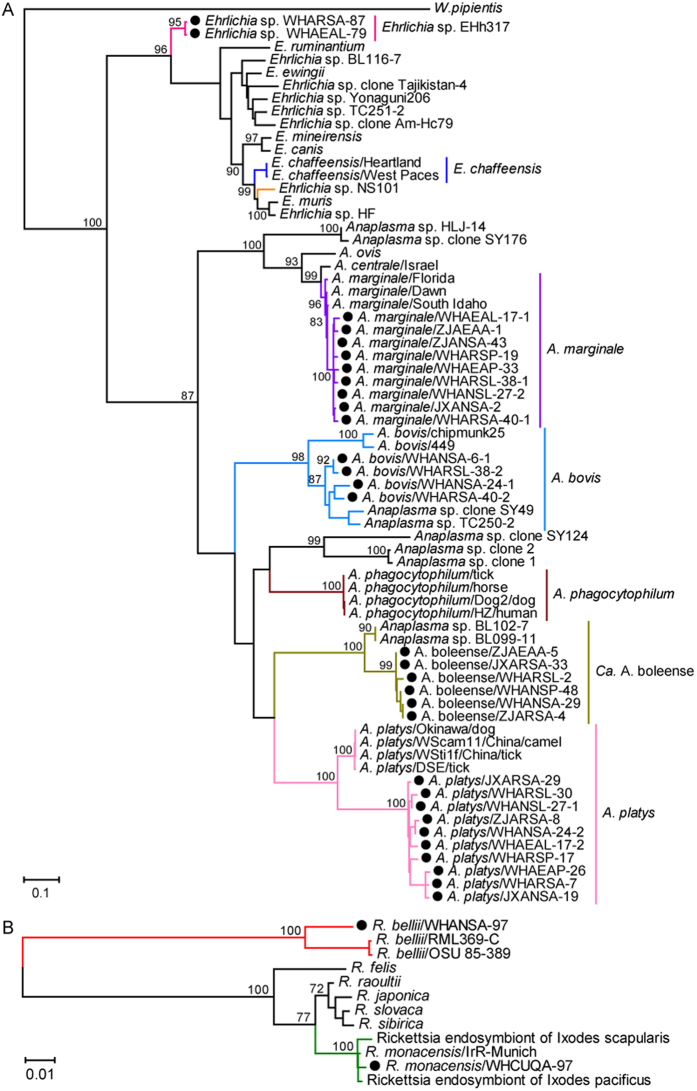
In the groEL gene tree (Fig. 2A), all sequences recovered from Anaplasma mosquitoes fell into four clusters corresponding to Candidatus A. boleense, A. bovis, A. marginale, and A. platys. Notably, within each of the clusters, the sequences from mosquitoes formed distinct lineages similar to those observed for rrs gene. The groEL gene sequences recovered from the Ehrlichia positive samples were also closely related each other and formed a distinct cluster. Finally, those mosquito sequences recovered from Rickettsia positive samples were closely related to those from R. belli and R. monacensis (Fig. 2B).
Figure 2.
Phylogenetic trees of the heat shock protein gene (groEL) of bacteria of the genera Anaplasma and Ehrlichia (A) and Rickettsia (B). The figure description follows that of Fig. 1.
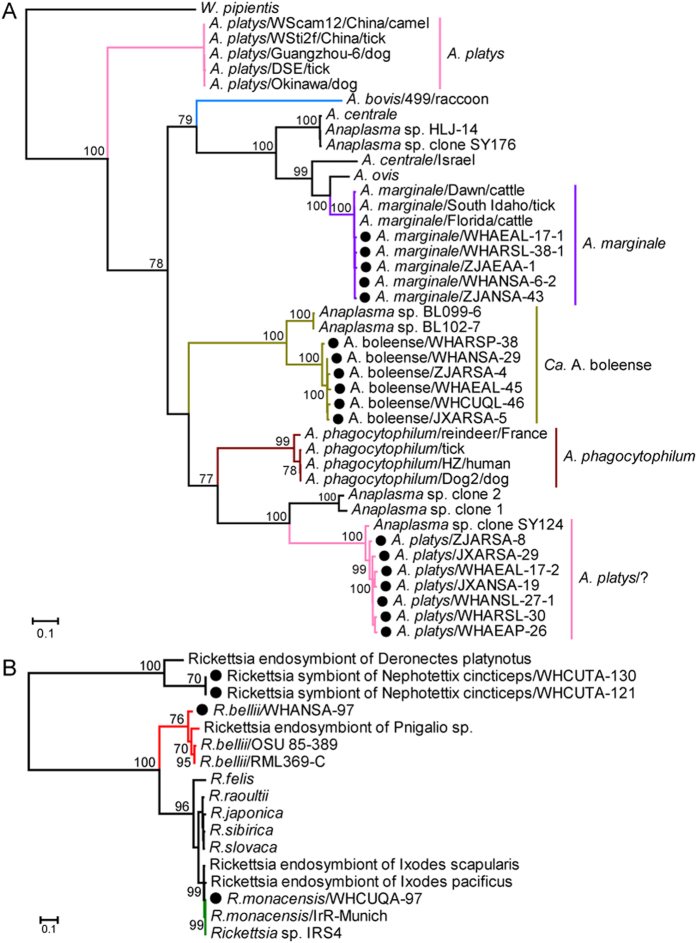
In the gltA gene tree, all Anaplasma sequences formed three clusters (Fig. 3A). Within the Candidatus A. boleense cluster, the bacteria formed a cluster clearly distinct from that previously documented in ticks. Strikingly, although the mosquito Anaplasma bacteria (ZJARSA-8, WHANSL-27–1, JXARSA-29, WHAEAL-17-2, JXANSA-19, WHARSL-30, WHAEAP-26) were most closely related to A. platys bacteria in both the rrs and groEL trees, in the gltA tree they grouped with strain Anaplasma sp. clone SY124 previously identified in ticks from Shenyang of China36. Further studies are needed to determine whether these bacteria belong to A. platys or represent co-infection (and/or recombinant event). Within the genus Rickettsia, the gltA gene sequences from mosquitoes were closely related to those from R. bellii and R. monacensis, respectively, while sequences WHCUTA-121 and WHCUTA-130 formed a distinct lineage (Fig. 3B). As no gltA sequences from the Rickettsia symbiont of Nephotettix cincticeps were available, we could not determine their phylogenetic relationship.
Figure 3.
Phylogenetic trees of the citrate synthase gene (gltA) of bacteria of the genera Anaplasma (A) and Rickettsia (B). The figure description follows that in Fig. 1.
Rickettsiales in different life stages of mosquitoes sampled in nature
The detection rate of Rickettsiales in adult mosquitoes from our three sampling sites was 19.78% in An. sinensis, 23.90% in Ar. subalbatus, 3.33% in Ae. albopictus, 4.86% in Cu. quinquefasciatus, and 6.25% in Cu. tritaeniorhynchus. Overall, Anaplasma bacteria showed the highest prevalence (10.2%), exhibiting species, geographic and annual variation (Table 1, Supplementary Table S2). In contrast, the detection rates of bacteria from the Ehrlichia, Candidatus Neoehrlichia, and Rickettsia genera were lower in adult mosquitoes (0.72%, 0.51%, and 1.13%, respectively), possible caused by bias in the PCR assay. Finally, among all the Rickettsiales identified in mosquitoes, A. marginale had the highest detection rate (4.43%), followed by A. phagocytophilum (2.16%).
To better understand the mosquito circulation of Rickettsiales, eggs, larvae, and pupae were collected from aquatic environments in Wuhan. Although only A. marginale and A. phagocytophilum were identified in two pools of An. sinensis eggs, more Rickettsiales were found in these five mosquito species at the larvae and pupae stages (Table 2, see Supplementary Table S3). In similar pattern to that observed in adult mosquitoes, Anaplasma bacteria were at higher prevalence in larvae and pupae than Ehrlichia, Candidatus Neoehrlichia, and Rickettsia. Notably, A. phagocytophilum and A. marginale were highly prevalent in larvae. As with the adults, three species of Ehrlichia (E. chaffeensis, Candidatus Ehrlichia sp. EHh317, and Candidatus Ehrlichia sp. NS101) were detected in the larvae and pupae stages. With the exception of Ar. subalbatus, Candidatus N. mikurensis were identified in four other species of mosquitoes. Finally, a novel candidate species (or symbiont/endosybionts) (Candidatus Rickettsia sp. Culex tritaeniorhynchus) was identified in Cu. tritaeniorhynchus larvae.
In juvenile mosquitoes, the prevalence of these bacteria was high in Ar. subalbatus (52.08% larvae and 12.50% in pupae), An. sinensis (27.08% in larvae and 4.55% in pupae), and Ae. albopictus (18.75% in larvae and 4.86% in pupae), but relatively low in Cu. quinquefasciatus (7.64% in larvae and 2.08% in pupae) and Cu. tritaeniorhynchus (2.78% in larvae and 2.31% in pupae). In sum, these data suggest that Rickettsiales may be transmitted through the transstadial and/or transovarian transmission routes in mosquitoes, although the number of observations in eggs was small (95% CI 0–24.8%).
More than one species of Anaplasma bacteria was detected in some individuals of both adult and juvenile mosquitoes (Table 4), indicative of co-infection. In adult mosquitoes, the most common combinations were A. marginale and A. bovis, and A. marginale with A. platys. The co-infection of three bacteria (A. bovis, A. marginale, and A. platys) was detected in one An. sinensis individual. Interestingly, co-infection with A. phagocytophilum and Ehrlichia sp. EHh317 was also detected in Ar. subalbatus. Notably, co-infection was relatively more common in An. sinensis mosquitoes, but absent in Cu. quinquefasciatus and Cu. tritaeniorhynchus mosquitoes. For juvenile mosquitoes, the co-infection of Anaplasma bacteria was observed within larvae and pupae of An. sinensis, Ar. subalbatus and Ae. albopictus mosquitoes, but not in Cu. quinquefasciatus and Cu. tritaeniorhynchus (Table 4). The most common combination was also A. marginale or A. phagocytophilum with other bacteria. Finally, co-infection with bacteria from the Ehrlichia and Anaplasma genera was also detected in juvenile mosquitoes.
Table 4. Co-infection of Rickettsiales in adult, larve and pupae mosquitoes collected in Hubei, Jiangxi, and Zhejiang provinces, China, during 2014–2015.
| Species | Location | Life stage | Bacteria | PCR positive/Mosquitoes collected (%) |
|---|---|---|---|---|
| Ae. albopictus | Zhejiang | Adult | C. A. boleense, A. marginale | 1/6 (16.67) |
| Hubei | Larvae | A. marginale, A. phagocytophilum | 1/144 (0.69) | |
| A. bovis, A. marginale | 1/144 (0.69) | |||
| A. marginale, Ehrlichia sp. EHh317 | 1/144 (0.69) | |||
| A. bovis, A. phagocytophilum, Ehrlichia sp. EHh317 | 1/144 (0.69) | |||
| An. sinensis | Hubei | Adult | C. A. boleense, A. marginale | 1/192 (0.52) |
| A. bovis, A. marginale | 3/192 (1.56) | |||
| A. bovis, A. platys | 1/192 (0.52) | |||
| A. marginale, A. platys | 3/192 (1.56) | |||
| A. bovis, A. marginale, A. platys | 1/192 (0.52) | |||
| Larvae | A. bovis, A. marginale | 2/48 (4.2) | ||
| A. marginale, A. phagocytophilum | 1/48 (2.1) | |||
| A. marginale, A. platys | 1/48 (2.1) | |||
| Pupae | C. A. boleense, A. phagocytophilum | 1/88 (1.0) | ||
| Jiangxi | A. bovis, A. platys | 2/38 (5.26) | ||
| Ar. subalbatus | Hubei | Adult | A. bovis, A. marginale | 2/103 (1.94) |
| A. marginale, A. platys | 2/103 (1.94) | |||
| A. phagocytophilum, Ehrlichia sp. EHh317 | 1/103 (0.97) | |||
| Larvae | A. marginale, A. phagocytophilum | 3/48 (6.25) | ||
| C. A. boleense, A. phagocytophilum | 1/48 (2.08) | |||
| A. bovis, A. phagocytophilum | 1/48 (2.08) | |||
| A. bovis, A. marginale | 2/48 (4.17) | |||
| Pupae | A. bovis, A. marginale | 1/48 (2.08) |
Rickettsiales in different life stages of laboratory reared mosquitoes
To confirm the transstadial and transovarian transmission of Rickettsiales, adult (and/or larvae for Cu. tritaeniorhynchus) mosquitoes collected in the field from Wuhan were reared in the laboratory for an entire life cycle (adult-egg-larva-pupae-adult) and tested for the presence of bacteria. From the adult (parent) mosquitoes collected in field, Ehrlichia and Rickettsia bacteria were identified; Ehrlichia sp. EHh317 in Ae. albopictus, R. japonica in all five species of mosquitoes, and R. monacensis and R. sibirica in Cu. tritaeniorhynchus (Table 3). Accordingly, these bacteria were also identified in their offspring. Notably, R. japonica was detected in all life stages of Ar. subalbatus and Cu. quinquefasciatus, and in three stages of the other three mosquito species. Additionally, both R. japonica and R. monacensis were successfully identified at each life stage in Cu. tritaeniorhynchus, again supporting the transstadial and transovarial transmission of Rickettsiales. As Anaplasma and Candidatus N. mikurensis were not identified in adult mosquitos sampled in the field for this experiment, they were similarly not found in laboratory reared eggs, larvae, pupae and adult mosquitoes.
Discussion
Rickettsiales are associated with a wide range of animals including diverse arthropods, mammals and birds3,6,14,15,16,17,18,19,20. High levels of genetic diversity in Rickettsiales have been identified in both ticks and vertebrates6,14,36, including multiple infection by distinct bacteria in a single tick species18,37. Our phylogenetic analysis revealed the co-circulation of nine documented and five tentative species bacteria, as well as three symbionts/endosybionts in five species of mosquitoes. Of particular note in this context was that two distinct Rickettsia species (or symbionts/endosybionts) were identified in mosquitoes and that a single mosquito can harbor two or more species from the genera Anaplasma and Ehrlichia. As more than 3,500 species of mosquitoes are distributed worldwide, it is likely that additional (and/or novel) mosquito-associated Rickettsiales (or symbiont/endosybionts) will be discovered in the future.
Ticks are considered the primary vectors for Rickettsiales, especially in the case of Anaplasma and Ehrlichia14,18,19. Although mosquitoes are both diverse and abundant, with the exception of the genus Wolbachia24,26 there is little evidence for mosquitoes serving as competent vectors or hosts of Rickettsiales bacteria. Previous studies showed that mosquito cell lines (i.e. Ae. albopictus and An. gambiae cells) could be used to propagate A. marginale, R. felis, R. montanensis, and R. peacockii bacteria38,39. Lindh and colleagues also reported the detection of the rrs gene of A. platys and A. ovis in the Anopheles midgut, but could not exclude that the Anaplasma bacteria were derived from ingested blood27. Recently, Dieme and colleagues reported the transmission potential of R. felis infection by An. gambiae mosquitoes28. Herein, we document the presence of diverse bacteria of the genera Anaplasma, Ehrlichia, Candidatus Neoehrlichia and Rickettsia in five species of adult and juvenile mosquitoes sampled from three Chinese provinces. In addition, Rickettsiales bacteria were identified in each mosquito life stage (egg, larvae, pupae, and adult). Hence, these data clearly show that diverse Rickettsiales are present in mosquitoes in nature, such that mosquitoes may have played an important role in the transmission, and likely evolution, of Rickettsiales bacteria. In addition, it was noteworthy that some of the Rickettsiales sequences (for example, WHCUTL-65, WHCUTA-121, WHCUTA-130 and WHANSA-146) recovered from mosquitoes were closely related to those of symbionts or endosymbionts. Finally, as only PCR was used here, which may result in some bias, it is possible that the diversity and the prevalence of Rickettsiales bacteria in mosquitoes may be higher than we report.
Intracellular parasites are transmitted by either transstadial and/or transovarial mechanisms (i.e. vertical transmission) in arthropods or by horizontal transmission via infected vertebrates. As horizontal transmission is dependent on the density of susceptible hosts and their intervals of patent infection, the transovarial and/or transstadial pathways may be more reliable routes to transmit intracellular parasites40,41. Although transovarial and/or transstadial transmission has been documented in members of the Rickettsiaceae15,17,42,43, this process is not thought to occur to a significant degree in those Anaplasma and Ehrlichia bacteria documented to date, which may in part be due to lack of the aldolase/adducing domain protein44. Hence, the known Anaplasma and Ehrlichia bacteria (excluding endosymbionts such as Midichloria) transmitted by ticks require feeding on an infected vertebrate14. Notably, we identified Anaplasma, Ehrlichia, Candidatus Neoehrlichia, and Rickettsia bacteria in eggs, larvae, and pupae, as well as adult mosquitoes collected in the field. Additionally, Ehrlichia and Rickettsia bacteria were identified in eggs, larvae, pupae, and adults reared in the laboratory. Together, these data suggest that Rickettsiales, including Anaplasma and Ehrlichia spp., may be transmitted transovarially and transstadially in mosquitoes in nature. However, as the detection rates of Ehrlichia and Rickettsia bacteria in laboratory reared mosquitoes were relatively low, further studies are needed to determine the efficiency of transovarial transmission.
The prevalence of Anaplasma, Ehrlichia, and Rickettsia bacteria in nature varies substantially with respect to vectors, hosts and geographic regions4,14,45. Similarly, ticks collected at different time points in the same locality can display different infection rates44. Such variation can be attributed to several factors, including host susceptibility and competence, the availability of different reservoir hosts, and geo-ecologic factors6,14,36,45,46,47. In addition, adult ticks have an additional blood meal compared to nymphs, such that the infection rates of A. phagocytophilum in adult ticks are higher than in nymphs36, and because of a lack of transovarial transmission, tick larvae are considered free of Anaplasmataceae bacteria45. Our analysis of mosquitoes also revealed variation in infection rate according to geographic location, mosquito species and their life stage, and sampling times. For example, infection rates were higher in adult mosquitoes of An. sinensis (20.50%) and Ar. subalbatus (23.90%), but lower in adult Ae. Albopictus (3.33%), Cu. quinquefasciatus (4.86%), and Cu. tritaeniorhynchus (6.25%), perhaps reflecting further differences in susceptibility. Furthermore, compared with Ehrlichia, Candidatus N. mikurensis, and Rickettsia, Anaplasma bacteria showed the highest infection rates in adult and juvenile mosquitoes collected in the field.
In all the gene trees inferred here, A. marginale from mosquitoes were closely related to strains sampled from cattle31. For Candidatus A. rodmosense, a close relationship between mosquito- and rodent-associated bacteria is observed in the rrs gene tree. Hence, these data are compatible with the inter-species transmission of bacteria among mosquitoes and mammals, such that mosquitoes may act as transmission vectors. However, for other Anaplasma bacteria (Candidatus A. boleense, A. bovis, and A. platys), the strains identified here were phylogenetically distinct in all three gene trees, such that additional studies are needed to determine whether they are specifically adapted to mosquitoes.
Bovine anaplasmosis caused by A. marginale is widely distributed in tropical and subtropical regions globally, and responsible for substantial economic losses4,48. Indeed, cattle can develop persistent infections and serve as reservoirs of A. marginale4. It was therefore notable that the A. marginale bacteria identified here were more common than other bacteria in An. sinensis and Ar. subalbatus mosquitoes, and that these strains are closely related to widely distributed ruminant strains31,49. Additionally, An. sinensis and Ar. subalbatus mosquitoes prefer to feed on large animals including cattle, and the geographic distribution of a variety of mosquito species (An. sinensis, Ar. subalbatus, Ae. albopictus, and Cu. quinquefasciatus) overlaps with the distribution of bovine anaplasmosis in China. Consequently, it is possible that mosquitoes are involved in the spread of bovine anaplasmosis, beyond serving as mechanical fomites.
The identification of extensive genetic diversity of Rickettsiales (especially Anaplasma) in adult and juvenile mosquitoes indicates that mosquitoes may have played an important role in the transmission of Rickettsiales bacteria, and that their role as vectors needs to be investigated further. These data are also compatible with the notion that Rickettsiales can be maintained in mosquitoes through transstadial and transovarial transmission. Due to the global distribution of these mosquitoes, greater efforts are clearly needed to determine their role in the evolution and natural transmission of Rickettsiales.
Material and methods
Mosquito collection and identification
During the summer (June to August) in 2014 and 2015, 971 adult mosquitoes were collected by ultraviolet light traps from sheep folds, cattle pens, ponds, creeks, and indoors at night in three regions of China: (i) Wuhan city in Hubei province, (ii) Yudu county in Jiangxi province, and (iii) Cixi city in Zhejiang province (see Supplementary Fig. S1). Similarly, mosquito eggs, larvae and pupae were collected from aquatic environments in Wuhan. All mosquitoes were identified to the species level and life stage based on morphologic criteria50 and further by molecular differentiation as described previously51. The main morphological characters distinguishing mosquito pupae from larvae are that the latter are comma-shaped in their lateral aspect and the head and thorax are merged into a cephalothorax. All samples collected were stored at −80 °C until DNA extraction.
Mosquito culture
For mosquito culture, adult mosquitoes (fed or unfed, Table 3), including Ae. albopictus, An. sinensis, Ar. subalbatus and Cu. tritaeniorhynchus collected from the natural aquatic environments in Wuhan were reared for a complete life cycle (adult-egg-larvae-pupae-adult) as described previously50,52. Larvae and adult Cu. quinquefasciatus were also collected for a complete life circle (adult-egg-larvae-pupae-adult or larvae-pupae-adult-egg-larvae-pupae-adult). Samples were collected from adult/larvae mosquitoes caught in the field and at each life stage of cultured mosquitoes.
DNA extraction, PCR amplification, and sequencing
After washing twice with PBS, adult mosquitoes, larvae and pupae were individually homogenized with a mortar and pestle. Approximately 450 An. sinensis and 500 Cu. tritaeniorhynchus eggs were pooled (a total of 19 pools) and homogenized. After homogenization, the suspension was incubated at 4 °C for 1 h and centrifuged at 2,500 g for 5 min, and the upper fraction collected. DNA was extracted from individual mosquitoes or mosquito pools with the QIAamp DNA Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer’s instructions, and then subjected to PCR for amplification of both bacterial gene sequences (rrs, groEL and gltA genes) and mosquito 18S rRNA genes51.
Rickettsial DNA was detected using nested PCR targeting a conserved sequence of the Rickettsiales rrs gene using the primers Eh-out1/Eh-out2 (outer primers) and Eh-gs1/Eh-gs2 (inner primers)53. Primers designed in-house were also used to amplify complete rrs gene sequences and partial groEL and gltA gene sequences by nested PCR. All primer sequemnces are described in Supplementary Table S4.
DNA samples of E. chaffeensis strain Arkansas were used as positive controls, with distilled water used as a negative control. Amplified DNA was purified by electrophoresis in low-melting point agarose and ligated into the cloning vector pMD19-T. Subsequently, each vector was transformed into E. coli and plated onto agarose culture dishes. Twenty clones were picked from each 20th dish and sent to the Sangon Biotechnology Company (Shanghai, China) for sequencing. To prevent contamination, the pre-PCR mix was prepared in a separate room and template DNA was added using dedicated pipets and tips.
Sequence data and genetic analyses
DNA sequences of the three bacterial genes obtained here were aligned with existing reference sequences in GenBank using ClustalW (default parameters) as implemented in the MEGA program, version 5.254. Nucleotide and amino acid sequence identities were calculated using DNAStar (DNASTAR, Inc., Madison, WI). Data sets of the following sizes were then used in an evolutionary analysis: (i) a 1425 bp rrs alignment (N = 131 sequences); (ii) a 1656 bp groEL gene alignment (N = 82); and (iii) a 1227 bp gltA gene alignment (N = 59). The sequences recovered in this study were named according to their relatedness to known bacteria, geographic origins, and sample numbers. All sequences obtained here have been submitted to GenBank and assigned accession numbers KU585921-KU586334.
Phylogenetic analyses
The best-fit evolutionary model for all sequence alignments was determined using jModelTest55 and found to be the General Time Reversible (GTR) nucleotide substitution model with a gamma (Γ)-distribution model of among-site rate variation and a proportion of invariable sites (i.e. GTR + Γ + I). Phylogenetic trees using this model were then estimated using the Maximum Likelihood (ML) method implemented in PhyML (version 3)56. All trees were mid-point rooted for clarity only.
Additional Information
How to cite this article: Guo, W.-P. et al. Extensive genetic diversity of Rickettsiales bacteria in multiple mosquito species. Sci. Rep. 6, 38770; doi: 10.1038/srep38770 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This study was supported by the Special National Project on Research and Development of Key Biosafety Technologies (2016YFC1201900, 2016YFC1200101), and the National Natural Science Foundation of China (Grants 81290343, 81672057). ECH is supported by an NHMRC Australia Fellowship (AF30). The opinions expressed herein are those of the author(s) and are not necessarily representative of those of the United Uniformed Services University of the Health Sciences (USUHS), the U.S. Department of Defense (DOD), or the United States Army, Navy, or Air Force.
Footnotes
Author Contributions Y.Z.Z. conceived the research project; J.H.T., X.D.L. and Y.L. collected the samples, W.P.G., X.B.N., X.P.C., and S.Y.Y. performed research; W.P.G. and Y.Z.Z. analyzed the data; Y.Z.Z., W.P.G., J.X., J.S.D. and E.C.H. wrote the manuscript.
References
- Dumler J. S. et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51, 2145–2165 (2001). [DOI] [PubMed] [Google Scholar]
- Montagna M. et al. “Candidatus Midichloriaceae” fam. nov. Rickettsiales, an ecologically widespread clade of intracellular Alphaproteobacteria. Appl. Environ. Microbiol. 79, 3241–3248 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raoult D. & Parola P. Rickettsial Diseases (ed.). (Informa Healthcare, 2007). [Google Scholar]
- Aubry P. & Geale D. W. A review of bovine anaplasmosis. Transbound Emerg. Dis. 58, 1–30 (2011). [DOI] [PubMed] [Google Scholar]
- Zhao G. Y., Chen X. P. & Zhang Y. Z. Ehrlichia bacteria and ehrlichiosis. Zhonghua Liu Xing Bing Xue Za Zhi. 34, 642–646 (2013). [PubMed] [Google Scholar]
- Eremeeva M. E. & Dasch G. A. Challenges posed by tick-borne rickettsiae: eco-epidemiology and public health implications. Front. Public. Health. 3, 55 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parola P. et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin. Microbiol. Rev. 26, 657–702 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda K. et al. Human infection with Ehrlichia canis, a leukocytic rickettsia. N. Engl. J. Med. 316, 853–856 (1987). [DOI] [PubMed] [Google Scholar]
- Chen S. M., Dumler J. S., Bakken J. S. & Walker D. H. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J. Clin. Microbiol. 32, 589–595 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken J. S. & Dumler J. S. Human granulocytic anaplasmosis. Infect. Dis. Clin. North. Am. 29, 341–355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail N., Bloch K. C. & McBride J. W. Human ehrlichiosis and anaplasmosis. Clin. Lab. Med. 30, 261–292 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritt B. S. et al. Emergence of a new pathogenic Ehrlichia species, Wisconsin and Minnesota, 2009. N. Engl. J. Med. 365, 422–429 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. Human infection with a novel tick-borne Anaplasma species in China: a surveillance study. Lancet Infect. Dis. 15, 663–70 (2015). [DOI] [PubMed] [Google Scholar]
- Rar V. & Golovljova I. Anaplasma, Ehrlichia, and “Candidatus Neoehrlichia” bacteria: pathogenicity, biodiversity, and molecular genetic characteristics, a review. Infect. Genet. Evol. 11, 1842–1861 (2011). [DOI] [PubMed] [Google Scholar]
- Merhej V. & Raoult D. Rickettsial evolution in the light of comparative genomics. Biol. Rev. Camb. Philos. Soc. 86, 379–405 (2011). [DOI] [PubMed] [Google Scholar]
- Chen X. P. et al. Detection of Wolbachia genes in a patient with non-Hodgkin’s lymphoma. Clin. Microbiol. Infect. 21, 182. e1–4 (2015). [DOI] [PubMed] [Google Scholar]
- Weinert L. A., Werren J. H., Aebi A., Stone G. N. & Jiggins F. M. Evolution and diversity of Rickettsia bacteria. BMC Biol. 7, 6 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y. J. et al. Extensive diversity of Rickettsiales bacteria in two species of ticks from China and the evolution of the Rickettsiales. BMC Evol. Biol. 14, 167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby A. C., Cho N. H., Fuxelius H. H., Westberg J. & Andersson S. G. Intracellular pathogens go extreme: genome evolution in the Rickettsiales. Trends Genet. 23, 511–520 (2007). [DOI] [PubMed] [Google Scholar]
- Sicard M., Dittmer J., Grève P., Bouchon D. & Braquart-Varnier C. A host as an ecosystem: Wolbachia coping with environmental constraints. Environ. Microbiol. 16, 3583–3607 (2014). [DOI] [PubMed] [Google Scholar]
- Zemtsova G., Killmaster L. F., Mumcuoglu K. Y. & Levin M. L. Co-feeding as a route for transmission of Rickettsia conorii israelensis between Rhipicephalus sanguineus ticks. Exp. Appl. Acarol. 52, 383–392 (2010). [DOI] [PubMed] [Google Scholar]
- Brown L. D., Christofferson R. C., Banajee K. H., Del Piero F., Foil L. D. & Macaluso K. R. Cofeeding intra- and interspecific transmission of an emerging insect-borne rickettsial pathogen. Mol. Ecol. 24, 5475–5489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolle M. A. Mosquito-borne diseases. Curr. Probl. Pediatr. Adolesc. Health. Care. 39, 97–140 (2009). [DOI] [PubMed] [Google Scholar]
- Iturbe-Ormaetxe I., Walker T. & O’ Neill S. L. Wolbachia and the biological control of mosquito-borne disease. EMBO Rep. 12, 508–518 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubálek Z., Rudolf I. & Nowotny N. Arboviruses pathogenic for domestic and wild animals. Adv. Virus Res. 89, 201–275 (2014). [DOI] [PubMed] [Google Scholar]
- Ricci I., Cancrini G., Gabrielli S., D’Amelio S. & Favi G. Searching for Wolbachia (Rickettsiales: Rickettsiaceae) in mosquitoes (Diptera: Culicidae): large polymerase chain reaction survey and new identifications. J. Med. Entomol. 39, 562–567 (2002). [DOI] [PubMed] [Google Scholar]
- Lindh J. M., Terenius O. & Faye I. 16S rRNA gene-based identification of midgut bacteria from field-caught Anopheles gambiae sensu lato and A. funestus mosquitoes reveals new species related to known insect symbionts. Appl. Environ. Microbiol. 71, 7217–7223 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieme C. et al. Transmission potential of Rickettsia felis infection by Anopheles gambiae mosquitoes. Proc. Natl. Acad. Sci. USA. 112, 8088–8093 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. et al. Molecular survey and genetic identification of Anaplasma species in goats from central and southern China. Appl. Environ. Microbiol. 78, 464–470 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X. G. et al. Dual infection with Anaplasma phagocytophilum and Babesia microti in a Rattus norvegicus, China. Ticks Tick Borne Dis. 4, 399–402 (2013). [DOI] [PubMed] [Google Scholar]
- Liu Z. et al. Amplification of 16S rRNA genes of Anaplasma species in China for phylogenetic analysis. Vet. Microbiol. 107, 145–148 (2005). [DOI] [PubMed] [Google Scholar]
- Cheng C., Paddock C. D. & Reddy Ganta R. Molecular heterogeneity of Ehrlichia chaffeensis isolates determined by sequence analysis of the 28-kilodalton outer membrane protein genes and other regions of the genome. Infect. Immun. 71, 187–195 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. Wide distribution and genetic diversity of “Candidatus Neoehrlichia mikurensis” in rodents from China. Appl. Environ. Microbiol. 79, 1024–1027 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Yukuhiro F., Matsuura Y., Fukatsu T. & Noda H. Intrasperm vertical symbiont transmission. Proc. Natl. Acad. Sci. USA. 111, 7433–7437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C. et al. Flagellar movement in two bacteria of the family Rickettsiaceae: a re-evaluation of motility in an evolutionary perspective. PLoS One. 9, e87718 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welc-Falęciak R. et al. Rickettsiaceae and Anaplasmataceae infections in Ixodes ricinus ticks from urban and natural forested areas of Poland. Parasit. Vectors. 7, 121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X., Chen X. P., Liu N., Dumler S. J. & Zhang Y. Z. Co-circulation of multiple species of Rickettsiales bacteria in one single species of hard ticks in Shenyang, China. Ticks Tick Borne Dis. 5, 727–733 (2014). [DOI] [PubMed] [Google Scholar]
- Mazzola V., Amerault T. E. & Roby T. O. Electron microscope studies of Anaplasma marginale in an Aedes albopictus culture system. Am. J. Vet. Res. 40, 1812–1815 (1979). [PubMed] [Google Scholar]
- Sakamoto J. M. & Azad A. F. Propagation of arthropod-borne Rickettsia spp. in two mosquito cell lines. Appl. Environ. Microbiol. 73, 6637–6643 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz L. C. et al. Natural transovarial transmission of dengue virus 4 in Aedes aegypti from Cuiabá, State of Mato Grosso, Brazil. Rev. Soc. Bras. Med. Trop. 48, 18–25 (2015). [DOI] [PubMed] [Google Scholar]
- Saiyasombat R., Bolling B. G., Brault A. C., Bartholomay L. C. & Blitvich B. J. Evidence of efficient transovarial transmission of Culex flavivirus by Culex pipiens (Diptera: Culicidae). J. Med. Entomol. 48, 1031–1038 (2011). [DOI] [PubMed] [Google Scholar]
- Brumin M., Levy M. & Ghanim M. Transovarial transmission of Rickettsia spp. and organ-specific infection of the whitefly Bemisia tabaci. Appl. Environ. Microbiol. 78, 5565–74 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C. L., Gaff H. D., Sonenshine D. E. & Hynes W. L. Experimental vertical transmission of Rickettsia parkeri in the Gulf Coast tick, Amblyomma maculatum. Ticks Tick Borne Dis. 6, 568–573 (2015). [DOI] [PubMed] [Google Scholar]
- Dunning Hotopp J. C. et al. Comparative genomics of emerging human ehrlichiosis agents. PLOS Genet. 2, e21 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuen S., Granquist E. G. & Silaghi C. Anaplasma phagocytophilum - a widespread multi-host pathogen with highly adaptive strategies. Front Cell Infect. Microbiol. 3, 31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massung R. F. et al. Genetic variants of Ehrlichia phagocytophila, Rhode Island and Connecticut. Emerg. Infect. Dis. 8, 467–472 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katargina O. et al. Identification of Anaplasma phagocytophilum in tick populations in Estonia, the European part of Russia and Belarus. Clin. Microbiol. Infect. 18, 40–46 (2012). [DOI] [PubMed] [Google Scholar]
- Kocan K. M., de la Fuente J., Blouin E. F., Coetzee J. F. & Ewing S. A. The natural history of Anaplasma marginale. Vet. Parasitol. 167, 95–107 (2010). [DOI] [PubMed] [Google Scholar]
- Pierlé S. A. et al. Genetic diversity of tick-borne rickettsial pathogens; insights gained from distant strains. Pathogens. 3, 57–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z. K., Zheng Z. M. & Wang Z. C. Management of Hygienic Pest (People’s Medical Publishing House Co., Ltd, 2011). [Google Scholar]
- Machida R. J. & Knowlton N. PCR primers for metazoan nuclear 18S and 28S ribosomal DNA sequences. PLOS ONE. 7, e46180 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict M. Q. Care and maintenance of anopheline mosquito colonies In Molecular Biology of Insect Disease Vectors: A Methods Manual. (ed. Crampton J. M., Beard C. B. & Louis C.) 3–12 (Springer, 1997). [Google Scholar]
- Wen B., Jian R., Zhang Y. & Chen R. Simultaneous detection of Anaplasma marginale and a new Ehrlichia species closely related to Ehrlichia chaffeensis by sequence analyses of 16S ribosomal DNA in Boophilus microplus ticks from Tibet. J. Clin. Microbiol. 40, 3286–3290 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A. & Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (2008). [DOI] [PubMed] [Google Scholar]
- Guindon S. et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.