Abstract
Most organisms are exposed to the genotoxic chemical formaldehyde, either from endogenous or environmental sources. Therefore, biology has evolved systems to perceive and detoxify formaldehyde. The frmRA(B) operon that is present in many bacteria represents one such system. The FrmR protein is a transcriptional repressor that is specifically inactivated in the presence of formaldehyde, permitting expression of the formaldehyde detoxification machinery (FrmA and FrmB, when the latter is present). The X-ray structure of the formaldehyde-treated Escherichia coli FrmR (EcFrmR) protein reveals the formation of methylene bridges that link adjacent Pro2 and Cys35 residues in the EcFrmR tetramer. Methylene bridge formation has profound effects on the pattern of surface charge of EcFrmR and combined with biochemical/biophysical data suggests a mechanistic model for formaldehyde-sensing and derepression of frmRA(B) expression in numerous bacterial species.
All organisms must adapt to withstand the detrimental effects of toxic chemicals. Some of these toxic compounds, such as reactive oxygen and nitrogen species are generated endogenously, as a result of metabolic processes; others are natural or synthetic products that are present in the environment. Many of these chemicals have broad reactivity, which presents biological systems with a challenge to specifically sense and then respond to their presence before the cell suffers irreversible damage. Formaldehyde is a toxic electrophilic chemical that is generated endogenously by many organisms1,2. For example, formaldehyde is generated: (i) as an intermediate in methylotrophic metabolism; (ii) in the degradation of glycine, either by the glycolytic byproduct methylgloxal or by Fenton chemistry; (iii) in the degradation of heme, during iron acquisition by some Gram-positive bacteria; (iv) by lipid peroxidation of sugars; (v) by the demethylation of histones; (vi) as a product of methylated-DNA repair by AlkB; and (vii) by the action of N-methyltryptophan oxidase (SolA)2,3,4. Consequently, biological systems are often exposed to both endogenous and exogenous sources of formaldehyde.
Formaldehyde mediates its toxic effects by chemically modifying vital cell components, including DNA and proteins, thereby leading to cellular dysfunction. Formaldehyde-mediated genotoxicity is caused by the formation DNA-DNA and DNA-protein cross-links, as well as covalent DNA monoadducts1,2,5,6,7. In addition, formaldehyde is able to covalently modify proteins, inhibiting their functions7,8. The life-threatening damage caused by the broad chemical reactivity of formaldehyde has driven the evolution of mechanisms to detoxify formaldehyde and counteract its detrimental effects2. In order to regulate expression of these detoxification systems, it is imperative to maintain specific response-regulators in the cell that can perceive the presence of formaldehyde and induce protective systems before significant damage to vital cell components occurs.
The starting point for the work reported here was the observation that formaldehyde is generated when the model bacterium Escherichia coli adapts to the presence of the alternative electron acceptor trimethylamine-N-oxide9. An inability to respond (by induction of the frmRAB operon) to this endogenous formaldehyde challenge resulted in growth inhibition, rather than growth promotion, when anaerobic E. coli cultures were provided with trimethylamine-N-oxide9. The frmRAB operon codes for: a regulator, FrmR (EcFrmR); a formaldehyde dehydrogenase, FrmA; and an S-formylglutathione hydrolase, FrmB10. In E. coli, and many other organisms, formaldehyde in the cytosol reacts with the major reductant glutathione, yielding S-hydroxymethylglutathione, which is then oxidized by FrmA to S-formylglutathione11,12. In E. coli S-formylglutathione is hydrolyzed to formate and glutathione by the product of the final gene of the frmRAB operon, FrmB, or by a second, less-efficient hydrolase YeiG11; some bacteria, such as the closely related Salmonella enterica serovar Typhimurium, lack frmB but possess yeiG. Thus, the proteins coded by the frmRAB operon and yeiG permit bacteria to sense and detoxify formaldehyde.
The first gene of the E. coli frmRAB operon is the regulatory protein EcFrmR. EcFrmR is a member of the CsoR/RcnR family of transcriptional repressors13,14. Most of the characterized members of this family are metal-ion sensors; the properties of Cu(I) (CsoR, RicR), Ni(II) (InrS) and Ni(II)/Co(II) (RcnR, DmeR) sensors in diverse bacterial species have been reported, as well as those of the sulfite/sulfide sensor CstR15,16,17,18,19. Amino acid sequence alignments of the metal-sensing members of the family revealed characteristic amino acid signatures (known as the W-X-Y-Z fingerprint) that are associated with metal-binding (Fig. 1a). The availability of structure-function data now allows the W-X-Y-Z fingerprint to be considered spatially, centered about the absolutely conserved Cys residue (position X, Fig. 1a; residue 35 in RcnR and FrmR), rather than as a primary sequence motif17,20. In addition to the absolute conservation at position X, there is a highly conserved His residue at position Y (Fig. 1a). RcnR and InrS use all four positions to co-ordinate their cognate metal, and in addition RcnR uses the N-terminal residue, which is hereafter regarded as spatially part of position W, for metal-coordination17,21. CsoR binds Cu(I) via the X-Y-Z positions (Fig. 1a). Amino acid residue variation at different positions within the fingerprint is implicated in signal specificity by coupling metal-coordination preferences to the allosteric network connecting the metal- and DNA-binding residues of the proteins21. Whilst the Ni(II)/Co(II) and Cu(I)-responsive family members are relatively well-characterized, little is known about signal perception and DNA-binding mechanisms of the remaining proteins (CstR and FrmR), although the Cys residues at positions X and Z in S. aureus CstR, and Pro2 and Cys35 (positions W and X) of Salmonella enterica serovar Typhimurium FrmR (hereafter StyFrmR) have been implicated in sulfide and formaldehyde sensing, respectively (Fig. 1a)19,22,23.
Figure 1. Amino acid fingerprints associated with signal perception by members of the CsoR/RcnR family and amino acid sequences of EcFrmR and StyFrmR.
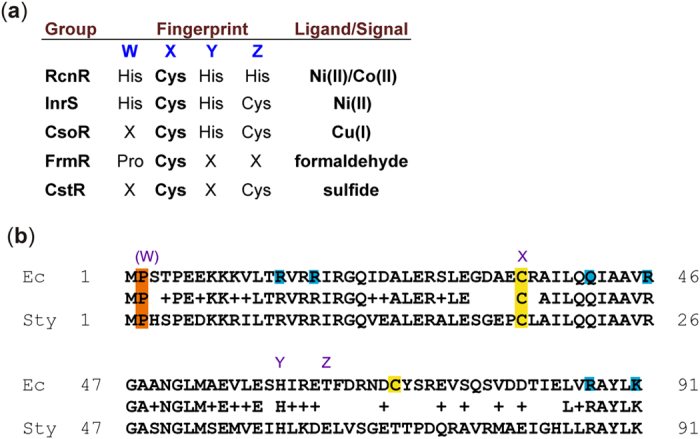
(a) Amino acid residues in the W-X-Y-Z fingerprint of CsoR/RcnR family proteins and the signals perceived by the indicated proteins. (b) Alignment of the E. coli (Ec) and S. enterica serovar Typhimurium (Sty) FrmR proteins. Identical (single letter code) and similar (+) residues, Pro2 (brown background), Cys35 and Cys70 (yellow background) are indicated. Residues of the W-X-Y-Z fingerprint (blue font) are indicated. Position W is shown to incorporate both Pro2 and Ser/His3 (as indicated by parentheses; see text for details). Residues on blue backgrounds have been implicated in DNA-binding in other CsoR/RcnR family members.
StyFrmR retains 3 of the metal-binding ligands (His3, Cys35 and His60, with Glu instead of His64) that are present in the paralogous Salmonella Ni(II)-binding regulator RcnR (Fig. 1a)22. Recent in vitro studies showed that StyFrmR binds Co(II), Cu(I) and Zn(II), but the binding affinities were weaker than those of the biological sensors of these metal ions in Salmonella, such that StyFrmR was outcompeted by CueR, RcnR, ZntR and ZurR for their cognate metals22. Thus in vivo, StyFrmR was able to act as a formaldehyde-responsive transcription regulator, but was unable to act as a metal ion sensor22. Further work showed that formaldehyde-sensing by StyFrmR is specific, direct and requires two residues of the W-X-Y-Z fingerprint (the N-terminal Pro at position W and Cys35 at position X; Fig. 1)23. The crystal structure of a metal-sensing StyFrmR variant (StyFrmR(E64H)), created by site-directed mutagenesis, revealed that it consists of a homotetrameric disc with a surface-exposed positively-charged region that is predicted to participate in DNA-binding23. Hence, the fold of StyFrmR resembles other members of the CsoR/RcnR family24,25.
Amino acid sequence alignment of the E. coli K-12 FrmR (EcFrmR) and StyFrmR proteins reveals strong conservation in the first 63 amino acids (67% identical, 89% similar) but weaker conservation thereafter (21% identical; 39% similar; Fig. 1b). There is limited conservation between the two proteins within the W-X-Y-Z fingerprint. Both have Cys and His at positions X and Y, as well as Pro at residue 2 (position W). However, EcFrmR differs with Ser not His at residue 3 (a component of position W; note that the equivalent of His3 is involved in metal-binding in RcnR and InrS proteins) and Thr not Glu at position Z. Notably, EcFrmR possesses an additional non-conserved Cys residue (Cys70) (Fig. 1b). The differences between StyFrmR and EcFrmR along with the biological imperative to mount an effective response to endogenous sources of formaldehyde, as evidenced by the observation that the frmRAB operon was essential for adaptation of E. coli to growth on trimethylamine-N-oxide, prompted an investigation of the EcFrmR protein9. This is worthy of investigation because, although the activities of detoxifying enzymes, such as FrmA and FrmB, have been established, the mechanism(s) used by regulatory proteins to perceive and respond to formaldehyde are poorly understood. Hence, the aim of the work reported here was to provide new insight into the mechanism used by biological systems to sense the presence of the highly reactive and toxic chemical, formaldehyde. In vivo and in vitro data show that EcFrmR senses formaldehyde directly, with no metal-dependence, via the formation of inter-subunit methylene bridges between adjacent Pro2 and Cys35 residues. This formaldehyde-mediated cross-linking remodels the surface of the tetrameric EcFrmR disc resulting in de-repression of frmRAB expression by promoting disassociation of the frmRAB promoter (Pfrm)-EcFrmR complex.
Results and Discussion
EcFrmR is a formaldehyde sensor
Herring and Blattner showed that EcFrmR is necessary for repression of the frmRAB promoter (Pfrm) in E. coli, and that the operon is induced in the presence of formaldehyde10. Similarly, StyFrmR responds to formaldehyde, but not acetaldehyde or organic alcohols23. A Pfrm-frmR-lacZ reporter fusion containing ~500 bp upstream of the frmA open reading frame, so as to include frmR, was constructed to determine the range of molecules that de-repress frmRAB expression (Table S1). β-Galactosidase activity (proxy for frmRAB expression) was induced 28-fold in the presence of 700 μM formaldehyde (Fig. 2a). The role of EcFrmR in this regulation was confirmed by mutation of codon six of frmR to a stop codon (Pfrm-frmRstop-lacZ), whereupon constitutive high expression was observed, consistent with EcFrmR-mediated repression of frmRAB in the absence of formaldehyde (Fig. 2a). The signal specificity of frmRAB induction was assessed by culturing the reporter strain in the presence of different aldehydes (Fig. 2b). Many of these compounds have been found to induce intracellular damage, suggesting that they can cross the cell membrane, and hence they could be directly or indirectly perceived by cytosolic FrmR26,27,28. Formaldehyde induced the highest β-galactosidase activity, but acetaldehyde, methylglyoxal and glyoxal also induced expression, albeit to lesser extents (Fig. 2b). This contrasts with StyFrmR which did not respond to acetaldehyde (in the same type of experiment), suggesting sequence differences between the two proteins around the sensory site could affect selectivity. Bulky aldehydes, such as furaldehyde and tribromoacetaldehyde, were unable enhance lacZ expression. Overall, these responses suggest that, although induction of frmRAB is not absolutely specific, formaldehyde is by far the most effective inducer, consistent with the specificity of the detoxification machinery (FrmA and FrmB/YeiG). However, it is possible that induction results from EcFrmR responding to the formation of S-hydroxymethylglutathione in the cytoplasm rather than formaldehyde per se.
Figure 2. EcFrmR-mediated repression of frmRAB expression is relieved by formaldehyde.
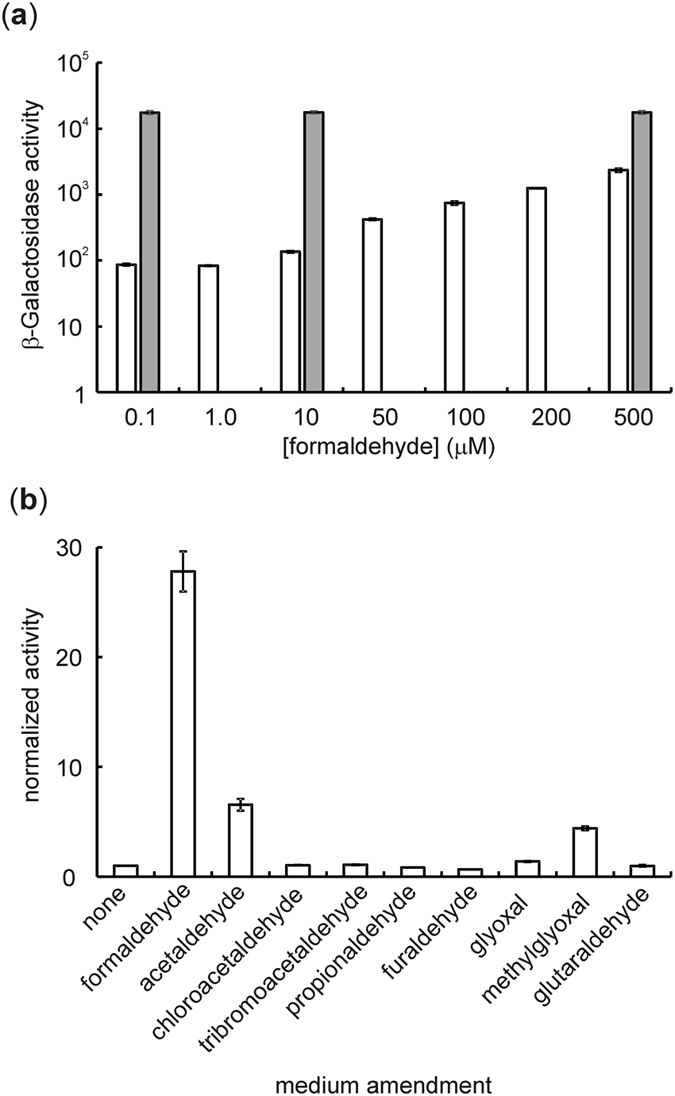
(a) Cultures of E. coli PC677 carrying Pfrm-frmR-lacZ (open bars) or Pfrm-frmRstop-lacZ (gray bars) were grown as described in Methods in the absence and presence of the indicated concentrations of formaldehyde. β-Galactosidase activities (Miller units plotted on a log scale) were measured as a proxy for in vivo transcription from the frmRAB promoter. (b) β-Galactosidase activities (Miller units) of cultures of E. coli PC677 carrying Pfrm-frmR-lacZ were measured after anaerobic cultivation in the presence of the indicated aldehydes as described in Methods. Activities were normalized to that measured in the absence of formaldehyde. For both panels, the error bars represent the standard deviation from the mean (n = 3).
EcFrmR binds directly to the frmRAB promoter (P frm ) to repress transcription
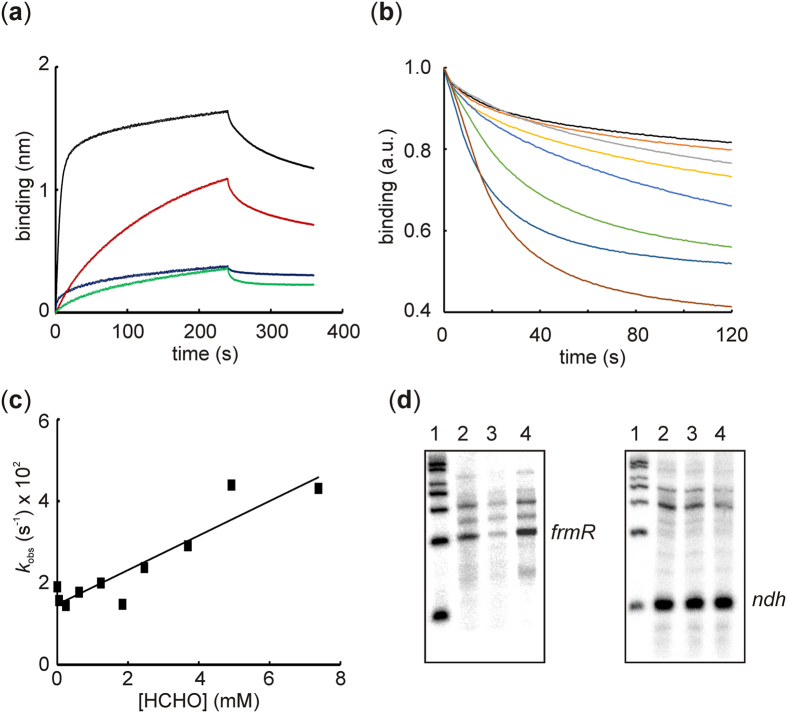
The effect of signal molecules on DNA-binding affinity and transcriptional regulation provides a sensitive measure of allosteric effectiveness. Bio-Layer Interferometry (BLItz) measurements were used to investigate interactions between the EcFrmR protein and frmRAB promoter DNA (Pfrm). Under these conditions a Kd for EcFrmR binding to immobilized Pfrm DNA was ~220 nM, determined from an overall on-rate constant (kf) of ~13000 M−1 s−1 and an overall off-rate constant (kr) of ~0.003 s−1 at 20 °C (Fig. 3a; Table 1). This Kd value is similar to that reported for StyFrmR (~100 nM)22, but should be considered as an upper limit because the potential effects of low level metal contamination and protein oxidation (see below) on DNA-binding are unknown. Nevertheless, this interaction was specific because the Kd for EcFrmR binding at an unrelated promoter DNA fragment (E. coli ydhY) was only ~3600 nM (Table 1). Pre-treatment of EcFrmR protein with excess formaldehyde for 3 min essentially abolished specific binding to Pfrm (Fig. 3a). Exposure of the pre-formed Pfrm-EcFrmR binary complex to increasing concentrations of formaldehyde resulted in disassociation of the complex (Fig. 3b). The Pfrm–EcFrmR disassociation curves were fitted to a single exponential function. Disassociation of the Pfrm-EcFrmR-complex exhibited a linear dependence on formaldehyde concentration, with a rate constant of ~4 M−1 s−1 at 20 °C (Fig. 3c; Table 1). Accordingly, in vitro transcription reactions showed that synthesis of the frmRAB transcript was inhibited in the presence of EcFrmR and that this inhibition was relieved when EcFrmR was treated with formaldehyde (Fig. 3d). EcFrmR did not affect transcription from the E. coli ndh promoter, indicating that the effects of EcFrmR and formaldehyde on frmRAB transcription were specific (Fig. 3d). Therefore, the in vivo and in vitro data showed that EcFrmR is a repressor of frmRAB expression that responds directly to formaldehyde by disassociation of the Pfrm–EcFrmR complex, allowing the expression of the formaldehyde detoxification system.
Figure 3. Formaldehyde enhances disassociation of the Pfrm-EcFrmR complex.
(a) Bio-Layer Interferometry (BLItz) assays. Reactions to evaluate the interaction of biotin-labeled Pfrm DNA, immobilized on a streptavidin probe, with EcFrmR were carried out with 10 different concentrations of EcFrmR (Table S4A). Representative traces for EcFrmR (6.16 μM tetramer, black line, 0.88 μM tetramer; red line), as well as EcFrmR pre-treated with 200-fold molar excess of formaldehyde (0.88 μM tetramer; blue line), and EcFrmR binding at a non-target DNA (PydhY, 0.88 μM EcFrmR tetramer; green line) are shown. (b) Pre-formed Pfrm-EcFrmR complexes were exposed to 10 different concentrations (Table S4C) of formaldehyde and disassociation curves were recorded. Traces for 0 (black); 0.05 mM (orange); 0.25 mM (gray); 0.62 mM (yellow); 1.25 mM (blue); 3.69 mM (green); 4.92 mM (dark blue); 7.38 mM (brown) are shown. (c) Single exponential fits to formaldehyde disassociation curves were used to obtain the observed rate constants (kobs) which were plotted against formaldehyde concentration to obtain the apparent second order rate constant. (d) Inhibition of frmRAB transcription by EcFrmR in vitro is relieved by formaldehyde. Reaction conditions are described in the Methods section. Left panel, Pfrm; right panel, Pndh. Lanes 1, RNA size markers, top to bottom: 600, 500, 400, 300, 200, 100 bases; Lanes 2, no EcFrmR; lanes 3, 1 nM EcFrmR tetramer; lane 4, 1 nM EcFrmR tetramer pre-treated with 200-molar excess formaldehyde. The locations of the frmR and ndh are indicated.
Table 1. Rate constants for EcFrmR DNA interactions.
| Reactiona | kf (M−1 s−1)b | kr (s−1)a | Kd (nM) | kapp (M−1 s−1) |
|---|---|---|---|---|
| EcFrmR + Pfrm ↔ Pfrm − EcFrmR | 13000 ± 390 | 0.0028 ± 0.000086 | 220 | — |
| EcFrmR − (Zn)4 + Pfrm ↔ Pfrm − EcFrmR − (Zn)4 | 5660 ± 165 | 0.003 ± 0.000078 | 520 | — |
| Pfrm − EcFrmR + HCHO → EcFrmR − HCHO + Pfrm | — | — | — | 4.2 |
| Pfrm − EcFrmR − (Zn)4 + HCHO → EcFrmR − (Zn)4 − HCHO + Pfrm | — | — | — | 0.7 |
| EcFrmR + PydhY ↔ PydhY − EcFrmR | 1950 ± 1000 | 0.007 ± 0.00017 | 3600 | — |
aThe BLItz data used to calculate the kinetic parameters shown are provided in Table S4. The data were fitted to a 1:1 binding model to derive kf, kr and Kd values using all the sample data simultaneously (Global fitting).
bValue and standard error.
Properties of isolated EcFrmR
Liquid chromatography-mass spectrometry (LC-MS) showed that EcFrmR lacked an N-terminal methionine (confirmed by N-terminal amino acid sequencing) and had the expected monomeric molecular mass of 10186.50 Da. EcFrmR eluted from a calibrated size exclusion chromatography column at a volume indicative of a tetramer and this assignment was confirmed by analytical ultracentrifugation, which yielded a mass of 44.9 kDa (Fig. S1a). Thus, like other members of the CsoR/RcnR family, EcFrmR is a homotetramer. The isolated protein was metal-free as judged by inductively coupled plasma mass spectrometry (ICP-MS) analysis and reaction with 5, 5′-dithiobis(2-nitrobenzoic acid) (DTNB) indicated the presence of 1.86 ± 0.21 reactive thiols per monomer. However, the number titratable thiol groups decreased to 1.02 ± 0.08 per monomer upon aerobic storage (>72 h), suggesting that protein can adopt an oxidized form with two disulfide bonds per tetramer during prolonged exposure to molecular oxygen.
As StyFrmR is able to bind Zn(II)22, the interaction of EcFrmR with Zn(II) was also examined. Zn(II) addition resulted in changes in intrinsic fluorescence at 304 nm (Fig. S1b), with saturation at 4 Zn(II) atoms per EcFrmR tetramer. In contrast, titrations with Mn(II) resulted in a linear non-specific binding response that did not saturate even at a [Mn(II)]:[EcFrmR tetramer] ratio of 19. The titration with Zn(II) suggested a sub-micromolar binding affinity, so a competition assay with mag-fura2 (Kd = 61.9 nM) was used to measure KZn (Fig. S1c). The data were best fit to a model of two pairs of two independent sites (K1 = K2 and K3 = K4), with K1 = 3.1 ± 0.3 nM and K3 = 219 ± 19 nM. The value for K1 and K2 represents an upper limit as the theoretical curve determined for K1 and K2 = 0.31 nM (i.e. 10-fold tighter binding at the first two sites) is not well-distinguished from the experimental data (Fig. S1c). This model is consistent with different stepwise binding affinities reported for other family members20. The Zn(II) affinity of EcFrmR was significantly weaker under non-reducing conditions (Kdapp = 146 ± 32 nM), suggesting that the thiolate of a Cys residue (most likely Cys35 at the X position of the W-X-Y-Z metal binding motif: Fig. 1) is important for Zn(II) binding. The affinity of EcFrmR for Zn(II) is similar to that reported for StyFrmR (0.17 ± nM), so Zn(II) could contribute to formaldehyde-sensing by both these proteins22. To gain structural insight into Zn(II) coordination by EcFrmR, Zn-saturated EcFrmR tetramers were analyzed by X-ray absorption spectroscopy (XAS). X-ray absorption near edge structure (XANES) data suggested a five-coordinate Zn-site (Fig. S1d). However, the best fit to the extended X-ray absorption fine structure (EXAFS) data was obtained with a tetrahedral model in which Zn(II) is coordinated by a His N atom at 2.00 Å (probably His60; see above), one thiolate ligand at 2.27 Å (probably Cys35; see above), an N/O ligand at 2.00 Å and an additional ligand from the solvent (Br- or Cl- from the buffer; Fig. S1d; Table S2). Thus, EcFrmR likely binds Zn(II) via three amino acid side-chains leaving the fourth co-ordination position free, possibly to participate in binding formaldehyde, by analogy to substrate coordination in Zn-dependent alcohol dehydrogenases29.
The ability of FrmR proteins to bind Zn(II) raises the possibility that formaldehyde sensing and/or DNA-binding is enhanced by metal ions. Therefore, the effect of Zn(II) on the ability of EcFrmR to bind to DNA (Pfrm) was assessed by BLItz (Table 1). Loading the EcFrmR tetramer with 4 molar equivalents of Zn(II) increased the Kd for binding at Pfrm ~2-fold as a result of a decrease in the rate constant for DNA-binding (Table 1). However, the disassociation rate constant of the pre-formed Zn(II)-loaded Pfrm–EcFrmR complex in response to formaldehyde exposure was ~7-fold lower than that observed in the absence of Zn(II), suggesting that Zn(II) blocks amino acid residues required for formaldehyde sensing, as determined by the XAS experiments, and would be antagonistic to the deployment of the detoxification machinery. Therefore, it was concluded that formaldehyde-sensing and DNA-binding are not metal- (Zn(II)-) dependent and, based on the affinity of EcFrmR (and StyFrmR) for Zn(II), it is likely that Zn(II) could only inhibit FrmR activity in vivo under conditions when Zn(II) homeostasis is severely perturbed (Fig. S2; Table 1)22.
Identification of EcFrmR residues necessary for formaldehyde sensing
Site-directed mutagenesis of the amino acids of the W-X-Y-Z fingerprint has revealed the importance of these residues for the function of CsoR/RcnR family proteins (Fig. 1a). Recently, Pro2 (position W) and Cys35 (position X) of StyFrmR have been shown to be required for formaldehyde-responsiveness in vivo and in vitro, whereas Glu64 (position Z) was not23. Therefore, to identify EcFrmR residues necessary for formaldehyde sensing, site-directed mutation of the Pfrm-frmR-lacZ construct (see above) was undertaken. β-Galactosidase activity measurements showed that, like StyFrmR, EcFrmR(P2A) and EcFrmR(C35A) failed to respond to formaldehyde, confirming that Pro2 and Cys35 (W and X positions in the CsoR/RcnR family fingerprint; Fig. 1a) are essential for perception of formaldehyde (Fig. 4a). Replacement of His60 (position Y) resulted in high basal activity that was further enhanced in the presence of formaldehyde, whereas replacement of Thr64 (position Z) had no effect on the function of EcFrmR (Fig. 4a). Additionally, by analogy to RcnR metal-sensing residues17, insertion of an Ala codon before Pro2 (EcFrmR(A2*)) also resulted in formaldehyde insensitivity, albeit with higher basal Pfrm activity, implicating the Pro2 imino group in formaldehyde sensing (Fig. 4a). Replacement of the only other cysteine residue (Cys70) in EcFrmR did not impair the response to formaldehyde (Fig. 4a). The conclusion that Pro2 and Cys35 are required for EcFrmR to respond to formaldehyde was supported by the formaldehyde-sensitivity of E. coli strains expressing EcFrmR(P2A) and EcFrmR(C35A) in place of EcFrmR. Cultures expressing these variants exhibited enhanced sensitivity to formaldehyde, consistent with the low frmRAB expression observed in the reporter fusion experiments, presumably arising from constitutive repression frmRAB expression (Fig. 4a and b; Table 2).
Figure 4. Identification of formaldehyde-insensitive EcFrmR protein variants.
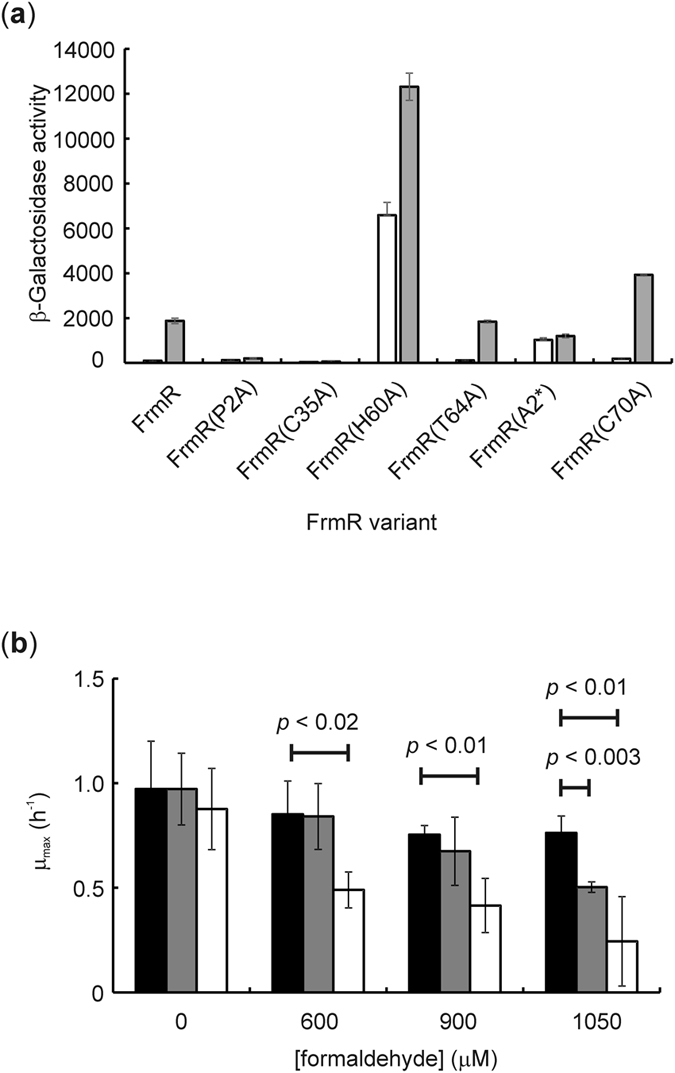
(a) The Pfrm-frmR-lacZ reporter was modified to encode EcFrmR variants with the indicated amino acid substitutions. Cultures of E. coli PC677 carrying these reporters were grown under anaerobic conditions in the absence (open bars) of presence (gray bars) of formaldehyde (250 μM) and β-galactosidase activities (Miller units) were measured as described in Methods. The error bars represent the standard deviation from the mean (n = 3). (b) Maximum growth rates (μmax) of E. coli MG1655 frmRAB mutant transformed with plasmids expressing the frmRAB operon from Pfrm under the control of wild-type EcFrmR (closed bars), EcFrmR(P2A) (gray bars) or EcFrmR(C35A) (open bars) cultured in the presence of the indicated initial concentrations of formaldehyde. The mean and standard deviations (n = 3) and the p values for one-tailed t-tests are shown.
Table 2. Liquid chromatography mass spectrometry analyses of EcFrmR proteins treated with formaldehyde.
| Protein sample | Measured mass (Da) | Relative abundance (a. u.) | Mass difference(Da)a | Commentb |
|---|---|---|---|---|
| EcFrmR | 10186.60 | 2.0 × 107 | 0.1 | FrmR monomer |
| 20371.02 | 4.4 × 106 | −2.18 | FrmR disulfide-linked dimer | |
| HCHO-treated EcFrmR | 10187.02 | 1.8 × 106 | 0.52 | FrmR monomer |
| 20396.87 | 2.0 × 105 | 23.67 | FrmR dimer with 2 methylene bridges | |
| 40768.73 | 2.0 × 105 | 22.30 | Two FrmR dimers, each with 1 methylene bridge, linked to form a tetramer by 1 disulfide bond | |
| HCHO-treated EcFrmR plus Zn(II) | 10187.01 | 1.8 × 106 | 0.51 | FrmR monomer |
| 20396.95 | 3.0 × 105 | 23.75 | FrmR dimer with 2 methylene bridges | |
| Zn(II)-treated EcFrmR plus HCHO | 10187.04 | 1.8 × 106 | 0.54 | FrmR monomer |
| 20396.99 | 4.0 × 105 | 23.79 | FrmR dimer with 2 methylene bridges | |
| HCHO-treated EcFrmR(P2A) | 20319.69 | 4.0 × 105 | −1.11 | FrmR(P2A) disulfide-linked dimer |
| HCHO-treated EcFrmR(P2A)c | 10190.71 | 6.8 × 105 | 29.91 | Hydroxymethylated FrmR(P2A) monomer |
| HCHO-treated EcFrmR(C35A) | 20307.40 | 2.8 × 106 | −1.40 | FrmR(C35A) disulfide-linked dimer |
| HCHO-treated EcFrmR(C35A)c | 20307.21 | 1.7 × 105 | −1.59 | FrmR(C35A) disulfide-linked dimer |
aDifference between predicted mass and measured mass.
bInterpretation of mass difference; disulfide bond, −2.2 Da; methylene bridge, net mass gain 12 Da; hydroxymethylation, net mass gain 30 Da.
cExposure to formaldehyde without quenching.
Reaction of EcFrmR with formaldehyde
The modification of EcFrmR by formaldehyde, and the competition with Zn(II), was analyzed by LC-MS. Without formaldehyde, the major species corresponded to the EcFrmR monomer lacking the N-terminal methionine (10186.60 Da; predicted mass 10186.50 Da) with lower amounts of a disulfide-linked dimer (20371.02 Da; predicted unmodified dimer mass 20373.20 Da) (Table 2). After exposure to formaldehyde (8-fold molar excess) for 3 min the EcFrmR monomer was still detected (10187.02 Da) along with new species of molecular mass 20396.87 Da, corresponding to an EcFrmR dimer plus an additional mass of 23.67 Da, and 40768.73 Da, corresponding to a tetramer with an additional mass of 22.30 Da (Table 2; Fig. S3). The former modified species represents EcFrmR dimers linked by two intermolecular methylene bridges (-CH2-) (net mass gain of 2 × 12 Da per EcFrmR dimer); the latter modified species can be accounted for by an EcFrmR tetramer in which each subunit participates in only one methylene bridge (total of two in the tetramer) and these cross-linked dimers are held together by one disulfide bond (Table 2; Fig. S3). Incubation with stoichiometric Zn(II) (4 Zn(II) per EcFrmR tetramer), before or after formaldehyde treatment, yielded dimeric species of molecular masses 20396.99 Da and 20396.95 Da, respectively, indicating that Zn(II) ions did not prevent formaldehyde-dependent methylene bridge formation under these conditions.
Formaldehyde modification of EcFrmR(P2A) and EcFrmR(C35A) was examined because they failed to respond to formaldehyde in vivo (Fig. 4). Neither mutation affected the oligomeric state of the protein, as both variants eluted as tetramers upon size exclusion chromatography. LC-MS showed the presence of disulfide-linked EcFrmR(P2A) dimers (20319.69 Da; predicted unmodified mass 20320.8 Da) in the absence or presence of formaldehyde (Table 2). However, exposure to formaldehyde without quenching permitted the detection of a monomeric EcFrmR(P2A) species (10190.71 Da) with additional mass 29.91 Da, equivalent to an hydroxymethyl adduct. In contrast for EcFrmR(C35A), a disulfide-linked dimer (20307.4 Da; predicted unmodified mass 20308.8 Da) that was not modified in the presence of formaldehyde was detected with or without quenching (Table 2). These data suggest that reaction with Cys35 is likely to be the first step in formaldehyde perception by EcFrmR (Table 2). Taken together, the in vivo data and the LC-MS data are consistent with a mechanism in which EcFrmR senses formaldehyde by the formation of methylene bridges between Pro2 and Cys35 residues of adjacent subunits; moreover, EcFrmR is oxidized in air, forming dimers linked by intermolecular disulfide bonds.
The structural response of EcFrmR to formaldehyde modification and a mechanism for formaldehyde sensing
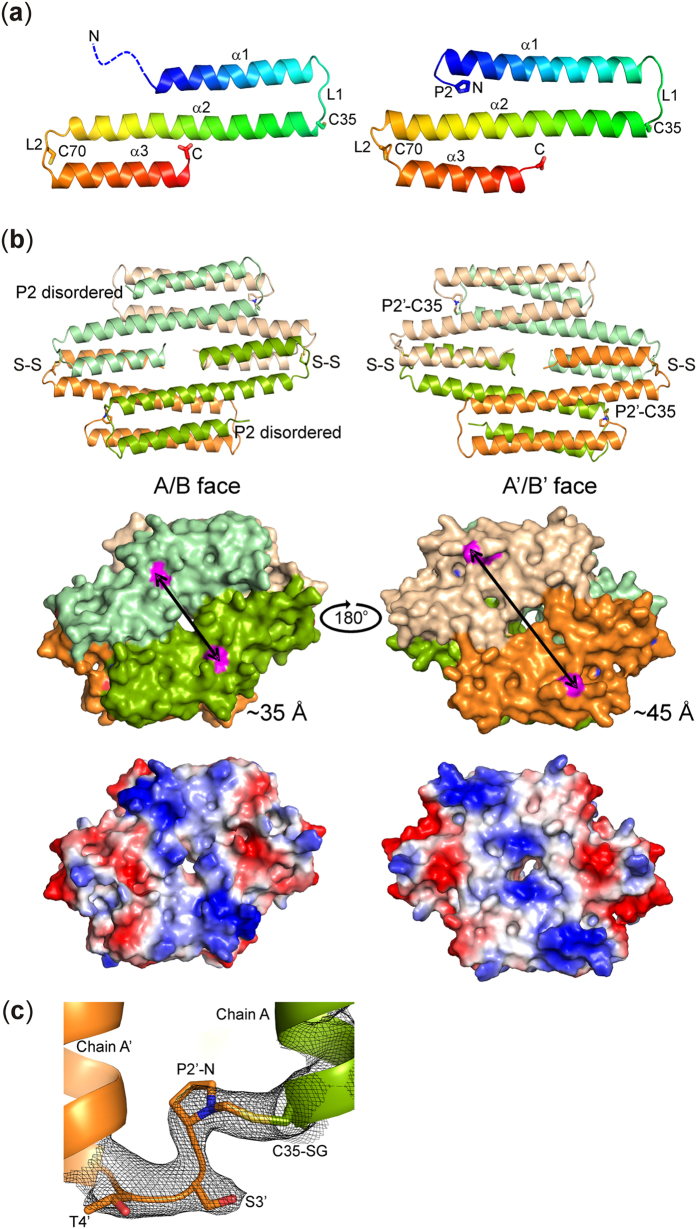
The crystal structure of formaldehyde-exposed EcFrmR was determined to a resolution of 2.7 Å (protein databank identifier PDB: 5LBM). Like other members of the CsoR/RcnR family, EcFrmR oligomerizes to form a disc-like tetramer, constructed from two homodimers, each of which forms one face of the disc. Each subunit consists of three helices (α1, residues 2–30; α2, residues 35–68; and α3, residues 73–91), linked by two short loops (L1, residues 31–34; and L2, residues 69–72), that are arranged as a flattened S-shape (Fig. 5a). The α3 helices slot together at the homodimer interface, such that each face of the tetramer is formed from a platform of five parallel helices with a hole at the centre (Fig. 5b). This arrangement differs from that seen in CsoR and StyFrmR(E64H) (PDB: 5LCY) where the equivalent α3 helix is domain swapped onto the opposite face of the tetramer (Fig. S4)23,24. This difference between EcFrmR and StyFrmR(E64H) is striking especially as the conserved regions of the α1 and α2 helices of these proteins superpose well; superposition of the Cα atoms of residues 10–63 (α1-α2) of the uncross-linked or cross-linked chains of EcFrmR with the uncross-linked StyFrmR yielded root mean square deviation (RMSD) values of ~1.0 Å for both. However, superposition of the Cα atoms from the full length chains (either cross-linked or uncross-linked) gave RMSD values of ~5 Å, reflecting the different organisation of α3 within the tetramer, which could be a consequence of the differences in amino acid sequence in the region spanning the terminus of α2, L2 and the beginning of α3 (Fig. 1b).
Figure 5. Structure of EcFrmR.
(a) Cartoon representations of uncross-linked (left) and cross-linked (right) EcFrmR monomers colored blue (N-terminal) to red (C-terminal). Secondary structure elements (α-helices, α1 to α3; loops, L1 and L2) are labeled and the amino acid residues (single letter code, P2, C35 and C70) involved in cross-linking and disulfide bond formation are shown as sticks. The disordered N-terminal region in the uncross-linked subunit is represented by the blue dashed line. (b) A comparison of the overall size and shape of the uncross-linked (left) and cross-linked (right) faces of the EcFrmR tetramer. The upper images show the arrangement of the helices on each face of the tetramer, the positions of the methylene bridges (P2′-C35) and the Cys70-Cys70′ disulfide bonds (S-S). The homodimer (A/B) on the uncross-linked face is drawn in shades of green and the cross-linked face (A′/B′) in shades of orange. The middle images show the expansion of the surface envelope upon cross-linking (black double headed arrow drawn between Arg14 Cα atoms, highlighted in pink). The lower images show the surface-charge on either side of the tetramer (red represents negative charge, blue positive charge and white neutral). (c) Section of the 2Fo-Fc map between chains A and A′ obtained when the coordinates for Pro2 and the methylene bridge were omitted from the refinement (black mesh, contoured at 1σ). Residues are indicated by their single letter codes.
The formaldehyde-exposed EcFrmR tetramer is asymmetric. One face of the tetramer (A/B face) is comprised of an unmodified homodimer with electron density visible for residues Lys9-Lys91 of both polypeptides with a disordered N-terminal region (residues Pro2-Lys8). Electron density corresponding to Zn(II) or other metal ions was not detected; however the W-X-Y-Z fingerprint residues in the unmodified homo-dimer are located in similar positions in 3D space to those of the CsoR proteins from Geobacillus thermodenitrificans and Thermus thermophilus and some of these residues are likely to constitute the EcFrmR Zn(II) binding site (Fig. S5)24,25. The XAS data supports the assignment of Cys35 and His60 as Zn(II) ligands (Fig. S1d). The residue at position Z (Thr64) does not interact with other residues of the W-X-Y-Z motif in either form of the EcFrmR dimer, consistent with the lack of effect of the T64A mutation on formaldehyde-dependent de-repression of Pfrm (Fig. 4a).
The homodimer forming the other face of the tetramer (A′/B′ face) is sandwiched against the first via a network of hydrophobic packing interactions between the secondary structural elements. In contrast to the A/B face, clear electron density is present for residues Pro2-Lys8 of both subunits on the A′/B′ face, resulting in an ordered extension to the N-terminal region of α1. The Pro2′-N atom is located within ~2.5 Å of the Cys35-SG atom of the corresponding subunit on the opposite face of the tetramer (Fig. 5b). There is extra density between these two atomic positions, indicating the presence of the formaldehyde cross-link, and a methylene bridge (-CH2-), as indicated by the LC-MS data, has been modeled at this location (Fig. 5c).
The crystal structure of EcFrmR reveals both the free and signal-triggered states of the protein. One face of the EcFrmR tetramer represents the conformation of the protein in the absence of formaldehyde (A/B face with a disordered Pro2), whilst the other face represents the conformation of the protein that has responded to formaldehyde and has formed the methylene bridge (A′/B′ face with an ordered Pro2) (Fig. 5b). ‘On’- and ‘off’-states of an RcnR/CsoR family member from a single source have not been observed previously and the structure presented here facilitates a detailed analysis of the conformational changes that occur upon reaction of EcFrmR with formaldehyde to inhibit DNA-binding. Unlike EcFrmR, the N-terminal region of StyFrmR(E64H) is visible in the absence of the methylene bridge23. This showed that the StyFrmR(E64H) N-terminal Pro2′ is located close to Cys35 (~3 Å between Pro2′-N and Cys35-SG atoms), poised to facilitate formaldehyde-specific cross-linking. With the exception of the domain swapped α3, the uncross-linked (A/B) face of EcFrmR superposes well on the StyFrmR structure (RSMD ~1.0 Å). This suggests that a remarkably subtle change in the locations of Pro2′ and Cys35 and the constraints imposed by the covalent methylene bridge cross-link contribute to creating a distinct difference in the relative conformation and orientation of the subunits in the cross-linked face compared to those of the uncross-linked face. Whilst the position of α3 is similar on both faces of the tetramer, there is a translational movement of α1 and α2 on the A′/B′ face, which slide across the equivalent helices on the opposite face by ~1.5 α-helical turns in response to formation of the methylene bridge. In addition, the C-terminal half of α2 twists and buckles towards α3, which alters the packing of these two helices, generating a 10° off-parallel angle between α3 and α2. This movement changes the overall size and shape of the A′/B′ face, which expands and elongates by ~10 Å on the diagonal in response to formaldehyde (Fig. 5b; Animation S1). Notably, expansion of the envelope of CsoR in response to Cu(I) was apparent in small angle X-ray scattering studies27. Furthermore, in addition to an H-bond network, involving His60-Tyr34-Glu80 that is thought to be involved in signal (Cu(I)) perception but not signal binding by M. tuberculosis and G. thermodenitrificans CsoR proteins, the N-terminal region of G. thermodenitrificans CsoR becomes ordered over the Cu(I)-binding site24. The stable ordering of the N-terminal regions of EcFrmR and CsoR proteins upon signal perception establishes new interactions between the α1 and α2 helices of these proteins (Fig. S6). Thus, signal perception by EcFrmR results in a H-bond interaction between Glu7 (α1) and Ser59 (α2), which is not present in the uncross-linked form of EcFrmR, StyFrmR(E64H) structure or in the Cu(I)-free form of S. lividans CsoR. The same two residue positions of Cu(I)-loaded G. thermodenitrificans CsoR (Glu22-Arg74) participate in an electrostatic interaction. Moreover, an equivalent interaction occurs in M. tuberculosis CsoR, but involving a slightly different residue position in α2 (Lys8-Glu63). Thus, it is suggested that N-terminal ordering and the establishment of interactions between α1 and α2 could be a common feature of signal perception and transduction in the CsoR/RcnR family that has only been revealed because both ‘on’ and ‘off’ states are captured in the EcFrmR structure.
The different conformational states of the two faces of formaldehyde-treated EcFrmR have a dramatic effect on the pattern of surface charge and the position of protrusions on each face of the tetramer (Fig. 5b). There are two patches of positive charge either side of the central hole on both faces of the tetramer, which contain several residues implicated in DNA-binding (Arg14, Arg16, Gln41, Arg46 and Lys91)24,30,31. The differences in size and shape of the two faces of EcFrmR in the crystal structure mean that these patches are separated by ~35 Å on the A/B face but by ~45 Å on the A′/B′ face. Whilst an asymmetric tetramer has been captured in the crystal structure, modeling suggests that both faces of EcFrmR could adopt the compact DNA-binding conformation in the absence of formaldehyde-induced cross-linking. However, models of a fully cross-linked tetramer, in which subunits A and A′ and B and B′ were both linked by two methylene bridges, as implied by the LC-MS data, contained many inter-subunit clashes, indicating that both faces of EcFrmR might not be able to adopt the extended conformation whilst retaining the tetrameric state. The presence of only two methylene bridges in the structure of the EcFrmR tetramer raises the possibility that, at least under some conditions, the formation of the third and fourth cross-links might be subject to negative cooperativity, as observed for binding the third and fourth Zn(II) ions to EcFrmR (see above) and that the fully cross-linked protein might disassociate into dimers incapable of DNA-binding. The negative cooperativity is also consistent with the effects of the H60A mutation on Pfrm-lacZ activity (Fig. 4a). This mutant shows high constitutive promoter activity, similar to the stop codon mutant (Fig. 2a). However, the maximum induction by formaldehyde is substantially lower. This contrasts with the ability of Ni(II) and Co(II) repress Prcn to the same extent as a stop codon mutant of RcnR16. The His60 residue of EcFrmR is within H-bond distance of Cys35 in the unmodified dimer (3.2 Å), and makes no obvious interactions with side chain or main chain residues in the methylene-bridged structure. Hence, the H60A mutation may mimic the modified form of the protein at all four sites, instead of two, resulting in greater de-repression.
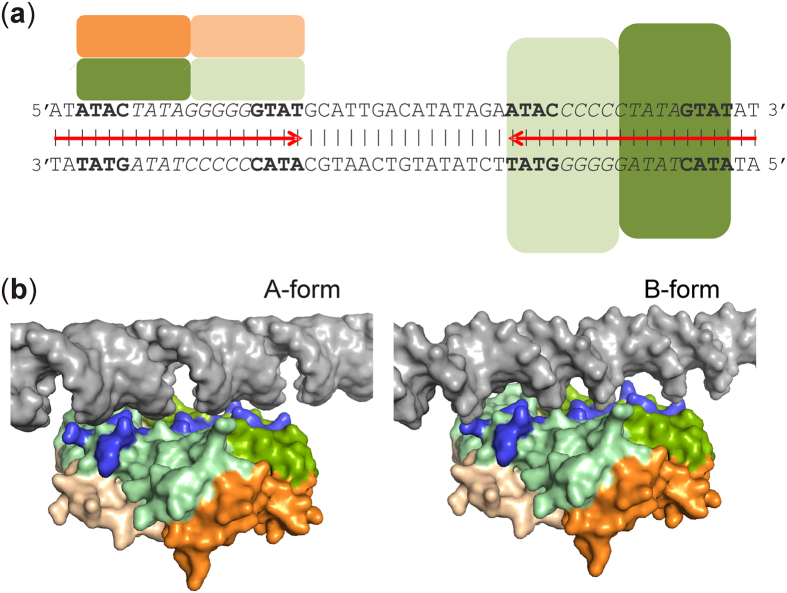
It has been suggested that CsoR/RcnR family members recognize their DNA target by a combination of: (i) shape selectivity, resulting from the propensity of the central GC tract to adopt the A-DNA form; (ii) specific interactions with the flanking inverted repeats; and (iii) non-specific binding to distant DNA that might result in DNA-wrapping30,31. Operator sequences for CsoR/RcnR family members have been classified into two groups: type I sites consist of GC tracts (3–8 bp) flanked by AT-rich inverted repeats; type II sites have shorter interrupted GC tracts30,31. The E. coli Pfrm contains a large inverted repeat centered at −29 relative to the predicted transcriptional start site (Fig. 6a). This region contains tandem type I FrmR-binding sites consisting of 9 bp GC-rich tracts flanked by ATAC/GTAT inverted repeats (Fig. 6a). The E. coli rcnR-A intergenic region also possesses tandem type I RcnR-binding sites containing a TACTGGGGGGNAGTA motif, which imparts some A-form DNA character on this region of DNA, and one RcnR tetramer binds at each site on the same face of the DNA helix31. The recently reported structure of the site-directed mutant StyFrmR(E64H), which responds to Co(II), Zn(II) and formaldehyde in vivo showed the presence of positively-charged surface patches for protein that had not been exposed to formaldehyde22,23. As noted above for the uncross-linked surface of EcFrmR (Fig. 5b), the StyFrmR(E64H) positively-charged patches were also separated by ~35 Å (distance between the Arg14 Cα atoms)23. These data were used to inform models of the Pfrm–EcFrmR complexes with A- and B-form DNA (Fig. 6). Models with both A- and B-form DNA suggested that residues (Lys10, Arg14, Arg16 and Arg17) forming the two positively-charged protrusions on the A/B face could interact with the major grooves of the DNA, with Lys91 from the central hole packing into the minor groove, but the better fit is with B-form DNA. It is noted that in the bacterial cell Pfrm DNA is unlikely to be exclusively in A- or B-form, but more likely a hybrid that will be further distorted upon binding of the FrmR tetramer. Nevertheless, the ~45 Å that separates the positively-charged patches on the cross-linked A′/B′ face precludes convincing interactions with either A- or B-form DNA. These differences suggest a mechanism for de-repression of the frmRAB promoter in response to formaldehyde. In the DNA bound state, Lys91 is held in a surface-exposed position by an ion pair interaction between Arg14 and the C-terminal carboxyl. Upon cross-linking, α1 and α2 are pulled away from the centre of the face of the tetramer, with loop 2 acting as a pivot point. This motion pulls Arg14 away from Lys91, breaking the interaction with the C-terminal carboxyl, which causes it to rotate by ~90° into the central hole, forming a new interaction with the guanidyl group of Arg46. The surface-exposed side-chain of Lys91 moves into the hole, burying the NZ atom so that it is no longer available to interact with DNA (Fig. S7). In addition, the motion of α1 and α2 increases the distance between the cluster of residues within the positively-charged protrusions by ~10 Å, such that Lys10, Arg14, Arg16 and Arg17 can no longer interact with the major groove, thus breaking the complex between EcFrmR and DNA. The centers of the tandem binding sites at Pfrm are separated by 31 bp compared to 19 bp for the rcnR-A intergenic region, raising the possibility that, unlike RcnR, both faces of a single EcFrmR tetramer could participate in binding to the tandem sites at Pfrm.
Figure 6. Modeling the Pfrm–EcFrmR complex.
(a) The DNA sequence of the frmRAB promoter region (Pfrm) contains tandem EcFrmR binding sites consisting of ATAC/GTAT inverted repeats (bold) separated by G/C-rich tracts (italic) that form a larger inverted repeat (convergent red arrows). The size of EcFrmR (subunits colored in shades of green and orange as in Fig. 5b) suggests that two tetramers could bind to the frmRAB promoter region. One EcFrmR tetramer (side view) is shown on the top face of the DNA sequence and the other (top view) behind the DNA sequence, offset by approximately a quarter turn relative to the first tetramer. (b) Models of binary complexes formed from EcFrmR and A- or B-form DNA. One of the tandem EcFrmR binding sites of Pfrm (dark gray) is modeled as A- (left) and B-form (right) DNA. EcFrmR is shown as surface representation with subunits colored in shades of green (uncross-linked A/B face) and orange (cross-linked A′/B′ face), with the amino acid side-chains on the A/B face that are implicated in DNA-binding highlighted in blue.
Concluding remarks
EcFrmR is the first example of a CsoR/RcnR family protein where the asymmetry of the tetramer in the crystal structure reveals the conformational changes induced by signal perception that lead to de-repression of target promoters. In vivo and in vitro experimental evidence show that the EcFrmR specifically reacts with the toxic chemical formaldehyde, resulting in the formation of inter-molecular methylene bridges between adjacent Pro2 and Cys35 residues. In the absence of formaldehyde the StyFrmR(E64H) structure indicates that the N-terminal regions are ordered, such that the N-terminal Pro residues are in close proximity to Cys35 of an adjacent subunit23. The conservation of His60 and its proximity to Cys35 in the adjacent subunit suggests that it could act to abstract a proton from the thiol group to facilitate the initial reaction with formaldehyde forming an S-hydroxymethyl adduct (LC-MS data Table 2). The initial hydroxymethylated Cys35 residues appear to be associated with one surface of the EcFrmR disc permiting nucleophilic attack by Pro2 residues associated with the opposite face of the EcFrmR disc resulting in methylene bridge formation (detected as the oxidized EcFrmR tetramer with additional mass 22.3 Da in LC-MS; Table 2, Fig. S3); an example of an N-terminal Pro residue acting as a nucleophile has been reported for the DNA-repair enzyme, formamidopyrimidine-DNA glycosylase32. The covalent capture of the Pro2 residues of one face of EcFrmR could contribute to the disordering of the uncross-linked N-terminal regions, which is apparent in the crystal structure reported here (Fig. 5). Alternatively, fully uncross-linked EcFrmR and StyFrmR could differ in the degree of flexibility in their N-terminal regions despite the high level of sequence conservation (Fig. 1b). The N-terminal disordering captured in the formaldehyde-treated EcFrmR crystal structure could account for the negative cooperativity observed for Zn(II) binding and perhaps in formaldehyde reactivity. Formaldehyde-induced methylene bridge formation (two per tetramer) expands the A′/B′ surface of EcFrmR resulting in a similar change in size to that observed for G. thermodenitrificans CsoR upon Cu(I) binding24. Interestingly, CsoR does not use direct coordination of the signal molecule via the N-terminus, unlike FrmR and RcnR. An allosteric network has been proposed for CsoR that involves an interaction between His60 and two other residues (Tyr34 and Glu80, numbered based on the N-terminus of FrmR and RcnR)33. However, these interactions are not present in FrmR and RcnR, suggesting that the allosteric network may initiate at the N-terminus upon signal perception. This notion is consistent with the observation that the G. thermodenitrificans CsoR structure shows the N-terminus capping the Cu(I)-binding site, but not directly coordinating the Cu(I) atom24. Thus, despite specific differences between RcnR, FrmR, and CsoR in the connectivity between signal perception residue(s) and the common DNA-binding interface of the proteins, an ordering of the N-terminal region and α1 may be a common feature of signal perception in this protein family. The inability of Zn(II) to drive EcFrmR off DNA is likely to be because Zn(II) binding cannot order the N-terminal region of α1. Ultimately, all four EcFrmR N-terminal Pro2 residues can be methylene bridged to their partner Cys35 residues resulting in derepression of frmRAB expression (detected as EcFrmR dimers +24 Da in LC-MS; Table 2; Fig. S3).
Finally, it is notable that the second EcFrmR Cys residue (Cys70), which is not conserved in StyFrmR, can form a disulfide bond linking loops 2 of the A/B′ and the B/A′ chains. The effect of this disulfide on the ability of all four EcFrmR subunits to undergo formaldehyde modification to fine-tune the EcFrmR response has not been assessed here, but the potential for EcFrmR to act as a redox sensor, through the formation of a disulfide bond, is of interest because formaldehyde exposure is associated with glutathione-depletion and oxidative stress in higher organisms34,35,36,37,38. Work is on-going to determine the possible role of glutathione (S-hydroxymethylglutathione is formed in E. coli exposed to formaldehyde) and oxidative stress (disulfide bond formation) in regulating EcFrmR activity in vivo as well as to determine the structure of the Pfrm-EcFrmRcomplex.
Methods
Bacterial strains, plasmids and oligonucleotides
These are listed in Supplementary Table S1. The Pfrm-frmR-lacZ plasmid was constructed from pPC163, which contains the Pnik promoter and lacZ ligated into pACYC18417. A fragment consisting of 499 bp of DNA located upstream of frmA, which included frmR, was ligated into the EagI and SalI sites of pPC163, replacing the Pnik cassette. For overproduction of EcFrmR, the frmR gene was amplified from E. coli K12 genomic DNA with primers JI174 and JI175, and ligated into pET22b at the NdeI and NcoI sites. Mutations in frmR were introduced by overlap PCR or by the Quikchange protocol (Stratagene) using the appropriate oligonucleotide primers (Table S1). Formaldehyde-sensitivities of E. coli JRG6703, harboring either pGS2497 (frmR wild-type), pGS2547 (frmR P2A) or pGS2548 (frmR C35A) were determined using aerobic, Luria-Bertani (LB) medium (200 μl), 96-well plate cultures. Formaldehyde (0–1650 μM) was added and the cultures were grown at 37 °C with shaking (250 rpm). Growth was monitored using a Sunrise absorbance reader (Tecan) at A595 for 500 min at intervals of 20 min. All cultures were grown in triplicate. The maximum growth rate (μmax h−1) for each strain was calculated.
Isolation of EcFrmR and selenomethionine (Se-Met) incorporated EcFrmR protein
For isolation of EcFrmR and the P2A and C35A variants, cultures of the E. coli expression strains (JRG6782, 6783 and 6784) were grown at 37 °C in LB medium containing ampicillin (100 μg ml−1), to an OD of ~0.6, at which point 1 mM IPTG was added and the cultures were incubated for a further 3 h. To obtain selenomethionine (Se-Met) incorporated EcFrmR, cultures were grown to OD ~0.6 in LB, then pelleted and resuspended into Se-Met over-expression medium (60 mM K2HPO4, 8 mM (NH4)2SO4, 33 mM KH2PO4, 2 mM tri-sodium citrate, 54 mM glycerol, 4 mM adenine, 2 mM guanosine, 4 mM thymine, 4 mM uracil, 4 mM MgSO4, 12 mM thiamine, L-lysine (100 mg l−1), L-phenylalanine (100 mg l−1), L-threonine (100 mg l−1) L-isoleucine (50 mg l−1), L-leucine (50 mg l−1), L-valine (50 mg l−1) and seleno –L –methionine (40 mg l−1)) before further incubation for 1 h at 37°C prior to induction of EcFrmR protein expression with IPTG. Cells were lysed after re-suspension in Buffer A (50 mM Tris, 0.1 M NaCl; pH 8.0) by sonication (Soniprep150 ultrasonic disintegrator) at ~16 microns for 2 cycles of 20 s. The lysate was cleared by centrifugation (10 min, 70000 g) and the resulting cell-free extract was applied to a Heparin-HP column (GE Healthcare) and EcFrmR was eluted using a NaCl gradient (0 to 0.5 M in 50 ml) in Buffer A. Fractions containing EcFrmR protein were combined and dialyzed overnight against 50 mM Tris-HCl buffer, pH 8.0 at 4 °C. Micro-crystals of EcFrmR were collected by centrifugation and dissolved in 1 ml of 1 M NaCl in buffer A (see above). The sample was then applied to a HiLoad Superdex 200 column (GE Healthcare) equilibrated with 50 mM Tris-HCl, 0.5 M NaCl (pH 8.0). EcFrmR-containing fractions were pooled and the purity of the samples was assessed by SDS-polyacrylamide gel electrophoresis. For crystallization, protein was concentrated to 10 mg ml−1 using a VivaSpin device with MWCO 30000 (GE Healthcare). The authenticity of EcFrmR was confirmed by total amino acid analysis (Alta Biosciences; University of Birmingham, UK) allowing calibration of EcFrmR protein concentrations measured by either the Bio-Rad protein reagent or using the calculated molar extinction coefficient for EcFrmR39,40.
Zn(II) binding Assays
Zn(II) titrations, XAS and EXAFS analyses are described in Supplementary material.
Analytical ultracentrifugation
Details are provided in the supplementary material.
Estimation of the number of reactive thiols in EcFrmR
Reactive protein thiols were estimated by the method of Thelander41. In a Quartz cuvette, a total reaction volume of 1 ml was prepared consisting of purified protein (100 μl) diluted with protein elution buffer (50 mM Tris, 0.5 M NaCl (pH 8.0) and 200 μl DTNB solution (0.2 ml 0.4% DTNB in ethanol and 1.8 ml 1 M Tris-HCl, pH 8.0). Absorbance at 412 nm was measured and the 2-nitrothiobenzoate concentration was used to calculate the number of reactive sulfydryl groups.
Mass spectrometry
Protein samples (~12 μM) for LC-MS were incubated with either an 8-fold or 40-fold molar excess of formaldehyde to EcFrmR tetramer concentration at room temperature. After 3 min, reactions were quenched with 10 mM glycine. Samples were loaded onto an Agilent 1260 Infinity liquid chromatograph fitted with an Agilent Extended C18 column (2.1 mm × 50 mm) and eluted with a gradient of 5-95% acetonitrile in 0.1% formic acid at 400 μl min−1 over 8 min. The eluate was directly coupled to an Agilent 6530 Q-ToF mass spectrometer fitted with an electrospray ionisation (ESI) source for determination of the masses of species detected in the chromatograph.
For ICP-MS, EcFrmR (200 μM) in 50 mM Tris (pH 8.0) buffer containing 0.5 M NaCl or buffer was incubated with concentrated nitric acid (1:1 ratio) at 60 °C for 1 h. Samples were cooled, diluted with dH2O (up to 10 ml) and filtered before analysis on a Perkin Elmer Nexlon ICP-MS system. Ions were quantified using a dilution series of certified multi-element reference standard (Sigma-Aldrich). Counts per second values for elements in the buffer and protein samples were then compared with the calibration curve to determine actual concentrations.
Bio-layer interferometry (BLItz)
Biotinylated-promoter DNA for frm (Pfrm) and ydhY (PydhY) was amplified from E. coli MG1655 genomic DNA by PCR using appropriate oligonucleotide primers (Table S1). Where indicated, EcFrmR tetramers were incubated with formaldehyde (up to an 800-fold molar excess) for 3 min (before quenching with 10 mM glycine) or Zn(II) (16-fold molar excess) at room temperature, prior to measurements. Analysis of the interaction between purified EcFrmR and biotinylated-promoter DNA was carried out using the BLItz system (FortéBio), at 20 °C. Streptavidin (SA) biosensors (FortéBio) were hydrated by soaking the tips in 250 μl protein elution buffer (50 mM Tris-HCl, pH 8.0 containing 0.5 M NaCl) for at least 10 min. The measurements were made using the Advanced Kinetics function of the BLItz Pro 1.1.0.31 software (FortéBio). The sequence for each run was as follows: (1) baseline step with protein elution buffer for 60 s, (2) loading step with 100 nM biotinylated DNA in buffer for 240 s, (3) baseline step with buffer for 60 s, (4) association step with various concentrations of purified EcFrmR in buffer for 240 s, (5) dissociation step with buffer (or formaldehyde-containing buffer) for 120 s. Baseline and dissociation steps were carried out by placing the biosensor in a black 500 μl Eppendorf tube filled with 250 μl buffer. Loading and association steps were carried out by placing the biosensor in the drop holder containing either 5 μl of Pfrm DNA or EcFrmR protein depending on the step. For each run a new biosensor was used. The BLItz Pro 1.1.0.31 software using global fitting and correcting both association and dissociation curves was used for analysis.
β-Galactosidase reporter experiments
Starter cultures of E. coli PC677 (ΔfrmR) containing the appropriate reporter plasmid (wild type or mutant) were grown aerobically in LB medium with chloramphenicol (0.1 mM) to an OD600 ~0.5. These cultures were then used to inoculate 1.87 ml of LB medium with 0.1 mM chloramphenicol in capped microcentrifuge tubes in triplicate with aldehyde compounds as indicated. Cultures were grown overnight (12–15 h) before measurement of β-galactosidase activity42. The maximal aldehyde concentrations that inhibited growth by <15% were: 700 μM formaldehyde, 7 mM acetaldehyde, 100 μM chloroacetaldehyde, 11 μM tribromoacetaldehyde, 4 mM propionaldehyde, 10 mM furaldehyde, 3.5 mM glyoxal, 350 μM methylglyoxal and 500 μM glutaraldehyde.
In vitro transcription reactions
The promoter and part of the coding regions of frmR and ndh (~200 bp upstream of the start codon to ~190 bp (frmR) or ~100 bp (ndh) into the gene) were amplified from E. coli MG1655 genomic DNA using appropriate oligonucleotides (Table S1). These DNA fragments (~0.1 pmol) were incubated for 30 min at 37 °C in a 10.5 μl reaction volume containing 40 mM Tris-HCl, pH 8.0, 10 mM MgCl2, 1 mM dithiothreitol, 75 mM KCl, 0.1 mM EDTA, 5% glycerol, 250 μg ml−1 bovine serum albumin, 20 units of RiboLoc RNase inhibitor (Fermentas), 1 pmol E. coli RNA polymerase holoenzyme (New England BioLabs, Inc.) and 0 or 1 nM EcFrmR tetramer. EcFrmR was reduced with 10 mM dithiothreitol and when required treated with 200-fold molar excess of formaldehyde for 5 min at room temperature, before quenching with 10 mM glycine. Transcription was initiated by the addition of 2 μl solution containing UTP at 50 μM; ATP, CTP and GTP at 1 mM; and 2.5 μCi of [α-32P]UTP (800 Ci mmol−1; PerkinElmer Life Sciences), followed by incubation for 15 min at 37 °C. Reactions were terminated by the addition of 12.5 μl Stop/Loading dye solution (95% formamide, 20 mM EDTA, pH 8, 0.05% bromophenol blue, 0.05% xylene cyanol). Samples (10 μl) of each reaction were loaded onto a 6% acrylamide, 1x TBE, 8 M urea gel and analysed using a phosphorimager (Typhoon 700; GE Healthcare). Markers (0.1–1 kb) were prepared using Perfect RNA Marker template mix (Novagen). A 20 μl reaction containing 0.75 μg of RNA template mix, 80 mM HEPES, pH 7.5, 12 mM MgCl2, 10 mM NaCl, 10 mM dithiothreitol, 2 mM ATP, 2 mM GTP, 2 mM CTP, 0.1 mM UTP, 5 μCi of [α-32P]UTP (800 Ci mmol−1, PerkinElmer Life Sciences), 20 units of RiboLoc RNase inhibitor (Fermentas) and 50 units of T7 RNA polymerase (Novagen), was incubated for 1 h at 37 °C, before storing at −20 °C. Markers from ~20 ng template were used for gel calibration.
Crystallization and structural determination of EcFrmR
Crystals of SeMet-labeled EcFrmR (in 50 mM Hepes pH 7.5 and 0.5 M NaCl) were grown in 0.2 M MgCl2, 0.1 M Na cacodylate pH 6.5 and 31% PEG 2000. Crystals were harvested and cryoprotected in their mother liquor with an additional 25% ethylene glycol, before a single-wavelength (λ 0.9798 Å) anomalous dispersion (SAD) experiment (100 K) was carried out on beamline i03 at the Diamond Light Source (Table S3). The data were auto-processed using Xia243 (XDS/Aimless) in space group P3112 and AutoSharp44 was used to locate eight selenium sites (four full occupancy and four half occupancy) and build the basic polypeptide structure within the asymmetric unit, before multiple rounds of structure building and refinement using COOT45, Buccaneer46 and Refmac547 from CCP4i48 were carried out to produce a poly-alanine model. The six subunits in the asymmetric unit comprised three half tetramers that were related by translational NCS, with poor electron density for one dimer. Subsequent higher resolution data (2.7 Å) were collected on beamline i03 (100 K) from a second SeMet-labeled crystal (λ = 0.9763 Å) that had been soaked in formaldehyde for 3 h before cryo-cooling. These data were in spacegroup P31, with a tetramer of EcFrmR in the asymmetric unit (Table S3). The poly-alanine model was used in molecular replacement48 as a starting point to build and refine the full structure of EcFrmR using COOT45, ArpWarp49and Refmac547 The final structure was refined to Rfactor/Rfree values of 0.21 and 0.27, respectively, and was validated using Molprobity50. Structure superpositions were made using the SuperPose software51.
Additional Information
Accession code: Structural data have been deposited in with the PDB; accession code 5LBM.
How to cite this article: Denby, K. J. et al. The mechanism of a formaldehyde-sensing transcriptional regulator. Sci. Rep. 6, 38879; doi: 10.1038/srep38879 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work was supported by the Biotechnology and Biological Sciences Research Council UK through the White Rose-BBSRC Mechanistic Biology Doctoral Training Programme (BB/J014443/1) (K.J.D) and project grant BB/L008114/1 (J.G) and the National Science Foundation (USA) grant MCB0520877 (PTC). We thank Mr. Simon Thorpe (The University of Sheffield Faculty of Science Mass Spectrometry Centre) for LC-MS analysis; Dr. Jamie Young (Sheffield Hallam University) for ICP-MS analysis; beam-line scientists from i03 at the Diamond Light Source for assistance with data collection; Professor Jon Sayers and Dr. Guta Vitovski for access to the BLItz equipment; and Professor Nigel Robinson and Dr. Deenah Osman (University of Durham) for their insightful intellectual input and critical comments during the course of this research project.
Footnotes
Author Contributions P.J.B., P.T.C. and J.G. conceived the study and supervised the experiments. K.J.D., J.I., M.D.R., S.E.S. and J.W. carried out the molecular genetics, purified and determined the biochemical properties of EcFrmR proteins. M.J.M. and K.H. were responsible for the XAS experimental design, data collection, and analysis. C.B. and P.J.B. solved the structure of EcFrmR; all the authors contributed to analyzing data and writing the manuscript.
References
- Yu R. et al. Formation, accumulation, and hydrolysis of endogenous and exogenous formaldehyde-induced DNA damage. Toxicol. Sci. 146, 170–182 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N. H., Djoko K. Y., Veyrier F. J. & McEwan A. G. Formaldehyde stress responses in bacterial pathogens. Front. Microbiol. 7, 257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trewick S. C., Henshaw T. F., Hausinger R. P., Lindahl T. & Sedgwick B. Oxidative demethylation by Escherichia coli AlkB directly reverts DNA base damage. Nature 419, 174–178 (2002). [DOI] [PubMed] [Google Scholar]
- Koyama Y. & Ohmori H. Nucleotide sequence of the Escherichia coli solA gene encoding a sarcosine oxidase-like protein and characterization of its product. Gene 181, 179–183 (1996). [DOI] [PubMed] [Google Scholar]
- Lai Y. et al. Measurement of endogenous versus exogenous formaldehyde-induced DNA-protein crosslinks in animal tissues by stable isotope labeling and ultrasensitive mass spectrometry. Cancer Res. 76, 2652–2661 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuykendall J. R. & Bogdanffy M. S. Efficiency of DNA-histone crosslinking induced by saturated and unsaturated aldehydes in vitro. Mutat. Res. 283, 131–136 (1992). [DOI] [PubMed] [Google Scholar]
- Xie M. Z. et al. Aldehydes with high and low toxicities inactivate cells by damaging distinct cellular targets. Mutat. Res. 786, 41–51 (2016). [DOI] [PubMed] [Google Scholar]
- Metz B. et al. Identification of formaldehyde-induced modifications in proteins: reactions with model peptides. J. Biol. Chem. 279, 6235–6243 (2004). [DOI] [PubMed] [Google Scholar]
- Denby K. J. et al. J. Adaptation of anaerobic cultures of Escherichia coli K-12 in response to environmental trimethylamine-N-oxide. Environ. Microbiol. 17, 2477–2491 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring C. D. & Blattner F. R. Global transcriptional effects of a suppressor tRNA and the inactivation of the regulator frmR. J. Bacteriol. 186, 6714–6720 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C. F. et al. Molecular basis of formaldehyde detoxification. Characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J. Biol. Chem 281, 14514–14522 (2006). [DOI] [PubMed] [Google Scholar]
- Harms N., Ras J., Reijnders W. N., van Spanning R. J. & Stouthamer A. H. S-formylglutathione hydrolase of Paracoccus denitrificans is homologous to human esterase D: a universal pathway for formaldehyde detoxification? J. Bacteriol. 178, 6296–6299 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc D. P. & Arunkumar A. I. Metal sensor proteins: nature’s metalloregulated allosteric switches. Dalton Trans. 7, 3107–3120 (2007). [DOI] [PubMed] [Google Scholar]
- Higgins K. A. & Giedroc D. Insights into protein allostery in the CsoR/RcnR family of transcriptional repressors. Chem, Lett. 43, 20–25 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. et al. CsoR is a novel Mycobacterium tuberculosis copper-sensing transcriptional regulator. Nat.Chem. Biol. 3, 60–68 (2007). [DOI] [PubMed] [Google Scholar]
- Iwig J. S., Rowe J. L. & Chivers P. T. Nickel homeostasis in Escherichia coli: the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol. Microbiol. 62, 252–262 (2006). [DOI] [PubMed] [Google Scholar]
- Iwig J. S., Leitch S., Herbst R. W., Maroney M. J. & Chivers P. T. Ni(II) and Co(II) sensing by Escherichia coli RcnR. J. Am. Chem. Soc. 130, 7592–7606 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster A. W., Pernil R., Patterson C. J. & Robinson N. J. Metal specificity of cyanobacterial nickel-responsive repressor InrS: cells maintain zinc and copper below the detection threshold for InrS. Mol. Microbiol. 92, 797–812 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossoehme N. et al. Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J. Biol. Chem. 286, 13522–13531 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z., Jacobsen F. E. & Giedroc D. P. Coordination chemistry of bacterial metal transport and sensing. Chem. Rev. 109, 4644–4681 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins K. A., Chivers P. T. & Maroney M. J. Role of the N-terminus in determining metal-specific responses in the Escherichia coli Ni- and Co-responsive metalloregulator, RcnR. J. Am. Chem. Soc. 134, 7081–7093 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D. et al. Generating a metal-responsive transcriptional regulator to test what confers metal sensing in cells. J. Biol. Chem. 290, 19806–19822 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman D. et al. The effectors and sensory sites of formaldehyde-responsive regulator FrmR and metal-sensing variant. J. Biol. Chem. doi: 10.1074/jbc.M116.745174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F. M. et al. Cu(I)-mediated allosteric switching in a copper-sensing operon repressor (CsoR). J. Biol. Chem. 289, 19204–19217 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K., Agari Y., Agari K., Kuramitsu S. & Shinkai A. Structural and functional characterization of the transcriptional repressor CsoR from Thermus thermophilus HB8. Microbiol. 156, 1993–2005 (2010). [DOI] [PubMed] [Google Scholar]
- Dillon D., Combes R. & Zeiger E. The effectiveness of Salmonella strains TA100, TA102 and TA104 for detecting mutagenicity of some aldehydes and peroxides. Mutagenesis 13, 19–26 (1998). [DOI] [PubMed] [Google Scholar]
- Mroczkowska M. M., Kolasa I. K. & Kusmierek J. T. Chloroacetaldehyde induced mutagenesis in Escherichia coli: specificity of mutations and modulation by induction of the adaptive response to alkylating agents. Mutagenesis 8, 341–348 (1993). [DOI] [PubMed] [Google Scholar]
- Zeiger E., Gollapudi B. & Spencer P. Genetic toxicity and carcinogenicity studies of glutaraldehyde–a review. Mutat. Res 589, 136–151 (2005). [DOI] [PubMed] [Google Scholar]
- Levin I., Meiri G., Peretz M., Burstein Y. & Frolow F. The ternary complex of Pseudomonas aeruginosa alcohol dehydrogenase with NADH and ethylene glycol. Protein Sci. 13, 1547–1556 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan B. G., Vijgenboom E. & Worrall J. A. R. Conformational and thermodynamic hallmarks of DNA operator site specificity in the copper sensitive operon repressor from Streptomyces lividans. Nucl. Acids Res. 42, 1326–1340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwig J. S. & Chivers P. T. DNA recognition and wrapping by Escherichia coli RcnR. J. Mol. Biol. 393, 514–526 (2009). [DOI] [PubMed] [Google Scholar]
- Zharkov D. O., Rieger R. A., Iden C. R. & Grollmann A. P. NH2-terminal proline acts as a nucleophile in the glycosylase/AP-lyase reaction catalyzed by Escherichia coli formamidopyrimidine-DNA glycosylase (Fpg) protein. J. Biol. Chem. 272, 5335–5341 (1997). [DOI] [PubMed] [Google Scholar]
- Ma Z. et al. Unnatural amino acid substitution as a probe of the allosteric coupling pathway in a mycobacterial Cu(I) sensor. J. Am. Chem. Soc. 131, 18044–18045 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levovich I. et al. Formaldehyde-releasing prodrugs specifically affect cancer cells by depletion of intracellular glutathione and augmentation of reactive oxygen species. Cancer Chemother. Pharmacol. 62, 471–482 (2008). [DOI] [PubMed] [Google Scholar]
- Tulpule K. & Dringen R. Formaldehyde stimulates Mrp1-mediated glutathione deprivation of cultured astrocytes. J. Neurochem. 116, 626–635 (2011). [DOI] [PubMed] [Google Scholar]
- Yano T., Takigami E., Yurimoto H. & Sakai Y. Yap1-regulated glutathione redox system curtails accumulation of formaldehyde and reactive oxygen species in methanol metabolism of Pichia pastoris. Eukaryot. Cell 8, 540–549 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerin T., Kim J. S., Gil H. W., Song H. Y. & Hong S. Y. Effects of formaldehyde on mitochondrial dysfunction and apoptosis in SK-N-SH neuroblastoma cells. Cell Biol. Toxicol. 31, 261–272 (2015). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Bone marrow injury induced via oxidative stress in mice by inhalation exposure to formaldehyde. PLoS One 8, e74974 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976). [DOI] [PubMed] [Google Scholar]
- Gasteiger E. et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucl. Acids Res. 31, 3784–3788 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thelander L. Studies on thioredoxin reductase from Escherichia coli B. The relation of structure and function. Eur. J. Biochem. 4, 407–419 (1968). [DOI] [PubMed] [Google Scholar]
- Miller J. H. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY (1992). [Google Scholar]
- Winter G. xia2: an expert system for macromolecular crystallography data reduction. J. Appl. Crystallogr. 43, 186–190 (2010). [Google Scholar]
- Vonrhein C., Blanc E., Roversi P. & Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 (2007). [DOI] [PubMed] [Google Scholar]
- Emsley P. & Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- Murshudov G. N., Vagin A. A. & Dodson E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D 53, 240–255 (1997). [DOI] [PubMed] [Google Scholar]
- Cowtan K. Fitting molecular fragments into electron density. Acta Crystallogr. Sect. D 64, 83–89 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer G., Cohen S. X., Lamzin V. S. & Perrakis A. Automated macromolecular model building for x-ray crystallography using ARP/wARP version. Nat. Protoc. 3, 1171–1179 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E. & Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. Sect. D 60, 2256–2268 (2004), [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.