Cadherin proteolysis reduces cell–cell adhesion and generates cleavage products that could possess independent functions. Here, Schiffmacher et al. reveal that the intracellular C-terminal fragment generated by Cadherin-6B proteolysis promotes chick cranial neural crest cell EMT through positive transcriptional feedback into the neural crest gene regulatory network.
Abstract
During epithelial-to-mesenchymal transitions (EMTs), cells disassemble cadherin-based junctions to segregate from the epithelia. Chick premigratory cranial neural crest cells reduce Cadherin-6B (Cad6B) levels through several mechanisms, including proteolysis, to permit their EMT and migration. Serial processing of Cad6B by a disintegrin and metalloproteinase (ADAM) proteins and γ-secretase generates intracellular C-terminal fragments (CTF2s) that could acquire additional functions. Here we report that Cad6B CTF2 possesses a novel pro-EMT role by up-regulating EMT effector genes in vivo. After proteolysis, CTF2 remains associated with β-catenin, which stabilizes and redistributes both proteins to the cytosol and nucleus, leading to up-regulation of β-catenin, CyclinD1, Snail2, and Snail2 promoter-based GFP expression in vivo. A CTF2 β-catenin–binding mutant, however, fails to alter gene expression, indicating that CTF2 modulates β-catenin–responsive EMT effector genes. Notably, CTF2 association with the endogenous Snail2 promoter in the neural crest is β-catenin dependent. Collectively, our data reveal how Cad6B proteolysis orchestrates multiple pro-EMT regulatory inputs, including CTF2-mediated up-regulation of the Cad6B repressor Snail2, to ensure proper cranial neural crest EMT.
Introduction
Epithelial-to-mesenchymal transition (EMT) is a highly coordinated process that integrates multiple mechanisms to generate a mesenchymal cell from one of epithelial origin. At the onset of EMT, junctional complexes such as adherens junctions are dismantled to facilitate loss of adhesion between cells, promote delamination, and enable motility (Gheldof and Berx, 2013). Cadherin-based adherens junctions consist of cis-interacting cadherin homodimers that associate in trans with cadherin ectodomains on neighboring cells. Intracellular cadherin C-terminal domains directly interact with catenins, including β-, γ-, and δ (p120)-catenin, and indirectly interact with α-catenin via β-catenin to affix the underlying F-actin network to the membrane (Nelson et al., 2013). Cytoskeletal anchoring of the cadherin–catenin complexes promotes stability of the adherens junction and maintains apicobasal polarity in epithelial cells (Hong et al., 2013; Nelson et al., 2013).
Cadherin expression and function have been well characterized throughout neural crest ontogeny (Taneyhill and Schiffmacher, 2013). Premigratory neural crest cells undergo EMT to become motile and later differentiate into numerous cell types including the craniofacial skeleton, glia and neurons, and pigment cells. Avian species have been extensively used as models for studying neural crest development and, specifically, the mechanisms underpinning EMT. Chicken premigratory neural crest cells initially express at least three cadherins: N-cadherin, E-cadherin, and Cadherin-6B (Cad6B; Duband and Thiery, 1982; Hatta et al., 1987; Duband et al., 1988; Akitaya and Bronner-Fraser, 1992; Nakagawa and Takeichi, 1995, 1998; Coles et al., 2007; Shoval et al., 2007; Lee et al., 2013). In the head, N-cadherin is reduced before EMT, whereas E-cadherin is maintained in early migratory neural crest cells. Cad6B expression in the midbrain, however, achieves maximal levels before EMT but is depleted within hours to allow en masse exit of emerging neural crest cells from the dorsal neural tube (Nakagawa and Takeichi, 1995; Coles et al., 2007). Loss of Cad6B during EMT occurs through various layers of regulation working in concert, including transcriptional repression of Cad6B by a Snail2–PHD12–Sin3A complex (Taneyhill et al., 2007; Strobl-Mazzulla and Bronner, 2012) and removal of membrane-bound Cad6B protein via a disintegrin and metalloproteinase (ADAM)–mediated and γ-secretase–mediated proteolysis (Schiffmacher et al., 2014) and clathrin-mediated endocytosis and macropinocytosis (Padmanabhan and Taneyhill, 2015).
The coordination of cadherin proteolysis by ADAMs and γ-secretase has been well characterized in vitro (Marambaud et al., 2002, 2003; Maretzky et al., 2005; Reiss et al., 2005; Uemura et al., 2006a,b). In many cell lines, N- and E-cadherin are substrates for metalloproteinases that cleave the cadherin ectodomain to generate a shed N-terminal fragment (NTF). The resultant membrane-bound C-terminal fragment (CTF1) is then subjected to intracellular proteolysis by γ-secretase to create a cytosolic C-terminal fragment (CTF2). Intriguingly, in many instances these cadherin NTFs and CTF2s possess unique functions independent of those associated with the full-length cadherin (McCusker and Alfandari, 2009). The generation of such functional cadherin fragments also occurs in vivo throughout neural crest ontogeny (Shoval et al., 2007; McCusker et al., 2009; Neuner et al., 2009; Schiffmacher et al., 2014; Abbruzzese et al., 2016). In the chicken trunk, ADAM10 processes N-cadherin to promote neural crest delamination during EMT (Shoval et al., 2007), and N-cadherin CTF2s then translocate to the nucleus and purportedly up-regulate EMT effector genes such as β-catenin and CyclinD1. In vitro studies provide some insight into how this CTF2-driven mechanism of transcriptional regulation may occur. Cadherin CTF2s can remain associated with bound catenins and enter the nucleus (Marambaud et al., 2003; Uemura et al., 2006a; Ferber et al., 2008), where they potentially modulate T-cell factor (TCF)/lymphoid enhancer-binding factor (LEF) target genes via an interaction with β-catenin (Sadot et al., 1998; Gottardi et al., 2001; Maretzky et al., 2005; Reiss et al., 2005; Uemura et al., 2006a; Shoval et al., 2007; Klinke et al., 2015), block CREB-mediated gene expression through degradation of CREB-binding protein (Marambaud et al., 2003), or associate with p120-catenin and the transcriptional repressor Kaiso to inhibit its function (Ferber et al., 2008).
During chick cranial neural crest EMT, ADAM10, ADAM19, and γ-secretase cleave Cad6B, facilitating removal of preexisting Cad6B pools and generating CTF2 (Schiffmacher et al., 2014). Importantly, ectopic expression of Cad6B CTF2 precociously clears Cad6B in premigratory cranial neural crest cells, suggesting that negative regulatory feedback on Cad6B exists after proteolysis. The role of Cad6B CTF2 during EMT, however, remains to be determined. In this study, we reveal a unique function for Cad6B CTF2 in augmenting gene expression during cranial neural crest EMT. We demonstrate that Cad6B CTF2 is generated before and during early EMT and associates with β-catenin in the cytosol and nucleus, both in vitro and in vivo. Strikingly, CTF2-mediated transactivation of several EMT effector genes including Snail2, which represses Cad6B transcription, occurs in the cranial neural crest in a manner dependent on β-catenin binding. Importantly, this interaction with β-catenin is required for CTF2 association with the Snail2 promoter in cranial neural crest cells. Altogether, these findings now provide a molecular mechanism by which Cad6B proteolysis orchestrates EMT through both concomitant repression of Cad6B transcription and up-regulation of other EMT effector genes.
Results
Generation of Cad6B CTFs is correlated with cranial neural crest EMT
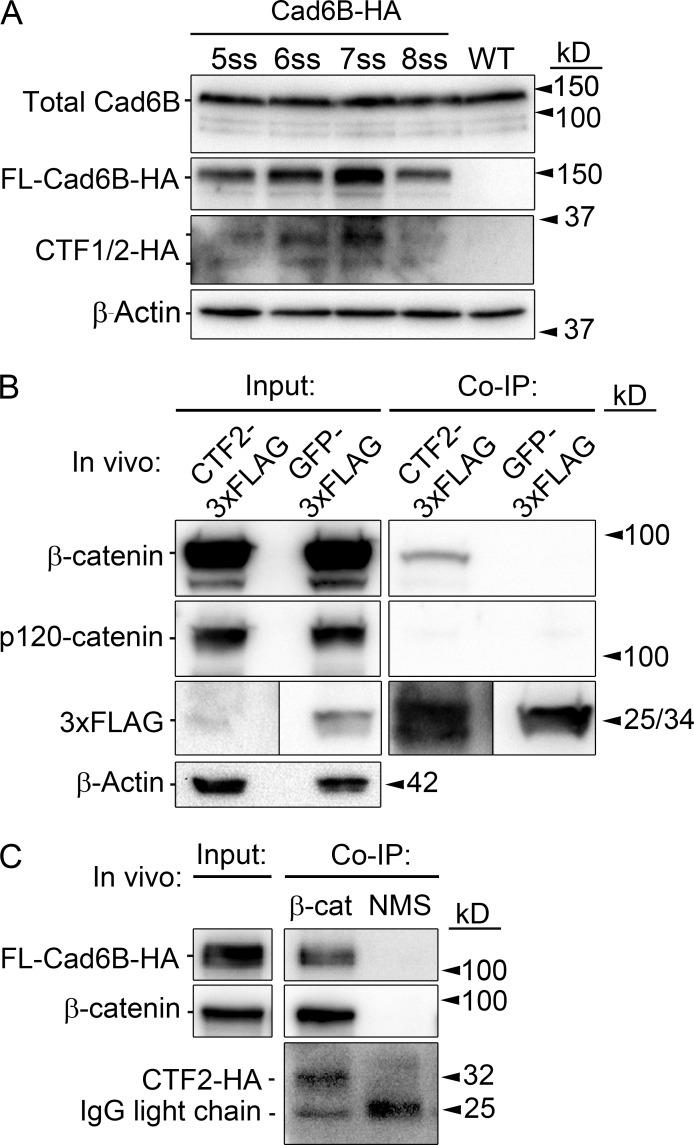
Our prior work revealed that proteolysis reduces Cad6B membrane pools before and during EMT, leading to the appearance of shed NTFs, the levels of which increased at each successive somite stage (ss; Schiffmacher et al., 2014). Although membrane-bound CTF1 and intracellular CTF2 were evident during EMT, it is unknown whether conversion of CTF1 to CTF2 is also a recurring process that is initiated before, and extends throughout, EMT. To address this, we evaluated the appearance of Cad6B CTFs pre-EMT (5ss, a stage at which robust Cad6B protein is first noted; Taneyhill et al., 2007; Schiffmacher et al., 2014) and during active stages of EMT (6–8ss) in the midbrain. Owing to the lack of an available antibody to the endogenous Cad6B C-terminal domain, we overexpressed full-length Cad6B tagged with HA at the C terminus (Cad6B-HA) in premigratory cranial neural crest cells using DNA concentrations previously validated (Schiffmacher et al., 2014) to not elicit a Cad6B overexpression phenotype (Coles et al., 2007). In line with our prior studies on the NTF (Schiffmacher et al., 2014), we detected HA-tagged CTF1 and CTF2 at all developmental stages examined (5–8ss; Fig. 1 A). These results suggest that Cad6B proteolysis is a continual two-step process that begins before EMT, liberating shed NTFs and cytosolic CTF2s.
Figure 1.
Cad6B CTF2s are generated before and throughout cranial neural crest EMT and remain associated with β-catenin. (A) Cad6B is subjected to γ-secretase–mediated proteolysis before, and during, EMT. Premigratory neural crest cells were electroporated at 2–3ss, and cranial neural folds were collected at specific stages for HA immunoblot analysis. (B) CTF2 physically associates with β-catenin, but not p120-catenin, in vivo. Embryo midbrains were electroporated at 4ss and incubated beyond the initiation of EMT. Dorsal neural tubes containing CTF2-3xFLAG– or GFP-3xFLAG–expressing neural crest cells were subjected to coimmunoprecipitation (co-IP) and immunoblotting. (C) HA-tagged CTF2s created via full-length Cad6B-HA proteolysis remain bound to β-catenin in cranial neural crest cells in vivo. Embryos were dorsal/ventrally electroporated at 4ss, and dorsal neural tubes were collected at 8ss. Normal mouse serum (NMS) immunoprecipitation with Cad6B-HA lysate served as a negative control. WT, wild type.
Cad6B CTF2 forms complexes with β-catenin in vitro and in vivo
In vitro studies indicate that cadherin CTF2 molecules likely function in association with other proteins, most notably catenin family members (Sadot et al., 1998; Marambaud et al., 2002; Uemura et al., 2006a; Ferber et al., 2008). To examine potential Cad6B CTF2 binding partners, we performed CTF2 coimmunoprecipitations using triple FLAG-tagged Cad6B CTF2 (CTF2-3xFLAG), or GFP-3xFLAG as a negative control, both in vitro in transfected LMH cells (Fig. S1) and in vivo in electroporated midbrain neural crest cells (Fig. 1 B). The CTF2-3xFLAG pull-down in vitro revealed abundant coimmunoprecipitation of β-catenin (Fig. S1). Interestingly, p120-catenin, which interacts with the juxtamembrane region of the cadherin cytoplasmic tail at a site that differs from the β-catenin–binding domain, does not coimmunoprecipitate with CTF2 in vitro (Fig. S1). Strikingly, coimmunoprecipitation of endogenous β-catenin was noted with minimal expression and pull-down of CTF2-3xFLAG in vivo (Fig. 1 B). As in our in vitro findings (Fig. S1), p120-catenin coimmunoprecipitation was not observed in vivo (Fig. 1 B). Importantly, the interaction between CTF2 and β-catenin in vivo is also evident upon immunoprecipitation of endogenous β-catenin from cranial neural crest cells (Fig. 1 C). Based on this interaction, we hypothesized that CTF2 may serve as a subunit in a functional complex with β-catenin and designed experiments to test this, as outlined here.
Cad6B CTF2 molecules are stabilized and protected from degradation when bound to β-catenin
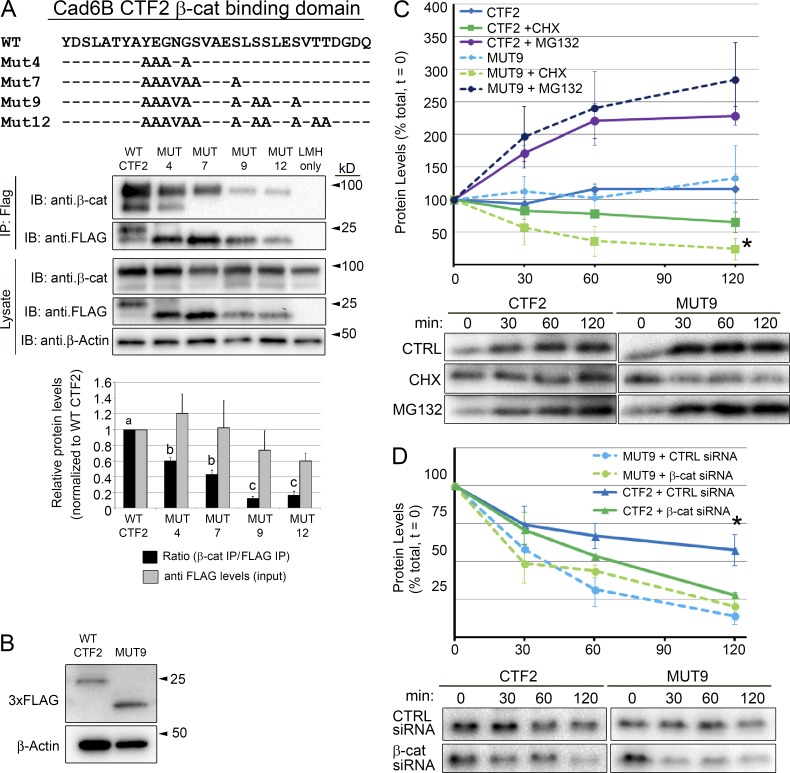
Cadherin CTF2 binding to β-catenin stabilizes β-catenin levels and protects β-catenin from proteasomal degradation (Sadot et al., 1998), but it is unclear whether the converse is true. To assess possible β-catenin–mediated CTF2 stabilization, 3xFLAG-tagged CTF2 mutant constructs were created by incorporating alanine substitutions into a minimal sequence (YEGNG) within the Cad6B β-catenin–binding domain (Huber et al., 2001; Simcha et al., 2001) as well as into key serines phosphorylated to increase β-catenin binding affinity (Fig. 2 A; Simcha et al., 2001; McEwen et al., 2014). These mutants were transiently transfected into LMH cells, and FLAG coimmunoprecipitations were performed. Although all mutants exhibited impaired binding to β-catenin, the CTF2 mutant MUT9, which contains five alanine substitutions within the YEGNG sequence and four serine-to-alanine substitutions, inhibited β-catenin binding by almost 90% (Fig. 2 A; MUT9, 87.5%; n = 3, P < 0.05). We next investigated the expression levels of all proteins in vitro and in vivo. Analysis of CTF2 mutants in vitro revealed no statistically significant change in expression levels compared with wild-type CTF2 (P < 0.49), although MUT9 and MUT12 levels tended to be somewhat reduced (Fig. 2 A). To corroborate this finding, wild-type CTF2 and MUT9 were expressed in neural crest cells in vivo for 5 h. Immunoblotting revealed no significant difference in overall expression level of each protein (Fig. 2 B). These data indicate that the mutations present in MUT9 do not significantly affect MUT9 expression levels.
Figure 2.
Cad6B CTF2s are stabilized through β-catenin binding. (A) Cad6B CTF2 mutants were generated through alanine substitutions within the β-catenin minimal binding domain and at key adjacent phosphorylation sites. Black bars represent the ratio of coimmunoprecipitated β-catenin to immunoprecipitated (IP) mutant CTF2 levels normalized to the wild-type (WT) ratio (set at 100%); gray bars represent the input levels of mutant CTF2 protein (normalized to β-actin levels) relative to WT CTF2 input levels (set at 100%). Means that share letter superscripts are not significantly different (P < 0.05). Immunoblots are representative of three independent experiments. (B) Introduction of CTF2 or MUT9 reveals no change in expression levels in vivo. Embryos were electroporated with triple FLAG-tagged constructs and incubated for 5 h before electroporated neural tubes/neural crest cells were dissected out for immunoblotting. Each lane represents 20 pooled electroporated neural folds. (C) WT CTF2s exhibit decreased degradation rates compared with MUT9 proteins in vitro. FLP-In CHO cell lines expressing either CTF2-HA or MUT9-HA were treated with CHX or MG132 over a 2-h time course (n = 3). Asterisk denotes a significant difference in protein between CHX-treated CTF2 and MUT9 at 120 min. (D) Depletion of β-catenin corroborates dependence of CTF2, but not MUT9, on β-catenin binding for its stability. A validated DsiRNA (#3) to CHO cell β-catenin (Fig. S2 A) or negative control (CTRL) DsiRNA was transiently transfected into Flp-In-Cad6B-CTF2-HA or Flp-In-Cad6B-MUT9-HA CHO cells. CTF2 and MUT9 levels were analyzed after a 2-h CHX chase by immunoblotting (as in C). Asterisk denotes a significant difference in protein in negative control DsiRNA-treated CTF2-HA cells compared with β-catenin DsiRNA-treated CTF2-HA cells or DsiRNA-treated MUT9 cells at 120 min (n = 3, P < 0.05). Error bars in all panels indicate SEMs.
Next, we examined the stability of wild-type CTF2 and MUT9 in vitro (Fig. 2 C). Flp-In-CHO cell lines constitutively expressing wild-type CTF2-HA or MUT9-HA from a single, stably integrated locus were treated with cycloheximide (CHX) or the proteasome inhibitor MG132, and lysates were collected at subsequent time points for immunoblotting analysis (Schiffmacher et al., 2014). After the CHX chase, wild-type CTF2 levels were extended compared with MUT9 levels (CTF2 t1/2 = 170.3 min; MUT9 t1/2 = 59.9 min). The decreased degradation rate for wild-type CTF2 leads to significantly higher CTF2 levels than MUT9 levels by 2 h (CTF2 65.6% vs. MUT9 24.4%; n = 3; P < 0.05). CTF2 and MUT9 are also subjected to proteasomal degradation, as both proteins responded similarly to MG132 supplementation compared with no treatment (n = 3; P < 0.05). To further confirm that the enhanced stabilization of wild-type CTF2 levels over MUT9 levels is dependent on β-catenin and not a result of increased turnover/proteolysis of MUT9 caused by the mutations MUT9 possesses, we depleted β-catenin from each Flp-In-CHO cell line using validated β-catenin (or negative control) Dicer-substrate short interfering RNAs (DsiRNAs; Fig. S2, A and B), achieving a >58% reduction in β-catenin (Fig. S2 C; P < 0.05). Upon examining CTF2 and MUT9 levels after a CHX chase, we noted that CTF2, in the absence of β-catenin depletion (CTF2 + CTRL DsiRNA), maintained a significantly reduced degradation rate (t1/2 = 123.2 min) compared with the other treatments (CTF2 + β-cat DsiRNA, t1/2 = 75.1 min; MUT9 + β-cat DsiRNA, t1/2 = 57.8 min; MUT9 + CTRL DsiRNA, t1/2 = 54.2 min; P < 0.05; Fig. 2 D). MUT9 levels, however, were unaffected by a reduction in β-catenin (P < 0.40). Collectively, these data reveal that cytosolic Cad6B CTF2s retain their key phosphorylated serines necessary to maintain an interaction with β-catenin. Notably, CTF2 binding to β-catenin stabilizes CTF2 and decreases its degradation rate, with complex formation mutually beneficial to both β-catenin and CTF2.
Elevated CTF2 levels result in β-catenin redistribution and accumulation within neural crest cells in vivo
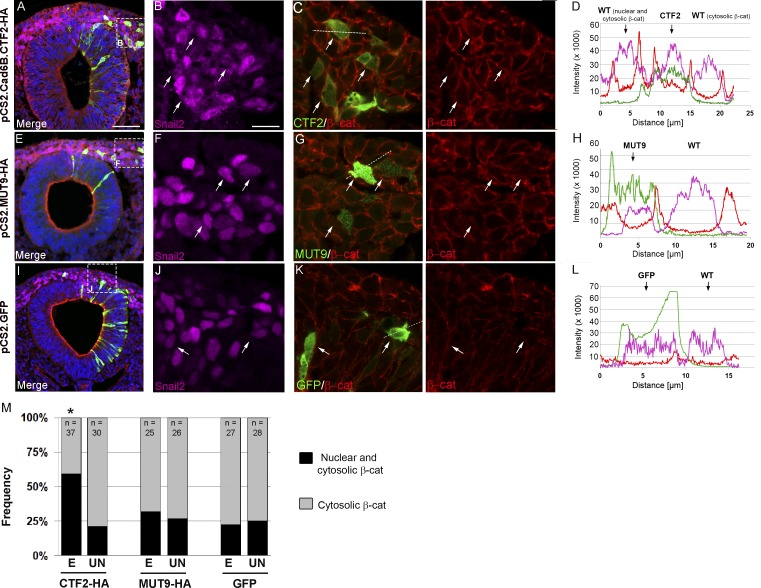
As cytosolic Cad6B CTF2s are generated during EMT, bound β-catenin molecules should exhibit altered distribution in the neural crest. To determine whether elevated CTF2 levels change β-catenin protein localization, we overexpressed HA-tagged wild-type CTF2 or MUT9 (or GFP as a control) in premigratory cranial neural crest cells and evaluated changes in β-catenin levels and distribution by immunohistochemistry (Fig. 3). Cells expressing CTF2 exhibited a redistribution/accumulation of β-catenin from the membrane to the cytosol and nucleus within 8 h after electroporation (Fig. 3, A–D, arrows). This β-catenin redistribution also occurs in a cell-autonomous manner (Fig. 3, B and C, arrows). Importantly, elevated MUT9 (Fig. 3, E–H, arrows) or GFP (Fig. 3, I–L, arrows) did not elicit this β-catenin phenotype. Cell-count analysis coupled with β-catenin fluorescence intensity profile line scans (Fig. 3, D, H, and L, red) in Snail2-positive (Fig. 3, D, H, and L, purple) electroporated (Fig. 3, D, H, and L, green) and adjacent unelectroporated cells revealed a 38% increase in cells showing cytosolic/nuclear β-catenin in the presence of CTF2 (59% total) compared with unelectroporated cells (24% averaged total, P < 0.05), MUT9 (32% total, P < 0.05), or GFP (22% total, P < 0.05; Fig. 3 M). Cells expressing MUT9 or GFP also did not cause any significant changes in β-catenin distribution (MUT9, P < 0.43; GFP, P < 0.62). Interestingly, cells with increased cytoplasmic β-catenin still retained membrane-associated β-catenin, suggesting that ectopic CTF2s are not outcompeting full-length cadherins for the available β-catenin pool. Collectively, our results indicate that Cad6B CTF2s can alter β-catenin distribution in vivo.
Figure 3.
Cad6B CTF2 overexpression results in the redistribution and accumulation of β-catenin in cranial neural crest cells in vivo. Representative transverse sections taken through the midbrain region of 8–9ss embryos expressing CTF2-HA (A–C), MUT9-HA (E–G), or GFP (I–K), followed by immunostaining for Snail2 (purple), HA or GFP (green), and β-catenin (red). Boxes in lower-magnification (20×) merge images (A, E, and I) mark respective magnified (63×) areas (B, C, F, G, J, and K). (D, H, and L) Line scans showing subcellular fluorescent intensity of recombinant protein (green), β-catenin (red), and Snail2 (purple) across cell diameters (C, G, and K, dotted lines) from representative electroporated and adjacent unelectroporated cells. (M) Quantification of β-catenin levels in electroporated (E) and unelectroporated (UN) Snail2-positive neural crest cells reveals a statistically significant increase in the number of cells containing elevated nuclear and cytosolic β-catenin levels (black bars) versus cytosolic-only β-catenin (gray bars) in the presence of CTF2 but not MUT9 or GFP (*, P < 0.05). DAPI (blue) labels nuclei in A, E, and I. Bars: (A, E, and I) 50 µm; (B and all remaining images) 10 µm. Samples sizes (n) for each group are shown in M. WT, wild type.
Cad6B CTF2 and β-catenin physically interact in the nucleus and cytosol both ex vivo and in vitro
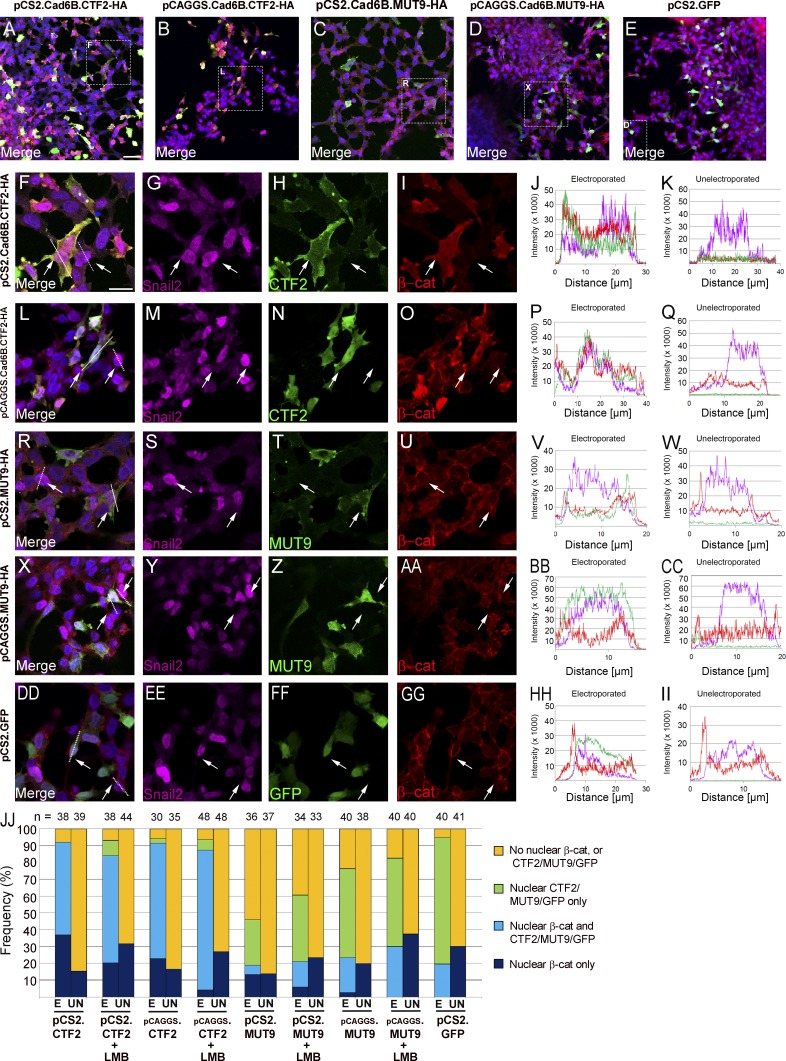
Prior in vitro studies have shown that cadherin CTF2 molecules often translocate to the nucleus in association with β-catenin (Sadot et al., 1998; Maretzky et al., 2005; Uemura et al., 2006a; Ferber et al., 2008). Furthermore, exogenous N-cadherin CTF2s localize within chicken trunk neural crest cell nuclei during EMT and migration ex vivo (Shoval et al., 2007). To determine whether Cad6B CTF2 and β-catenin cotranslocate into the nucleus, explants of premigratory cranial neural crest cells electroporated with either pCS2- or pCAGGs-based expression constructs were treated with (Fig. S3) or without (Fig. 4) the nuclear export inhibitor leptomycin B (LMB). Both electroporated and unelectroporated cells treated with LMB exhibited an overall 11% increase in β-catenin nuclear localization (P < 0.05). In the absence (or presence) of LMB, CTF2 altered β-catenin distribution as previously observed in vivo (Fig. 3). Increased levels of CTF2 and β-catenin were noted within the nuclei of neural crest cells expressing CTF2 from either expression vector (Fig. 4, F–Q, arrows), with detected levels correlating with the strength of the vector (Fig. 4 JJ: pCAGGs [stronger promoter] shows a 13% [LMB−] and 20% [LMB+] increase versus pCS2). This enhanced β-catenin nuclear localization, however, was not readily detectable in cells overexpressing MUT9 from either expression vector (Fig. 4, R–CC, arrows) or GFP (Fig. 4, DD–II, arrows). To quantify effects on β-catenin distribution, we analyzed electroporated and unelectroporated Snail2-positive migratory neural crest cells in at least six ex vivo neural crest explants per plasmid for the presence of nuclear β-catenin (Fig. 4 JJ). Our results indicate that 92% of electroporated versus 15% of unelectroporated cells exhibit nuclear β-catenin upon CTF2 overexpression (pCS2, P < 0.05; 91% vs. 17% for pCAGGs, P < 0.05). Conversely, cells expressing MUT9 show nuclear β-catenin in 19% of electroporated cells versus 14% of unelectroporated cells (P < 0.76). In cells expressing MUT9, LMB supplementation or increased promoter strength significantly increases MUT9 nuclear distribution but does not affect β-catenin nuclear localization (Fig. 4 JJ; P < 0.70). These data demonstrate that increased levels of Cad6B CTF2 enhance β-catenin nuclear localization, but not in CTF2 mutants deficient in β-catenin binding (MUT9).
Figure 4.
Cad6B CTF2 and β-catenin coimport into the nucleus in cranial neural crest cells ex vivo. Representative explants of neural crest cells overexpressing CTF2-HA (pCS2-CTF2-HA, A and F–I; pCAGGs-CTF2-HA, B and L–O), MUT9-HA (pCS2-MUT9-HA, C and R–U; pCAGGs-MUT9-HA, D and X–AA), or GFP (E and DD–GG), followed by immunostaining for Snail2 (purple), HA or GFP (green), and β-catenin (red). Boxes in lower-magnification 20× merge images (A–E) mark respective magnified (63×) areas (F–I, L–O, R–U, X–AA, and DD–GG). Bar: (A–E) 20 µm; (F–GG) 10 µm. (JJ) Quantification of electroporated (E) and unelectroporated (UN) neural crest cells exhibiting β-catenin nuclear localization reveals a statistically significant increase in the presence of CTF2 but not MUT9. Protein subcellular distribution was determined by fluorescence intensity line profiling (J, K, P, Q, V, W, BB, CC, HH, and II) across candidate cell diameters (arrows and dotted lines in F, L, R, X, and DD). Cell counts were performed based on protein distribution pattern (orange, no nuclear β-catenin or CTF2/MUT9/GFP; green, nuclear CTF2/MUT9/GFP only; light blue, both nuclear β-catenin and CTF2/MUT9/GFP; dark blue, nuclear β-catenin only). Samples sizes (n) for each group shown in JJ.
Nuclear colocalization of both β-catenin and Cad6B CTF2 could suggest that their physical association is maintained after nuclear import. To address this hypothesis, coimmunoprecipitations were performed with cytoplasmic and nuclear fractions prepared from transfected LMH cells (Fig. S4). The coimmunoprecipitation of β-catenin with CTF2 in the nuclear fraction indicates that CTF2 remains associated with β-catenin after nuclear coimportation. Collectively, these results confirm that stabilized, cytosolic Cad6B CTF2–β-catenin complexes also translocate to the nucleus.
Cad6B CTF2 up-regulates EMT effector gene expression through a β-catenin–dependent mechanism in vivo
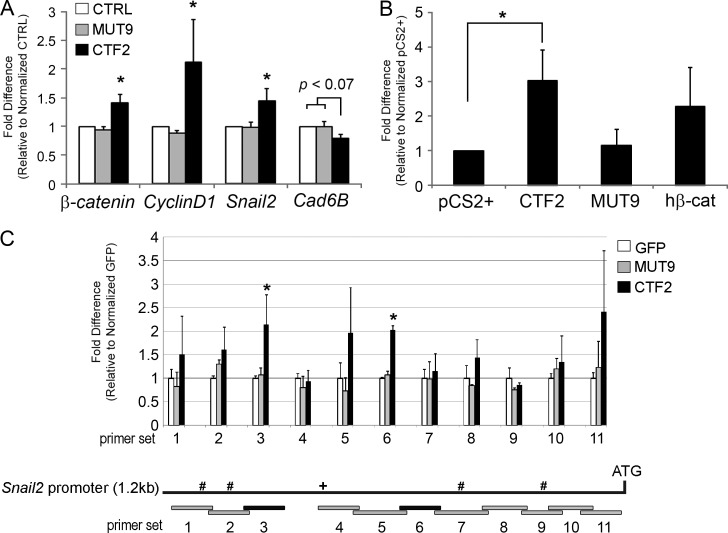
Because the generation of CTF2 molecules depends on full-length Cad6B expression and is correlated with EMT, we hypothesize that Cad6B CTF2 may regulate EMT itself. This is supported by our prior work demonstrating that CTF2 overexpression results in premature clearance of full-length Cad6B in cranial neural crest cells undergoing EMT (Schiffmacher et al., 2014). Given a previously described role for cadherin CTF2 molecules in regulating gene expression, we investigated the potential for the Cad6B CTF2–β-catenin complex to function similarly. Whereas earlier studies detected changes in gene expression only semiquantitatively in vitro and in vivo after long cadherin CTF2 expression times (12–24 h; Uemura et al., 2006a; Shoval et al., 2007), we limited CTF2 expression to 5 h, which is the minimal duration to achieve detectable levels. Premigratory cranial neural crest cells overexpressing CTF2, MUT9, or GFP were analyzed for changes in expression of known downstream targets of β-catenin transactivation important for EMT (CyclinD1 and β-catenin) and an independent regulator of EMT (Snail2) by quantitative PCR (QPCR; Fig. 5 A). Transient expression of CTF2, but not MUT9 or GFP, resulted in a statistically significant 41%, 110%, and 44% increase in β-catenin, CyclinD1, and Snail2 transcripts, respectively, relative to MUT9 and GFP (P < 0.05). No significant gene expression changes were noted in MUT9 and GFP samples, suggesting that Cad6B CTF2 regulates expression of these genes through a β-catenin–dependent mechanism. Conversely, transient expression of CTF2 led to a 20% decrease in Cad6B transcripts (P < 0.07). Although not statistically significant, this reduction is likely associated with the observed rise in Snail2 and subsequent increased Snail2 repression of Cad6B. These results are further supported by our findings that CTF2 and MUT9 are expressed at comparable levels in vivo and in vitro (Figs. 2, B and C), indicating that the inability of MUT9 to activate EMT effector genes is not caused by differences in expression levels that could occur independently of β-catenin binding, such as by altering the susceptibility of MUT9 to proteases.
Figure 5.
Transient elevation of Cad6B CTF2 in the cranial neural crest up-regulates multiple EMT effector genes and controls endogenous Snail2 expression via an association with chromatin. (A) Embryos were unilaterally electroporated with GFP (white), MUT9 (gray), or CTF2 (black) expression vectors at 3ss and incubated for an additional 5 h (6–7ss). Excised neural crest cells were pooled and lysed for RNA extraction, followed by generation of cDNA. Gene levels were normalized to 18S ribosomal RNA, and the graph shows differences in gene expression as determined by calculating the fold-change from the control group (CTRL; arbitrarily set to 1; n = 3). (B) Evaluation of the Snail2-GFP reporter to increased nuclear β-catenin as a result of CTF2 overexpression. Embryos were electroporated as described in A with a GFP reporter driven by a minimal Snail2 promoter, along with either empty vector, CTF2, MUT9, or human β-catenin. Differences in GFP were assessed and depicted graphically, with GFP levels for CTF2 arbitrarily set to 1 (n = 3). (C) In vivo ChIP-QPCR demonstrates β-catenin–dependent association of CTF2, but not MUT9 or GFP, to chromatin regions within the endogenous Snail2 promoter. Embryos were electroporated as described in A, and fold enrichment of amplicon expression between treatments per primer set was assessed after normalization to GFP group means (n = 2). The Snail2 promoter schematic (not to scale) depicts the 1.2-kb sequence upstream of the translational start site that was tested. Gray bars represent amplicons not significantly enriched; black bars represent amplicons from primer sets 3 and 6 showing CTF2 association. +, relative position of a validated TCF/LEF site (Sakai et al., 2005) covered by primer set 4; #, putative TCF/LEF site. Data in all graphs show means and SEM, and all asterisks indicate a significant difference in gene expression (A and B) or enrichment (C; P < 0.05).
To corroborate our results, we coelectroporated a Snail2-GFP reporter (possessing 1.2 kb of the chicken Snail2 promoter, including one validated [D1.2; Sakai et al., 2005] and four computationally predicted TCF/LEF binding sites) with empty vector, CTF2, MUT9, or human β-catenin (positive control) into premigratory cranial neural crest cells and then analyzed GFP expression after 5 h by QPCR (Fig. 5 B). Transient CTF2 expression induced a threefold increase in GFP transcripts over the empty vector (P < 0.05) in a manner similar to β-catenin overexpression (2.3-fold increase), showing that reporter activation occurs only in the presence of wild-type CTF2 and is thus dependent on β-catenin binding. Altogether, these data reveal that Cad6B CTF2 facilitates β-catenin–mediated transactivation within premigratory cranial neural crest cells and that proteolysis of Cad6B can feedback directly on Cad6B transcriptional repression during EMT.
Cad6B CTF2 associates with chromatin in a β-catenin–dependent manner in vivo
The mechanism by which CTF2 activates EMT effector genes could be indirect through its capacity to shuttle β-catenin to the nucleus or direct via an association with chromatin. E-cadherin CTF2 binds DNA in vitro (Ferber et al., 2008), but to our knowledge, no studies to date have endeavored to detect cadherin CTF2 association to chromatin in vivo. To this end, we examined the ability of wild-type CTF2 and MUT9 (or GFP) to associate with specific chromatin regions within the 1.2-kb upstream sequence of the endogenous Snail2 promoter identified previously to be CTF2 responsive (Fig. 5 B) using in vivo chromatin immunoprecipitation (ChIP) coupled with QPCR, as in our prior work (Taneyhill et al., 2007; Taneyhill and Adams, 2008; Jhingory et al., 2010). Chromatin immunoprecipitated from neural crest cells expressing CTF2, MUT9, or GFP (all 3xFLAG-tagged) was amplified by QPCR using primer sets spanning this 1.2-kb region (Fig. 5 C). Although no primer set significantly amplified DNA from the MUT9-expressing samples relative to GFP controls, we observed significant twofold increases in association of CTF2 to DNA in two of 11 primer sets (Fig. 5 C; primer sets 3 and 6, black bars; P < 0.05). Interestingly, these two regions do not contain either validated (+, primer set 4) or putative (#, primer sets 1, 2, 7, and 9) TCF/LEF binding sites (Fig. 5 C). Altogether, our data reveal that CTF2 association with the Snail2 promoter is dependent on an intact β-catenin–binding domain at these analyzed chromatin regions.
Discussion
The molecular changes underscoring EMT must be efficiently integrated and temporally coordinated for its successful implementation. Although cells acquire a gene regulatory network that will direct the new mesenchymal identity, post-transcriptional mechanisms are simultaneously occurring to depolarize the cell, disassemble intercellular junctions, and alter the cytoskeletal architecture (Acloque et al., 2009). At the onset of EMT, Cad6B is transcriptionally repressed (Taneyhill et al., 2007; Strobl-Mazzulla and Bronner, 2012), and membrane-bound Cad6B protein is cleared post-translationally (Yap et al., 2007; Schiffmacher et al., 2014; Padmanabhan and Taneyhill, 2015). Tight spatiotemporal regulation of Cad6B is critical, as Cad6B knockdown or overexpression accelerates or impedes cranial neural crest emigration, respectively (Coles et al., 2007). Together, these studies demonstrate a vital role for Cad6B, and the crucial requirement for Cad6B reduction during EMT, to ensure a successful transition to a migratory neural crest cell.
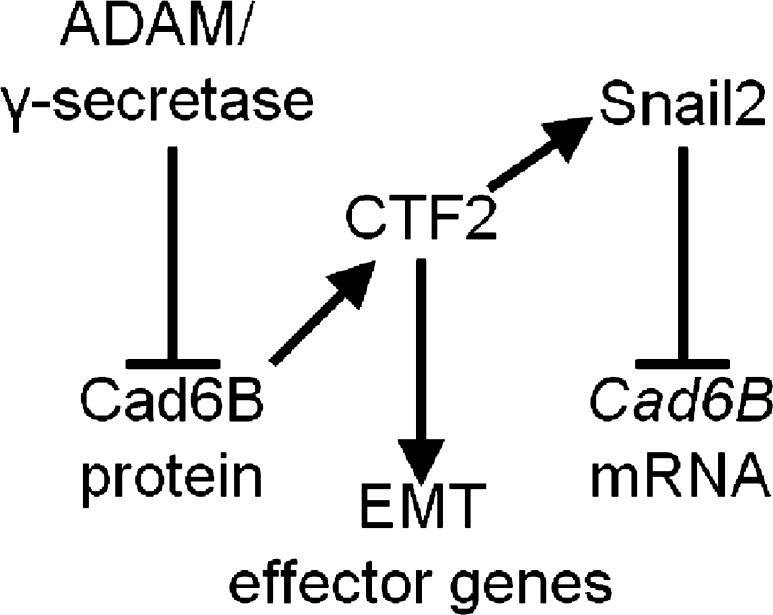
This current study further substantiates the importance of Cad6B by demonstrating that Cad6B proteolysis not only dismantles adherens junctions to promote delamination, but also provides transcriptional regulatory feedback into the neural crest gene regulatory network. This transcriptional regulation depends on the presence of an intact β-catenin–binding domain in Cad6B CTF2 and leads to the up-regulation of pro-EMT genes including β-catenin, CyclinD1, and most notably Snail2 (Fig. 6). Collectively, our results show how a single cellular mechanism can cascade into multiple, coordinated regulatory inputs that direct cranial neural crest EMT.
Figure 6.
Cad6B proteolysis provides transcriptional regulatory feedback to positively impact cranial neural crest EMT. Cad6B proteolysis reduces full-length Cad6B protein levels and generates shed NTFs and CTF2s. CTF2 transactivates multiple EMT effector genes, including Snail2, which in turn represses Cad6B transcription. Altogether, this regulatory circuit promotes cranial neural crest EMT.
Cad6B CTF2 is generated throughout neural crest EMT
Our previous work demonstrated that serial proteolysis of Cad6B yielded Cad6B CTFs at a specific stage of EMT, but did not provide information on the duration of CTF2 generation during chick development (Schiffmacher et al., 2014). Our current study reveals that Cad6B CTFs are present before (5ss) and throughout (6–8ss) EMT (Fig. 1 A). Lacking an antibody that binds endogenous Cad6B CTFs, we are unable to determine whether CTF2 levels are positively correlated with rising full-length Cad6B levels within premigratory cranial neural crest cells (up to 6ss). We surmise, though, that this is the case, given that endogenous Cad6B NTFs increase during neural crest EMT (Schiffmacher et al., 2014). At later stages of EMT, we would still expect to detect CTFs, albeit at much lower levels as a result of the considerable reduction of Cad6B protein after 8ss (Taneyhill et al., 2007; Schiffmacher et al., 2014). These data are further supported by our prior analysis of presenilin-1, the catalytic subunit of γ-secretase, which possesses an overlapping spatiotemporal expression profile with Cad6B (Schiffmacher et al., 2014). This distribution of presenilin-1 and its Cad6B substrate, together with the detection of Cad6B CTFs before and during EMT, suggest that Cad6B proteolysis is an ongoing event within premigratory cranial neural crest cells and is predominantly limited by existing full-length Cad6B protein. Therefore, the onset of EMT may be triggered, in part, by the accumulation of Cad6B CTF2–β-catenin complexes to a defined level, allowing for the up-regulation of EMT effector genes, and is not contingent on the initial detection of CTF2 itself.
Cad6B CTF2–β-catenin complex formation is important to stabilize both proteins and modulate gene expression during neural crest EMT
Cytosolic cadherin CTFs possess novel roles in multiple in vitro and in vivo systems, most often functioning via an interaction with catenins (Sadot et al., 1998; Marambaud et al., 2002). For example, cytosolic E-cadherin CTF2 forms a complex with p120-catenin that coimports into the nucleus and influences nuclear p120-catenin activity by interacting with Kaiso to control gene expression (Ferber et al., 2008). In the current study, we evaluated potential catenin interactors for Cad6B CTF2 both in vitro and in vivo (Fig. 1, B and C; and Fig. S1). Typically, β- and p120-catenin remain bound to cadherin CTF2 after γ-secretase proteolysis, at least in vitro (Marambaud et al., 2002; Ferber et al., 2008). Our data demonstrate that this also occurs in vivo, because a pool of β-catenin remains bound to Cad6B CTF2-HA derived solely from full-length Cad6B-HA proteolysis (Fig. 1 C); however, p120-catenin pull-down was not evident. This disparity could reflect actual events occurring in vivo or be unique to chickens. Alternatively, although our CTF2 construct was designed to mimic a CTF2 peptide generated via γ-secretase, it is possible that in both LMH cells and the cranial neural crest, recombinant CTF2 is subjected to caspase-3–mediated proteolysis to remove the CTF2 N terminus containing the p120-binding domain, as noted for other cadherin CTF2s (Steinhusen et al., 2001; Marambaud et al., 2002; Wolfe and Kopan, 2004; Uemura et al., 2006a). Because of the appreciable affinity with which β-catenin binds to Cad6B CTF2 in vivo, we explored the function of Cad6B CTF2 through its interaction with β-catenin.
To evaluate potential β-catenin–dependent or –independent roles for Cad6B CTF2 in the neural crest, we designed a Cad6B CTF2 mutant deficient in β-catenin binding (MUT9, Fig. 2 A) that is expressed at comparable levels to wild-type CTF2. MUT9 was then used to determine whether β-catenin binding affords any biochemical “advantage” to CTF2. Here, we show that the Cad6B CTF2–β-catenin interaction stabilizes CTF2 and protects CTF2 from proteasomal degradation in a manner dependent on β-catenin binding (Fig. 2, C and D). Interestingly, formation of N-cadherin CTF2–β-catenin complexes in vitro stabilizes endogenous β-catenin pools by inhibiting glycogen synthase kinase-3β phosphorylation (Uemura et al., 2006a). Mutual stabilization of β-catenin and Cad6B CTF2 implies that complex formation may be essential to maintain and elevate levels of CTF2 beyond a threshold and allow for sufficient accumulation of the complex to influence neural crest EMT, as supported by our data. Our results are also in line with those showing that cadherin cytoplasmic tails are highly unstructured and contain PEST domains that mediate protein degradation (Huber et al., 2001). β-Catenin binding to full-length cadherins and CTF2s, however, masks this PEST sequence, thereby preventing cadherin (or CTF2) degradation (Huber et al., 2001).
Nuclear translocation of cadherin CTF2 is a common hallmark of recombinant CTF2 expression and appears to depend on catenin binding (Sadot et al., 1998; Uemura et al., 2006a; Shoval et al., 2007; Ferber et al., 2008). Although it cannot be ruled out that cadherin CTFs possess a cryptic nuclear localization signal, Cad6B CTF2 does not appear to have any importin-α nuclear localization signal observed in other catenins (Kosugi et al., 2009; Klinke et al., 2015), which is critical for nuclear coimport of both the catenin and CTF2 (Sadot et al., 1998; Uemura et al., 2006a; Ferber et al., 2008; Klinke et al., 2015). Conversely, nuclear localization of catenins is not dependent on binding to cadherin CTF2s. Our results, however, reveal that increased Cad6B CTF2, but not MUT9, stabilizes, redistributes, and enhances nuclear import of the CTF2–β-catenin complex within cranial neural crest cells (Figs. 3 and 4). Our in vivo data are in agreement with previous in vitro findings that ascribe a β-catenin transport scaffold function to cytosolic E-cadherin CTF2s (Uemura et al., 2006a; Klinke et al., 2015). As such, mutual stabilization, and the resultant accumulation of both proteins, may lead to increased nuclear accumulation of the complex as well.
Multiple studies contend that nuclear cadherin CTF2s, through various mechanisms, regulate expression of CyclinD1 and other β-catenin–responsive genes (Maretzky et al., 2005; Reiss et al., 2005; Uemura et al., 2006a; Shoval et al., 2007; Klinke et al., 2015). The majority of these studies, however, semiquantitatively assessed changes in gene expression after long incubations that followed CTF2 introduction. To more directly address the involvement of CTF2 in transcriptional regulation, we allowed only 5 h of CTF2 expression in vivo. Within this short time frame, we still detected statistically significant up-regulation of CyclinD1, β-catenin, and Snail2 (Fig. 5 A). Introduction of MUT9, however, abrogated this increase, revealing that conuclear imported β-catenin is likely responsible for this up-regulation. This is further supported by our data indicating that CTF2 and MUT9 are expressed at comparable levels in vivo (Fig. 2 B), arguing against the notion that the absence of EMT effector gene up-regulation is caused by differences in MUT9 expression levels and/or stability. Elevated CTF2, but not MUT9, also augments GFP driven by an ectopically expressed minimal chick Snail2 promoter in vivo (Fig. 5 B), further validating Snail2 as a bona fide target of CTF2 in vivo, and likely through β-catenin–mediated transactivation.
The specific mechanism outlined now provides additional evidence for a universal role for nuclear cadherin CTF2 molecules, as up-regulation of the Wnt target CyclinD1 has been documented in vitro and in vivo after manipulation of other cadherin CTF2s (Maretzky et al., 2005; Reiss et al., 2005; Uemura et al., 2006a; Shoval et al., 2007; Klinke et al., 2015). CyclinD1 is a known Wnt target in the chicken trunk neural crest (Burstyn-Cohen et al., 2004), and our data now show it is possibly regulated by β-catenin-TCF/LEF signaling in the cranial neural crest as well. Although it is uncertain whether β-catenin autoregulates via canonical Wnt signaling in the neural crest, our results, together with the noted β-catenin autoregulation after N-cadherin CTF2 overexpression in the chick trunk neural crest and in vitro, support this hypothesis (Uemura et al., 2006a; Shoval et al., 2007). It also remains to be determined whether Snail2 is up-regulated in response to β-catenin-TCF/LEF signaling, although our ChIP-QPCR data (Fig. 5 C) suggest that this (via CTF2-mediated nuclear shuttling of β-catenin), together with other mechanisms, is likely. These findings in the chicken are also consistent with the observation that three alleles of the Xenopus laevis Snail2 ortholog Slug possess Wnt- and β-catenin–responsive promoters (Vallin et al., 2001).
Our findings do not exclude the possibility that only β-catenin-TCF/LEF targets are activated in response to elevated nuclear Cad6B CTF2 levels. Indeed, prior studies have shown that β-catenin binding to cadherin CTF2s and TCF/LEF proteins is mutually exclusive because of the overlap in the CTF2 and TCF/LEF binding sites on β-catenin (Sadot et al., 1998; Orsulic et al., 1999). Thus, β-catenin–bound CTF2 cannot interact with TCF/LEF proteins, suggesting that other β-catenin targets exist that do not rely on TCF/LEF proteins for their regulation. Given that our ChIP-QPCR data revealed that the Snail2 promoter region possessing the greatest enrichment of CTF2 lacks any putative TCF/LEF binding sites (Fig. 5 C), CTF2–β-catenin complexes may in fact be interacting with other proteins (e.g., DNA binding proteins) to regulate Snail2. Moreover, CTF2, in a complex with other proteins that bind CTF2 within the β-catenin binding domain (given the MUT9 data), could associate directly with chromatin to regulate Snail2. With respect to the regulation of Snail2 expression, illustrated here in chickens and as shown in Xenopus, these findings demonstrate that CTF2–β-catenin complex formation is important to mediate nuclear shuttling of β-catenin for binding to TCF/LEF or other proteins (Valenta et al., 2012) or to allow chromatin-associated CTF2–β-catenin (or other protein) complexes to participate in transactivation, with CTF2 performing scaffolding (Ortiz et al., 2015) or other functions outside of β-catenin binding (Marambaud et al., 2003). As such, CTF2 may mediate transactivation of EMT effector genes through both indirect and direct methods. We hypothesize that this is likely a universal mechanism by which CTF2 transactivates other genes, which will be borne out in future experiments.
ADAM-mediated proteolysis of cadherins has been implicated during disease, particularly cancer cell EMT/metastasis (Duffy et al., 2011). Elevated levels of ADAMs and E-cadherin often serve as biomarkers for cancer and can lead to increased shed cadherin NTFs (Duffy et al., 2011; David and Rajasekaran, 2012). Although there is growing insight on NTF function during cancer metastasis (Najy et al., 2008; David and Rajasekaran, 2012; Patil et al., 2015; Hu et al., 2016), how CTF2s mediate metastasis remains unexplored. Our results suggest a transcriptional role for a cadherin CTF2 during an in vivo developmental EMT that may be directly translatable to EMTs underlying disease and, in turn, may shed light on the repertoire of genes activated as cancer cells become invasive.
Our current data and previous findings (Schiffmacher et al., 2014) reveal how proteolysis during EMT establishes negative feedback on Cad6B expression, at both post-translational and transcriptional levels, in a temporally and spatially restricted manner. The exclusive expression of Cad6B within the premigratory cranial neural crest limits subsequent creation and function of Cad6B CTF2 to cells initiating and undergoing EMT. While membrane Cad6B protein is being reduced through proteolysis, Cad6B CTF2–β-catenin complexes are contributing positive feedback on the expression of Snail2, a potent activator of chicken cranial neural crest EMT and delamination (Nieto et al., 1994; del Barrio and Nieto, 2002), and other EMT effectors, and possibly indirect negative feedback on Snail2 targets such as Cad6B and αN-catenin (Taneyhill et al., 2007; Jhingory et al., 2010; Strobl-Mazzulla and Bronner, 2012). Because of the transient, EMT-specific nature of this mechanism, this feedback may be in place to ensure that Snail2 accumulates to appropriate levels at this critical stage in development. Collectively, our results not only underscore the need to proteolytically process Cad6B in the premigratory cranial neural crest, but also highlight the complexity of positive regulation on other EMT effectors, thereby ensuring that neural crest cells acquire their proper mesenchymal identity and, later, correctly pattern the vertebrate embryo.
Materials and methods
Chicken embryo culture
Fertilized chicken eggs were obtained from B & E Farms and incubated at 38°C in humidified incubators (EggCartons.com). Embryos were staged according to the number of pairs of somites (somite stage).
Cad6B CTF2 expression constructs
The coding sequence corresponding to theoretical CTF2 (produced by γ-secretase–mediated proteolysis (Uemura et al., 2006b; Cad6B; NCBI accession number NP_001001758.1; aa 637–790) was PCR-amplified from pCIG.Cad6B (Coles et al., 2007) and cloned into the pCS2+ and pCIG vectors. C-terminal HA-tagged CTF2 was PCR-amplified and cloned into pCS2+ from pCIG.CAD6B-HA (Schiffmacher et al., 2014). Triple FLAG-tagged CTF2 constructs (epitope: DYKDHDGDYKDHDIDYKDDDDK) were generated via fusion PCR combining CTF2 templates and 3xFLAG sequence from p3xFLAG-CMV-7 (Sigma-Aldrich) as previously described (Hobert, 2002). All alanine substitutions generated within epitope-tagged pCS2.CTF2 mutant constructs were created by incorporating nucleotide changes into internal amplicon overhang primers used with fusion PCR. All constructs were verified for sequence accuracy (Genewiz). The IRES-GFP sequence was removed from pCIG during cloning to generate HA-tagged pCAGGs.CAD6B.CTF2 and MUT9 constructs.
Cell culture and transfection
Chicken LMH cells (CRL-2117; ATCC) were cultured according to ATCC instructions. Transient transfection assays were performed in LMH cells using Lipofectamine 2000 (Thermo Fisher Scientific). Cells were grown to 90% confluence, and transfections were performed according to the manufacturer’s protocol. FLP-In-CHO cells were cultured in complete medium supplemented with 100 µg/ml Zeocin (Thermo Fisher Scientific) and transfected according to the manufacturer’s instructions (Thermo Fisher Scientific).
Protein extraction, fractionation, coimmunoprecipitations, and immunoblotting
48 h after transfection, LMH cells were harvested by gentle mechanical scraping in ice-cold PBS and centrifuged at 4°C for 5 min at 500 g. Pellets were then flash-frozen in liquid nitrogen and stored at −80°C until needed for immunoblot analysis. Embryo protein extracts were collected as previously described (Schiffmacher et al., 2014). In brief, whole midbrains or dorsal neural tubes of midbrains were extracted from embryos, rinsed in Ringer’s solution, centrifuged at 500 g for 5 min at 4°C, and snap-frozen in liquid nitrogen. Cytoplasmic and nuclear fractionation was performed as described (Folco et al., 2012). Triple-FLAG coimmunoprecipitations were performed using FLAG M2 antibody-coated magnetic beads (M8823; Sigma-Aldrich). Transfected cells growing on 100-mm plates were harvested as previously described (Schiffmacher et al., 2014). Cell pellets were lysed in FLAG buffer (100 mM Tris-HCl, pH 8.0, 150 mM NaCl, 5 mM EDTA, 5% glycerol, and 0.1% IGEPAL CA-630) supplemented with 1 mM PMSF, complete protease inhibitor cocktail (Roche), 10 mM NaF, and 1 mM sodium orthovanadate. Lysates were incubated for 30 min at 4°C with periodic mixing and clarified by centrifugation at maximum speed for 15 min at 4°C. Input protein concentrations were quantified by Bradford assay (Thermo Fisher Scientific). Equivalent amounts of protein lysates were incubated with washed M2 beads overnight at 4°C with constant rotating. Beads were magnetically separated from supernatant and washed four times with FLAG buffer. Beads were then washed twice with FLAG buffer containing no IGEPAL CA-360. Bound proteins were eluted with FLAG elution buffer (FLAG buffer without IGEPAL CA-360 supplemented with 125 µg/ml 3xFLAG peptide [F4799; Sigma-Aldrich]). Equivalent volumes of eluates were boiled at 100°C for 5 min in 4× reducing Laemmli sample buffer and then centrifuged at maximum speed for 5 min at RT. β-Catenin immunoprecipitations were performed as described earlier with a monoclonal β-catenin antibody (sc-393501; Santa Cruz Biotechnology, Inc.) and protein A/G magnetic beads (Thermo Fisher Scientific). Western blots were performed as previously described (Schiffmacher et al., 2014). Primary antibodies used for immunoblotting were Cad6B (CC6DB-1, 1:80; Developmental Studies Hybridoma Bank), β-actin (1:1,000, sc-47778; Santa Cruz Biotechnology, Inc.), HA (1:1,000, 3F10; Roche; or 12CA5; Thermo Fisher Scientific), β-catenin (1:1,000, sc-53483, or 1:3,000, sc-393501; Santa Cruz Biotechnology, Inc.), p53 (1:1,000, sc-99; Santa Cruz Biotechnology, Inc.), and FLAG (1:2,000, M2, F1804; Sigma-Aldrich). Immunoblots were serially probed between antibody stripping with Restore Western Blot Stripping Buffer according to the manufacturer’s instructions (Thermo Fisher Scientific). Immunoblot images for figures were gamma-modified and processed using Photoshop 9.0 (Adobe Systems). Immunoblot band volumes (intensities) were calculated from unmodified immunoblot images using Image Lab software (Bio-Rad Laboratories), and relative protein levels were calculated by normalizing Cad6B CTF2 and β-catenin band volumes to β-actin band volumes. Differences in the amount of CTF2–β-catenin coimmunoprecipitation between mutant and wild-type CTF2 were assessed by comparing the ratios of normalized β-catenin band volumes to normalized CTF2 (FLAG) band volumes. Fold changes in mutant CTF2 coimmunoprecipitation amounts are presented as means and SEM of these ratios relative to the wild-type ratio set at 1. Immunoblots were analyzed by ANOVA using the PROC MIXED model in SAS statistical software (SAS Institute), and levels were deemed significantly different P < 0.05 using Tukey’s method.
CTF2 protein stability assays with Flp-In-Cad6B-CTF2 stable cell lines
Flp-In-CHO clonal cell lines expressing Cad6B CTF2 or MUT9 from a single genomic integration site were created according to modified manufacturer’s instructions (K6010-02, R75807; Thermo Fisher Scientific; Schiffmacher et al., 2014; Padmanabhan and Taneyhill, 2015). Cells were cultured in six-well plates in Ham’s F12 medium (10-080; Mediatech) supplemented with 10% FBS and 500 µg/ml hygromycin (Thermo Fisher Scientific). Confluent Flp-In-Cad6B-CTF2-HA CHO cells and Flp-In-Cad6B-MUT9-HA CHO cells were treated with 100 µg/ml cycloheximide (Thermo Fisher Scientific; dissolved in DMSO), 20 µM MG132 dissolved in DMSO (Cayman Chemicals), or both chemicals and incubated at 37°C for 0.5, 1, or 2 h. Cells were then lysed in lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, and 5% IGEPAL CA-630) supplemented with complete protease inhibitor cocktail (Roche) and 1 mM PMSF for 30 min at 4°C with periodic mixing. β-Catenin knockdown in Flp-In CHO cell lines was performed by transfecting 50 nM 27mer duplex DsiRNAs (Integrated DNA Technologies) with Lipofectamine 2000 for 48 h before CHX chase for 2 h. DsiRNA efficiencies were optimized using Cy3 Transfection Control DsiRNA (Integrated DNA Technologies), and nontargeting Universal Negative Control DsiRNAs (Integrated DNA Technologies) were used as negative controls for all DsiRNA experiments (sequences available in Fig. S2). Relative protein levels at each time point and across time were analyzed by ANOVA using the PROC MIXED model in SAS software. Degradation rates (t1/2, measured as time at which degradation results in 50% of total protein at t = 0) were calculated by linear regression using Excel software (Microsoft).
In ovo electroporation
Expression constructs were unilaterally electroporated into premigratory midbrain neural crest cells in developing 2–3ss chick embryos using a modified version of in ovo electroporation (Itasaki et al., 1999; Schiffmacher et al., 2014). Constructs were electroporated at 3.0 µg/µl unless otherwise stated. Reincubation times varied according to the experiment. Immunohistochemistry images are representative of a minimum of five embryos, with five to seven sections taken through the midbrain. To ensure sufficient electroporation efficiency, electroporated embryos were randomly selected for 5- or 8-h incubations. Ventral/dorsal electroporation was performed as previously described (Schiffmacher et al., 2014).
Neural crest cell explants
Neural crest cells were explanted in vitro as previously described (Coles et al., 2007; Padmanabhan and Taneyhill, 2015). Embryos were reincubated for 3 h postelectroporation before midbrain dorsal neural folds (containing premigratory neural crest cells) were dissected out in PB-1 standard medium and placed into chamber slides (Nunc Lab-tek II; Thermo Fisher Scientific) coated with poly-l-lysine (P5899; Sigma-Aldrich) and fibronectin (356008; Corning). Explants were incubated in serum-free DMEM (10-013-CV; Corning) supplemented with 10% FBS for 10 h at 37°C. Where indicated, explants were treated with 20 nM LMB (sc-358688; Santa Cruz Biotechnology, Inc.) for the remaining 2 h of the incubation.
Immunohistochemistry
Immunohistochemical detection of various proteins was performed in whole-mount embryos, transverse sections, or neural crest explants after 4% PFA fixation and cryostat-sectioning as in Schiffmacher et al. (2014). The following primary antibodies and concentrations were used: GFP (1:750, clone 3E6; Thermo Fisher Scientific), HA (1:1,000, 3F10; Roche; or 12CA5; Thermo Fisher Scientific), β-catenin (1:250, sc-393501; Santa Cruz Biotechnology, Inc.), Snail2 (1:100, C19G7; Cell Signaling Technology), and FLAG (1:1,000 M2, F1804; Sigma-Aldrich). Appropriate fluorescence-conjugated secondary antibodies (Alexa Fluor 488, 594, 647; Thermo Fisher Scientific) were used at a concentration of 1:500. Sections or explants were stained with DAPI to mark cell nuclei and mounted using Fluoromount G. Images from five to seven serial transverse sections through the midbrain of a minimum of three embryos, or at least six explants, were collected using an LSM 800 microscope (ZEISS) with a 63× oil objective (NA 1.4; ZEISS) at RT and analyzed with Zen Blue software (ZEISS). All exported files were processed (cropping, minor gamma adjustments) in Photoshop CS6, and a representative image was chosen for applicable figures. Subcellular fluorescent intensity was performed using the profile line intensity tool in Zen Blue software, and exported datasets were rendered into graphs using Excel software. Cell counts were evaluated by χ2 analysis using the PROC FREQ model in SAS software.
QPCR
Overexpression of wild-type CTF2, MUT9, and GFP followed by QPCR was performed as previously described (Taneyhill et al., 2007). In brief, 2–3ss embryos were electroporated in ovo with 3.0 µg/µl DNA. To evaluate Snail2 promoter activity caused by CTF2 overexpression, an additional 0.5 µg/µl D1.2-EGFP was coelectroporated with the other constructs described earlier or human β-catenin. The D1.2-EGFP construct contains an EGFP reporter driven by 1.2 kb of chicken Snail2 promoter upstream of the coding sequence (a gift from Y. Wakamatsu, Tohoku University, Sendai, Japan; Sakai et al., 2005). After 5 h at 37°C, midbrain neural folds were excised, pooled, and lysed in RNA lysis buffer, with total RNA isolated using the RNAqueous Total RNA Isolation kit (AM1912; Thermo Fisher Scientific). cDNAs were synthesized using random hexamers and the Superscript II RT-PCR system (Thermo Fisher Scientific) according to the manufacturer’s instructions. QPCR was performed using the ABI 7000 in TaqMan or SYBR Green (Thermo Fisher Scientific) assays as previously described (Taneyhill and Bronner-Fraser, 2005). In brief, 25-µl QPCR reactions for CyclinD1, Snail2, β-catenin, and GFP included 2× SYBR Green master mix, cDNA, and 75 nM of each primer. All reactions produced single amplicons that were verified for correct sequence. Cad6B QPCR reactions included TaqMan master mix, cDNA, 150 nM of each primer, and 150 nM of each Cad6B TaqMan probe, as described (Taneyhill et al., 2007). After normalization to a standard (chick 18S rRNA), fold up-regulation or down-regulation for each of three or more replicates was determined by dividing the relative expression value for the gene of interest in the presence of the elevated CTF2 or mutant CTF2 by that obtained for the gene of interest in the presence of GFP (negative control). To reduce the overall variance in Cad6B QPCR, unelectroporated contralateral excised neural folds collected from electroporated embryos were used to generate negative control cDNA, because Cad6B expression is highly variable among embryos at 6–7ss. To account for potential genomic DNA contamination amplification during reverse transcription, QPCR was also performed on identical sample cDNA synthesis reactions that lacked reverse transcription. Primers were also designed to span exon–exon junctions to reduce background amplification. Data were analyzed by analysis of variance (ANOVA) using the PROC MIXED model (SAS), and levels were deemed significantly different (P < 0.05) using the PDIFF procedure. Data were transformed to meet assumptions of normality and homogeneity of variance.
ChIP-QPCR assays
ChIP-QPCR was performed as described previously (Taneyhill et al., 2007; Taneyhill and Adams, 2008; Jhingory et al., 2010) to demonstrate association of 3xFLAG-tagged CTF2 with regions of the Snail2 promoter. In brief, embryos were electroporated with pCS2-CTF2-HA, pCS2-MUT9-HA, or pCS2-GFP-HA, and neural folds were dissected out from the midbrain region using tungsten needles 8 h postelectroporation. Tissue was fixed for 10 min in 4% PFA and pooled for use in lysate preparation and sonication to achieve a chromatin size of 200–1,000 bp. Chromatin was immunoprecipitated after overnight incubation at 4°C using M2 FLAG antibody-coated magnetic beads. Captured chromatin-3xFLAG–tagged protein complexes were eluted from the beads, followed by cross-link reversal, proteinase K treatment, and DNA precipitation. QPCR was then performed on the chromatin as described earlier using primer sets spanning 1.2 kb upstream of the Snail2 translational start site (Table S1). Data were transformed and analyzed as described in the QPCR methods. Putative TCF/LEF sites within the Snail2 promoter were computationally predicted using the JASPAR 2016 server.
Online supplemental material
Fig. S1 demonstrates that Cad6B CTF2 physically associates with β-catenin, but not p120-catenin, in vitro. Fig. S2 shows the sequence and efficacy of the β-catenin DsiRNAs used to deplete β-catenin from Flp-In parental, Cad6B-CTF2-HA, and Cad6B-MUT9-HA CHO cells. Fig. S3 reveals Cad6B CTF2 and β-catenin colocalization within the cytosol and nucleus of cranial neural crest cells after treatment with LMB. Fig. S4 illustrates β-catenin coimmunoprecipitation with CTF2-3xFLAG, but not GFP-3xFLAG, from cytosolic and nuclear protein fractions of transfected LMH cells. Table S1 outlines the primer sequences used in the Snail2 promoter ChIP-QPCR experiments.
Supplementary Material
Acknowledgments
We thank Ms. Karishma Sharma for technical assistance and Dr. Yoshio Wakamatsu for the D1.2-EGFP construct.
This work was supported by grants to L.A. Taneyhill (National Institutes of Health R01DE024217 and American Cancer Society Research Scholar Grant RSG-15-023-01-CSM) and A.T. Schiffmacher (National Institutes of Health F32DE022990).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ADAM
- a disintegrin and metalloproteinase
- ANOVA
- analysis of variance
- ChIP
- chromatin immunoprecipitation
- CHX
- cycloheximide
- CTF
- C-terminal fragment
- DsiRNA
- Dicer-substrate short interfering RNA
- EMT
- epithelial-to-mesenchymal transition
- LEF
- lymphoid enhancer-binding factor
- LMB
- leptomycin B
- NTF
- N-terminal fragment
- QPCR
- quantitative PCR
- ss
- somite stage
- TCF
- T-cell factor
References
- Abbruzzese G., Becker S.F., Kashef J., and Alfandari D.. 2016. ADAM13 cleavage of cadherin-11 promotes CNC migration independently of the homophilic binding site. Dev. Biol. 415:383–390. 10.1016/j.ydbio.2015.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acloque H., Adams M.S., Fishwick K., Bronner-Fraser M., and Nieto M.A.. 2009. Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J. Clin. Invest. 119:1438–1449. 10.1172/JCI38019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akitaya T., and Bronner-Fraser M.. 1992. Expression of cell adhesion molecules during initiation and cessation of neural crest cell migration. Dev. Dyn. 194:12–20. 10.1002/aja.1001940103 [DOI] [PubMed] [Google Scholar]
- Burstyn-Cohen T., Stanleigh J., Sela-Donenfeld D., and Kalcheim C.. 2004. Canonical Wnt activity regulates trunk neural crest delamination linking BMP/noggin signaling with G1/S transition. Development. 131:5327–5339. 10.1242/dev.01424 [DOI] [PubMed] [Google Scholar]
- Coles E.G., Taneyhill L.A., and Bronner-Fraser M.. 2007. A critical role for Cadherin6B in regulating avian neural crest emigration. Dev. Biol. 312:533–544. 10.1016/j.ydbio.2007.09.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David J.M., and Rajasekaran A.K.. 2012. Dishonorable discharge: The oncogenic roles of cleaved E-cadherin fragments. Cancer Res. 72:2917–2923. 10.1158/0008-5472.CAN-11-3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Barrio M.G., and Nieto M.A.. 2002. Overexpression of Snail family members highlights their ability to promote chick neural crest formation. Development. 129:1583–1593. [DOI] [PubMed] [Google Scholar]
- Duband J.L., and Thiery J.P.. 1982. Distribution of fibronectin in the early phase of avian cephalic neural crest cell migration. Dev. Biol. 93:308–323. 10.1016/0012-1606(82)90120-8 [DOI] [PubMed] [Google Scholar]
- Duband J.L., Volberg T., Sabanay I., Thiery J.P., and Geiger B.. 1988. Spatial and temporal distribution of the adherens-junction-associated adhesion molecule A-CAM during avian embryogenesis. Development. 103:325–344. [DOI] [PubMed] [Google Scholar]
- Duffy M.J., Mullooly M., O’Donovan N., Sukor S., Crown J., Pierce A., and McGowan P.M.. 2011. The ADAMs family of proteases: New biomarkers and therapeutic targets for cancer? Clin. Proteomics. 8:9 10.1186/1559-0275-8-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferber E.C., Kajita M., Wadlow A., Tobiansky L., Niessen C., Ariga H., Daniel J., and Fujita Y.. 2008. A role for the cleaved cytoplasmic domain of E-cadherin in the nucleus. J. Biol. Chem. 283:12691–12700. 10.1074/jbc.M708887200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco E.G., Lei H., Hsu J.L., and Reed R.. 2012. Small-scale nuclear extracts for functional assays of gene-expression machineries. J. Vis. Exp. 4140 10.3791/4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheldof A., and Berx G.. 2013. Cadherins and epithelial-to-mesenchymal transition. Prog. Mol. Biol. Transl. Sci. 116:317–336. 10.1016/B978-0-12-394311-8.00014-5 [DOI] [PubMed] [Google Scholar]
- Gottardi C.J., Wong E., and Gumbiner B.M.. 2001. E-cadherin suppresses cellular transformation by inhibiting beta-catenin signaling in an adhesion-independent manner. J. Cell Biol. 153:1049–1060. 10.1083/jcb.153.5.1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K., Takagi S., Fujisawa H., and Takeichi M.. 1987. Spatial and temporal expression pattern of N-cadherin cell adhesion molecules correlated with morphogenetic processes of chicken embryos. Dev. Biol. 120:215–227. 10.1016/0012-1606(87)90119-9 [DOI] [PubMed] [Google Scholar]
- Hobert O. 2002. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 32:728–730. [DOI] [PubMed] [Google Scholar]
- Hong S., Troyanovsky R.B., and Troyanovsky S.M.. 2013. Binding to F-actin guides cadherin cluster assembly, stability, and movement. J. Cell Biol. 201:131–143. 10.1083/jcb.201211054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Q.P., Kuang J.Y., Yang Q.K., Bian X.W., and Yu S.C.. 2016. Beyond a tumor suppressor: Soluble E-cadherin promotes the progression of cancer. Int. J. Cancer 138:2804–2812. 10.1002/ijc.29982 [DOI] [PubMed] [Google Scholar]
- Huber A.H., Stewart D.B., Laurents D.V., Nelson W.J., and Weis W.I.. 2001. The cadherin cytoplasmic domain is unstructured in the absence of beta-catenin. A possible mechanism for regulating cadherin turnover. J. Biol. Chem. 276:12301–12309. 10.1074/jbc.M010377200 [DOI] [PubMed] [Google Scholar]
- Itasaki N., Bel-Vialar S., and Krumlauf R.. 1999. ‘Shocking’ developments in chick embryology: Electroporation and in ovo gene expression. Nat. Cell Biol. 1:E203–E207. 10.1038/70231 [DOI] [PubMed] [Google Scholar]
- Jhingory S., Wu C.Y., and Taneyhill L.A.. 2010. Novel insight into the function and regulation of alphaN-catenin by Snail2 during chick neural crest cell migration. Dev. Biol. 344:896–910. 10.1016/j.ydbio.2010.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinke D.J. II, Horvath N., Cuppett V., Wu Y., Deng W., and Kanj R.. 2015. Interlocked positive and negative feedback network motifs regulate β-catenin activity in the adherens junction pathway. Mol. Biol. Cell. 26:4135–4148. 10.1091/mbc.E15-02-0083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S., Hasebe M., Tomita M., and Yanagawa H.. 2009. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc. Natl. Acad. Sci. USA. 106:10171–10176. 10.1073/pnas.0900604106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R.T., Nagai H., Nakaya Y., Sheng G., Trainor P.A., Weston J.A., and Thiery J.P.. 2013. Cell delamination in the mesencephalic neural fold and its implication for the origin of ectomesenchyme. Development. 140:4890–4902. 10.1242/dev.094680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud P., Shioi J., Serban G., Georgakopoulos A., Sarner S., Nagy V., Baki L., Wen P., Efthimiopoulos S., Shao Z., et al. . 2002. A presenilin-1/gamma-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 21:1948–1956. 10.1093/emboj/21.8.1948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marambaud P., Wen P.H., Dutt A., Shioi J., Takashima A., Siman R., and Robakis N.K.. 2003. A CBP binding transcriptional repressor produced by the PS1/epsilon-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell. 114:635–645. 10.1016/j.cell.2003.08.008 [DOI] [PubMed] [Google Scholar]
- Maretzky T., Reiss K., Ludwig A., Buchholz J., Scholz F., Proksch E., de Strooper B., Hartmann D., and Saftig P.. 2005. ADAM10 mediates E-cadherin shedding and regulates epithelial cell-cell adhesion, migration, and beta-catenin translocation. Proc. Natl. Acad. Sci. USA. 102:9182–9187. 10.1073/pnas.0500918102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker C.D., and Alfandari D.. 2009. Life after proteolysis: Exploring the signaling capabilities of classical cadherin cleavage fragments. Commun. Integr. Biol. 2:155–157. 10.4161/cib.7700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCusker C., Cousin H., Neuner R., and Alfandari D.. 2009. Extracellular cleavage of cadherin-11 by ADAM metalloproteases is essential for Xenopus cranial neural crest cell migration. Mol. Biol. Cell. 20:78–89. 10.1091/mbc.E08-05-0535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen A.E., Maher M.T., Mo R., and Gottardi C.J.. 2014. E-cadherin phosphorylation occurs during its biosynthesis to promote its cell surface stability and adhesion. Mol. Biol. Cell. 25:2365–2374. 10.1091/mbc.E14-01-0690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najy A.J., Day K.C., and Day M.L.. 2008. The ectodomain shedding of E-cadherin by ADAM15 supports ErbB receptor activation. J. Biol. Chem. 283:18393–18401. 10.1074/jbc.M801329200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S., and Takeichi M.. 1995. Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development. 121:1321–1332. [DOI] [PubMed] [Google Scholar]
- Nakagawa S., and Takeichi M.. 1998. Neural crest emigration from the neural tube depends on regulated cadherin expression. Development. 125:2963–2971. [DOI] [PubMed] [Google Scholar]
- Nelson W.J., Dickinson D.J., and Weis W.I.. 2013. Roles of cadherins and catenins in cell-cell adhesion and epithelial cell polarity. Prog. Mol. Biol. Transl. Sci. 116:3–23. 10.1016/B978-0-12-394311-8.00001-7 [DOI] [PubMed] [Google Scholar]
- Neuner R., Cousin H., McCusker C., Coyne M., and Alfandari D.. 2009. Xenopus ADAM19 is involved in neural, neural crest and muscle development. Mech. Dev. 126:240–255. 10.1016/j.mod.2008.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M.A., Sargent M.G., Wilkinson D.G., and Cooke J.. 1994. Control of cell behavior during vertebrate development by Slug, a zinc finger gene. Science. 264:835–839. 10.1126/science.7513443 [DOI] [PubMed] [Google Scholar]
- Orsulic S., Huber O., Aberle H., Arnold S., and Kemler R.. 1999. E-cadherin binding prevents beta-catenin nuclear localization and beta-catenin/LEF-1-mediated transactivation. J. Cell Sci. 112:1237–1245. [DOI] [PubMed] [Google Scholar]
- Ortiz A., Lee Y.C., Yu G., Liu H.C., Lin S.C., Bilen M.A., Cho H., Yu-Lee L.Y., and Lin S.H.. 2015. Angiomotin is a novel component of cadherin-11/β-catenin/p120 complex and is critical for cadherin-11-mediated cell migration. FASEB J. 29:1080–1091. 10.1096/fj.14-261594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan R., and Taneyhill L.A.. 2015. Cadherin-6B undergoes macropinocytosis and clathrin-mediated endocytosis during cranial neural crest cell EMT. J. Cell Sci. 128:1773–1786. 10.1242/jcs.164426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil P.U., D’Ambrosio J., Inge L.J., Mason R.W., and Rajasekaran A.K.. 2015. Carcinoma cells induce lumen filling and EMT in epithelial cells through soluble E-cadherin-mediated activation of EGFR. J. Cell Sci. 128:4366–4379. 10.1242/jcs.173518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss K., Maretzky T., Ludwig A., Tousseyn T., de Strooper B., Hartmann D., and Saftig P.. 2005. ADAM10 cleavage of N-cadherin and regulation of cell-cell adhesion and beta-catenin nuclear signalling. EMBO J. 24:742–752. 10.1038/sj.emboj.7600548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadot E., Simcha I., Shtutman M., Ben-Ze’ev A., and Geiger B.. 1998. Inhibition of beta-catenin-mediated transactivation by cadherin derivatives. Proc. Natl. Acad. Sci. USA. 95:15339–15344. 10.1073/pnas.95.26.15339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D., Tanaka Y., Endo Y., Osumi N., Okamoto H., and Wakamatsu Y.. 2005. Regulation of Slug transcription in embryonic ectoderm by beta-catenin-Lef/Tcf and BMP-Smad signaling. Dev. Growth Differ. 47:471–482. 10.1111/j.1440-169X.2005.00821.x [DOI] [PubMed] [Google Scholar]
- Schiffmacher A.T., Padmanabhan R., Jhingory S., and Taneyhill L.A.. 2014. Cadherin-6B is proteolytically processed during epithelial-to-mesenchymal transitions of the cranial neural crest. Mol. Biol. Cell. 25:41–54. 10.1091/mbc.E13-08-0459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoval I., Ludwig A., and Kalcheim C.. 2007. Antagonistic roles of full-length N-cadherin and its soluble BMP cleavage product in neural crest delamination. Development. 134:491–501. 10.1242/dev.02742 [DOI] [PubMed] [Google Scholar]
- Simcha I., Kirkpatrick C., Sadot E., Shtutman M., Polevoy G., Geiger B., Peifer M., and Ben-Ze’ev A.. 2001. Cadherin sequences that inhibit beta-catenin signaling: A study in yeast and mammalian cells. Mol. Biol. Cell. 12:1177–1188. 10.1091/mbc.12.4.1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhusen U., Weiske J., Badock V., Tauber R., Bommert K., and Huber O.. 2001. Cleavage and shedding of E-cadherin after induction of apoptosis. J. Biol. Chem. 276:4972–4980. 10.1074/jbc.M006102200 [DOI] [PubMed] [Google Scholar]
- Strobl-Mazzulla P.H., and Bronner M.E.. 2012. A PHD12-Snail2 repressive complex epigenetically mediates neural crest epithelial-to-mesenchymal transition. J. Cell Biol. 198:999–1010. 10.1083/jcb.201203098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill L.A., and Adams M.S.. 2008. Investigating regulatory factors and their DNA binding affinities through real time quantitative PCR (RT-QPCR) and chromatin immunoprecipitation (ChIP) assays. Methods Cell Biol. 87:367–389. 10.1016/S0091-679X(08)00219-7 [DOI] [PubMed] [Google Scholar]
- Taneyhill L.A., and Bronner-Fraser M.. 2005. Dynamic alterations in gene expression after Wnt-mediated induction of avian neural crest. Mol. Biol. Cell. 16:5283–5293. 10.1091/mbc.E05-03-0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill L.A., and Schiffmacher A.T.. 2013. Cadherin dynamics during neural crest cell ontogeny. Prog. Mol. Biol. Transl. Sci. 116:291–315. 10.1016/B978-0-12-394311-8.00013-3 [DOI] [PubMed] [Google Scholar]
- Taneyhill L.A., Coles E.G., and Bronner-Fraser M.. 2007. Snail2 directly represses cadherin6B during epithelial-to-mesenchymal transitions of the neural crest. Development. 134:1481–1490. 10.1242/dev.02834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura K., Kihara T., Kuzuya A., Okawa K., Nishimoto T., Bito H., Ninomiya H., Sugimoto H., Kinoshita A., and Shimohama S.. 2006a Activity-dependent regulation of beta-catenin via epsilon-cleavage of N-cadherin. Biochem. Biophys. Res. Commun. 345:951–958. 10.1016/j.bbrc.2006.04.157 [DOI] [PubMed] [Google Scholar]
- Uemura K., Kihara T., Kuzuya A., Okawa K., Nishimoto T., Ninomiya H., Sugimoto H., Kinoshita A., and Shimohama S.. 2006b Characterization of sequential N-cadherin cleavage by ADAM10 and PS1. Neurosci. Lett. 402:278–283. 10.1016/j.neulet.2006.04.018 [DOI] [PubMed] [Google Scholar]
- Valenta T., Hausmann G., and Basler K.. 2012. The many faces and functions of β-catenin. EMBO J. 31:2714–2736. 10.1038/emboj.2012.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallin J., Thuret R., Giacomello E., Faraldo M.M., Thiery J.P., and Broders F.. 2001. Cloning and characterization of three Xenopus slug promoters reveal direct regulation by Lef/beta-catenin signaling. J. Biol. Chem. 276:30350–30358. 10.1074/jbc.M103167200 [DOI] [PubMed] [Google Scholar]
- Wolfe M.S., and Kopan R.. 2004. Intramembrane proteolysis: Theme and variations. Science. 305:1119–1123. 10.1126/science.1096187 [DOI] [PubMed] [Google Scholar]
- Yap A.S., Crampton M.S., and Hardin J.. 2007. Making and breaking contacts: The cellular biology of cadherin regulation. Curr. Opin. Cell Biol. 19:508–514. 10.1016/j.ceb.2007.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.