Centrosomes play a key role in organizing the microtubule spindle that separates chromosomes during mitosis. Bennabi et al. review how microtubule spindle formation and chromosomal segregation also occur in oocytes during cell division by meiosis despite the absence of centrosomes.
Abstract
Oocytes accumulate maternal stores (proteins, mRNAs, metabolites, etc.) during their growth in the ovary to support development after fertilization. To preserve this cytoplasmic maternal inheritance, they accomplish the difficult task of partitioning their cytoplasm unequally while dividing their chromosomes equally. Added to this complexity, most oocytes, for reasons still speculative, lack the major microtubule organizing centers that most cells use to assemble and position their spindles, namely canonical centrosomes. In this review, we will address recent work on the mechanisms of meiotic spindle assembly and chromosome alignment/segregation in female gametes to try to understand the origin of errors of oocyte meiotic divisions. The challenge of oocyte divisions appears indeed not trivial because in both mice and humans oocyte meiotic divisions are prone to chromosome segregation errors, a leading cause of frequent miscarriages and congenital defects.
Introduction
Sexual reproduction relies on the fusion of paternal and maternal haploid gametes—the sperm and the extremely large oocyte, respectively—forming a new diploid organism. Meiotic divisions contribute solely to the formation of haploid gametes. They consist of two successive divisions, without intervening DNA replication, meiosis I and II, which reduce the genetic content by half. It has been known for over a decade that female meiosis is highly prone to chromosome segregation errors, especially in humans (Hassold and Hunt, 2001; Hassold et al., 2007; Nagaoka et al., 2012). At least 10% of human pregnancies produce aneuploid embryos (presenting a gain or loss of entire chromosomes), inducing spontaneous abortions and congenital defects such as trisomies, for which incidence increases with maternal age (Nagaoka et al., 2012). In eukaryotes, the structure orchestrating chromosome alignment and segregation during cell division is the microtubule spindle. In mitotic cells, the microtubules that compose the spindle are mostly nucleated from centrosomes acting as major microtubule organizing centers (MTOCs). Canonical centrosomes are composed of a pair of centrioles surrounded by pericentriolar material (PCM) that possesses microtubule nucleation activity. The microtubule slow-growing end (minus end) is tethered to the PCM of the centrosome, whereas the fast growing (plus end) is directed toward chromosomes. At mitosis entry, centrosomes separate on opposite sides of the nuclear envelope, defining the future spindle poles and allowing bipolar spindle formation. Whereas the majority of male gametes retain centrosomes containing centrioles, in oocytes of most metazoan species, centrioles are eliminated before meiotic divisions (Szollosi et al., 1972; Manandhar et al., 2005). Thus, spindle morphogenesis and positioning are atypical in these cells. The lack of centrosomes could favor the asymmetric partitioning of the cytoplasm by reducing the distance between the pole of the spindle that is anchored to the cortex and the cell cortex. Indeed, astral microtubules, a subpopulation of microtubules connecting the spindle pole to the cortex in most mitotic cells, are absent in most oocytes because of the lack of centrioles. However, as a result of the large size of oocytes, even when centrosomes are retained, oocytes can still divide extremely asymmetrically, as in starfish. In these oocytes, centriole-containing centrosomes participate in chromosome capture once chromosomes are close enough to be reached by microtubules. Chromosome gathering is, however, achieved by a contractile actin mesh that delivers chromosomes to the spindle (Lénárt et al., 2005). Interestingly, the lack of centrioles imposes atypical modes of spindle assembly in oocytes that we are going to review in this study.
Centrosome-independent microtubule nucleation
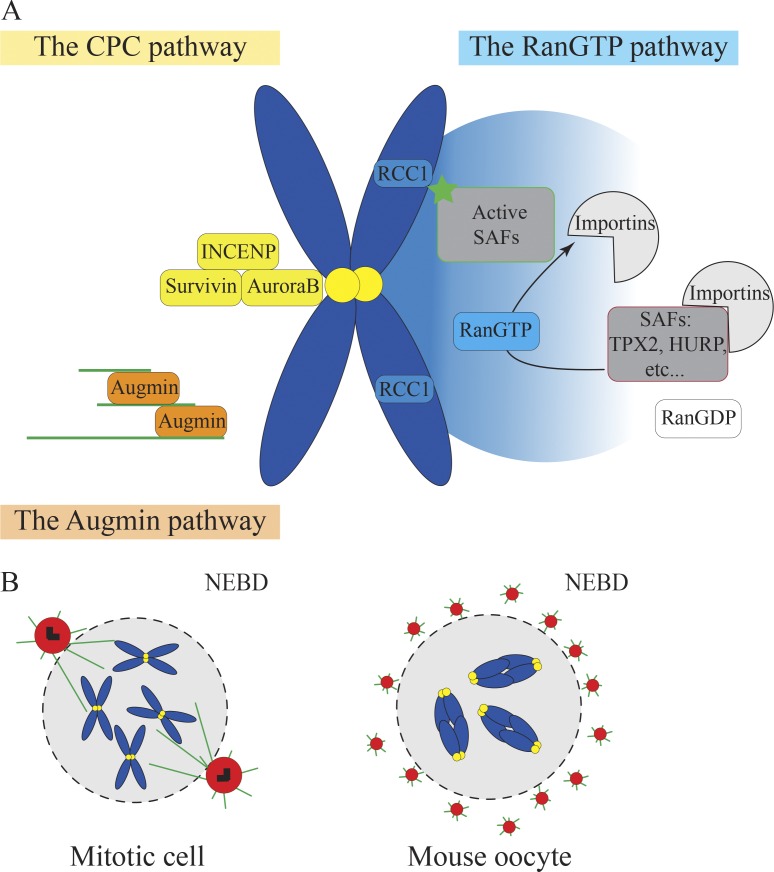
In mitosis, the spindle is formed by microtubules that are nucleated from canonical centrosomes. Although centrosome-mediated spindle formation is dominant in most mitotic cells, mitosis can still take place in the absence of centrosomes, showing that other centrosome-independent pathways can participate in spindle formation (Khodjakov et al., 2000; Basto et al., 2006; Azimzadeh et al., 2012; Bazzi and Anderson, 2014). These centrosome-independent pathways become dominant in cells lacking centrosomes such as oocytes. Indeed, because most oocytes lack canonical centrosomes, they use alternative pathways to nucleate microtubules (Fig. 1 A). Among them, the RanGTP pathway has been very well described (Fig. 1 A). The small Ran GTPase (Ras-like nuclear protein) is present in a gradient around chromosomes both in mitotic and meiotic cells. The RanGTP active form is produced by the Ran guanosine exchange factor regulator of chromosome condensation 1 (RCC1) that is localized on chromosomes (Kalab et al., 1999). This gradient locally activates spindle assembly factors (SAFs), such as, for example, targeting protein for Xklp2 (Tpx2), that participate in microtubule nucleation, interaction, and stabilization as well as motor activities (Meunier and Vernos, 2016). These SAFs interact with importins via their NLS and are kept inhibited. The RanGTP gradient is proposed to promote the dissociation of SAFs from their inhibitory binding to importins, causing their local activation and release (Gruss et al., 2001; Nachury et al., 2001). In human oocytes, RanGTP inhibition seems to delay microtubule nucleation and impair spindle formation (Holubcová et al., 2015). However, human oocytes used in this study were atretic (oocytes from patients receiving in vitro fertilization that did not spontaneously resume meiosis in response to hormonal treatment), and thus, they might not behave similarly to healthy human oocytes. Differently, inhibition of RanGTP delays but does not impair spindle assembly in mouse and Drosophila melanogaster oocytes (Dumont et al., 2007; Cesario and McKim, 2011). This suggests that although the RanGTP pathway is involved in microtubule nucleation for spindle assembly in the absence of centrosomes, other pathways seem important. Among these, the Augmin pathway (Fig. 1 A) generates new microtubules along preexisting microtubules (Sánchez-Huertas and Lüders, 2015). The Augmin complex is composed of eight proteins (named HAUS 1–8) able to recruit γ-tubulin to the sides of microtubules within the spindle (Goshima et al., 2008; Lawo et al., 2009; Uehara et al., 2009). In Xenopus laevis egg extracts, Augmin depletion results in reduced microtubule nucleation and multipolar spindle formation, suggesting a role of the Augmin complex in spindle bipolarization (Petry et al., 2011). In fruit flies, Augmin compensates for the lack of centrosomes by promoting microtubules nucleation at meiotic spindle poles (Colombié et al., 2013). Similarly, the chromosomal passenger complex (CPC) pathway (Fig. 1 A) is also involved in microtubule stabilization and spindle assembly in Xenopus egg extracts and Drosophila oocytes (Sampath et al., 2004; Kelly et al., 2007; Tseng et al., 2010; Radford et al., 2012; Das et al., 2016). The CPC is associated with kinetochores and is composed of the Aurora B/C kinase, the inner centromeric protein (INCENP), Survivin, and Borealin (Dumont and Desai, 2012). In Caenorhabditis elegans oocytes, Katanin increases the density of small microtubules by severing preexisting ones and could thus contribute to microtubule formation by amplifying microtubule nucleation via other pathways (Srayko et al., 2006).
Figure 1.
Pathways replacing centrosomes for microtubule nucleation in oocytes. (A) The three microtubule nucleation pathways: the RanGTP pathway, CPC pathway, and Augmin pathway. (B) Microtubule nucleation by centrosomes in mitotic cells (left) and by multiple aMTOCs in mouse oocytes (right). For A and B, DNA is in blue, microtubules in green, kinetochores in yellow, pericentriolar material in red, and centrioles in black.
In addition to these microtubule nucleation pathways, mouse oocytes contain acentriolar MTOCs (aMTOCs) capable of nucleating microtubules (Fig. 1 B). At nuclear envelope breakdown (NEBD), the nucleation capacity of these aMTOCs is low, but it increases throughout meiosis I. Indeed, levels of the RanGTPase effector TPX2 (Wittmann et al., 2000) rise progressively during meiosis I (Brunet et al., 2008), which intensifies the extent of phosphorylation of the aMTOC protein transforming acidic coiled coil 3 (TACC) and increases microtubule nucleation activity at aMTOCs (Still et al., 1999; Bayliss et al., 2003; Eyers et al., 2003; Tsai et al., 2003; Kinoshita et al., 2005; Brunet et al., 2008). These aMTOCs are perinuclear before meiotic divisions (Fig. 1 B) so that they can be readily distributed around the chromatin when NEBD occurs (Łuksza et al., 2013). Although the exact composition of these structures is not exhaustively known, they contain classical PCMs such as γ-tubulin and pericentrin and are likely bona fide PCMs (Gueth-Hallonet et al., 1993; Carabatsos et al., 2000). In mitotic cells, the PCM size is regulated by centrioles such that microtubule nucleation is carefully regulated (Kirkham et al., 2003; Conduit and Raff, 2010; Gopalakrishnan et al., 2012; Woodruff et al., 2015). In mouse oocytes, the size of the PCM seems to scale with the cell volume, but the regulatory mechanisms at play are unknown (Łuksza et al., 2013). Surprisingly, such acentriolar MTOCs are not detected on the nuclear envelope in prophase I or at later stages in spindle poles from Xenopus, C. elegans, Drosophila, and human atretic oocytes (Gard, 1991; Matthies et al., 1996; Srayko et al., 2006; Holubcová et al., 2015). Although all of these microtubule nucleation pathways are essential for spindle assembly in the absence of nucleation by centrosomes, little is known about their relative contribution in oocytes and how they interact.
Spindle bipolarization
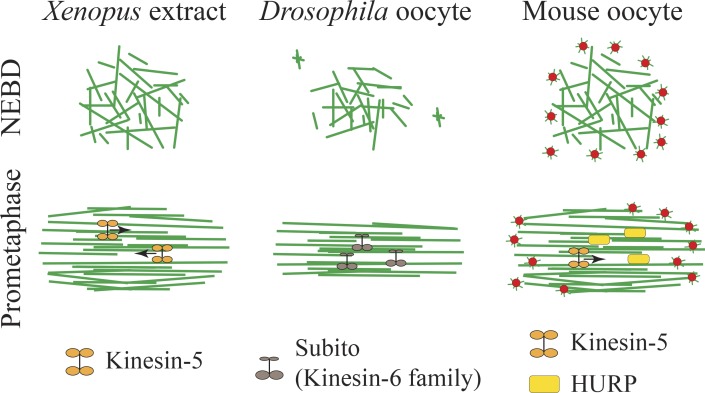
Once microtubules are formed, the spindle must assemble in a bipolar fashion to accurately segregate chromosomes in two distinct groups. In mitotic cells, centrosomes are duplicated during interphase of the cell cycle, and cells enter mitosis with two centrosomes. At the onset of mitosis, the two centrosomes separate and nucleate microtubules (Fig. 1 B). Duplicated centrosomes thus form the spindle axis and promote rapid spindle bipolarization (Toso et al., 2009; Tanenbaum and Medema, 2010). In oocytes, spindle bipolarization does not rely on a bipolar axis predefined by the two separated centrosomes. Instead, spindle bipolarization is a sequential and slow process. It can take up to 12 min in C. elegans, 4 h in mouse, and 6.5 h in human atretic oocytes, which corresponds to around half of the transition time from NEBD to anaphase in these species (Dumont et al., 2007; Schuh and Ellenberg, 2007; Holubcová et al., 2015; Sumiyoshi et al., 2015) and 40 min in Drosophila oocytes (Sköld et al., 2005). In the absence of centrosomes, the establishment of a bipolar spindle depends on the sorting and stabilization of microtubules into a central array via microtubule motors and microtubule-associated proteins (Heald et al., 1996; Walczak et al., 1998). A crucial step in this process is the transformation of an unorganized ball of microtubules into a bipolar array presenting antiparallel microtubules in opposite orientations. This is achieved via the sorting and bundling of microtubules by plus end–directed microtubule motors (Fig. 2). Among them, Kinesin-5 (Eg5) was shown to be essential for the establishment and maintenance of spindle bipolarity in Xenopus extracts and mouse oocytes (Fig. 2) because its inhibition results in monopolar spindles (Walczak et al., 1998; Kapoor et al., 2000; Mailhes et al., 2004; Schuh and Ellenberg, 2007; Fitzharris, 2009). In Drosophila, the Kinesin-6 family member Subito facilitates spindle bipolarization (Fig. 2) by promoting the formation of a central microtubule array (Jang et al., 2005, 2007). In particular, CPC central spindle proteins such as INCENP and Aurora B fail to localize to this central region in subito mutants. In mice, where oocytes assemble a meiotic spindle in the presence of multiple aMTOCs, these aMTOCs have to be properly organized to ensure correct spindle bipolarization. Before NEBD, aMTOCs are decondensed by Polo-like kinase 1 (PLK1); upon NEBD, they are spread along the nuclear envelope by a microtubule- and dynein-dependent mechanism; and after NEBD, aMTOCs are fragmented in smaller structures by Kinesin-5 (Łuksza et al., 2013; Clift and Schuh, 2015). This fragmentation process is essential for bipolar spindle formation, as a failure to fragment aMTOCs induces defects in bipolar spindle assembly and chromosome alignment. Next, concomitant to the formation of a central microtubule array, aMTOCs are progressively sorted along the central spindle into distinct poles between NEBD and 4 h after (Schuh and Ellenberg, 2007; Breuer et al., 2010). A key player in this process is the microtubule-associated protein and RanGTPase factor hepatoma up-regulated protein (HURP), which has a role very comparable to the one of Subito in Drosophila (Tsou et al., 2003). HURP is recruited by Kinesin-5 to the central spindle (Fig. 2) and permits aMTOCs sorting by facilitating microtubule stability in this region (Breuer et al., 2010). The stabilization of microtubules in the region of overlap of antiparallel microtubules provides tracks on which motors can bind aMTOCs as their cargo and transport them to spindle poles.
Figure 2.
Spindle bipolarization. Organization of microtubules into a bipolar array via microtubule motors and microtubule-associated proteins in Xenopus egg extracts, Drosophila, and mouse oocytes between NEBD and prometaphase. Microtubules are in green and aMTOCs in red.
Interestingly, in human atretic oocytes in which spindle bipolarization is extremely slow, most spindles fail to maintain a bipolar shape but instead go through phases of multipolarity (Holubcová et al., 2015). Such unstable spindles are rarely observed in mitotic spindles or meiotic spindles from other species, except in oocytes from the hurp−/− strain (Breuer et al., 2010), thus raising the question of the nature of the regulatory mechanisms at play in this type of human oocyte favoring this instability.
Spindle pole formation
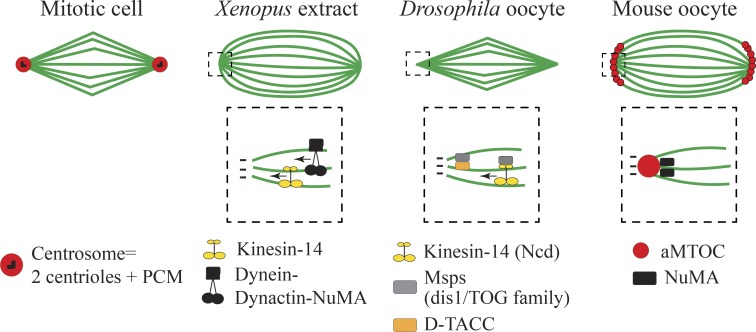
Spindle poles in mitosis are organized by a single centrosome (Fig. 3). Pole formation in oocytes is differentbecause it is not organized by a single entity. The formation of spindle poles, which is the region where microtubule minus ends are converging, relies on the activity of microtubule motors and microtubule-associated proteins (Fig. 3). Drosophila excepted, most oocytes present spindle poles that are less focused than in mitosis, having this typical barrel-shape aspect. Studies in Xenopus egg extracts have shown that Dynein and Kinesin-14 minus-end motors (Fig. 3) shape the poles by focusing microtubule minus ends in these regions (Heald et al., 1996; Walczak et al., 1998). In Drosophila oocytes, nonclaret disjunctional (Ncd; Kinesin-14) prevents pole splitting and multipolar spindle formation (Fig. 3; Endow and Komma, 1997; Sköld et al., 2005). Furthermore, Dynein in a complex with Dynactin and nuclear mitotic apparatus (NuMa) are essential to cross-link parallel microtubules (in the same orientation) and thus tether together microtubule minus ends at meiotic spindle poles in Xenopus egg extracts (Fig. 3; Merdes et al., 1996). Acentrosomal poles in Drosophila oocytes contain the microtubule-associated protein mini-spindles (Msps), which is a member of the defect in sister chromatid disjoining 1/tumor overexpression gene (dis1/TOG) family conserved in C. elegans, Xenopus, and humans. Msps is recruited to spindle poles by Kinesin-14 (Ncd) and D-TACC (Fig. 3), where it prevents loss of bipolarity possibly by stabilization of microtubules ends (Cullen and Ohkura, 2001). The C. elegans homologue ZYG-9 is enriched at spindle poles and required for spindle assembly (Matthews et al., 1998). Remarkably, the function of NuMa in tethering microtubule minus ends is conserved in acentriolar spindles. Indeed, the microtubule-associated protein NuMa accumulates at the poles in rabbit, human, and mouse oocytes (Yan et al., 2006; Alvarez Sedó et al., 2011; Kolano et al., 2012). In mouse oocytes, NuMa is required for the formation of barrel-shaped spindle poles as well as microtubule minus-end cohesion (Fig. 3) because its impairment causes hyperfocused poles that often lose microtubule connection (Kolano et al., 2012).
Figure 3.
Spindle pole formation and final spindle shape. Top rows show spindle shape in metaphase in mitotic cells, Xenopus egg extracts, Drosophila, and mouse oocytes. The dashed square shows magnification of the spindle pole where microtubule motors and microtubule-associated proteins organize microtubule minus ends. Microtubules are in green.
In mouse oocytes, the discrete aMTOCs organize spindle poles (Fig. 3). After their bipolar sorting, aMTOCs progressively cluster together between 4 and 7 h after NEBD and will contribute to the cohesion and integrity of spindle poles (Kolano et al., 2012). Even though not addressed so far, if the sorting of aMTOCs fails to be optimal, the number of aMTOCs at each pole might not be identical and could thus favor force imbalance within the spindle compared with mitotic spindles where the poles are formed by equivalent centrosomes. This would resemble the process of clustering of extra-centrosomes in cancer cells in which unbalanced poles favor chromosome missegregation (Kwon et al., 2008; Breuer et al., 2010). In C. elegans, Drosophila, Xenopus, and humans, microtubule minus ends do not seem to be anchored to discrete aMTOC entities (Fig. 3). Although they are not anchored to detectable structures, their poles are shaped by a combination of factors as described above (in this section). In addition, most meiotic spindle poles, with the exception of Drosophila, have a broad shape compared with the more focused mitotic spindle poles, which could be related to the lack of tight organizers, the centrosomes (Fig. 3). Thus, meiotic spindle poles could possibly be less robust than the mitotic ones that are anchored to distinct centrosomes.
Chromosome alignment
After a bipolar spindle is formed, chromosomes align in the spindle equator. In mitosis, the “search and capture” model states that microtubules growing toward the chromosomes are rapidly captured and stabilized by the kinetochores, establishing stable kinetochore–microtubule attachments (Kirschner and Mitchison, 1986; Wollman et al., 2005). In oocytes, chromosome alignment is a much slower and progressive process that depends on the interaction of microtubules with chromosome arms and kinetochores. The interaction of chromosome arms with microtubules and microtubule motors, which also exist in the short prometaphase of mitotic cells, are thought to generate forces pushing chromosomes toward the spindle equator (Mazumdar and Misteli, 2005; Cheeseman and Desai, 2008; Cai et al., 2009; Wandke et al., 2012). In C. elegans, the kinesin-like protein KPL-19 localizes to a nonkinetochore chromatin region where microtubules contact chromosomes and could promote the motion of chromosomes toward the equator (Wignall and Villeneuve, 2009).
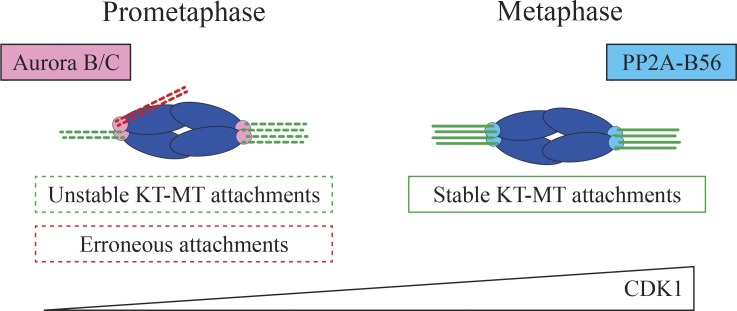
An EM study has suggested that mouse oocytes establish extremely delayed kinetochore–microtubule attachments (K-fibers), 1 to 2 h before anaphase (Brunet et al., 1999). However, even though stable K-fibers appear to be formed late in mouse oocytes, this does not exclude the possibility that microtubules could establish earlier contacts with kinetochores. Indeed, kinetochore–microtubule attachments are observed after calcium or cold treatment 3 to 4 h before anaphase (Lane et al., 2012). Yet K-fiber stability varies until late metaphase I (Fig. 4). A study with high-resolution live microscopy revealed that almost all kinetochores undergo multiple steps of error correction before engaging into stable bipolar attachments (Kitajima et al., 2011). Thus, K-fibers may not have been well preserved during EM fixation procedures and failed to be detected at earlier stages (Brunet et al., 1999). It may be interesting to reanalyze in more detail the timing of apparition of K-fibers by EM. The delay in K-fiber formation depends on cyclin-dependent kinase 1 (CDK1) activity (Fig. 4), which increases very gradually throughout meiosis I (Davydenko et al., 2013). A precocious increase in CDK1 activity leads to premature stable kinetochore–microtubule attachments and lagging chromosomes at anaphase. Aurora B/C phosphorylation activity destabilizes the attachments, whereas protein phosphatase 2A-B56 (PP2A-B56), recruited at kinetochores by an increase in CDK1 activity, stabilizes kinetochore–microtubule attachments (Fig. 4; Yoshida et al., 2015). Using a genetic approach, it has been shown in mouse oocytes that Aurora C corrects erroneous kinetochore attachments (Balboula and Schindler, 2014). In addition, kinetochore microtubule stability is regulated by their position within the spindle, as they can undergo Aurora A–dependent destabilization near spindle poles (Chmátal et al., 2015). It is thought that a delay in K-fiber formation would prevent the stabilization of erroneous attachments before bipolar spindle formation, a very slow and unsteady process in meiosis I.
Figure 4.
Establishment of stable kinetochore–microtubule attachments in mouse oocytes. Mouse oocytes form stable kinetochore–microtubule (KT-MT) attachments only at late metaphase I. Aurora B/C phosphorylation destabilizes kinetochore–microtubule attachments, whereas PP2A-B56 dephosphorylation activity stabilizes the attachments. This process is regulated by a progressive increase in CDK1 activity. DNA is in blue.
A recent study has shown that stable K-fibers formation is also slow in Drosophila oocytes (Głuszek et al., 2015) but depends on an alternative mechanism. The catastrophe-promoting complex Sentin–EB1 (end-binding protein 1) is responsible for delaying stable K-fiber attachments by regulating microtubule end dynamics. Mutant oocytes for sentin present more stable K-fibers early on in meiosis I, which is deleterious for bivalent segregation. Thus, one could speculate that slow K-fiber formation might be beneficial in the context of spindles organized from multiple aMTOCs or from chromosomes that might produce more merotelic attachments than spindles organized from centrosomes (when one kinetochore is attached to the two spindle poles).
Chromosome segregation
Once chromosomes are aligned on the spindle equator, pulling by K-fibers drives chromosome separation. In mitotic cells, chromosome separation is driven first by shortening of the kinetochore–microtubule attachments (anaphase A) and then by spindle elongation (anaphase B). In mouse oocytes, the opposite happens: first, the spindle elongates by a Kinesin-5–dependent mechanism, and then kinetochore–microtubule attachments shorten (FitzHarris, 2012). Interestingly, in nematodes, K-fibers align chromosomes but are not required for chromosome separation at anaphase (Dumont et al., 2010). Instead, it is proposed that microtubule assembly between chromosomes promotes their separation. This is consistent with the fact that spindle poles almost completely disappear at anaphase in this species. In addition, C. elegans chromosomes are holocentric presenting kinetochores ensheathing the entire chromosome length (Oegema et al., 2001). Although the presence of holocentric chromosomes could favor microtubule nucleation between chromosomes at anaphase, it could also promote the formation of merotelic attachments. Whether this kinetochore-independent separation mechanism is conserved in mammalian oocytes is still unknown, even though spindles lacking K-fibers are still able to undergo anaphase in mouse oocytes (Deng et al., 2009).
In mitosis, sister kinetochores are attached to opposite poles before segregation (bi-oriented), and cohesins (protein complexes holding the sister chromatid together) are cleaved at anaphase, leading to separation. In meiosis, sister kinetochores are attached to the same pole (mono-orientation), whereas homologous chromosomes are attached to opposite poles (Watanabe, 2012). At anaphase I, the meiotic-specific cohesin Rec8 is protected from cleavage at centromeres, permitting the separation of homologous chromosomes but not the separation of sister chromatids. Loss of cohesion is a leading cause of age-associated chromosome segregation errors (Chiang et al., 2010; Lister et al., 2010). The recently discovered kinetochore factor meiotic kinetochore factor (MEIKIN) is conserved from yeast to humans and required for both mono-orientation and cohesion protection (Kim et al., 2015). This suggests that MEIKIN could be a novel candidate implicated in age-associated chromosome segregation errors.
Why lose centrioles?
Lack of centrioles in oocytes imposes atypical modes of spindle formation that might contribute to the inherent high rate of chromosome segregation errors observed in meiosis. A puzzling observation is that whereas centrioles and PCMs are lost in oocytes of most metazoan species, mouse oocytes still retain multiple discrete PCMs or aMTOCs that can participate in bipolar spindle formation. In contrast to most species, sperm centrioles degenerate in rodents during spermatogenesis and thus are not contributed by the sperm at fertilization (Woolley and Fawcett, 1973; Manandhar et al., 1998). Instead, centrioles progressively assemble de novo in early embryos (Gueth-Hallonet et al., 1993). How centrioles are generated in rodent early embryos is not known. Nevertheless, these discrete PCMs could serve as templates for a later generation of centriole-containing centrosomes in the embryo.
Whether they possess discrete PCM foci at their poles or not, oocyte meiotic spindles appear to be fragile with steps of assembly that are slow and even unstable, as in humans. In addition, their shapes are often peculiar. Female meiotic spindles of many species are small and do not closely scale to cell size unlike mitotic spindles (Crowder et al., 2015). In mouse after fertilization and until centrioles assemble de novo at the 64-cell stage (Gueth-Hallonet et al., 1993), the spindle transitions from a meiotic shape to a mitotic one: the aMTOCs number sequentially decreases, poles become more focused, and the length of the spindle scales with the size of the cell (Courtois et al., 2012). This raises the question of the contribution of the centrosome in spindle size scaling. Furthermore, the large size of oocytes could dilute some components required for spindle morphogenesis and thus contribute to the fact that spindle size does not strictly correlate with cell size.
Still, very little is known about why and how centrioles are eliminated in oocytes of most species. One hypothesis is that centriole elimination prevents multipolar spindle formation in the first embryonic division after introduction of the sperm centrioles upon fertilization. However, in rodents, the sperm does not contribute with a centriole. Another hypothesis would be that it prevents parthenogenesis (egg activation in the absence of fertilization) because injection of centrosomes in Xenopus eggs induces activation without fertilization (Tournier et al., 1989). Recent studies have started to unravel how centrioles are removed in oocytes. In starfish, meiotic divisions take place in the presence of centriole-containing centrosomes. Mother centrioles are eliminated by extrusion into polar bodies, and the remaining daughter centriole is degraded in the cytoplasm (Borrego-Pinto et al., 2016). In the fruit fly, centriole elimination is a progressive process that ends up just before meiotic spindle assembly. It is dependent on PLK1 because its loss triggers PCM down-regulation, which leads to centriole removal. Centriole maintenance by perturbing this process results in spindle assembly defects in oocytes and early embryos and thus to female sterility (Pimenta-Marques et al., 2016). The absence of canonical centrosomes constitutes one of the many factors that could contribute to the innate susceptibility of oocyte to produce errors in chromosome segregation. However, despite its contribution to oocyte aneuploidy, centriole elimination must likely be crucial for gamete fitness of most metazoan species.
Conclusion
Recent advances have shed light on the mechanisms of spindle assembly in both mitosis and meiosis. It appears that oocytes use the same nucleation pathways as mitotic cells, namely the RanGTP, Augmin, and CPC pathways, with the exception that they are dominant in this study, in the absence of a centrosome pathway. Although they share common pathways, meiotic spindles are not just mitotic spindles without centrosomes, and these pathways are likely regulated in a meiosis-specific manner. Yet one can speculate that in the absence of centrosomes, the initial conditions might be key parameters influencing the entire process of spindle assembly with consequences on chromosome segregation. Oocytes have to face circumstances in which the critical mass of microtubules to capture and gather chromosomes could be limiting early on when they are polymerized only locally around chromosomes. This effect could be amplified by the fact that oocytes present huge nuclei (30 µm wide in the mouse and up to 450 µm in Xenopus), such that the volume at which the spindle starts assembling is gigantic compared with one of mitotic cells. It might be so that the critical concentration for tubulin to polymerize might be much more difficult to reach than in somatic cells when the nucleus breaks down, reinforcing the importance of pathways acting as catalyzers/amplifiers of tubulin polymerization locally around chromosomes. How these pathways, and yet-to-be-discovered ones, interact to promote early stages of spindle assembly has not been thoroughly addressed and remains an important question for future studies.
Acknowledgments
This work was supported by grants from the Agence Nationale de la Recherche (ANR-14-CE11-0002 to M.-H. Verlhac) and from the Fondation pour la Recherche Médicale (Team FRM 2015 to M.-H. Verlhac). This work has received support under the program Investissements d'Avenir launched by the French government and implemented by the Agence Nationale de la Recherche, with references ANR-10-LABX-54 MEMO LIFE and ANR-11-IDEX-0001-02 PSL* Research University.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- aMTOC
- acentriolar microtubule organizing center
- CDK1
- cyclin-dependent kinase 1
- CPC
- chromosomal passenger complex
- MTOC
- microtubule organizing center
- NEBD
- nuclear envelope breakdown
- NuMa
- nuclear mitotic apparatus
- PCM
- pericentriolar material
- SAF
- spindle assembly factor
- TACC
- transforming acidic coiled coil
References
- Alvarez Sedó C., Schatten H., Combelles C.M., and Rawe V.Y.. 2011. The nuclear mitotic apparatus (NuMA) protein: Localization and dynamics in human oocytes, fertilization and early embryos. Mol. Hum. Reprod. 17:392–398. 10.1093/molehr/gar009 [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Wong M.L., Downhour D.M., Sánchez Alvarado A., and Marshall W.F.. 2012. Centrosome loss in the evolution of planarians. Science. 335:461–463. 10.1126/science.1214457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balboula A.Z., and Schindler K.. 2014. Selective disruption of aurora C kinase reveals distinct functions from aurora B kinase during meiosis in mouse oocytes. PLoS Genet. 10:e1004194 10.1371/journal.pgen.1004194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., and Raff J.W.. 2006. Flies without centrioles. Cell. 125:1375–1386. 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Bayliss R., Sardon T., Vernos I., and Conti E.. 2003. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol. Cell. 12:851–862. 10.1016/S1097-2765(03)00392-7 [DOI] [PubMed] [Google Scholar]
- Bazzi H., and Anderson K.V.. 2014. Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc. Natl. Acad. Sci. USA. 111:E1491–E1500. 10.1073/pnas.1400568111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrego-Pinto J., Somogyi K., Karreman M.A., König J., Müller-Reichert T., Bettencourt-Dias M., Gönczy P., Schwab Y., and Lénárt P.. 2016. Distinct mechanisms eliminate mother and daughter centrioles in meiosis of starfish oocytes. J. Cell Biol. 212:815–827. 10.1083/jcb.201510083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breuer M., Kolano A., Kwon M., Li C.-C., Tsai T.-F., Pellman D., Brunet S., and Verlhac M.-H.. 2010. HURP permits MTOC sorting for robust meiotic spindle bipolarity, similar to extra centrosome clustering in cancer cells. J. Cell Biol. 191:1251–1260. 10.1083/jcb.201005065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S., Maria A.S., Guillaud P., Dujardin D., Kubiak J.Z., and Maro B.. 1999. Kinetochore fibers are not involved in the formation of the first meiotic spindle in mouse oocytes, but control the exit from the first meiotic M phase. J. Cell Biol. 146:1–12. 10.1083/jcb.146.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet S., Dumont J., Lee K.W., Kinoshita K., Hikal P., Gruss O.J., Maro B., and Verlhac M.-H.. 2008. Meiotic regulation of TPX2 protein levels governs cell cycle progression in mouse oocytes. PLoS One. 3:e3338 10.1371/journal.pone.0003338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., O’Connell C.B., Khodjakov A., and Walczak C.E.. 2009. Chromosome congression in the absence of kinetochore fibres. Nat. Cell Biol. 11:832–838. 10.1038/ncb1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabatsos M.J., Combelles C.M., Messinger S.M., and Albertini D.F.. 2000. Sorting and reorganization of centrosomes during oocyte maturation in the mouse. Microsc. Res. Tech. 49:435–444. [DOI] [PubMed] [Google Scholar]
- Cesario J., and McKim K.S.. 2011. RanGTP is required for meiotic spindle organization and the initiation of embryonic development in Drosophila. J. Cell Sci. 124:3797–3810. 10.1242/jcs.084855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman I.M., and Desai A.. 2008. Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9:33–46. 10.1038/nrm2310 [DOI] [PubMed] [Google Scholar]
- Chiang T., Duncan F.E., Schindler K., Schultz R.M., and Lampson M.A.. 2010. Evidence that weakened centromere cohesion is a leading cause of age-related aneuploidy in oocytes. Curr. Biol. 20:1522–1528. 10.1016/j.cub.2010.06.069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmátal L., Yang K., Schultz R.M., and Lampson M.A.. 2015. Spatial regulation of kinetochore microtubule attachments by destabilization at spindle poles in meiosis I. Curr. Biol. 25:1835–1841. 10.1016/j.cub.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clift D., and Schuh M.. 2015. A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat. Commun. 6:7217 10.1038/ncomms8217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombié N., Głuszek A.A., Meireles A.M., and Ohkura H.. 2013. Meiosis-specific stable binding of augmin to acentrosomal spindle poles promotes biased microtubule assembly in oocytes. PLoS Genet. 9:e1003562 10.1371/journal.pgen.1003562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conduit P.T., and Raff J.W.. 2010. Cnn dynamics drive centrosome size asymmetry to ensure daughter centriole retention in Drosophila neuroblasts. Curr. Biol. 20:2187–2192. 10.1016/j.cub.2010.11.055 [DOI] [PubMed] [Google Scholar]
- Courtois A., Schuh M., Ellenberg J., and Hiiragi T.. 2012. The transition from meiotic to mitotic spindle assembly is gradual during early mammalian development. J. Cell Biol. 198:357–370. 10.1083/jcb.201202135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder M.E., Strzelecka M., Wilbur J.D., Good M.C., von Dassow G., and Heald R.. 2015. A comparative analysis of spindle morphometrics across metazoans. Curr. Biol. 25:1542–1550. 10.1016/j.cub.2015.04.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen C.F., and Ohkura H.. 2001. Msps protein is localized to acentrosomal poles to ensure bipolarity of Drosophila meiotic spindles. Nat. Cell Biol. 3:637–642. 10.1038/35083025 [DOI] [PubMed] [Google Scholar]
- Das A., Shah S.J., Fan B., Paik D., DiSanto D.J., Hinman A.M., Cesario J.M., Battaglia R.A., Demos N., and McKim K.S.. 2016. Spindle assembly and chromosome segregation requires central spindle proteins in Drosophila oocytes. Genetics. 202:61–75. 10.1534/genetics.115.181081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydenko O., Schultz R.M., and Lampson M.A.. 2013. Increased CDK1 activity determines the timing of kinetochore-microtubule attachments in meiosis I. J. Cell Biol. 202:221–229. 10.1083/jcb.201303019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng M., Gao J., Suraneni P., and Li R.. 2009. Kinetochore-independent chromosome poleward movement during anaphase of meiosis II in mouse eggs. PLoS One. 4:e5249 10.1371/journal.pone.0005249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., and Desai A.. 2012. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 22:241–249. 10.1016/j.tcb.2012.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., Petri S., Pellegrin F., Terret M.-E., Bohnsack M.T., Rassinier P., Georget V., Kalab P., Gruss O.J., and Verlhac M.-H.. 2007. A centriole- and RanGTP-independent spindle assembly pathway in meiosis I of vertebrate oocytes. J. Cell Biol. 176:295–305. 10.1083/jcb.200605199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J., Oegema K., and Desai A.. 2010. A kinetochore-independent mechanism drives anaphase chromosome separation during acentrosomal meiosis. Nat. Cell Biol. 12:894–901. 10.1038/ncb2093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S.A., and Komma D.J.. 1997. Spindle dynamics during meiosis in Drosophila oocytes. J. Cell Biol. 137:1321–1336. 10.1083/jcb.137.6.1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyers P.A., Erikson E., Chen L.G., and Maller J.L.. 2003. A novel mechanism for activation of the protein kinase Aurora A. Curr. Biol. 13:691–697. 10.1016/S0960-9822(03)00166-0 [DOI] [PubMed] [Google Scholar]
- Fitzharris G. 2009. A shift from kinesin 5-dependent metaphase spindle function during preimplantation development in mouse. Development. 136:2111–2119. 10.1242/dev.035089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzHarris G. 2012. Anaphase B precedes anaphase A in the mouse egg. Curr. Biol. 22:437–444. 10.1016/j.cub.2012.01.041 [DOI] [PubMed] [Google Scholar]
- Gard D.L. 1991. Organization, nucleation, and acetylation of microtubules in Xenopus laevis oocytes: A study by confocal immunofluorescence microscopy. Dev. Biol. 143:346–362. 10.1016/0012-1606(91)90085-H [DOI] [PubMed] [Google Scholar]
- Głuszek A.A., Cullen C.F., Li W., Battaglia R.A., Radford S.J., Costa M.F., McKim K.S., Goshima G., and Ohkura H.. 2015. The microtubule catastrophe promoter Sentin delays stable kinetochore-microtubule attachment in oocytes. J. Cell Biol. 211:1113–1120. 10.1083/jcb.201507006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan J., Chim Y.-C.F., Ha A., Basiri M.L., Lerit D.A., Rusan N.M., and Avidor-Reiss T.. 2012. Tubulin nucleotide status controls Sas-4-dependent pericentriolar material recruitment. Nat. Cell Biol. 14:865–873. 10.1038/ncb2527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Mayer M., Zhang N., Stuurman N., and Vale R.D.. 2008. Augmin: A protein complex required for centrosome-independent microtubule generation within the spindle. J. Cell Biol. 181:421–429. 10.1083/jcb.200711053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O.J., Carazo-Salas R.E., Schatz C.A., Guarguaglini G., Kast J., Wilm M., Le Bot N., Vernos I., Karsenti E., and Mattaj I.W.. 2001. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 104:83–93. 10.1016/S0092-8674(01)00193-3 [DOI] [PubMed] [Google Scholar]
- Gueth-Hallonet C., Antony C., Aghion J., Santa-Maria A., Lajoie-Mazenc I., Wright M., and Maro B.. 1993. gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J. Cell Sci. 105:157–166. [DOI] [PubMed] [Google Scholar]
- Hassold T., and Hunt P.. 2001. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2:280–291. 10.1038/35066065 [DOI] [PubMed] [Google Scholar]
- Hassold T., Hall H., and Hunt P.. 2007. The origin of human aneuploidy: Where we have been, where we are going. Hum. Mol. Genet. 16 Spec No. 2:R203–R208. 10.1093/hmg/ddm243 [DOI] [PubMed] [Google Scholar]
- Heald R., Tournebize R., Blank T., Sandaltzopoulos R., Becker P., Hyman A., and Karsenti E.. 1996. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 382:420–425. 10.1038/382420a0 [DOI] [PubMed] [Google Scholar]
- Holubcová Z., Blayney M., Elder K., and Schuh M.. 2015. Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science. 348:1143–1147. 10.1126/science.aaa9529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J.K., Rahman T., and McKim K.S.. 2005. The kinesinlike protein Subito contributes to central spindle assembly and organization of the meiotic spindle in Drosophila oocytes. Mol. Biol. Cell. 16:4684–4694. 10.1091/mbc.E04-11-0964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J.K., Rahman T., Kober V.S., Cesario J., and McKim K.S.. 2007. Misregulation of the kinesin-like protein Subito induces meiotic spindle formation in the absence of chromosomes and centrosomes. Genetics. 177:267–280. 10.1534/genetics.107.076091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalab P., Pu R.T., and Dasso M.. 1999. The ran GTPase regulates mitotic spindle assembly. Curr. Biol. 9:481–484. 10.1016/S0960-9822(99)80213-9 [DOI] [PubMed] [Google Scholar]
- Kapoor T.M., Mayer T.U., Coughlin M.L., and Mitchison T.J.. 2000. Probing spindle assembly mechanisms with monastrol, a small molecule inhibitor of the mitotic kinesin, Eg5. J. Cell Biol. 150:975–988. 10.1083/jcb.150.5.975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly A.E., Sampath S.C., Maniar T.A., Woo E.M., Chait B.T., and Funabiki H.. 2007. Chromosomal enrichment and activation of the aurora B pathway are coupled to spatially regulate spindle assembly. Dev. Cell. 12:31–43. 10.1016/j.devcel.2006.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A., Cole R.W., Oakley B.R., and Rieder C.L.. 2000. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 10:59–67. 10.1016/S0960-9822(99)00276-6 [DOI] [PubMed] [Google Scholar]
- Kim J., Ishiguro K., Nambu A., Akiyoshi B., Yokobayashi S., Kagami A., Ishiguro T., Pendas A.M., Takeda N., Sakakibara Y., et al. 2015. Meikin is a conserved regulator of meiosis-I-specific kinetochore function. Nature. 517:466–471. 10.1038/nature14097 [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Noetzel T.L., Pelletier L., Mechtler K., Drechsel D.N., Schwager A., Lee M., Raff J.W., and Hyman A.A.. 2005. Aurora A phosphorylation of TACC3/maskin is required for centrosome-dependent microtubule assembly in mitosis. J. Cell Biol. 170:1047–1055. 10.1083/jcb.200503023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M., Müller-Reichert T., Oegema K., Grill S., and Hyman A.A.. 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 112:575–587. 10.1016/S0092-8674(03)00117-X [DOI] [PubMed] [Google Scholar]
- Kirschner M., and Mitchison T.. 1986. Beyond self-assembly: From microtubules to morphogenesis. Cell. 45:329–342. 10.1016/0092-8674(86)90318-1 [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Ohsugi M., and Ellenberg J.. 2011. Complete kinetochore tracking reveals error-prone homologous chromosome biorientation in mammalian oocytes. Cell. 146:568–581. 10.1016/j.cell.2011.07.031 [DOI] [PubMed] [Google Scholar]
- Kolano A., Brunet S., Silk A.D., Cleveland D.W., and Verlhac M.-H.. 2012. Error-prone mammalian female meiosis from silencing the spindle assembly checkpoint without normal interkinetochore tension. Proc. Natl. Acad. Sci. USA. 109:E1858–E1867. 10.1073/pnas.1204686109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M., Godinho S.A., Chandhok N.S., Ganem N.J., Azioune A., Thery M., and Pellman D.. 2008. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 22:2189–2203. 10.1101/gad.1700908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane S.I.R., Yun Y., and Jones K.T.. 2012. Timing of anaphase-promoting complex activation in mouse oocytes is predicted by microtubule-kinetochore attachment but not by bivalent alignment or tension. Development. 139:1947–1955. 10.1242/dev.077040 [DOI] [PubMed] [Google Scholar]
- Lawo S., Bashkurov M., Mullin M., Ferreria M.G., Kittler R., Habermann B., Tagliaferro A., Poser I., Hutchins J.R.A., Hegemann B., et al. 2009. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19:816–826. 10.1016/j.cub.2009.04.033 [DOI] [PubMed] [Google Scholar]
- Lénárt P., Bacher C.P., Daigle N., Hand A.R., Eils R., Terasaki M., and Ellenberg J.. 2005. A contractile nuclear actin network drives chromosome congression in oocytes. Nature. 436:812–818. 10.1038/nature03810 [DOI] [PubMed] [Google Scholar]
- Lister L.M., Kouznetsova A., Hyslop L.A., Kalleas D., Pace S.L., Barel J.C., Nathan A., Floros V., Adelfalk C., Watanabe Y., et al. 2010. Age-related meiotic segregation errors in mammalian oocytes are preceded by depletion of cohesin and Sgo2. Curr. Biol. 20:1511–1521. 10.1016/j.cub.2010.08.023 [DOI] [PubMed] [Google Scholar]
- Łuksza M., Queguigner I., Verlhac M.-H., and Brunet S.. 2013. Rebuilding MTOCs upon centriole loss during mouse oogenesis. Dev. Biol. 382:48–56. 10.1016/j.ydbio.2013.07.029 [DOI] [PubMed] [Google Scholar]
- Mailhes J.B., Mastromatteo C., and Fuseler J.W.. 2004. Transient exposure to the Eg5 kinesin inhibitor monastrol leads to syntelic orientation of chromosomes and aneuploidy in mouse oocytes. Mutat. Res. 559:153–167. 10.1016/j.mrgentox.2004.01.001 [DOI] [PubMed] [Google Scholar]
- Manandhar G., Sutovsky P., Joshi H.C., Stearns T., and Schatten G.. 1998. Centrosome reduction during mouse spermiogenesis. Dev. Biol. 203:424–434. 10.1006/dbio.1998.8947 [DOI] [PubMed] [Google Scholar]
- Manandhar G., Schatten H., and Sutovsky P.. 2005. Centrosome reduction during gametogenesis and its significance. Biol. Reprod. 72:2–13. 10.1095/biolreprod.104.031245 [DOI] [PubMed] [Google Scholar]
- Matthews L.R., Carter P., Thierry-Mieg D., and Kemphues K.. 1998. ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J. Cell Biol. 141:1159–1168. 10.1083/jcb.141.5.1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthies H.J., McDonald H.B., Goldstein L.S., and Theurkauf W.E.. 1996. Anastral meiotic spindle morphogenesis: Role of the non-claret disjunctional kinesin-like protein. J. Cell Biol. 134:455–464. 10.1083/jcb.134.2.455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumdar M., and Misteli T.. 2005. Chromokinesins: Multitalented players in mitosis. Trends Cell Biol. 15:349–355. 10.1016/j.tcb.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Merdes A., Ramyar K., Vechio J.D., and Cleveland D.W.. 1996. A complex of NuMA and cytoplasmic dynein is essential for mitotic spindle assembly. Cell. 87:447–458. 10.1016/S0092-8674(00)81365-3 [DOI] [PubMed] [Google Scholar]
- Meunier S., and Vernos I.. 2016. Acentrosomal microtubule assembly in mitosis: the where, when, and how. Trends Cell Biol. 26:80–87. 10.1016/j.tcb.2015.09.001 [DOI] [PubMed] [Google Scholar]
- Nachury M.V., Maresca T.J., Salmon W.C., Waterman-Storer C.M., Heald R., and Weis K.. 2001. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 104:95–106. 10.1016/S0092-8674(01)00194-5 [DOI] [PubMed] [Google Scholar]
- Nagaoka S.I., Hassold T.J., and Hunt P.A.. 2012. Human aneuploidy: Mechanisms and new insights into an age-old problem. Nat. Rev. Genet. 13:493–504. 10.1038/nrg3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K., Desai A., Rybina S., Kirkham M., and Hyman A.A.. 2001. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J. Cell Biol. 153:1209–1226. 10.1083/jcb.153.6.1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petry S., Pugieux C., Nédélec F.J., and Vale R.D.. 2011. Augmin promotes meiotic spindle formation and bipolarity in Xenopus egg extracts. Proc. Natl. Acad. Sci. USA. 108:14473–14478. 10.1073/pnas.1110412108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta-Marques A., Bento I., Lopes C.A., Duarte P., Jana S.C., and Bettencourt-Dias M.. 2016. A mechanism for the elimination of the female gamete centrosome in Drosophila melanogaster. Science. 353:aaf4866 10.1126/science.aaf4866 [DOI] [PubMed] [Google Scholar]
- Radford S.J., Jang J.K., and McKim K.S.. 2012. The chromosomal passenger complex is required for meiotic acentrosomal spindle assembly and chromosome biorientation. Genetics. 192:417–429. 10.1534/genetics.112.143495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath S.C., Ohi R., Leismann O., Salic A., Pozniakovski A., and Funabiki H.. 2004. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 118:187–202. 10.1016/j.cell.2004.06.026 [DOI] [PubMed] [Google Scholar]
- Sánchez-Huertas C., and Lüders J.. 2015. The augmin connection in the geometry of microtubule networks. Curr. Biol. 25:R294–R299. 10.1016/j.cub.2015.02.006 [DOI] [PubMed] [Google Scholar]
- Schuh M., and Ellenberg J.. 2007. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 130:484–498. 10.1016/j.cell.2007.06.025 [DOI] [PubMed] [Google Scholar]
- Sköld H.N., Komma D.J., and Endow S.A.. 2005. Assembly pathway of the anastral Drosophila oocyte meiosis I spindle. J. Cell Sci. 118:1745–1755. 10.1242/jcs.02304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M., O’toole E.T., Hyman A.A., and Müller-Reichert T.. 2006. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr. Biol. 16:1944–1949. 10.1016/j.cub.2006.08.029 [DOI] [PubMed] [Google Scholar]
- Still I.H., Vince P., and Cowell J.K.. 1999. The third member of the transforming acidic coiled coil-containing gene family, TACC3, maps in 4p16, close to translocation breakpoints in multiple myeloma, and is upregulated in various cancer cell lines. Genomics. 58:165–170. 10.1006/geno.1999.5829 [DOI] [PubMed] [Google Scholar]
- Sumiyoshi E., Fukata Y., Namai S., and Sugimoto A.. 2015. Caenorhabditis elegans Aurora A kinase is required for the formation of spindle microtubules in female meiosis. Mol. Biol. Cell. 26:4187–4196. 10.1091/mbc.E15-05-0258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szollosi D., Calarco P., and Donahue R.P.. 1972. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J. Cell Sci. 11:521–541. [DOI] [PubMed] [Google Scholar]
- Tanenbaum M.E., and Medema R.H.. 2010. Mechanisms of centrosome separation and bipolar spindle assembly. Dev. Cell. 19:797–806. 10.1016/j.devcel.2010.11.011 [DOI] [PubMed] [Google Scholar]
- Toso A., Winter J.R., Garrod A.J., Amaro A.C., Meraldi P., and McAinsh A.D.. 2009. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J. Cell Biol. 184:365–372. 10.1083/jcb.200809055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier F., Karsenti E., and Bornens M.. 1989. Parthenogenesis in Xenopus eggs injected with centrosomes from synchronized human lymphoid cells. Dev. Biol. 136:321–329. 10.1016/0012-1606(89)90259-5 [DOI] [PubMed] [Google Scholar]
- Tsai M.-Y., Wiese C., Cao K., Martin O., Donovan P., Ruderman J., Prigent C., and Zheng Y.. 2003. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat. Cell Biol. 5:242–248. 10.1038/ncb936 [DOI] [PubMed] [Google Scholar]
- Tseng B.S., Tan L., Kapoor T.M., and Funabiki H.. 2010. Dual detection of chromosomes and microtubules by the chromosomal passenger complex drives spindle assembly. Dev. Cell. 18:903–912. 10.1016/j.devcel.2010.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou A.-P., Yang C.-W., Huang C.-Y.F., Yu R.C.-T., Lee Y.-C.G., Chang C.-W., Chen B.-R., Chung Y.-F., Fann M.-J., Chi C.-W., et al. 2003. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 22:298–307. 10.1038/sj.onc.1206129 [DOI] [PubMed] [Google Scholar]
- Uehara R., Nozawa R.S., Tomioka A., Petry S., Vale R.D., Obuse C., and Goshima G.. 2009. The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl. Acad. Sci. USA. 106:6998–7003. 10.1073/pnas.0901587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak C.E., Vernos I., Mitchison T.J., Karsenti E., and Heald R.. 1998. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr. Biol. 8:903–913. 10.1016/S0960-9822(07)00370-3 [DOI] [PubMed] [Google Scholar]
- Wandke C., Barisic M., Sigl R., Rauch V., Wolf F., Amaro A.C., Tan C.H., Pereira A.J., Kutay U., Maiato H., et al. 2012. Human chromokinesins promote chromosome congression and spindle microtubule dynamics during mitosis. J. Cell Biol. 198:847–863. 10.1083/jcb.201110060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y. 2012. Geometry and force behind kinetochore orientation: Lessons from meiosis. Nat. Rev. Mol. Cell Biol. 13:370–382. 10.1038/nrm3349 [DOI] [PubMed] [Google Scholar]
- Wignall S.M., and Villeneuve A.M.. 2009. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat. Cell Biol. 11:839–844. 10.1038/ncb1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T., Wilm M., Karsenti E., and Vernos I.. 2000. TPX2, A novel Xenopus MAP involved in spindle pole organization. J. Cell Biol. 149:1405–1418. 10.1083/jcb.149.7.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman R., Cytrynbaum E.N., Jones J.T., Meyer T., Scholey J.M., and Mogilner A.. 2005. Efficient chromosome capture requires a bias in the ‘search-and-capture’ process during mitotic-spindle assembly. Curr. Biol. 15:828–832. 10.1016/j.cub.2005.03.019 [DOI] [PubMed] [Google Scholar]
- Woodruff J.B., Wueseke O., Viscardi V., Mahamid J., Ochoa S.D., Bunkenborg J., Widlund P.O., Pozniakovsky A., Zanin E., Bahmanyar S., et al. 2015. Centrosomes. Regulated assembly of a supramolecular centrosome scaffold in vitro. Science. 348:808–812. 10.1126/science.aaa3923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolley D.M., and Fawcett D.W.. 1973. The degeneration and disappearance of the centrioles during the development of the rat spermatozoon. Anat. Rec. 177:289–301. 10.1002/ar.1091770209 [DOI] [PubMed] [Google Scholar]
- Yan L.-Y., Huang J.-C., Zhu Z.-Y., Lei Z.-L., Shi L.-H., Nan C.-L., Zhao Z.-J., Ouyang Y.-C., Song X.-F., Sun Q.-Y., and Chen D.-Y.. 2006. NuMA distribution and microtubule configuration in rabbit oocytes and cloned embryos. Reproduction. 132:869–876. 10.1530/rep.1.01224 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Kaido M., and Kitajima T.S.. 2015. Inherent instability of correct kinetochore-microtubule attachments during meiosis I in oocytes. Dev. Cell. 33:589–602. 10.1016/j.devcel.2015.04.020 [DOI] [PubMed] [Google Scholar]