Mitofusin 1 (MFN1) mediates mitochondrial fusion, but the mechanisms involved are unclear. Qi et al. present the crystal structures of a minimal GTPase domain of human MFN1, which suggest that MFN1 tethers apposing membranes through nucleotide-dependent dimerization.
Abstract
Mitochondria undergo fusion and fission. The merging of outer mitochondrial membranes requires mitofusin (MFN), a dynamin-like GTPase. How exactly MFN mediates membrane fusion is poorly understood. Here, we determined crystal structures of a minimal GTPase domain (MGD) of human MFN1, including the predicted GTPase and the distal part of the C-terminal tail (CT). The structures revealed that a helix bundle (HB) formed by three helices extending from the GTPase and one extending from the CT closely attaches to the GTPase domain, resembling the configuration of bacterial dynamin-like protein. We show that the nucleotide-binding pocket is shallow and narrow, rendering weak hydrolysis and less dependence on magnesium ion, and that association of HB affects GTPase activity. MFN1 forms a dimer when GTP or GDP/BeF3−, but not GDP or other analogs, is added. In addition, clustering of vesicles containing membrane-anchored MGD requires continuous GTP hydrolysis. These results suggest that MFN tethers apposing membranes, likely through nucleotide-dependent dimerization.
Introduction
The double-membraned organelle mitochondria constantly remodels themselves by membrane fusion and fission (Hoppins and Nunnari, 2009; Lackner and Nunnari, 2009; Youle and van der Bliek, 2012; Mishra and Chan, 2016; Pernas and Scorrano, 2016). Fusion of the outer mitochondrial membranes is mediated by a class of dynamin-like GTPases: mitofusin (MFN) in mammals, fuzzy onions in Drosophila melanogaster, and Fzo1p in yeast (Hales and Fuller, 1997; Hermann et al., 1998; Rapaport et al., 1998). Two MFNs have been identified in mammals (Santel and Fuller, 2001; Rojo et al., 2002). Deletion of either MFN in mice causes embryonic lethality and mitochondrial fragmentation indicative of a lack of fusion and continued fission (Chen et al., 2003). Human mutations in MFN2 are associated with Charcot–Marie–Tooth neuropathy type 2A (CMT2A; Züchner et al., 2004), a classic axonal peripheral sensorimotor neuropathy. The fusion of viral and cellular membranes and of intracellular transport vesicles with target membranes (i.e., heterotypic fusion) has been studied extensively (Jahn and Scheller, 2006; Harrison, 2008; Martens and McMahon, 2008; Wickner and Schekman, 2008; Südhof and Rothman, 2009). However, how homotypic fusion such as MFN-mediated mitochondrial fusion occurs is poorly understood.
MFN is composed of an N-terminal GTPase followed by a predicted helix bundle (HB) region, two closely spaced transmembrane (TM) segments, and the C-terminal tail (CT). The crystal structure of a heptad repeat (HR) in the MFN-CT revealed an antiparallel coiled coil (Koshiba et al., 2004). Thus, MFN may tether apposing membranes via the homotypic interactions of the CT.
MFN shares some sequence similarities with bacterial dynamin-like protein (BDLP; Daumke and Praefcke, 2016). Structural studies of BDLP have revealed that the GTPase forms a dimer (Low and Löwe, 2006; Low et al., 2009). In the GMPPNP-bound state, the HB regions following the GTPase domains are straight and parallel in dimers, and BDLP oligomers may attach to lipid tubes in a helical pattern using their paddle domains (a helix hairpin equivalent to the TMs of MFN; Low et al., 2009). In the GDP-bound state, the HB rotates relative to the GTPase and bends drastically in the middle so that the paddle domains of the pairing molecules are close to the GTPase domain and associated with each other at the dimer interface (Low and Löwe, 2006). The membrane instability caused by such conformational changes could be used for fusion (Daumke and Praefcke, 2016; Schrepfer and Scorrano, 2016). However, the fusogenic activity of BDLP has yet to be demonstrated, making the mechanistic link between MFN and BDLP uncertain.
Another related member of the dynamin family, called atlastin (ATL) in mammals, Sey1p in yeast, and RHD3 in plants, mediates homotypic fusion; it has the same membrane topology as MFNs and fuses ER membranes (Hu and Rapoport, 2016). Structural and biochemical analysis of human ATL1 and Sey1p indicated that GTP-binding–induced dimerization of the GTPase promotes membrane tethering. GTP hydrolysis induces conformational changes in the HB, forcing the membranes to merge (Bian et al., 2011; Byrnes and Sondermann, 2011; Yan et al., 2015). The mechanistic analogy between MFN and ATL is supported by similar fusion-promoting amphipathic helices in the CT (Liu et al., 2012). In addition, ATL-mediated membrane tethering requires continuous GTP hydrolysis (Liu et al., 2015), whereas the cytosolic domain of MFN only self-associates when GTP is present at a hydrolyzable temperature (Ishihara et al., 2004), and MFN-dependent mitochondrial docking has recently been shown to be GTP hydrolysis dependent (Brandt et al., 2016).
How the cytosolic domain of MFN is configured at the atomic level is a long-standing question. Here, we determined the structures of a cytosolic module of human MFN1 and unveiled unique properties of the nucleotide pocket of MFN. Structural and biochemical analysis revealed that the HR in the MFN-CT may form part of the HB domain and that membrane tethering by MFN needs active hydrolysis of GTP.
Results and discussion
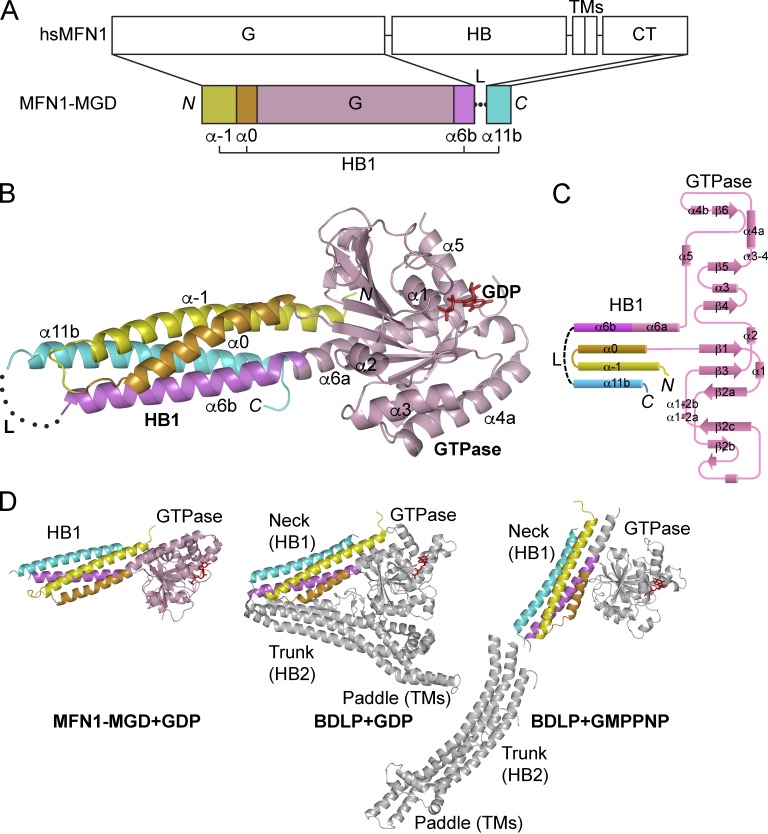
To obtain soluble human MFN1, we started with expression of the N-terminal cytosolic domain (residues 1–593). Serial truncations at the C terminus (constructs 1–558, 1–525, 1–429, and 1–364) were then attempted until only the predicted GTPase domain (residues 1–364) remained. We also engineered the full-length protein (residues 1–741) by replacing TM-containing portions (including residues 594–624, 567–645, 525–694, and 434–638) with an artificial linker (GSGSGSGGS). Unfortunately, all of these constructs either expressed poorly or appeared insoluble. In the end, we found that a fragment of human MFN1 containing the predicted GTPase (residues 1–364) and second half of the HR2 (residues 694–741) in the CT (Fig. 1 A) yields purifiable recombinant proteins in Escherichia coli. The N- and C-terminal regions are connected by the artificial linker. Because the GTPase can only be expressed with the addition of a CT portion, we termed the resulting construct minimal GTPase domain (MGD). Purified MGD was crystallized in the presence of GDP and the structure determined at 2.6-Å resolution by single anomalous diffraction (Table 1).
Figure 1.
Crystal structure of MFN1–MGD. (A) Scheme showing the domains of human MFN1 and the MGD construct used for crystallization. Regions of MGD are colored and secondary structure elements that form helix bundle 1 (HB1) are labeled. G, GTPase; TMs, transmembrane segments; CT, cytosolic tail; L, linker. (B) Structure of the GDP-bound form of MGD. As in A, the GTPase is colored in pink and the helices in HB1 yellow (α-1), orange (α0), purple (α6b), and cyan (α11b), respectively. GDP is shown as red sticks and the disordered linker by a dotted line. The main helices are labeled. (C) Topology plots of the MGD. Colored as in A. (D) Comparison of MFN and BDLP. The GTPase domains of MFN1–MGD and BDLP in GDP- (PDB 2J68) or GMPPNP-bound (PDB 2W6D) states are oriented similarly. The nucleotides are shown as red sticks. The part in BDLP that is equivalent to MGD is colored in the same way; the remaining part is shown in gray. Major domains are labeled.
Table 1. Data collection and refinement statistics.
| Data | MGD•GDP (SeMet-SAD) 5GNT | MGD 5GNU | MGD K88A•GTP 5GNS | MGD K88A•GDP 5GNR |
|---|---|---|---|---|
| Space group | P212121 | P212121 | P212121 | P212121 |
| Cell parameters | ||||
| a (Å) | 70.36 | 73.0 | 71.66 | 71.34 |
| b (Å) | 72.53 | 76.49 | 74.67 | 74.98 |
| c (Å) | 94.07 | 95.36 | 94.76 | 94.45 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Wavelength used (Å) | 0.9785 | 0.9785 | 0.9785 | 0.9785 |
| Resolution (Å) | 50 (2.71)–2.66 | 50 (4.19)–4.10 | 50 (2.42)–2.30 | 50 (2.74)a–2.65 |
| No. of all reflections | 204,360 (9397) | 19,796 (1112) | 122,150 (18,308) | 119,107 (12,080) |
| No. of unique reflections | 14,293 (681) | 4318 (227) | 40,802 (5930) | 15,269 (1510) |
| Completeness (%) | 99.9 (100) | 97.9 (99.6) | 99.1 (99.4) | 99.8 (100) |
| Average I/σ(I) | 56.7 (7.0) | 13.5 (1.34) | 10.2 (2.3) | 21.38 (1.74) |
| Rmergeb (%) | 8.4 (62.3) | 8.9 (78.0) | 5.3 (44.3) | 8.6 (81.5) |
| Refinement | ||||
| No. of reflections | 14,251 | 3371 | 14,425 | 13,579 |
| Rworkc (%) | 21.0 | 28.9 | 21.2 | 25.4 |
| Rfreec (%) | 27.3 | 31.3 | 28.3 | 29.3 |
| rmsd bond distance (Å) | 0.010 | 0.008 | 0.010 | 0.012 |
| rmsd bond angle (°) | 1.486 | 1.653 | 1.388 | 1.483 |
| Average B-value (Å2) | 46.3 | 120.3 | 56.23 | 41.8 |
| No. of protein atoms | 3,021 | 2,985 | 3,048 | 3,025 |
| No. of ligand atoms | 28 | 0 | 32 | 28 |
| No. of solvent atoms | 18 | 0 | 8 | 10 |
| Ramachandran plot | ||||
| Residues in favored regions (%) | 92.39 | 83.15 | 92.88 | 90.72 |
| Residues in generously allowed regions (%) | 7.07 | 14.95 | 7.12 | 8.75 |
| Residues in disallowed regions (%) | 0.54 | 1.9 | 0 | 0.53 |
rmsd, root mean square deviation.
Numbers in parentheses are corresponding values for the highest-resolution shell.
Rmerge=ΣhΣi|Ih,i − Ih|/ΣhΣiIh,i, where Ih is the mean intensity of the i observations of symmetry related reflections of h.
Rwork =Σ|Fobs − Fcalc|/ΣFobs, where Fcalc is the calculated protein structure factor from the atomic model; Rfree is an R factor for a preselected subset (5%) of reflections that was not included in refinement.
MGD appears as a monomer, consisting of a typical large GTPase domain and a four-helix bundle (Fig. 1 B) composed by two helices from an N-terminal extension of the GTPase (α-1 and α0), the second half of α6 (α6b), and the second half of α11 (α11b) from the CT (Fig. 1 C). Given that additional HBs may exist in full-length MFN, we designated the HB in MGD HB1. The configuration of MGD is most reminiscent of BDLP (Fig. 1 D). The N-terminal extension is observed in the neck region of BDLP (Low and Löwe, 2006), which also contains a helix extending from the last helix of the GTPase, and the last helix of the entire molecule. When BDLP is in the GDP-bound state, the orientation of the neck relative to the GTPase resembles that of MFN1–MGD in complex with GDP (Fig. 1 D).
MFN–HB1 is formed mainly through hydrophobic bundling. The residues in α11b previously proposed to form an antiparallel coiled coil are involved extensively in helical zippering of HB1. For example, F733 packs against L8 in the N-terminal α-1, and L705 interacts with L45 and I48 (Fig. S1 A). The CT of MFN1 contains three helices (α10, α11a, and α11b), all of which exhibit an amphipathic nature and have potential in helical zippering. When α11b was removed or replaced with α10 or α11a in MGD, the constructs were poorly expressed in E. coli and the resulting recombinant proteins precipitated heavily (Fig. S1 B), suggesting specific pairing of α11b with HB1. Substitution of L705 with Pro increased the trypsin sensitivity of the protein, and F733P yielded very little soluble protein (Fig. S1 C). These results suggest that α11b from the CT may complement the N-terminal helices and maintains the integrity of HB1 via hydrophobic interactions.
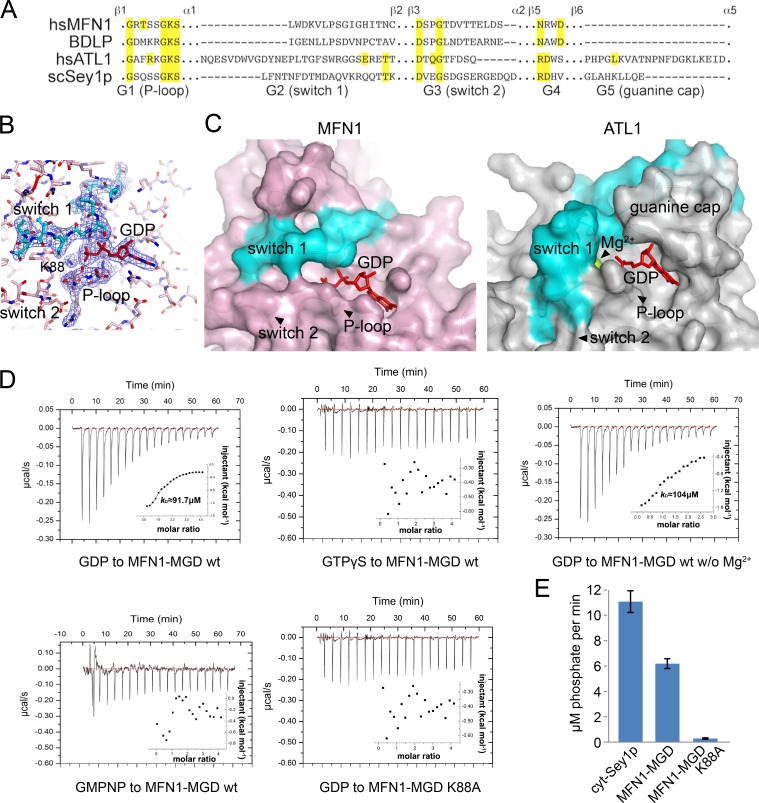
The GTPase of MFN1 is composed of a central β sheet with three α helices on each side (α1, 5, and 6 on the GDP-binding side and α2, 3, and 4 on the other). The active site of MFN1 is coordinated mainly by four signature motifs of dynamin-like GTPase (Fig. 2, A and B): G1/P-loop (β1–α1), G2/switch 1 (α1–β2), G3/switch 2 (β3–α2), and G4 (following β5). A fifth motif termed a “guanine cap” is found between β5 and α5 in other dynamin family members but is completely missing in MFN and BDLP (Fig. 2 A), rendering a rather exposed active site (Fig. 2 C). In addition, magnesium ion, which is commonly found in GTPase to facilitate nucleotide binding, is absent from the MFN1 structure.
Figure 2.
GTP binding and hydrolysis of MFN1. (A) Sequence alignment of the conserved GTPase motifs. Key residues are highlighted in yellow. Neighboring secondary structure elements of the motifs are labeled. (B) The active site of MFN1 is shown in sticks. The 2Fo − Fc electron density maps (1.0-σ contour) of switch 1, P-loop, and GDP are shown as wire mesh (blue). (C) Comparison of the active sites of MFN1 and ATL1. A surface representation is shown. The switch 1 region is highlighted in cyan, GDP as red sticks, and Mg2+ ion as a lime sphere. Key motifs are labeled. (D) Binding affinity of various nucleotides for wild-type (wt) MFN1 and the K88A mutant measured by ITC. 2 mM nucleotide solution was injected stepwise into 0.1 mM protein. The dissociation constant, Kd, if calculable, is given in the inlet. The data are representative of at least three repetitions. (E) GTPase activity of MFN1 and Sey1p. 2 µM protein was used for each sample. The activities were measured by phosphate release at saturating GTP concentrations (0.5 mM) using the cytosolic domains. Each bar is the mean and SD of four measurements. The data are representative of at least three repetitions.
To better understand the nucleotide-binding property of MFN1, we determined the structures of MGD without GDP (Apo form), with GTP, or with GTPγS (a nonhydrolyzable GTP analog; Fig. S2 A). In all cases, structures very similar to the GDP form were obtained. In the GTP-added structure, GDP was observed, presumably by continuous hydrolysis during crystallization. In the GTPγS-added structure, GDP was still seen, likely because of low levels of GDP contamination in GTPγS. In the Apo form, the active site did not change much, as expected. In particular, the two switches that normally move apart in the absence of nucleotide were positioned similar to the GDP-bound state (Fig. S2 B). These observations suggest that MFN possesses an exposed and less flexible nucleotide-binding pocket and is predicted to have low affinity for nucleotide and a weak GTPase.
To test nucleotide binding by MFN1, we performed isothermal titration calorimetry (ITC). Wild-type protein binds to GDP at a Kd of 91.7 µM (Fig. 2 D), 50 times lower than that of ATL1 (Bian et al., 2011). In addition, MFN1 exhibited no detectable interactions with GTPγS or GMPPNP, but with GDP even in the absence of Mg2+ (Fig. 2 D). Different dynamin GTPases have been shown to have different preferences for GTP analogs (Yan et al., 2015). The active site of MFN1 does not appear to tolerate subtle atomic modifications in the nucleotide. To capture MFN1 in a GTP-bound state, we mutated the highly conserved K88 in the P-loop to alanine, which decreased the GTPase activity (Fig. 2 E). Surprisingly, MFN1 adapted to the loss of K88 and used K99 from G2/switch 1 to coordinate γ-phosphate (Fig. S2, C–E). The phosphate chain of GTP bent drastically to present the γ-phosphate toward K99. These results confirm that MFN interacts poorly with nucleotides. Consistently, the GTPase activity of MFN1 was lower than that of Sey1p (Fig. 2 E).
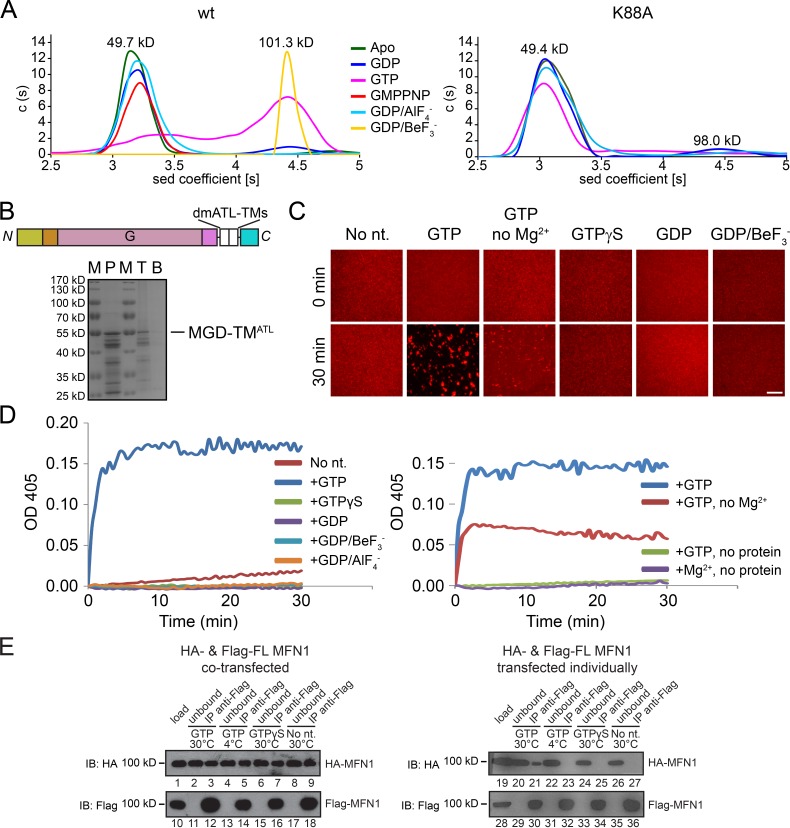
Nucleotide-dependent dimer formation is a common feature of dynamin-like GTPases. In cell lysates, the cytosolic domains of MFN self-associate only when GTP is added at an enzyme-active temperature (Ishihara et al., 2004). To test whether purified MFN forms a nucleotide-dependent dimer, we performed analytical ultracentrifugation (AUC). As expected, MGD behaved as a monomer in the absence of nucleotide or in the presence of GDP, forming dimers in the presence of GTP (Fig. 3 A). Unlike ATL or Sey1p, MFN is still a monomer when GDP/AlF4− (a commonly used transition state mimic) was added (Fig. 3 A). Considering the tightness of the active center, we replaced AlF4− with a smaller phosphate analog, BeF3−, and then observed strong dimer formation (Fig. 3 A). Consistent with these results, a trypsin protection assay showed that MGD is more resistant with GTP or GDP/BeF3− than GDP or GDP/AlF4− (Fig. S2 F), judging by the disappearance of the full-length band and the appearance of protected bands of ∼35 kD. Dimerization was not seen when GTPase-defective mutant K88A was used (Fig. 3 A). To further test whether MGD is sufficient for membrane tethering, we substituted the linker with the TMs of Drosophila ATL (MGD-TMATL) and purified and reconstituted the chimera into proteoliposomes (Fig. 3 B). Vesicle tethering, as visualized by confocal microscopy, was only observed with the presence of GTP (Fig. 3 C). Tethering was decreased, but still detectable, when GTP was added but Mg2+ was omitted (Fig. 3 C). The same results were obtained when tethering was measured as increasing solution turbidity (absorbance at 405 nm; Fig. 3 D). These findings suggest that MFN mediates membrane tethering by continuous GTP hydrolysis and is less dependent on Mg2+.
Figure 3.
Homotypic interactions of MFN1. (A) The sizes of wild-type (wt) MFN1-MGD (theoretical molecular mass, 49.5 kD) and the K88A mutant (both at 0.4 mM) were determined by analytical ultracentrifugation in the presence of 2 mM indicated nucleotides. The estimated molecular masses are given above the peaks (in kilodaltons). The data are representative of at least three repetitions. (B) Purified (P) and reconstituted dmATL TM-containing MGD (domain structure shown above) was subjected to floatation analysis. Top (T) and bottom (B) fractions were analyzed by SDS-PAGE and Coomassie staining. M, molecular marker shown in kilodaltons. (C) Membrane tethering by MGD-TMATL followed by a visual assay. Proteoliposomes containing MFN1 (protein/lipid ratio 1:2,000) and rhodamine-labeled lipids were analyzed by confocal microscopy. One aliquot was sampled immediately, and a second was taken after incubation at 37°C with 10 mM of the indicated nucleotide for 30 min. The data are representative of at least three repetitions. Bar, 50 µm. (D) As in C, but measured by absorbance at 405 nm. (E) HA-tagged and Flag-tagged full-length (FL) human MFN1 were cotransfected into HEK293T cells and solubilized in digitonin or transfected individually followed by mixing of the digitonin-solubilized cell extracts. Immunoprecipitation (IP) was performed with anti-HA or anti-Flag agarose beads. When indicated, 1 mM nucleotides was added and incubated. The samples were analyzed by SDS-PAGE and immunoblotting (IB). 10% of the starting material (load) and the material not bound to the antibodies (unbound) was also analyzed.
To test whether the CT of MFN is also involved in tethering as previously suggested (Koshiba et al., 2004), we performed coimmunoprecipitation experiments. When HA- and Flag-tagged full-length human MFN1 were cotransfected into HEK293T cells, Flag antibodies could precipitate HA-MFN1 (Fig. 3 E, lanes 3, 5, 7, and 9), even when nucleotide was not added. Thus, MFN could form nucleotide-independent homotypic interactions. However, when HA-tagged and Flag-tagged proteins were transfected individually into HEK293T cells and the extracts mixed, excluding TM-based interaction, no coimmunoprecipitation was observed in the absence of nucleotide (Fig. 3 E, lane 27). The same results were obtained after the addition of GTP and MgCl2 at 4°C or GTPγS and MgCl2 at 30°C (Fig. 3 E, lanes 23 and 25). Consistent with previous study (Ishihara et al., 2004), the interaction between MFN1 molecules was only restored when GTP and MgCl2 were added and incubated at 30°C (Fig. 3 E, lane 21), presumably allowing efficient GTP-dependent dimerization of MFN molecules. These results confirm that GTP-dependent self-association of MFN requires active GTP hydrolysis. It also suggests that MFN undergoes GTP-independent oligomerization, possibly mediated by the TM.
The overexpression of an MFN1 fragment containing no GTPase domain (residues 331–741) has been shown to cause mitochondria aggregation, and soluble HR2 fragments can self-associate (Koshiba et al., 2004). In agreement with this previous study, we found that HA-tagged MFN1 331–741 and Flag-tagged MFN1 331–741 (Fig. S1 D), which contain α6b and the remaining molecule, associate with one another when cotransfected (Fig. S1 E, lanes 13 and 20). However, the interaction was not detected when these proteins were transfected individually (Fig. S1 E, lanes 3 and 10). In contrast to the findings of (Koshiba et al., 2004), we found very weak binding between HA-tagged and Flag-tagged soluble MFN1-CT (residues 629–741; Fig. S1 F, lanes 3 and 10), and it only occurred when the constructs were cotransfected. These results support the notion that the TM domain can mediate nucleotide-independent oligomerization of MFN molecules. Under the conditions tested here, the MFN-CT fails to form homotypic interactions.
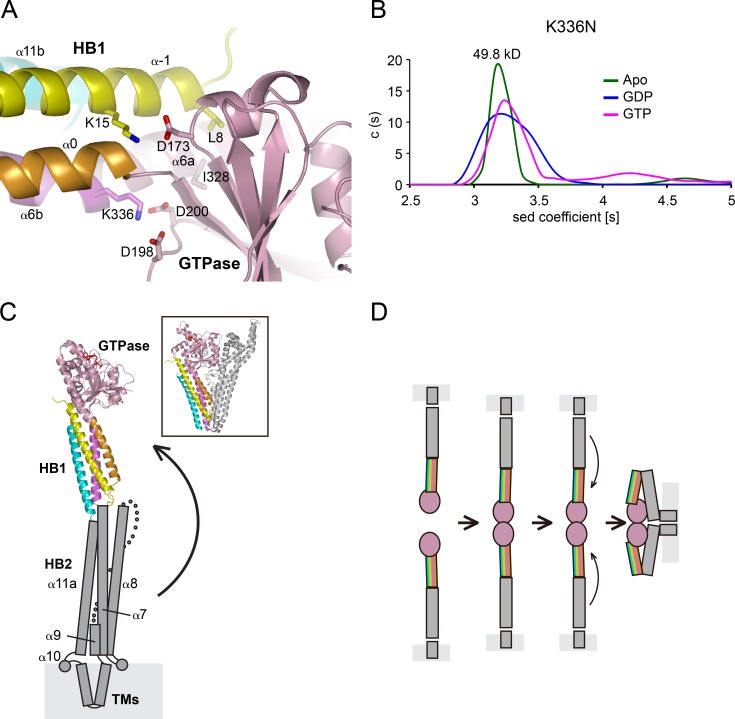
To gain further mechanistic insight, we positioned MFN2 mutations causing CMT2A (Liesa et al., 2009) in MFN1–MGD. Sequence similarity between MFN1 and 2 is high (Fig. S3 A), even though it was reported that these GTPases have different activity (Ishihara et al., 2004). The modeled mutations in MFN1–MGD can be categorized into three types (Fig. S3 B): directly involved in GTPase activity or GTPasa-HB1 interaction; maintenance of molecule folding, especially in α3/α4 and HB1; and on the molecule surface with functions yet to be identified. For the first category, T84 (T105 in MFN2) in the G1/P-loop, when mutated to Met, may block the entrance of nucleotide. P102 and G106 (P123 and G127) reside in G2/switch 1, and mutations may affect the flexibility of the switch. We found that P102L decreased nucleotide binding (Fig. S3, C and E) and GTPase activity (Fig. S3 D), and T84M expressed poorly. K336 (K357) connects the GTPase and HB1 as a kink in α6 (between pink and purple in Fig. 1 B) and forms salt bridges with D198 and D200 in the α2-β4 loop of the GTPase (Fig. 4 A). Additional GTPase–HB1 interactions include K15 from α-1 reaching D173 between β2′′ and β3, and L8 engaging I328 in α6a (Fig. 4 A). Movement of α6a in the GTPase could conveniently guide the rearrangement of HB1 and vice versa. Mutation of K336 to asparagine caused defects in nucleotide binding, GTPase activity, and dimerization (Fig. 4 B and Fig. S3, C and D), even though the residue is not part of the GTPase. Similar changes, including K15A, D173A, and I328A, also affected GTPase activity (Fig. S3 D). Finally, when tested in a cellular context, K336N failed to rescue mitochondrial morphology defects in MFN1-deleted MEF cells (Fig. S3 F). Collectively, these results suggest an important role of the GTPase-HB1 interaction.
Figure 4.
Association between GTPase and HBs of MFN1. (A) The interface between GTPase and HB1. The cartoon representation is colored as in Fig. 1 B. Key residues are indicated as sticks. (B) The size of MFN1–MGD K336N (theoretical molecular mass, 49.5 kD) at 0.4 mM was determined by analytical ultracentrifugation in the presence of the indicated nucleotides. The estimated molecular masses are given above the peaks (in kilodaltons). The data are representative of at least three repetitions. (C) A structural model of full-length MFN1. Predicted HB2 is shown in gray, with secondary structure elements labeled. A possible movement of the HB2 is indicated by an arrow, and the resulting folded configuration may resemble that of BDLP (shown in box for comparison). (D) A simple fusion model for MFN. First, the HBs are extended. Then, upon GTP hydrolysis, HB1 rotates and allows HB2 to bend over and attach to the GTPase, bringing apposing membranes together for fusion.
Our structural analysis showed that a GTPase-containing fragment of MFN1 folds like BDLP. The GTPase and HB1 of MFN1-MGD (Apo or plus GDP) superimpose with the GTPase and neck domain of BDLP in corresponding nucleotide states. The remaining part of the N-terminal cytosolic domain of MFN1 (residues 365–593) is predicted to be mostly α helical (Fig. S3 A) and likely adopts a similar configuration as in BDLP. Following α6b, the predicted α7, 8, and 9 could organize into a second HB (HB2) that is equivalent to the trunk domain of BDLP (Fig. 4 C). Similarly, α11a in the CT would complement HB2. Between α9 and α11a, a transmembrane hairpin of MFN1 (residues 596–628) corresponds to the paddle domain of BDLP, whereas the potential fusion-promoting amphipathic helix (α10) of MFN1 is not found in BDLP. α11a and b (collectively as HR2) have been proposed to tether membranes as an antiparallel coiled coil (Koshiba et al., 2004). Our structural model and biochemical analysis predicts that these helices serve as building blocks for the stalk-like domain, consistent with HR1 interacting with HR2 (Huang et al., 2011). In full-length MFN, whether HR2 folds stably with the N-terminal cytosolic domain or participates as a tethering factor during mitochondrial fusion remains to be tested, preferably in a cellular context.
Our results provide important insights into how MFN mediates homotypic fusion of mitochondrial outer membranes. MFN forms dimers upon GTP binding and/or hydrolysis. The association of HB1 with the GTPase affects its activity, as implicated by the role of K336. In Fzo1p, fusion activity is regulated by ubiquitination of the equivalent K464 (Anton et al., 2013). One simple scenario is that MFN tethers membranes through dimerization of the GTPase domain; during the GTP cycle, the HBs of MFN would rearrange to achieve fusion (Fig. 4 G). The HB2–GTPase interactions could occur near α3/α4 of the GTPase, suggested by the heavy cluster of CMT2A mutations in α3/α4. More possibilities for assembly and reassembly would emerge considering nucleotide-dependent and independent oligomerization. Indeed, MFN-containing docking rings were observed during mitochondrial fusion in vitro (Brandt et al., 2016). MFN folds like BDLP, acts like ATL in some aspects, and may mediate homotypic membrane fusion in a manner that is yet to be discovered.
Materials and methods
Protein purification
MFN1–MGD was produced from a MFN1 N-terminal (1–364) and C-terminal (694–741) fusion protein with a 9-amino-acid linker (GSGSGSGGS) and cloned into a modified pET-28a vector with an N-terminal 6×His. The plasmids were transformed into E. coli strain BL21 (DE3) and the transformed cells cultured at 37°C in LB media containing 100 mg/l kanamycin. After reaching OD600 0.8, the culture was cooled to 25°C and supplemented with 0.5 mM IPTG. After overnight induction, the cells were harvested by centrifugation, the pellets resuspended in the lysis buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, and 4 mM MgCl2), and the suspension homogenized with an ultra–high-pressure cell disrupter (JNBIO) at 4°C. The fusion protein was purified first by Ni-NTA affinity chromatography and elution with lysis buffer and then further purified by passage through a Mono-Q ion-exchange column (GE Healthcare), followed by a Superdex 200 gel filtration column (GE Healthcare) using the lysis buffer. All other MGD mutants were purified using the same protocol described in this paragraph.
Selenomethionyl (SeMet)-labeled MGD was expressed in minimal medium that inhibits methionine synthesis. The E. coli strain BL21was incubated overnight in LB medium at 37°C and harvested at 5,000 rpm (10 min, 4°C). The pellet was inoculated into 1 l of M9 medium (supplemented with 100 mg/l kanamycin and 3% glucose) at 37°C until an OD600 of 0.6. Next, 100 mg each of Lys, Phe, and Thr and 50 mg each of Ile, Leu, Val, and SeMet were added to the M9 medium, and the mixture was incubated for 15 min at 37°C. After induction with 1 mM IPTG, the cells were grown at 16°C for an additional 16 h. The SeMet-labeled protein was purified using the same procedure as described for the native protein.
For TM-containing MGD, the 9–amino acid linker (GSGSGSGGS) was substituted by TMs of D. melanogaster ATL (residues 422–470) by overlap PCR, and the chimeric fragment was subcloned into pGEX-6p-1 vector. The construct was transformed into E. coli BL21 (DE3). Cells were grown to an OD600 of 0.8, induced with 0.3 mM IPTG for 24 h at 16°C, and harvested by centrifugation. The pellets were resuspended in the A100 buffer (25 mM Hepes, pH 7.4, 100 mM KCl, 10% glycerol, 1 mM EDTA, and 1 mM BME). Membranes were then pelleted by centrifugation at 30,000 rpm for 1 h, dissolved by 1% FOS-CHOLINE-12 (Anatrace) in A100 buffer, and insoluble components were cleared by centrifugation. The recombinant protein was isolated by glutathione Sepharose (GE Healthcare), washed twice with A100 buffer containing 0.1% FOS-CHOLINE-12, and eluted by cleavage of 3C protease overnight at 4°C.
Crystallization and data collection
All native and SeMet-substituted proteins were concentrated to 12.5 mg/ml and crystallized by vapor diffusion at 16°C. SeMet-derived MGD with GDP crystals was obtained in a reservoir solution containing 200 mM sodium sulfate decahydrate and 20% (wt/vol) PEG 3350. MGD apo crystal appeared after ∼1 mo in 200 mM ammonium sulfate, 100 mM Tris, pH 8.5, and 25% (wt/vol) PEG 3350. The crystals of MGD K88A with GTP were obtained in a reservoir solution containing 200 mM sodium phosphate monobasic monohydrate and 20% (wt/vol) PEG 3350. Crystals of MGD K88A with GDP complex appeared in 200 mM ammonium fluoride and 20% (wt/vol) PEG 3350.
All crystals were gradually transferred into a cryoprotectant solution consisting of reservoir solution supplemented with 20% (vol/vol) glycerol before being flash frozen in liquid nitrogen for storage. The SAD and native datasets were collected on beamline BL17U and BL19U at the Shanghai Synchrotron Radiation Facility. Data were processed and scaled using the HKL2000 package (Otwinowski and Minor, 1997). Ten selenium atoms belonging to SeMet-labeled MGD in the asymmetric unit (one MGD per asymmetric unit) were located and refined, and the SAD data phases were calculated and substantially improved by solvent flattening using the PHENIX program (Afonine et al., 2012). A model was manually built into the modified experimental electron density using COOT (Emsley and Cowtan, 2004). Initially, large hydrophobic residues were assigned to facilitate the register of the long helices, and the model was further refined in PHENIX (Afonine et al., 2012) with rigid body, twin lattice symmetry, and secondary structure restraints. Native datasets were determined by molecular replacement using the initial searching model. The final refinement statistics are summarized in Table 1. Structural figures were drawn using the program PyMOL.
GTPase activity
GTPase assays were performed using the Enzchek phosphate assay kit (Invitrogen). Reactions were performed in a 100-µl volume with 5 µl 20× reaction buffer (1 M Tris-HCl, 20 mM MgCl2, pH 7.5, and 2 mM sodium azide), 200 µM 2-amino-6-mercapto-7-methylpurine riboside, 0.1 U purine nucleoside phosphorylase, and 0–5 µM MGD protein or the indicated mutants and incubated for 30 min at 37°C in a 96-well plate (Corning). Reactions were initiated by the addition of 0.5 mM GTP. The absorbance at 360 nm was measured every 1 min over 40 min at 37°C by using a microplate reader (Synergy 4; BioTek). The rate of phosphate release was then calculated based on a standard curve.
Trypsin protection assay
2 µg of MGD protein was incubated in the presence or absence of nucleotide (GTP, GDP·AlF4−, GDP·BeF3−, or GDP) and treated with 2 mM indicated amounts of trypsin at 37°C for 10 min. SDS buffer was added to stop the reaction. The samples were analyzed by SDS-PAGE and stained with Coomassie blue.
ITC
ITC was performed at 10°C with a MicroCal iTC200 instrument (GE Healthcare). MGD and mutant proteins were exchanged in ITC buffer containing 20 mM Tris, pH 8.0, 150 mM NaCl, and 4 mM MgCl2. GDP and GMPPNP were directly dissolved in ITC buffer. The concentrations of MGD protein and GDP or GMPPNP used for titration were 100 µM and 2 mM, respectively. Protein concentrations were determined by the absorbance at 280 nm. Acquired ITC data were analyzed by the program Origin 7.0 (GE Healthcare) using the “One Set of Binding Sites” fitting model.
AUC
Purified MGD and mutant proteins were used for AUC in a buffer containing 20 mM Tris, pH 8.0, 150 mM NaCl, and 4 mM MgCl2. Sedimentation velocity experiments were performed at 4°C in a proteomeLab XL-1 Protein Characterization System (Beckman Coulter). Before centrifugation, indicated nucleotide (2 mM GDP; 2 mM GMPPNP; 2 mM GDP, 2.5 mM AlCl3, and 25 mM NaF; or 2 mM GDP, 2.5 mM BeSO4, and 25 mM NaF) was added to 400 µM protein. All interference data were collected at 42,000 rpm using an An-60 Ti rotor (Beckman Coulter). The AUC data were processed according to a c(M) distribution model.
Tethering assays
Wild-type MGD containing the TMs of dmATL was purified in FOS-CHOLINE-12 (Anatrace) and reconstituted into preformed liposomes with Rhodamine-DPPE (Avanti Polar Lipids, Inc.) as previously described (Bian et al., 2011). To check reconstitution efficiency, 30 µl proteoliposomes was mixed with 100 µl of 1.9 M sucrose and overlaid with 100 µl of 1.25 M sucrose and 20 µl of 0.25 M sucrose. The samples were centrifuged in a Beckman TLS 55 rotor at 55,000 rpm for 80 min at 4°C. The gradient was fractionated into five 50-µl fractions, and the samples were analyzed by SDS-PAGE.
For visual assays, different nucleotides at 10 mM were added and the samples diluted 1:50 in reaction buffer. The samples were spotted onto a glass coverslip and visualized by confocal microscopy. Rhodamine dye was excited with 561-nm lasers, and their emissions selected with 620/60–nm filters. For OD405 analysis, absorbance at 405 nm was measured on a Tecan Microplate Reader. The absorbance before nucleotide addition was set to zero.
Mammalian cell culture, transfections, coimmunoprecipitation, and microscopy
The MFN1-deleted mouse embryonic fibroblast (MEF) cell line was obtained from D. Chan’s laboratory (California Institute of Technology, Pasadena, CA). In brief, MEFs were derived from E10.5 embryos of MFN1-knockout mice and infected with a retrovirus expressing SV40 large T antigen for immortalization (Chen et al., 2003). MFN1-deleted MEF or HEK293T were maintained in DMEM plus 10% FBS (Gibco) at 37°C in 5% CO2. Before transfection, cells were trypsinized, seeded on a cover glass, and grown to 50% confluence. Transfections were performed using X-tremeGENE HP DNA Transfection Reagent (Roche) according to the manufacturer’s instructions.
For coimmunoprecipitation experiments, 70% confluent HEK293T cells were transfected with indicated constructs and harvested 24 h later in HKM buffer (25 mM Hepes, pH 7.4, 150 mM potassium acetate, 2 mM magnesium acetate, and protease inhibitors) containing 1% digitonin (EMD Millipore). Cell lysates were incubated with anti-Flag or anti-HA agarose (Sigma-Aldrich) for 2 h at 4°C. Washed precipitates were separated by SDS-PAGE and immunoblotted with anti-Flag or anti-HA antibodies (Sigma-Aldrich).
For immunofluorescence experiments, 24 h after transfection, cells were fixed with 4% PFA in 1x PBS. Primary antibody against Myc tag (Abcam) and Alexa Fluor 488 donkey anti–mouse secondary antibody (Thermo Fisher Scientific) were applied. Mitochondria were stained by 100 nM prewarmed MitoTracker red CMXRos (Invitrogen) for 30 min in 37°C. Images were acquired with a laser scanning confocal microscope (TCS SP5; Leica Biosystems) with a 63×/1.4 NA Plan-Apochromat oil-immersion objective lens and controlled by LAS AF version 1.3.1 build 525 software. Image brightness and contrast were adjusted in Adobe Photoshop.
Accession numbers
The coordinates and structure factors have been deposited in the Protein Data Bank under accession numbers 5GNT, 5GNU, 5GNS, and 5GNR.
Online supplemental material
Fig. S1 shows the CT of MFN1 as part of the N-terminal domain. Fig. S2 shows comparisons of different MFN1 structures. Fig. S3 shows a comparison of MFN1, MFN2, and BDLP.
Supplementary Material
Acknowledgments
We thank T. Rapoport for critically reading the manuscript, A. Prater for proofreading, and the Tsinghua University Branch of China National Center for Protein Sciences (Beijing) for technical support.
L. Yu is a Tsinghua-Peking Center for Life Sciences postdoctoral fellow and is supported by the China Postdoctoral Science Foundation (grant 2014M550713). Z. Lou is supported by the National Natural Science Foundation of China (grant 81322023) and the National Basic Research Program of China (973 Program; grants 2013CB911103 and 2014CB542802). J. Hu is supported by the National Key Research and Development Program (grant 2016YFA0500201), the National Natural Science Foundation of China (grant 31225006), and an International Early Career Scientist grant from the Howard Hughes Medical Institute.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ATL
- atlastin
- AUC
- analytical ultracentrifugation
- BDLP
- bacterial dynamin-like protein
- CMT2A
- Charcot–Marie–Tooth neuropathy type 2A
- CT
- C-terminal tail
- HB
- helix bundle
- HR
- heptad repeat
- ITC
- isothermal titration calorimetry
- MEF
- mouse embryonic fibroblast
- MFN
- mitofusin
- MGD
- minimal GTPase domain
- SeMet
- selenomethionyl
- TM
- transmembrane
References
- Afonine P.V., Grosse-Kunstleve R.W., Echols N., Headd J.J., Moriarty N.W., Mustyakimov M., Terwilliger T.C., Urzhumtsev A., Zwart P.H., and Adams P.D.. 2012. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 68:352–367. 10.1107/S0907444912001308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton F., Dittmar G., Langer T., and Escobar-Henriques M.. 2013. Two deubiquitylases act on mitofusin and regulate mitochondrial fusion along independent pathways. Mol. Cell. 49:487–498. 10.1016/j.molcel.2012.12.003 [DOI] [PubMed] [Google Scholar]
- Bian X., Klemm R.W., Liu T.Y., Zhang M., Sun S., Sui X., Liu X., Rapoport T.A., and Hu J.. 2011. Structures of the atlastin GTPase provide insight into homotypic fusion of endoplasmic reticulum membranes. Proc. Natl. Acad. Sci. USA. 108:3976–3981. 10.1073/pnas.1101643108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt T., Cavellini L., Kühlbrandt W., and Cohen M.M.. 2016. A mitofusin-dependent docking ring complex triggers mitochondrial fusion in vitro. eLife. 5:e14618 10.7554/eLife.14618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrnes L.J., and Sondermann H.. 2011. Structural basis for the nucleotide-dependent dimerization of the large G protein atlastin-1/SPG3A. Proc. Natl. Acad. Sci. USA. 108:2216–2221. 10.1073/pnas.1012792108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Detmer S.A., Ewald A.J., Griffin E.E., Fraser S.E., and Chan D.C.. 2003. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J. Cell Biol. 160:189–200. 10.1083/jcb.200211046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daumke O., and Praefcke G.J.. 2016. Invited review: Mechanisms of GTP hydrolysis and conformational transitions in the dynamin superfamily. Biopolymers. 105:580–593. 10.1002/bip.22855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., and Cowtan K.. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- Hales K.G., and Fuller M.T.. 1997. Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell. 90:121–129. 10.1016/S0092-8674(00)80319-0 [DOI] [PubMed] [Google Scholar]
- Harrison S.C. 2008. Viral membrane fusion. Nat. Struct. Mol. Biol. 15:690–698. 10.1038/nsmb.1456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann G.J., Thatcher J.W., Mills J.P., Hales K.G., Fuller M.T., Nunnari J., and Shaw J.M.. 1998. Mitochondrial fusion in yeast requires the transmembrane GTPase Fzo1p. J. Cell Biol. 143:359–373. 10.1083/jcb.143.2.359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., and Nunnari J.. 2009. The molecular mechanism of mitochondrial fusion. Biochim. Biophys. Acta. 1793:20–26. 10.1016/j.bbamcr.2008.07.005 [DOI] [PubMed] [Google Scholar]
- Hu J., and Rapoport T.A.. 2016. Fusion of the endoplasmic reticulum by membrane-bound GTPases. Semin. Cell Dev. Biol. 10.1016/j.semcdb.2016.06.001 [DOI] [PubMed] [Google Scholar]
- Huang P., Galloway C.A., and Yoon Y.. 2011. Control of mitochondrial morphology through differential interactions of mitochondrial fusion and fission proteins. PLoS One. 6:e20655 10.1371/journal.pone.0020655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N., Eura Y., and Mihara K.. 2004. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. J. Cell Sci. 117:6535–6546. 10.1242/jcs.01565 [DOI] [PubMed] [Google Scholar]
- Jahn R., and Scheller R.H.. 2006. SNAREs--engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7:631–643. 10.1038/nrm2002 [DOI] [PubMed] [Google Scholar]
- Koshiba T., Detmer S.A., Kaiser J.T., Chen H., McCaffery J.M., and Chan D.C.. 2004. Structural basis of mitochondrial tethering by mitofusin complexes. Science. 305:858–862. 10.1126/science.1099793 [DOI] [PubMed] [Google Scholar]
- Lackner L.L., and Nunnari J.M.. 2009. The molecular mechanism and cellular functions of mitochondrial division. Biochim. Biophys. Acta. 1792:1138–1144. 10.1016/j.bbadis.2008.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liesa M., Palacín M., and Zorzano A.. 2009. Mitochondrial dynamics in mammalian health and disease. Physiol. Rev. 89:799–845. 10.1152/physrev.00030.2008 [DOI] [PubMed] [Google Scholar]
- Liu T.Y., Bian X., Sun S., Hu X., Klemm R.W., Prinz W.A., Rapoport T.A., and Hu J.. 2012. Lipid interaction of the C terminus and association of the transmembrane segments facilitate atlastin-mediated homotypic endoplasmic reticulum fusion. Proc. Natl. Acad. Sci. USA. 109:E2146–E2154. 10.1073/pnas.1208385109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.Y., Bian X., Romano F.B., Shemesh T., Rapoport T.A., and Hu J.. 2015. Cis and trans interactions between atlastin molecules during membrane fusion. Proc. Natl. Acad. Sci. USA. 112:E1851–E1860. 10.1073/pnas.1504368112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low H.H., and Löwe J.. 2006. A bacterial dynamin-like protein. Nature. 444:766–769. 10.1038/nature05312 [DOI] [PubMed] [Google Scholar]
- Low H.H., Sachse C., Amos L.A., and Löwe J.. 2009. Structure of a bacterial dynamin-like protein lipid tube provides a mechanism for assembly and membrane curving. Cell. 139:1342–1352. 10.1016/j.cell.2009.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S., and McMahon H.T.. 2008. Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9:543–556. 10.1038/nrm2417 [DOI] [PubMed] [Google Scholar]
- Mishra P., and Chan D.C.. 2016. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 212:379–387. 10.1083/jcb.201511036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., and Minor W.. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Pernas L., and Scorrano L.. 2016. Mito-morphosis: mitochondrial fusion, fission, and cristae remodeling as key mediators of cellular function. Annu. Rev. Physiol. 78:505–531. 10.1146/annurev-physiol-021115-105011 [DOI] [PubMed] [Google Scholar]
- Rapaport D., Brunner M., Neupert W., and Westermann B.. 1998. Fzo1p is a mitochondrial outer membrane protein essential for the biogenesis of functional mitochondria in Saccharomyces cerevisiae. J. Biol. Chem. 273:20150–20155. 10.1074/jbc.273.32.20150 [DOI] [PubMed] [Google Scholar]
- Rojo M., Legros F., Chateau D., and Lombès A.. 2002. Membrane topology and mitochondrial targeting of mitofusins, ubiquitous mammalian homologs of the transmembrane GTPase Fzo. J. Cell Sci. 115:1663–1674. [DOI] [PubMed] [Google Scholar]
- Santel A., and Fuller M.T.. 2001. Control of mitochondrial morphology by a human mitofusin. J. Cell Sci. 114:867–874. [DOI] [PubMed] [Google Scholar]
- Schrepfer E., and Scorrano L.. 2016. Mitofusins, from mitochondria to metabolism. Mol. Cell. 61:683–694. 10.1016/j.molcel.2016.02.022 [DOI] [PubMed] [Google Scholar]
- Südhof T.C., and Rothman J.E.. 2009. Membrane fusion: grappling with SNARE and SM proteins. Science. 323:474–477. 10.1126/science.1161748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W., and Schekman R.. 2008. Membrane fusion. Nat. Struct. Mol. Biol. 15:658–664. 10.1038/nsmb.1451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Sun S., Wang W., Shi J., Hu X., Wang S., Su D., Rao Z., Hu J., and Lou Z.. 2015. Structures of the yeast dynamin-like GTPase Sey1p provide insight into homotypic ER fusion. J. Cell Biol. 210:961–972. 10.1083/jcb.201502078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle R.J., and van der Bliek A.M.. 2012. Mitochondrial fission, fusion, and stress. Science. 337:1062–1065. 10.1126/science.1219855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züchner S., Mersiyanova I.V., Muglia M., Bissar-Tadmouri N., Rochelle J., Dadali E.L., Zappia M., Nelis E., Patitucci A., Senderek J., et al. . 2004. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet. 36:449–451. 10.1038/ng1341 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.