Key Points
Matching for MICA significantly reduces the incidence of acute and chronic GVHD in otherwise HLA 10/10-matched unrelated-donor HCT.
Our results formally define MICA as a novel major histocompatibility complex-encoded human transplantation antigen.
Abstract
Graft-versus-host disease (GVHD) is among the most challenging complications in unrelated donor hematopoietic cell transplantation (HCT). The highly polymorphic MHC class I chain–related gene A, MICA, encodes a stress-induced glycoprotein expressed primarily on epithelia. MICA interacts with the invariant activating receptor NKG2D, expressed by cytotoxic lymphocytes, and is located in the MHC, next to HLA-B. Hence, MICA has the requisite attributes of a bona fide transplantation antigen. Using high-resolution sequence-based genotyping of MICA, we retrospectively analyzed the clinical effect of MICA mismatches in a multicenter cohort of 922 unrelated donor HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 10/10 allele-matched HCT pairs. Among the 922 pairs, 113 (12.3%) were mismatched in MICA. MICA mismatches were significantly associated with an increased incidence of grade III-IV acute GVHD (hazard ratio [HR], 1.83; 95% confidence interval [CI], 1.50-2.23; P < .001), chronic GVHD (HR, 1.50; 95% CI, 1.45-1.55; P < .001), and nonelapse mortality (HR, 1.35; 95% CI, 1.24-1.46; P < .001). The increased risk for GVHD was mirrored by a lower risk for relapse (HR, 0.50; 95% CI, 0.43-0.59; P < .001), indicating a possible graft-versus-leukemia effect. In conclusion, when possible, selecting a MICA-matched donor significantly influences key clinical outcomes of HCT in which a marked reduction of GVHD is paramount. The tight linkage disequilibrium between MICA and HLA-B renders identifying a MICA-matched donor readily feasible in clinical practice.
Introduction
Allogeneic hematopoietic cell transplantation (HCT) is a well-established treatment of a large variety of diseases affecting the immune and/or hematopoietic systems.1 Well over a million HCTs have been performed to date.2 More than 40% of these were allogeneic, of which about half were from unrelated donors. Despite these encouraging developments, post-HCT adverse clinical outcomes are still frequent and remain a daily challenge. Among these, graft-versus-host disease (GVHD) is of chief concern, as it remains the most common life-threatening post-HCT complication and single handedly hampers the final outcome of this powerful and often unique curative option.3-5
Increasing the degree of HLA matching between the donor and the recipient is one of the most important paths to decrease the risk for post-HCT complications, and for GVHD in particular.6,7 That is why, in the absence of an HLA-identical sibling donor, an 8/8 or preferably a 10/10 allele-matched (HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1) unrelated donor is the best alternative option. But even under these stringent matching conditions, the incidence of GVHD remains quite high (ie, 50% of patients develop grade II-IV acute GVHD, up to 35% grade III-IV acute GVHD, and 40% to 50% chronic GVHD).8-10 The greater GVHD incidence in HLA allele-matched (ie, unrelated) vs haplotype-matched (ie, sibling) HCT hints at the existence of intra–major histocompatibility complex (MHC)–encoded, yet HLA-A-, HLA-B-, HLA-C-, HLA-DRB1-, and HLA-DQB1-independent (and hence hitherto unidentified), histocompatibility loci.11,12
Other than HLA-DPB1,13-17 the most promising MHC-encoded candidate is the MHC class I chain–related gene A (MICA). MICA is a highly polymorphic nonconventional MHC class I molecule, with 105 alleles reported to date. It encodes a single-chain (β2-microglobulin-independent), cargo (peptide)-free cell surface glycoprotein that is upregulated by cell stress.18 The MICA gene was discovered by the senior author (S.B.) 20 years ago and is located within the MHC, 46 kb centromeric to the HLA-B locus.19 The MICA glycoprotein is expressed on a restricted number of cell types, mainly epithelial. MICA binds NKG2D, an activating receptor expressed on the surface of cytotoxic CD8+ αβ and γδ T lymphocytes, as well as natural killer (NK) cells.18 Within the intestinal epithelium, MICA is also recognized by the T-cell receptor of Vδ1-bearing γδ T cells.20 Previous studies have hinted at a role for MICA in GVHD, but these analyzed MICA diversity only in patients (not donors) and/or in small, single-center cohorts, which engendered controversial, if not opposite, conclusions.21-25 Here we evaluate the MICA sequence in a retrospective multicenter European cohort of 922 patients having undergone HCT with HLA 10/10-allele-matched unrelated donors. All known covariables relevant to HCT were included in the final analysis. The results uncover the relevance of MICA matching in unrelated donor HCT and provide the rationale for including MICA typing in the donor selection process.
Methods
Study design and oversight
This retrospective study aimed to test whether donor/patient matching at the MICA locus improves the outcomes of unrelated donor HCT. Patients (and their donors) from 6 French and 2 Dutch centers were included. Genomic DNA and high-resolution HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-DPB1 genotyping data were collected for all donor–recipient pairs. Clinical information was made available by the Francophone Society of Bone Marrow Transplantation and Cell Therapies (SFGM-TC) and the Haemato Oncology Foundation for Adults in the Netherlands through the European Society for Blood and Marrow Transplantation ProMISe database. All authors vouch for the accuracy and completeness of the results. The funding agencies did not play any role in study design, data collection, analysis, decision as to whether to submit the manuscript for publication, or its content. The study, conducted under the auspices of SFGM-TC and the Haemato Oncology Foundation for Adults in the Netherlands, was approved by institutional review boards of participating centers and performed according to the principles of the Helsinki declaration. Written informed consent was obtained from all participants.
Patients and donors
The study population consisted of 922 patients (and donors) who underwent unrelated donor HCT for the treatment of blood disorders between 1992 and 2013. All patients received a first unrelated donor HCT, using bone marrow or peripheral blood stem cells from donors who were matched for 10 of the 10 possible alleles at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci (Table 1). HLA-DPB1 genotyping data were available for all donors and recipients. Further inclusion requirements were thus the availability of DNA from both donors and recipients, as well as access to HLA-DPB1 typing data.
Table 1.
Demographics of the study population
| Total transplants (n = 922) | MICA-matched transplants (n = 809) | MICA-mismatched transplants (n = 113) | P value* | |
|---|---|---|---|---|
| Transplantation centers† | .09 | |||
| 1 | 103 (11%) | 83 (10%) | 20 (18%) | |
| 2 | 211 (23%) | 180 (22%) | 31 (27%) | |
| 3 | 128 (14%) | 113 (14%) | 15 (13%) | |
| 4 | 166 (18%) | 144 (18%) | 22 (20%) | |
| 5 | 56 (6%) | 52 (6%) | 4 (4%) | |
| 6 | 90 (10%) | 82 (10%) | 8 (7%) | |
| 7 | 100 (11%) | 94 (12%) | 6 (5%) | |
| 8 | 68 (7%) | 61 (8%) | 7 (6%) | |
| Age at transplant, years | .02 | |||
| 0-17 | 78 (9%) | 71 (9%) | 7 (6%) | |
| 18-49 | 387 (42%) | 324 (40%) | 63 (56%) | |
| 50-64 | 400 (43%) | 363 (45%) | 37 (33%) | |
| 65 or older | 57 (6%) | 51 (6%) | 6 (5%) | |
| Year of transplantation | .81 | |||
| 1992-2003 | 88 (9%) | 79 (10%) | 9 (8%) | |
| 2004-2007 | 256 (28%) | 225 (28%) | 31 (27%) | |
| 2008-2013 | 578 (63%) | 505 (62%) | 73 (65%) | |
| Patient–donor sex | .07 | |||
| Male–Female | 159 (17%) | 132 (16%) | 27 (24%) | |
| Other combinations | 755 (82%) | 669 (83%) | 86 (76%) | |
| Missing | 8 (< 1%) | 8 (1%) | 0 (0%) | |
| Patient–donor serological status for cytomegalovirus | 1.00 | |||
| Negative–negative | 363 (39%) | 319 (39%) | 44 (39%) | |
| Other combinations | 543 (59%) | 476 (59%) | 67 (59%) | |
| Missing | 16 (< 2%) | 14 (2%) | 2 (2%) | |
| Source of cells | .86 | |||
| Bone marrow | 247 (27%) | 218 (27%) | 29 (26%) | |
| Peripheral blood stem cells | 675 (73%) | 591 (73%) | 84 (74%) | |
| Conditioning regimen | .45 | |||
| Nonmyeloablative/reduced-intensity | 563 (61%) | 496 (61%) | 67 (59%) | |
| Myeloablative without total-body irradiation | 145 (16%) | 123 (15%) | 22 (19%) | |
| Myeloablative with total-body irradiation | 211 (23%) | 188 (23%) | 23 (20%) | |
| Missing | 3 (< 1%) | 2 (< 1%) | 1 (< 1%) | |
| GVHD prophylaxis | .42 | |||
| Cyclosporin only | 198 (21%) | 172 (21%) | 26 (23%) | |
| Cyclosporin and methotrexate | 305 (33%) | 269 (33%) | 36 (32%) | |
| Cyclosporin and mycophenolate | 282 (31%) | 243 (30%) | 39 (35%) | |
| Other combinations | 123 (13%) | 113 (14%) | 10 (9%) | |
| Missing | 14 (< 2%) | 12 (< 2%) | 2 (< 2%) | |
| In vivo T-cell depletion ‡ | .39 | |||
| No | 283 (31%) | 244 (30%) | 39 (35%) | |
| Yes | 625 (68%) | 553 (68%) | 72 (64%) | |
| Missing | 14 (< 2%) | 12 (< 2%) | 2 (< 2%) | |
| Disease | .51 | |||
| Acute myeloid leukemia | 235 (25%) | 203 (25%) | 32 (28%) | |
| Chronic myeloid leukemia | 53 (6%) | 49 (6%) | 4 (4%) | |
| Acute lymphoblastic leukemia | 135 (15%) | 120 (15%) | 15 (13%) | |
| Myelodysplastic syndrome | 140 (15%) | 128 (16%) | 12 (11%) | |
| Non-Hodgkin lymphoma | 118 (13%) | 101 (12%) | 17 (15%) | |
| Others¶ | 241 (26%) | 208 (26%) | 33 (29%) | |
| Disease stage at transplantation§ | .54 | |||
| Early | 385 (42%) | 340 (42%) | 45 (40%) | |
| Late | 480 (52%) | 420 (52%) | 60 (53%) | |
| Not applicable | 34 (4%) | 28 (3%) | 6 (5%) | |
| Unknown | 23 (2%) | 21 (3%) | 2 (2%) | |
| Time from diagnosis until transplantation | .64 | |||
| <12 mo | 431 (47%) | 381 (47%) | 50 (44%) | |
| >12 mo | 491 (53%) | 428 (53%) | 63 (56%) | |
| HLA-DPB1 matching‖ | .69 | |||
| Matched | 100 (11%) | 86 (11%) | 14 (12%) | |
| Mismatched | 822 (89%) | 723 (89%) | 99 (88%) |
Results are presented as number of patients and corresponding percentages of the study population. All clinical variables of the table were used for adjustment in the multivariate models.
HLA, human leukocyte antigen.
P-values were determined with the Pearson’s χ square test.
Patients received their transplant in 6 centers of the Francophone Society of Bone Marrow Transplantation and Cell Therapies (SFGM-TC) (1 to 6; N = 754) and in 2 Dutch centers part of the Europdonor operated by Matchis Foundation network (7 and 8; N=168).
In vivo T-cell depletion was performed by addition of antithymocyte globulin or Alemtuzumab to the conditioning regimen.
Other diseases include multiple myeloma, Hodgkin lymphoma, Fanconi anemia, aplastic anemia, chronic lymphocytic leukemia, plasma cell leukemia, other acute leukemias, solid tumors (not breast), hemophagocytosis, and inherited disorders.
Early corresponds to diseases in first complete remission or in chronic phase. Late corresponds to second or higher complete remissions, accelerated phases, partial remissions, progressions, primary induction failures, relapses, or stable diseases. Not applicable corresponds to bone marrow failure (aplastic anemia, Fanconi anemia), inherited disorders, hemophagocytosis, and solid tumors. The Disease Risk index, as defined by Armand et al,49 was also evenly distributed in the MICA-matched and MICA-mismatched patients (P = 0.423) (supplemental Table 7).
HLA-DPB1 matching was defined with typing data at second-field resolution following the World Health Organization official nomenclature.
MICA genotyping
MICA was genotyped in all donors and recipients by sequenced-based typing: exons 2, 3, and 4 were Sanger sequenced bidirectionally, and the transmembrane microsatellite polymorphism was genotyped, following our previously established protocols.26,27 The sequences and the transmembrane microsatellite length were analyzed using SeqScape v2.6 and GeneMapper v4.0 software (ThermoFisher Scientific), respectively. Final MICA genotypes were assigned using an in-house-developed software that compiled sequence data and transmembrane genotypes. Ambiguous results were resolved by polymerase chain reaction amplification with sequence-specific primers. After this procedure, analysis of matching/mismatching between recipients and donors was performed at an identical level of resolution (second field in the HLA nomenclature) for both HLA and MIC genes.
Definitions
MICA matching was dissected using 4 different categories of mismatches: all types of mismatches, independent of their directions; mismatches in the GvH direction (the recipient, but not the donor, is mismatched), mismatches in the host-versus-graft (HvG) direction (the donor, but not the recipient, is mismatched), and bidirectional mismatches (all combinations except GvH and HvG mismatches). An HLA 10/10 match refers to a donor–recipient pair matched for HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 genes at the allele level. For HLA-DPB1 and MICA loci, a mismatched donor–recipient pair refers to a pair with 1 or 2 mismatched alleles. For the analysis presented in supplemental Table 5, available on the Blood Web site, HLA-DPB1 mismatches were classified into permissive and nonpermissive mismatches according to the latest version (2.0) of the DPB1 T-Cell Epitope Algorithm available at the IPD-IMGT/HLA web site (https://www.ebi.ac.uk/ipd/imgt/hla/dpb.html).28 Grading of acute and chronic GVHD was performed according to the classification of Glucksberg et al.29 For acute GVHD, severe corresponds to grades III and IV. Overall survival (OS) was defined as time from transplantation to death by any cause. Relapse-free survival (RFS) was defined as time to relapse of primary disease or death by any cause, whichever came first. Nonrelapse mortality (NRM) corresponds to mortality within the first complete remission of disease. The causes of NRM are summarized in supplemental Table 1. Causes of death unrelated to transplantation included deaths related to relapse, progression of original disease, secondary malignancy, and cell therapy (non-HCT). OS, RFS, and NRM were censored at the time of the last follow-up. Incidences of clinical outcomes were defined as the cumulative probability of the outcomes at any given point.
Statistical analysis
The primary endpoints of the study were severe (ie, grades III-IV) acute and chronic GVHD. Relapse, OS, RFS, and NRM were also tested as secondary endpoints. Multivariate analysis of OS and RFS was performed, using extended Cox models.30 For acute GVHD III-IV, chronic GVHD, relapse, and NRM, competing risks analysis was used using an extended Fine and Gray model.31-33 Death without GVHD and relapse were considered competing events for the GVHD endpoints (acute III-IV and chronic GVHD). Death from any other cause than transplantation was the competing event for NRM. Death from any cause and GVHD (acute and chronic) were the competing events for relapse. All statistical models were adjusted for center effect34 and covariates defining the European Society for Blood and Marrow Transplantation risk score: patient age, disease stage at transplantation, time to transplantation, and donor–recipient sex combination.35 In addition to these, the following relevant variables were included: HLA-DPB1 matching (all and nonpermissive alleles, respectively), patient–donor serological status for cytomegalovirus, year of transplantation, source of stem cells, conditioning regimen, GVHD prophylaxis, treatment with antithymocyte globulin or Alemtuzumab, and disease category. All models were evaluated for interactions and proportional hazards assumption. Violations of the proportional hazards assumption were adjusted using time-dependent covariates. No adjustments were made for multiple comparisons. All reported P values were 2-sided.
Results
The demographics of the study population are shown in Table 1. The median posttransplant follow-up was 32 months (mean, 37 months; 95% confidence interval [CI], 34-40 months), and the median patient age was 50 years (mean, 45 years; 95% CI, 44-47 years). Patients suffered from both malignant and nonmalignant diseases. Most transplants were performed with nonmyeloablative/reduced-intensity conditioning regimens (61%); in vivo T-cell depletion was performed in the majority of cases (68%), and peripheral blood was the main source for stem cells (73%). All donor–patient pairs were fully typed at high resolution (second field) for HLA-A, HLA-B, HLA-C, HLA-DRB1, HLA-DQB1, and HLA-DPB1 and were matched for 10 of 10 alleles at HLA-A, HLA-B, HLA-C, HLA-DRB1, and HLA-DQB1 loci (HLA 10/10 matched). In all 1844 samples, exons encoding MICA’s extracellular α1, α2, and α3 domains were completely sequenced, and the genotype of the trans-membrane exon encoded microsatellite was established, resulting in high-resolution allele-typing of MICA. Of these 922 transplantations, 822 (89.2%) and 113 (12.3%) were mismatched at the HLA-DPB1 and MICA loci, respectively. Only 26 of the 105 known MICA alleles were identified in the study population, and their distribution was similar in both patients and donors (supplemental Table 2). The observed allele frequencies were in line with data obtained from other European and European-American populations for this locus.18 All relevant covariates for the analyzed clinical outcomes were equally distributed in the MICA-matched and MICA-mismatched patients, with the exception of patients’ age category at the time of transplant, which reflects the fact that older patients (age > 49 years) were slightly (P = .02) better matched for MICA (Table 1).
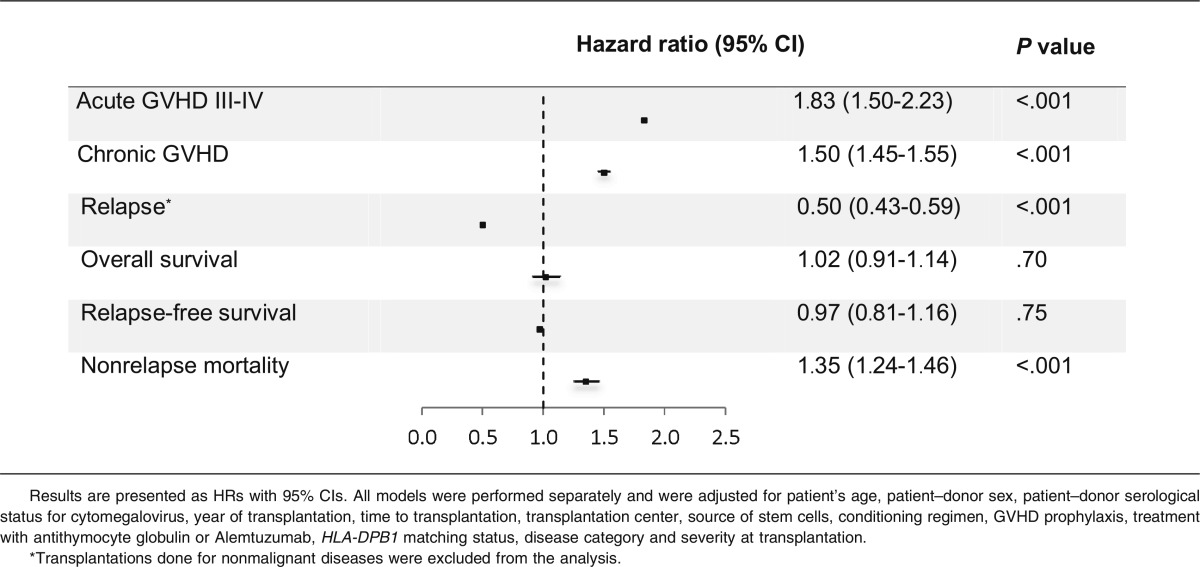
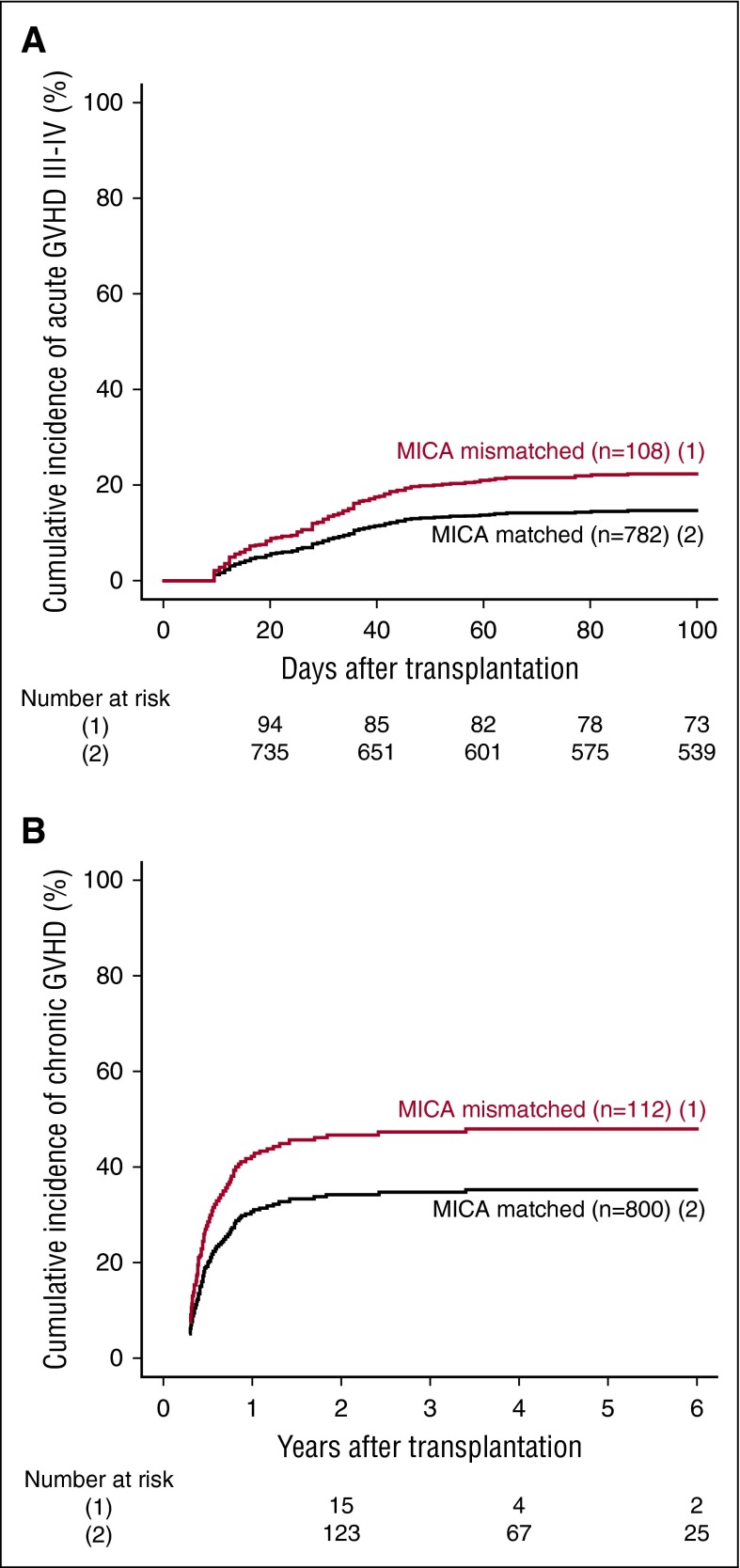
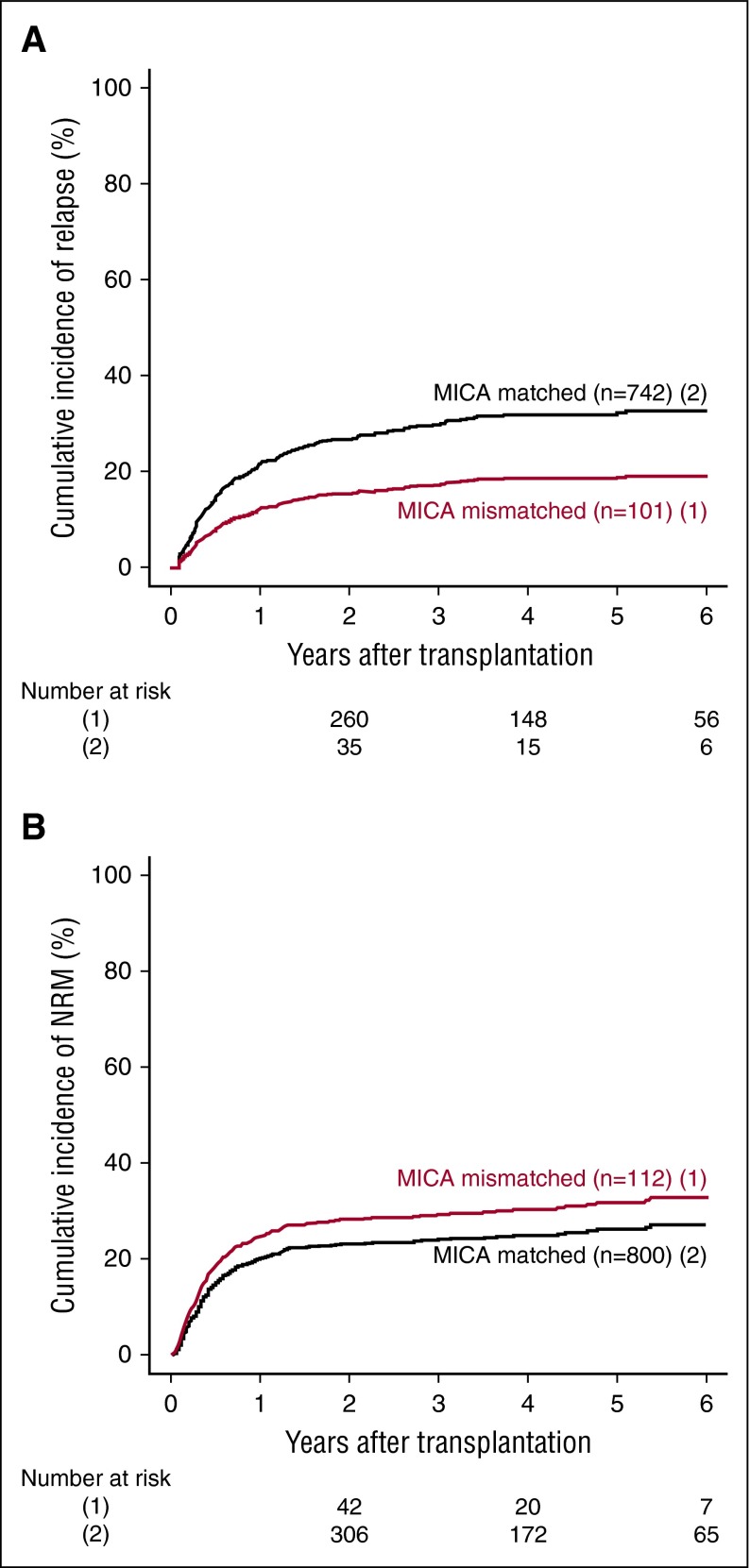
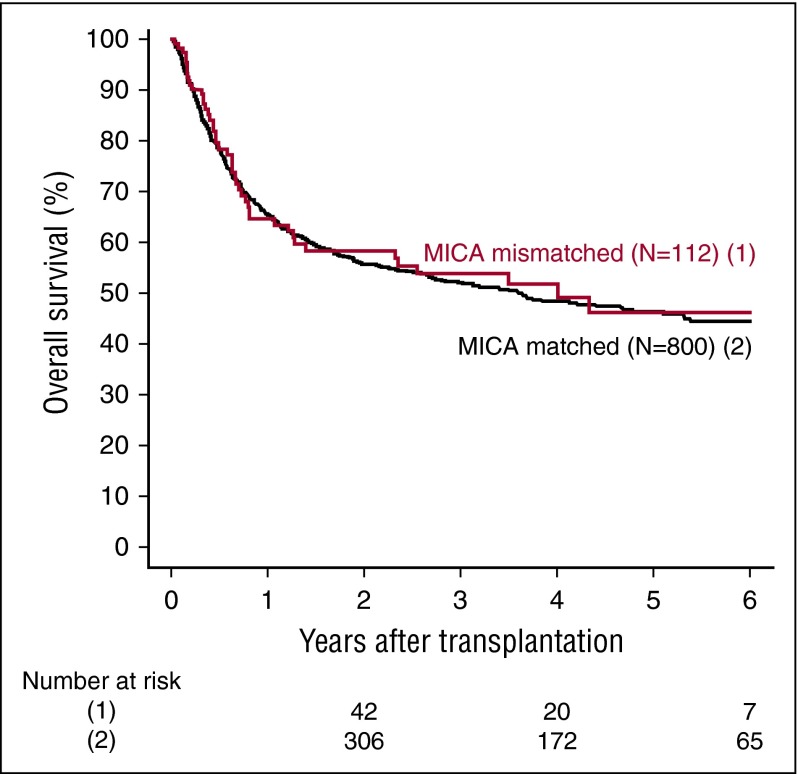
MICA mismatches were significantly associated with an increased incidence of severe (grades III-IV) acute GVHD (hazard ratio [HR], 1.83; 95% CI, 1.50-2.23; P < .001) (Table 2). At day 100 post-HCT, the cumulative incidences of severe acute GVHD in mismatched vs matched transplantations were 22.3% vs 14.7% (Figure 1A). Organ-specific subanalyses showed that this effect was more important in the gut (P = .004) and the skin (P = .006) than in the liver (P = .09; supplemental Figure 1). MICA mismatches showed also a significant effect on acute GVHD grades II-IV (HR, 1.27; 95% CI, 1.10-1.52; P = .009) and grades I-IV (HR, 1.47; 95% CI, 1.38-1.57; P < .001; supplemental Table 3). Chronic GVHD was also associated with MICA mismatches (HR, 1.50; 95% CI, 1.45-1.55; P < .001) (Table 2) and showed a 12.7% lower incidence at 6 years post-HCT in the MICA-matched vs mismatched groups (Figure 1B). Only the limited form of chronic GVHD was affected by MICA mismatches (HR, 1.99; 95% CI, 1.63-2.49; P < .001; supplemental Table 3). Interestingly, MICA-mismatched patients had a statistically significant lower hazard of disease relapse compared with MICA-matched patients (HR, 0.50, 95% CI 0.43-0.59; P < .001) (Table 2). Six years post-HCT, the estimated incidence of relapse was 10.2% in the mismatch group vs 18.3% in the matched group (Figure 2A). Analysis of patient survival, through assessment of OS, RFS, and NRM, revealed a significant difference in adjusted HRs in the MICA-matched (27.2% incidence at 6 years post-HCT) vs mismatched (32.3% incidence at 6 years post-HCT) groups solely for NRM (Figure 2B; Table 2). Total MICA-matching (not taking into account the mismatch direction; cf. infra) had no effect on OS and on RFS (Table 2; Figure 3).
Table 2.
Analysis of the effect of MICA mismatches on clinical outcomes after multivariate modeling
Figure 1.
Effect of MICA matching on GVHD. The cumulative incidences of grades III-IV acute GVHD (A) and chronic GVHD (B) are shown for mismatched (1) vs matched (2) patients at the MICA locus.
Figure 2.
Effect of MICA matching on relapse and NRM. The cumulative incidence of relapse (A) and NRM (B) for mismatched (1) vs matched (2) patients at the MICA locus are shown.
Figure 3.
Effect of MICA matching on OS. Kaplan Meier estimates of OS are shown.
The analysis of the effect of the direction of MICA mismatches revealed that acute and chronic GVHD were globally independent from the mismatch direction (Table 3). Except mismatches in the HvG direction; all types of mismatches showed a statistically significant protective effect on relapse (GvH direction: HR, 0.49; 95% CI 0.38-0.62; P < .001; bidirectional: HR, 0.46; 95% CI, 0.34-0.61; P < .001). Patients with GvH mismatches had a higher mortality compared with patients without GvH mismatches (HR, 1.63; 95% CI, 1.09-2.43; P = .017), whereas patients with HvG mismatches had a lower mortality compared with patients without HvG mismatches (HR, 0.62; 95% CI, 0.40-0.96; P = .033). Finally, all types of mismatches except HvG mismatches were associated with a higher rate of NRM (Table 3).
Table 3.
Effect of the direction of MICA mismatches on relevant clinical outcomes
| All mismatches (n = 113) | GvH mismatches (n = 11) | HvG mismatches (n = 17) | Bidirectional mismatches (n = 85) | |||||
|---|---|---|---|---|---|---|---|---|
| HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | HR (95% CI) | P value | |
| Acute GVHD III-IV | 1.83 (1.50-2.23) | <.001 | 1.64 (1.39-1.93) | <.001 | 1.86 (1.18-2.93) | .008 | 1.78 (1.45-2.19) | <.001 |
| Chronic GVHD | 1.50 (1.45-1.55) | <.001 | 1.60 (1.08-2.37) | .019 | 1.83 (0.80-4.15) | .150 | 1.38 (1.12-1.70) | .002 |
| Relapse* | 0.50 (0.43-0.59) | <.001 | 0.49 (0.38-0.62) | <.001 | 0.99 (0.66-1.47) | .96 | 0.46 (0.34-0.61) | <.001 |
| Overall survival | 1.02 (0.91-1.14) | .70 | 1.63 (1.09-2.43) | .017 | 0.62 (0.40-0.96) | .033 | 0.99 (0.88-1.11) | .864 |
| Relapse-free survival | 0.97 (0.81-1.16) | .75 | 1.40 (0.97-2.02) | .075 | 1.02 (0.66-1.56) | .944 | 0.85 (0.69-1.06) | .145 |
| Nonrelapse mortality | 1.35 (1.24-1.46) | <.001 | 2.18 (1.41-3.37) | <.001 | 1.07 (0.97-1.18) | .151 | 1.19 (1.05-1.34) | .005 |
N represents the number of mismatched patients considered. Results are presented as HRs with 95% CIs. All models were performed separately and were adjusted for patient’s age, patient–donor sex, patient–donor serological status for cytomegalovirus, year of transplantation, time to transplantation, transplantation center, source of stem cells, conditioning regimen, GVHD prophylaxis, treatment with antithymocyte globulin or Alemtuzumab, HLA-DPB1 matching status, disease category, and severity at transplantation.
Transplantations done for nonmalignant diseases were excluded from this analysis.
It is further of note that all factors previously shown to be associated with the outcomes analyzed here are replicated in our cohort, with the exception of plain HLA-DPB1 mismatches (supplemental Table 4). We further analyzed HLA-DPB1 mismatches through their classification into permissive vs nonpermissive, according to their belonging to different T-cell epitope groups.36 Although, as previously reported, HLA-DPB1 nonpermissive mismatches were associated with an increased incidence of acute GVHD III-IV and NRM (supplemental Table 5), they did not affect our conclusions (ie, the effect of MICA mismatches on acute GVHD III-IV, chronic GVHD, relapse, and NRM; supplemental Table 5). Finally, we found no association of mismatches at MICA amino acid position 129 (known to affect binding affinity for NKG2D) with any of the tested clinical endpoints (supplemental Table 6).
Discussion
Here we report that HCT from a MICA-mismatched, but otherwise fully HLA 10/10-matched donor, carries a significantly increased risk for acute severe (grade III-IV) GVHD, chronic GVHD, and NRM. The increased GVHD incidence was accompanied by a decrease in the relapse rate. This decrease could be a simple consequence of the higher GVHD incidences, or possibly a graft-versus-leukemia effect mediated by MICA (see the third paragraph of “Discussion”).37 The fact that the sole survival endpoint showing an increase was NRM (HR, 1.35) is consistent with a masking effect by the lower rate of relapse. This is further corroborated by the detailed analysis of MICA mismatch directions, which shows that the abovementioned decreased relapse rate is not observed when considering mismatches in the HvG direction. The absence of effect of global and bidirectional mismatches on OS can be explained by opposite effects of HvG and GvH mismatches. Finally, MICA influence on HCT outcome is independent of HLA-DPB1. These data formally define MICA as a bona fide transplantation antigen and provide the rationale for including MICA typing in the donor selection process.
Several publications have previously analyzed a role for MICA in HCT.21-25 In 2 independent small cohorts of sibling and unrelated donor HCT, respectively, the recipients (but not the donors) were genotyped for the SNP encoding p.M129V,38 which was found to be significantly associated with clinical outcome, albeit in opposite trends.21,25 No significant mismatch in this SNP was found to be associated with any clinical outcome in our cohort (supplemental Table 6). Parmar et al22 reported a higher rate of grade II-IV acute GVHD in MICA-mismatched vs matched patients. These findings were, however, immediately challenged.23 Finally, Askar et al24 reported a link between MICA mismatch (in conjunction with HLA-DPB1) and grade II-IV acute GVHD, but in univariate analysis only. The fact that these studies analyzed only recipients (and not donors) in small (<200 pairs), single-center cohorts that lacked full HLA matching/data and/or critical covariates or were restricted to unidirectional mismatch analyses, and so on, raised more questions than provided answers. Thus, the present study is by far the largest reported MICA full-sequence analysis in HCT (or any organ transplant). Moreover, it includes all relevant GVHD clinical covariates and demonstrates an independent effect of MICA mismatching on GVHD.
Recent data in murine models of allogeneic HCT corroborate our findings by showing that NKG2D activation enhances cytotoxicity and survival of CD8+ T cells and critically contributes to GVHD.39 Given that MICA expression is upregulated in intestinal tissues of patients with GVHD,40 and that the effects of MICA mismatches on severe acute GVHD are more significant in skin and gut, both epithelial tissues known to express MICA (supplemental Figure 1), it is plausible that a donor/recipient MICA mismatch directly affects the cytotoxicity of NKG2D (which is monomorphic) bearing T/NK cells and/or the intestinal Vδ1 T-cell receptor bearing γδ T cells. Indeed, it is well-established that certain MICA alleles/amino acids affect its binding affinity for NKG2D (for review, see Carapito and Bahram18; for an example, see Mellergaard et al41). Furthermore, and in accordance with a rather restricted expression pattern for the MICA glycoprotein, it is perhaps not surprising that MICA mismatches are associated with limited (and not extensive; ie, multiorgan) chronic GVHD (supplemental Table 3). Alternatively, MICA can act as a minor histocompatibility locus (ie, be a source of polymorphic antigenic peptides presented by cognate and/or donor classical MHC molecules, similar to H60 and H-Y in mouse or H-Y and HA-1 in man42), and hence participate in GVHD pathophysiology through its contribution to alloreactivity. Although this second possibility is theoretically feasible43 (and, interestingly, there is a peculiar precedent for this: the murine minor histocompatibility locus, H60, encodes an MHC-I-like structure that is a NKG2D ligand43,44), we much favor the first option, especially given that both NK and γδ T are considered important effector cells that mediate GVL reactivity within the first weeks after transplantation.45,46 In addition, although only NK and γδ T cells have been shown to detect MICA through NKG2D and/or TCR, it remains possible that other T-cell subsets detect MICA polymorphisms, whether through NKG2D and/or the TCR. In this respect, it will be important to isolate alloreactive, including αβ, T cells and determine whether any of these cells detect MIC polymorphisms in vivo. We believe the present work mandates these studies.
Collectively, these results suggest that the pretransplantation MICA typing may help in risk assessment/management of GVHD. Fortunately, because of the high degree of linkage disequilibrium with HLA-B, up to 88% of HLA 10/10 matched donor–patient pairs are also matched for MICA. Moreover, MICA displays a comparatively lower allelic variability than classical HLA genes (∼100 alleles for MICA vs ∼4100 for HLA-B; supplemental Table 2). Therefore, finding a MICA-matched donor should be relatively easy in clinical practice and could provide a straightforward mean to lower the incidence of both acute and chronic GVHD. In this respect, analogy with HLA-C is instructive, as this is the most recent HLA gene to be part of the search criteria for an unrelated donor.47 Indeed, 68% (vs 88% for MICA) of donors matched at high resolution for HLA-A, HLA-B, HLA-DRB1, and HLA-DQB1 were found to be also a full match at HLA-C.48 Matching at this locus diminishes the risk for acute GVHD (HR, 1.19-2.02)47 in near-identical proportions as that for MICA. Finally, for patients at high risk for relapse because of advanced or aggressive malignancy, the risk for GVHD associated with the use of a MICA-mismatched donor could be balanced against the potential benefits of lowered disease recurrence via the observed graft-versus-leukemia effect.
In conclusion, the present study identifies MICA as an additional human HLA-encoded transplantation antigen. It further supports the inclusion of pretransplantation typing of MICA in the search for a donor. Given the tight linkage disequilibrium between MICA and HLA-B, the enormous imbalance in allele numbers, and the relatively small number of MICA alleles covering diverse human populations, finding a 10/10 matched donor who is also matched for MICA could be readily achieved through presently established donor search protocols. To definitively assess the practical feasibility of the inclusion of MICA-matching data in the donor selection process, a randomized prospective study is necessary at present.
Acknowledgments
The authors thank Alain Fischer (Imagine Institute, Necker Hospital, Paris, France), Susan Gilfillan and Marco Colonna (both at Washington University School of Medicine, St. Louis, MO), Andreas Diefenbach (University of Mainz, Germany), Yasuo Morishima (Aichi Cancer Center Research Institute, Nagoya, Japan), and Effie Petersdorf (Fred Hutchinson Cancer Research Center, Seattle, WA) for critical reading of this manuscript. The authors also thank Nicole Raus (SFGM-TC, Lyon, France) for retrieving clinical data from the ProMISe database.
Supported by the National Research Agency (Agence Nationale de Recherche) and the Investment for the Future Program (Programme des Investissements d’Avenir) through a Laboratoire d’Excellence TRANSPLANTEX (ANR-11-LABX-0070_TRANSPLANTEX) grant (S.B.). Additional funding was provided by INSERM UMR_S 1109, the Institut Universitaire de France , and the Franco-Japanese NextGen HLA laboratory (all S.B.).
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: R.C. performed experiments, designed the study, analyzed the data, and wrote the manuscript. S.B. designed the study, analyzed the data, and wrote the manuscript. G.G., W.I., I.K., C.M., S.M., A.M., A. Pichot, and M.U. performed experiments and analyzed the data. M.K. and N.J. performed statistics. P.A.v.d.B., D.B., A.C., D.C., F. Claas, V.D., K.G., J.K., J.J.C., M. Labalette, X.L., P.L., M. Michallet, N.M., P.M., M. Oudshoorn, A. Parissiadis, R.P.d.L., C. Picard, G.S., E.S., R.T., A.T., and I.Y.-A. provided samples and clinical data, interpreted clinical data, and discussed results. P.A., BL, M. Mohty, A.N., C. Paillard, S.Q., A.S.-G., J.S., L.V., R.Z., F. Ciceri, and K.F. interpreted clinical data and discussed results. F.B., H.I., Y.K., M. Labopin, M.M.-B., M. Ota, and B.v.d.H. analyzed data and reviewed statistics. All authors contributed to the writing of the report and approved the final version of the manuscript.
Conflict-of-interest disclosure: S.B. is the scientific founder and a (minority) shareholder in BIOMICA SAS. J.K. is cofounder and chief scientific officer of Gadeta and received personal fees from Gadeta. In addition, J.K. has a patent issued/pending. E.S. is inventor of a patent application filed by the University Medical Center Utrecht on the prediction of an alloimmune response against mismatched HLA (PCT/EPT2013/073386). The remaining authors declare no competing financial interests.
Correspondence: Seiamak Bahram, Centre de Recherche d’Immunologie et d’Hématologie, 4 rue Kirschleger, 67085 Strasbourg Cedex, France; e-mail: siamak@unistra.fr; or Raphael Carapito, Centre de Recherche d’Immunologie et d’Hématologie, 4 rue Kirschleger, 67085 Strasbourg Cedex, France; e-mail: carapito@unistra.fr.
References
- 1.Forman SJ, Negrin RS, Antin JH, Appelbaum FR. Thomas' Hematopoietic Cell Transplantation. 5th ed. Hoboken, NJ: Wiley-Blackwell; 2016. [Google Scholar]
- 2.Gratwohl A, Pasquini MC, Aljurf M, et al. Worldwide Network for Blood and Marrow Transplantation (WBMT) One million haemopoietic stem-cell transplants: a retrospective observational study. Lancet Haematol. 2015;2(3):e91–e100. doi: 10.1016/S2352-3026(15)00028-9. [DOI] [PubMed] [Google Scholar]
- 3.Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nat Rev Immunol. 2012;12(6):443–458. doi: 10.1038/nri3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373(9674):1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Socié G, Ritz J. Current issues in chronic graft-versus-host disease. Blood. 2014;124(3):374–384. doi: 10.1182/blood-2014-01-514752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Loiseau P, Busson M, Balere ML, et al. HLA Association with hematopoietic stem cell transplantation outcome: the number of mismatches at HLA-A, -B, -C, -DRB1, or -DQB1 is strongly associated with overall survival. Biol Blood Marrow Transplant. 2007;13(8):965–974. doi: 10.1016/j.bbmt.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 7.Sasazuki T, Juji T, Morishima Y, et al. Effect of matching of class I HLA alleles on clinical outcome after transplantation of hematopoietic stem cells from an unrelated donor. Japan Marrow Donor Program. N Engl J Med. 1998;339(17):1177–1185. doi: 10.1056/NEJM199810223391701. [DOI] [PubMed] [Google Scholar]
- 8.Gooley TA, Chien JW, Pergam SA, et al. Reduced mortality after allogeneic hematopoietic-cell transplantation. N Engl J Med. 2010;363(22):2091–2101. doi: 10.1056/NEJMoa1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasquini M, Zhu X. Current uses and outcomes of hematopoietic stem cell transplantation: 2014 CIBMTR Summary Slides. http://www.cibmtr.org. Accessed December 2015.
- 10.Anasetti C, Logan BR, Lee SJ, et al. Blood and Marrow Transplant Clinical Trials Network. Peripheral-blood stem cells versus bone marrow from unrelated donors. N Engl J Med. 2012;367(16):1487–1496. doi: 10.1056/NEJMoa1203517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Warren EH, Zhang XC, Li S, et al. Effect of MHC and non-MHC donor/recipient genetic disparity on the outcome of allogeneic HCT. Blood. 2012;120(14):2796–2806. doi: 10.1182/blood-2012-04-347286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersdorf EW. The major histocompatibility complex: a model for understanding graft-versus-host disease. Blood. 2013;122(11):1863–1872. doi: 10.1182/blood-2013-05-355982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleischhauer K. Immunogenetics of HLA-DP--A New View of Permissible Mismatches. N Engl J Med. 2015;373(7):669–672. doi: 10.1056/NEJMe1505539. [DOI] [PubMed] [Google Scholar]
- 14.Petersdorf EW, Malkki M, O’hUigin C, et al. High HLA-DP Expression and Graft-versus-Host Disease. N Engl J Med. 2015;373(7):599–609. doi: 10.1056/NEJMoa1500140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersdorf EW. HLA mismatching in transplantation. Blood. 2015;125(7):1058–1059. doi: 10.1182/blood-2014-12-619015. [DOI] [PubMed] [Google Scholar]
- 16.Shaw BE, Arguello R, Garcia-Sepulveda CA, Madrigal JA. The impact of HLA genotyping on survival following unrelated donor haematopoietic stem cell transplantation. Br J Haematol. 2010;150(3):251–258. doi: 10.1111/j.1365-2141.2010.08224.x. [DOI] [PubMed] [Google Scholar]
- 17.Petersdorf EW. Optimal HLA matching in hematopoietic cell transplantation. Curr Opin Immunol. 2008;20(5):588–593. doi: 10.1016/j.coi.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carapito R, Bahram S. Genetics, genomics, and evolutionary biology of NKG2D ligands. Immunol Rev. 2015;267(1):88–116. doi: 10.1111/imr.12328. [DOI] [PubMed] [Google Scholar]
- 19.Bahram S, Bresnahan M, Geraghty DE, Spies T. A second lineage of mammalian major histocompatibility complex class I genes. Proc Natl Acad Sci USA. 1994;91(14):6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu B, Pizarro JC, Holmes MA, et al. Crystal structure of a gammadelta T-cell receptor specific for the human MHC class I homolog MICA. Proc Natl Acad Sci USA. 2011;108(6):2414–2419. doi: 10.1073/pnas.1015433108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boukouaci W, Busson M, Peffault de Latour R, et al. MICA-129 genotype, soluble MICA, and anti-MICA antibodies as biomarkers of chronic graft-versus-host disease. Blood. 2009;114(25):5216–5224. doi: 10.1182/blood-2009-04-217430. [DOI] [PubMed] [Google Scholar]
- 22.Parmar S, Del Lima M, Zou Y, et al. Donor-recipient mismatches in MHC class I chain-related gene A in unrelated donor transplantation lead to increased incidence of acute graft-versus-host disease. Blood. 2009;114(14):2884–2887. doi: 10.1182/blood-2009-05-223172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson E, Grzywacz B, Wang H, et al. Limited role of MHC class I chain-related gene A (MICA) typing in assessing graft-versus-host disease risk after fully human leukocyte antigen-matched unrelated donor transplantation. Blood. 2009;114(21):4753–4754, author reply 4754-4755. doi: 10.1182/blood-2009-08-239301. [DOI] [PubMed] [Google Scholar]
- 24.Askar M, Sun Y, Rybicki L, et al. Synergistic effect of major histocompatibility complex class I-related chain a and human leukocyte antigen-DPB1 mismatches in association with acute graft-versus-host disease after unrelated donor hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2014;20(11):1835–1840. doi: 10.1016/j.bbmt.2014.07.019. [DOI] [PubMed] [Google Scholar]
- 25.Isernhagen A, Malzahn D, Viktorova E, et al. The MICA-129 dimorphism affects NKG2D signaling and outcome of hematopoietic stem cell transplantation. EMBO Mol Med. 2015;7(11):1480–1502. doi: 10.15252/emmm.201505246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fodil N, Laloux L, Wanner V, et al. Allelic repertoire of the human MHC class I MICA gene. Immunogenetics. 1996;44(5):351–357. doi: 10.1007/BF02602779. [DOI] [PubMed] [Google Scholar]
- 27.Fodil N, Pellet P, Laloux L, Hauptmann G, Theodorou I, Bahram S. MICA haplotypic diversity. Immunogenetics. 1999;49(6):557–560. doi: 10.1007/s002510050536. [DOI] [PubMed] [Google Scholar]
- 28.Crivello P, Zito L, Sizzano F, et al. The impact of amino acid variability on alloreactivity defines a functional distance predictive of permissive HLA-DPB1 mismatches in hematopoietic stem cell transplantation. Biol Blood Marrow Transplant. 2015;21(2):233–241. doi: 10.1016/j.bbmt.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 29.Glucksberg H, Storb R, Fefer A, et al. Clinical manifestations of graft-versus-host disease in human recipients of marrow from HL-A-matched sibling donors. Transplantation. 1974;18(4):295–304. doi: 10.1097/00007890-197410000-00001. [DOI] [PubMed] [Google Scholar]
- 30.Therneau T, Grambsch P. Modeling Survival Data: Extending the Cox Model. New York: Springer; 2000. [Google Scholar]
- 31.Scheike TH, Zhang MJ. Analyzing competing risk data using the R timereg package. J Stat Softw. 2011;38(2):38. [PMC free article] [PubMed] [Google Scholar]
- 32.Scheike TH, Zhang MJ. Flexible competing risks regression modeling and goodness-of-fit. Lifetime Data Anal. 2008;14(4):464–483. doi: 10.1007/s10985-008-9094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheike TH, Zhang MJ, Gerds TA. Predicting cumulative incidence probability by direct binomial regression. Biometrika. 2008;95(1):205–220. [Google Scholar]
- 34.Zhou B, Latouche A, Rocha V, Fine J. Competing risks regression for stratified data. Biometrics. 2011;67(2):661–670. doi: 10.1111/j.1541-0420.2010.01493.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gratwohl A, Stern M, Brand R, et al. European Group for Blood and Marrow Transplantation and the European Leukemia Net. Risk score for outcome after allogeneic hematopoietic stem cell transplantation: a retrospective analysis. Cancer. 2009;115(20):4715–4726. doi: 10.1002/cncr.24531. [DOI] [PubMed] [Google Scholar]
- 36.Fleischhauer K, Shaw BE, Gooley T, et al. International Histocompatibility Working Group in Hematopoietic Cell Transplantation. Effect of T-cell-epitope matching at HLA-DPB1 in recipients of unrelated-donor haemopoietic-cell transplantation: a retrospective study. Lancet Oncol. 2012;13(4):366–374. doi: 10.1016/S1470-2045(12)70004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horowitz MM, Gale RP, Sondel PM, et al. Graft-versus-leukemia reactions after bone marrow transplantation. Blood. 1990;75(3):555–562. [PubMed] [Google Scholar]
- 38.Steinle A, Li P, Morris DL, et al. Interactions of human NKG2D with its ligands MICA, MICB, and homologs of the mouse RAE-1 protein family. Immunogenetics. 2001;53(4):279–287. doi: 10.1007/s002510100325. [DOI] [PubMed] [Google Scholar]
- 39.Karimi MA, Bryson JL, Richman LP, et al. NKG2D expression by CD8+ T cells contributes to GVHD and GVT effects in a murine model of allogeneic HSCT. Blood. 2015;125(23):3655–3663. doi: 10.1182/blood-2015-02-629006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gannagé M, Buzyn A, Bogiatzi SI, et al. Induction of NKG2D ligands by gamma radiation and tumor necrosis factor-alpha may participate in the tissue damage during acute graft-versus-host disease. Transplantation. 2008;85(6):911–915. doi: 10.1097/TP.0b013e31816691ef. [DOI] [PubMed] [Google Scholar]
- 41.Mellergaard M, Skovbakke SL, Schneider CL, et al. N-glycosylation of asparagine 8 regulates surface expression of major histocompatibility complex class I chain-related protein A (MICA) alleles dependent on threonine 24. J Biol Chem. 2014;289(29):20078–20091. doi: 10.1074/jbc.M114.573238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spierings E. Minor histocompatibility antigens: past, present, and future. Tissue Antigens. 2014;84(4):374–360. doi: 10.1111/tan.12445. [DOI] [PubMed] [Google Scholar]
- 43.Diefenbach A, Jamieson AM, Liu SD, Shastri N, Raulet DH. Ligands for the murine NKG2D receptor: expression by tumor cells and activation of NK cells and macrophages. Nat Immunol. 2000;1(2):119–126. doi: 10.1038/77793. [DOI] [PubMed] [Google Scholar]
- 44.Cerwenka A, Bakker AB, McClanahan T, et al. Retinoic acid early inducible genes define a ligand family for the activating NKG2D receptor in mice. Immunity. 2000;12(6):721–727. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- 45.Ruggeri L, Capanni M, Urbani E, et al. Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science. 2002;295(5562):2097–2100. doi: 10.1126/science.1068440. [DOI] [PubMed] [Google Scholar]
- 46.Minculescu L, Sengeløv H. The role of gamma delta T cells in haematopoietic stem cell transplantation. Scand J Immunol. 2015;81(6):459–468. doi: 10.1111/sji.12289. [DOI] [PubMed] [Google Scholar]
- 47.Tiercy JM. HLA-C Incompatibilities in Allogeneic Unrelated Hematopoietic Stem Cell Transplantation. Front Immunol. 2014;5:216. doi: 10.3389/fimmu.2014.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eberhard HP, Müller CR. The Impact of HLA-C Matching on Donor Identification Rates in a European-Caucasian Population. Front Immunol. 2014;5:501. doi: 10.3389/fimmu.2014.00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armand P, Gibson CJ, Cutler C, et al. A disease risk index for patients undergoing allogeneic stem cell transplantation. Blood. 2012;120(4):905–913. doi: 10.1182/blood-2012-03-418202. [DOI] [PMC free article] [PubMed] [Google Scholar]