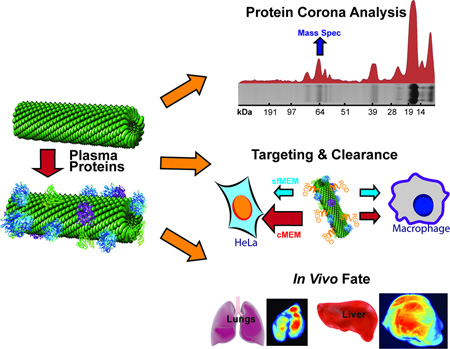
Abstract
Biomolecules in bodily fluids such as plasma can adsorb to the surface of nanoparticles and influence their biological properties. This phenomenon, known as the protein corona, is well established in the field of synthetic nanotechnology but has not been described in the context of plant virus nanoparticles (VNPs). We investigated the interaction between VNPs derived from Tobacco mosaic virus (TMV) and plasma proteins, and found that the VNP protein corona was significantly less abundant compared to the corona of synthetic particles. The formed corona was dominated by complement proteins and immunoglobulins, the binding of which could be reduced by PEGylating the VNP surface. We investigated the impact of the VNP protein corona on molecular recognition and cell targeting in the context of cancer and thrombosis. A library of functionalized TMV rods with PEG and peptide ligands targeting integrins or fibrin(ogen) showed different dispersion properties, cellular interactions and in vivo fates depending on the properties of the protein corona, influencing target specificity and non-specific scavenging by macrophages. Our results provide insight into the in vivo properties of VNPs and suggest that the protein corona effect should be considered during the development of efficacious, targeted VNP formulations.
Keywords: nanomedicine, drug/contrast agent delivery, RGD, GPRPP, CREKA
Graphical Abstract
The interactions between targeted plant virus-based nanoparticles (VNPs) and blood plasma proteins and their consequences are investigated. The amount of proteins associated with VNPs in situ is lower than for synthetic nanoparticles; complement proteins and immunoglobulins dominate. The consequences of protein adsorption for molecular targeting and in vivo biodistribution are demonstrated.
1 Introduction
Targeted nanoparticle formulations hold great promise for the diagnosis and treatment of cancer and cardiovascular disease because they enable the site-specific delivery of imaging agents and drugs, therefore reducing systemic toxicity while improving imaging contrast and pharmacological efficacy[1–5]. However, nanoparticles must overcome several biological barriers on their way to the target site including adsorption of blood proteins, which interact with nanoparticles to form a protein corona, a layer of proteins that becomes an interface between the nanomaterial and the surrounding biological milieu. The composition and structure of the corona, the latter reflecting which proteins and epitopes are exposed on the surface of the corona and which are sequestered in deeper layers, can influence molecular recognition of the nanoparticle by target and non-target cells, and thus determine its fate[6–9]. The protein corona effect has been extensively studied using different types of synthetic nanoparticles, including those composed of polystyrene, SiO2 and gold[10–12], but the role of the protein corona on protein-based nanoparticles such as plant virus nanoparticles (VNPs) has not been elucidated yet.
VNPs derived from plant viruses have several advantages for medical applications. They come in a variety of well-defined sizes and shapes (tubes, filaments or sphere-like icosahedra), they are genetically encoded and therefore identical (i.e. no batch to batch variations, which is a disadvantage of synthetic nanoparticles)[13], and they are highly biocompatible and biodegradable[14,15]. VNPs can be modified by genetic engineering, e.g. to incorporate targeting motifs or conjugation sites, or chemical synthesis, e.g. to introduce synthetic materials such as polyethylene glycol (PEG) stealth coatings, contrast agents, or drugs[16–18]. Furthermore, many VNPs can be disassembled into monomer capsid proteins and subsequently re-assembled, allowing the encapsulation of cargo molecules[19–22] and shape engineering[23]. VNPs therefore offer a highly versatile and tunable platform for unique biomedical applications such as the molecular magnetic resonance imaging of atherosclerotic plaques[16] and targeted drug delivery to cancer cells[18].
Some design principles for VNPs have been established: shape engineering allows the avoidance of non-specific sequestration in cells of the mononuclear phagocyte system (MPS)[23], polymer coatings reduce the immunogenicity of VNPs, and the addition of targeting ligands can increase cell specificity[24,25]. Nevertheless, the fundamental interactions between native or functionalized VNPs and plasma proteins, and the impact of such interactions on biodistribution, clearance, and molecular target recognition, are poorly understood. We therefore investigated the composition of the protein corona surrounding VNPs based on Tobacco mosaic virus (TMV). We considered different shapes, i.e. rod-like TMV structures vs. spherical TMV nanoparticles (SNPs), as well as different surface charges and chemistries (unmodified TMV particles vs. PEGylated and targeted formulations). We investigated how the different VNP formulations interacted with target cancer cells and non-target phagocytes using in vitro assays, and additionally studied in vivo VNP biodistribution and clearance using mouse models of thrombosis and cancer.
2 Results and discussion
2.1 Plant viral nanoparticles (VNPs) and the protein corona
The quantity and identity of plasma proteins interacting with VNPs with different shapes and surface chemistries was investigated by producing a diverse panel of TMV-derived VNP formulations. We compared wild-type TMV (TMV-wt), a mutant version containing a lysine residue at position 158 in the capsid protein (TMV-lys)[26], and spherical versions of both variants generated by heat-mediated shape switching, as previously described[27,28]. TMV-wt particles have a negative surface charge (ζ = −25.3 ± 2.3 mV)[29], whereas the additional lysine in the TMV-lys mutant is exposed to the solvent and reduces the surface charge accordingly (ζ = −14.5 ± 0.6 mV) as shown in Supplementary Figure S1. Spherical SiO2 nanoparticles with positive and negative surface charges were used as synthetic nanoparticle controls. The nominal “50 nm” SiO2 and TMV-based SNPs were comparable in size. The majority of SNPs generated by the thermal transition of a single TMV rod are 50–60 nm in diameter[27,28], although the presence of a small population of larger SNPs (reflecting the fusion of multiple rods during transition) increased the hydrodynamic diameters measured by dynamic light scattering (Supplementary Table S1 and Supplementary Figure S2A+B).
The nanoparticle formulations were incubated in ~100% human plasma for 1 h to allow the corona to form. The samples were then purified and washed to remove excess plasma proteins and the loosely-bound proteins defined as dynamic or soft corona, allowing us to focus on the tightly-bound proteins of the hard corona (Figure 1A)[6,8,30] which has the most significant impact on nanoparticle behavior because the proteins remain adsorbed for prolonged periods. TMV-derived rod-shaped and spherical nanoparticles remained stable when exposed to and incubated in human plasma. Particle fragmentation or aggregation was not apparent, attesting to the biocompatible nature of the protein-based nanoparticles in biological media (Figure 1A and Supplementary Figure S3+S4).
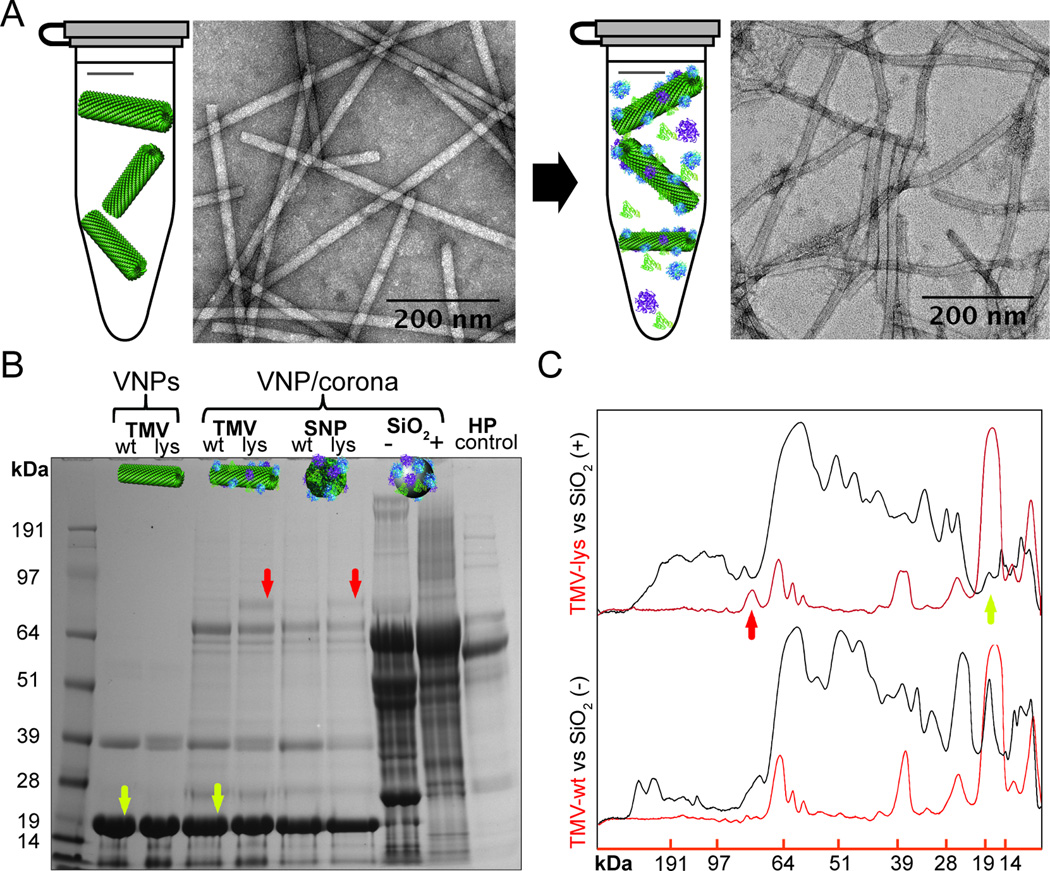
Figure 1. The protein corona of plant virus nanoparticles.
A, TEM images of TMV-lys before and after incubation in human plasma. Particle fragmentation or pronounced aggregation was not apparent. B, SDS-PAGE analysis of the protein corona formed on TMV and SiO2 particles after incubation for 1 h in ~100% human plasma. ‘Bare’ (not exposed to human plasma) TMV-wt, TMV-lys and diluted human plasma (HP) were used as references. Red arrows indicate a coronal component that is characteristic of TMV-lys particles. Yellow arrows indicate the 17.5-kDa TMV capsid protein. C, Quantitative densitometric analysis of selected bands from the gel in panel “B” showing the quantity of protein in the corona on the surface of VNPs and synthetic SiO2 particles. Additional data are shown in Supplementary Figure S4.
Quantitative analysis of the protein corona by SDS-PAGE (Figure 1B+C) indicated that although the TMV-based VNPs did acquire a protein corona, the amount of bound protein was significantly lower than that associated with the synthetic nanoparticles (Figure 1B+C). SDS-PAGE and densitometry indicated that the total quantity of corona-forming proteins on the surface of the VNPs (mass of protein bound per unit of surface area) was ~6 times lower than that found on the synthetic SiO2 particles (Supplementary Figure S4D). The less efficient adsorption of proteins to the TMV-based nanoparticles may reflect its surface properties, which feature patches of positive and negative charges and heterogeneous hydrophobic and hydrophilic domains, therefore providing a higher degree of biocompatibility than synthetic nanoparticles. In contrast, the SiO2 particles present a uniform surface charge, and a smooth morphology, helping to attract a rich protein corona[6,10].
The SDS-PAGE experiments did not reveal any differences in the amount of protein bound to the TMV-wt and TMV-lys rods or their corresponding SNPs (Figure 1B and Supplementary Figure S4) but the formulations with different surface charges attracted coronas with different compositions (it should be noted that SDS-PAGE only provides limited insight into protein corona composition; more detailed information is provided by mass spectrometry analysis below). The main difference was a 70-kDa band that was only present in the corona surrounding the TMV-lys VNPs and SNPs. Mass spectrometry showed that this band comprised complement protein C3/C4, plasminogen and IgM (see highlighted results in Table 1). These amino-acid sequence or surface chemistry-dependent differences in coronal composition are consistent with earlier reports showing that several parameters influence the corona of synthetic nanoparticles, including the base nanomaterial, particle size, surface curvature, particle shape, surface morphology, and surface charge[10,31,32]. We found that while the VNP surface chemistry (TMV/SNP-wt vs TMV/SNP-lys) had a moderate effect on the coronal composition (differences in 70-kDa band), shape had little impact on the components of the corona (as far as detectable using SDS-PAGE). The TMV and SNP particles consist of the same coat proteins, however the structure of the SNPs remains to be elucidated. Initial work has shown that while the coat proteins do not denature during the thermal transition, considerable conformational changes occur within the coat proteins during the TMV-to-SNP transition[33]. Our data indicate that the protein corona formation on TMV vs SNP does not show differences. Nevertheless, more sensitive methods are required to rule out subtle differences in protein corona magnitude or composition comparing the differently shaped VNP platforms.
Table 1.
The 20 most abundant proteins in the corona surrounding TMV-lys in human plasma determined by mass spectrometry.
| Protein name | Protein accession number |
Protein molecular weight (Da) |
Total Spectrum count |
|
|---|---|---|---|---|
| 1* | Complement C3 OS=Homo sapiens GN=C3 PE=1 SV=2 | CO3_HUMAN | 187,149.10 | 1696 |
| 2 | Ig kappa chain C region OS=Homo sapiens GN=IGKC PE=1 SV=1 | IGKC_HUMAN | 11,608.60 | 577 |
| 3 | Complement C4-B OS=Homo sapiens GN=C4B PE=1 SV=2 | CO4B_HUMAN | 192,752.80 | 387 |
| 4* | Complement C4-A OS=Homo sapiens GN=C4A PE=1 SV=2 | CO4A_HUMAN | 192,786.80 | 384 |
| 5* | Plasminogen OS=Homo sapiens GN=PLG PE=1 SV=2 | PLMN_HUMAN | 90,567.40 | 383 |
| 6* | Ig mu chain C region OS=Homo sapiens GN=IGHM PE=1 SV=3 | IGHM_HUMAN | 49,305.70 | 370 |
| 7 | Apolipoprotein B-100 OS=Homo sapiens GN=APOB PE=1 SV=2 | APOB_HUMAN | 515,614.80 | 316 |
| 8 | Ig mu heavy chain disease protein OS=Homo sapiens PE=1 SV=1 | MUCB_HUMAN | 43,056.70 | 189 |
| 9 | Serum albumin OS=Homo sapiens GN=ALB PE=1 SV=2 | ALBU_HUMAN | 69,366.90 | 117 |
| 10 | Huntingtin OS=Homo sapiens GN=HTT PE=1 SV=2 | HD_HUMAN | 347,603.10 | 90 |
| 11 | Immunoglobulin lambda-like polypeptide 5 OS=Homo sapiens GN=IGLL5 PE=2 SV=2 |
IGLL5_HUMAN | 23,062.90 | 86 |
| 12 | C4b-binding protein alpha chain OS=Homo sapiens GN=C4BPA PE=1 SV=2 |
C4BPA_HUMAN | 67,033.00 | 84 |
| 13 | Ig lambda-2 chain C regions OS=Homo sapiens GN=IGLC2 PE=1 SV=1 |
LAC2_HUMAN | 11,293.20 | 83 |
| 14 | Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 | FIBA_HUMAN | 94,973.40 | 64 |
| 15 | Alpha-2-macroglobulin OS=Homo sapiens GN=A2M PE=1 SV=3 | A2MG_HUMAN | 163,289.90 | 63 |
| 16 | Ig alpha-1 chain C region OS=Homo sapiens GN=IGHA1 PE=1 SV=2 |
IGHA1_HUMAN | 37,653.80 | 62 |
| 17 | Apolipoprotein(a) OS=Homo sapiens GN=LPA PE=1 SV=1 | APOA_HUMAN | 501,288.90 | 53 |
| 18 | Ig gamma-2 chain C region OS=Homo sapiens GN=IGHG2 PE=1 SV=2 |
IGHG2_HUMAN | 35,899.20 | 51 |
| 19 | Histidine-rich glycoprotein OS=Homo sapiens GN=HRG PE=1 SV=1 | HRG_HUMAN | 59,576.60 | 50 |
| 20 | Ig gamma-1 chain C region OS=Homo sapiens GN=IGHG1 PE=1 SV=1 |
IGHG1_HUMAN | 36,105.00 | 47 |
Proteins in the ~70-kDa band distinguishing TMV-lys from TMV-wt.
The identities of the coronal proteins were determined by mass spectrometry using the TMV-lys rods as a representative formulation (Table 1 and Supplementary Table S2). This revealed the presence of approximately 140 plasma proteins bound to TMV (Table 1 lists the 20 most abundant proteins; the complete list is shown in Supplementary Table S2). Overall, complement proteins accounted for ~42% of the total protein mass, and ~28% of the proteins were identified as immunoglobulins. This was anticipated because ‘naked’ (non-PEGylated or otherwise shielded) plant-derived VNPs are known to be immunogenic[24]. In contrast, the protein corona associated with SiO2, polystyrene and gold nanoparticles has been reported to contain predominantly fibrinogen and/or apolipoproteins[8,12]. Our data therefore suggest that the protein coronas formed around synthetic nanoparticles and VNPs are distinct.
2.2 The impact of protein corona on molecular recognition of targeted TMV particles
Next we investigated the effect of the protein corona on the interactions of targeted TMV formulations and cells. These studies were motivated by recent reports showing that the protein corona may cause steric hindrance between targeting components on the surface of nanoparticles and their receptors[34,35]. In some cases this has reduced the targeting efficiency of nanoparticles, but in other cases the functionality of the particles has been preserved, at least in in vitro[36,37].
We tested TMV formulations labeled with PEG (of various PEG chain lengths) and selected targeting peptides: cyclic RGD, CREKA or GPRPP. The cyclic RGD motif binds specifically to integrins that are overexpressed on cancer cells, making this peptide a popular choice for the development of cancer-targeting nanoformulations and contrast agents[38,39]. GPRPP[40,41] and CREKA[42,43] bind to fibrin(ogen), and have therefore been integrated into nanoparticle formulations that target fibrin-rich atherosclerotic plaques and thrombi. The PEG chains and peptides were conjugated to the TMV exterior using NHS, diazonium and click chemistries (see Materials and Methods, Figure 2).
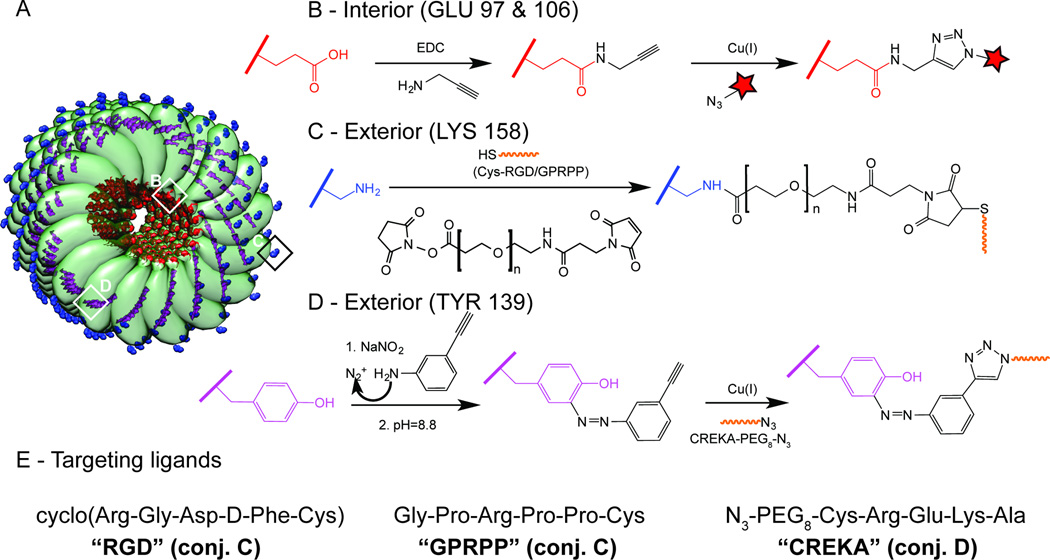
Figure 2. Chemical modification of TMV.
A, Schematic representation of TMV comprising 100 capsid protein subunits (the full length rod has 2,130 identical copies of the coat protein unit), with surface-exposed residues highlighted in color: glutamic acid (red), lysine (blue) and tyrosine (purple). B, Internal labeling with Cy5 fluorophores via EDC conjugation of alkyne molecules to the carboxyl group of glutamic acid and subsequent alkyne/azide-Cy5 click reaction. C, External conjugation of cysteine-tagged RGD and GPRPP peptides to lysine residues via a NHS ester–maleimide heterobifunctional PEG linker (–NH2 to –SH conjugation). D, External conjugation of azide-tagged CREKA peptide to tyrosine residues via reaction with diazonium salt and subsequent alkyne-azide click reaction. E, The peptide sequences of the targeting ligands.
The RGD targeting ligand (cyclic cRGDfC) was conjugated to TMV using a heterobifunctional linker with or without an intervening PEG spacer (4, 12, or 24 PEG units, corresponding to molecular weights of 513.5, 865.9 and 1394.6 Da). TMV-lys and TMV-PEG12 were used as controls. Successful conjugation was confirmed by SDS-PAGE (Figure 3A); the RGD ligand and PEG linkers increase the molecular weight of the modified capsid proteins. Ligand density was estimated by densitometry. On average, TMV was modified with ~550–600 PEG-RGD ligands per particle, corresponding to the occupation of ~25% of the available TMV capsid proteins. Structural integrity was confirmed by transmission electron microscopy (not shown).
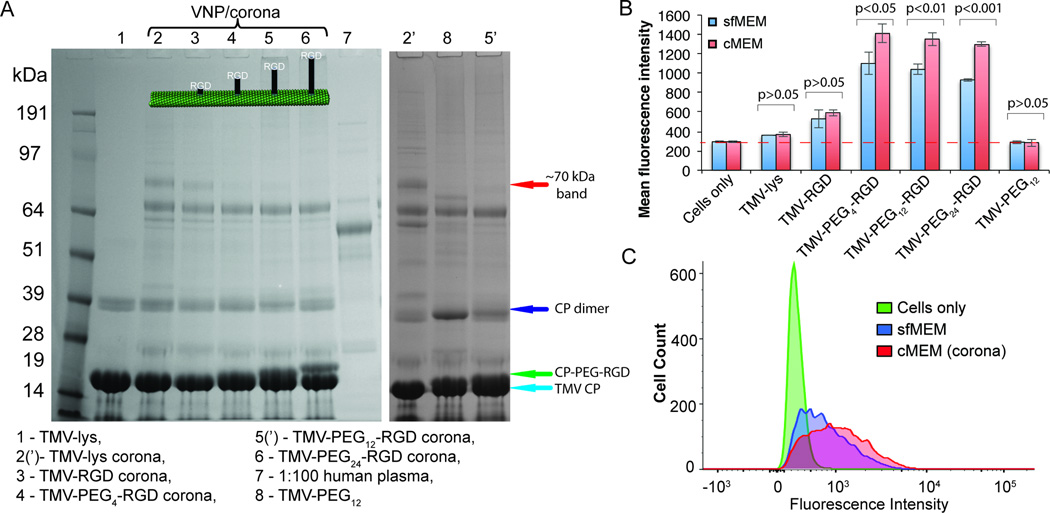
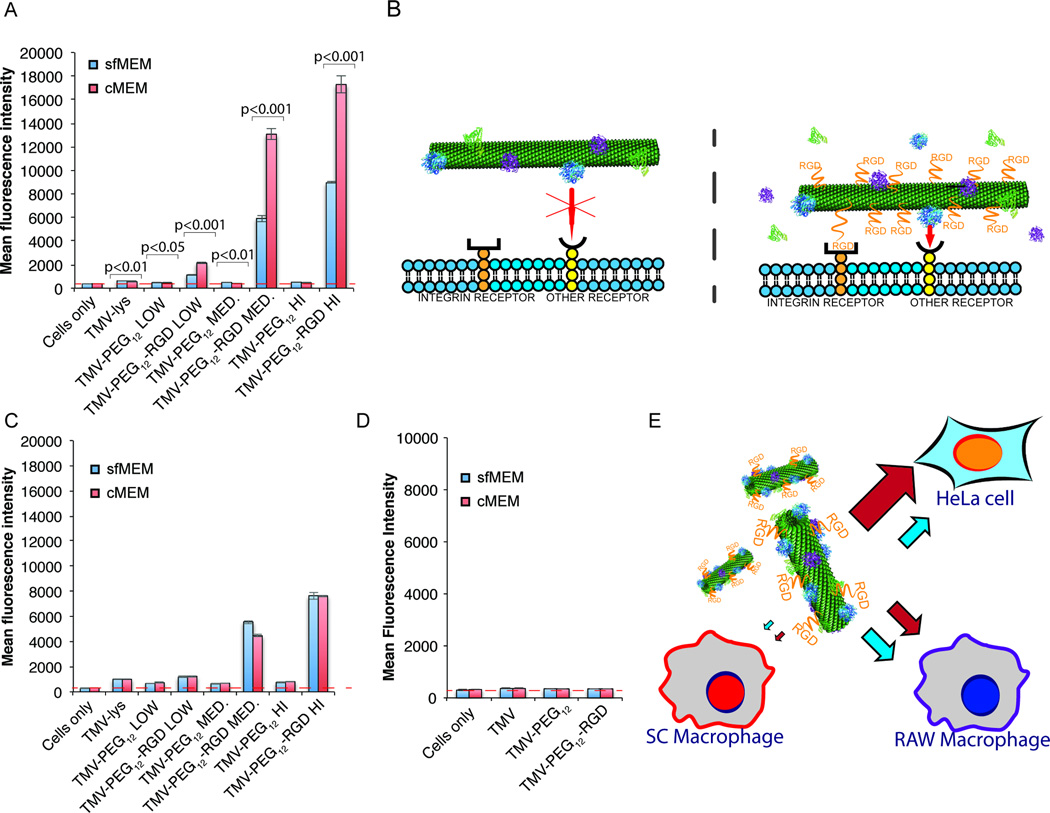
Figure 3. Influence of the PEG linker length of RGD-targeted TMV on protein corona formation and consequence for targeting of integrin-expressing cancer cells.
A, SDS-PAGE analysis of the protein corona surrounding TMV-lys and TMV-PEG0–24-RGD VNPs after 1 h incubation in ~100% human plasma. ‘Bare’ (not exposed to human plasma) TMV-lys and diluted plasma were used as references. B, FACS analysis of the interactions between RGD-targeted TMV VNPs and HeLa cells, which overexpress integrins, in serum-free (sfMEM) and complete (cMEM) media. Statistically significant differences between serum-free and complete media are indicated above the histograms. C, FACS histograms showing the promotion of TMV-PEG12-RGD uptake in the presence of a protein corona.
We confirmed the presence of a protein corona as described above using the preparation with human plasma followed by SDS-PAGE analysis (Figure 3A). Although the TMV-lys and TMV-PEG0–24-RGD particles were surrounded by protein coronas with a similar composition, we observed a monotonic reduction in the quantity of the ~70-kDa protein band (complement C3/C4, plasminogen and IgM) with increasing PEG spacer length. These data indicate that PEGylation is an effective method to reduce the immunogenicity of TMV and its interaction with plasma proteins, consistent with other reports[44–46]. PEG can adopt two conformations depending on the density of PEG grafting on the surface: the mushroom conformation (for low grafting density) and the brush conformation (for high grafting density)[24,45]. In our case, PEG was conjugated, on average, to every fourth capsid protein, equating to a low grafting density, which should yield the mushroom conformation. Longer PEG chains would therefore occupy more of the surface and cause greater steric hindrance, producing a more effective polymer cloak.
Next, we investigated the uptake of the targeted TMV formulations by cells in vitro. The non-specific uptake of nanomaterials by the MPS causes rapid clearance, and plasma proteins in the protein corona may tag the nanoparticles for macrophage recognition and thus prevent interactions with target cells. At the same time, the protein corona can block ligand–receptor interactions, thus reducing the efficiency of targeting[34,35]. We used HeLa cells, a cervical cancer cell line known to express the αVβ3 and αVβ5 integrins[39] as a proof-of-concept target cell line. SC human blood and RAW264.7 murine blood monocyte macrophages were used to model MPS clearance. The VNP–cell interactions were analyzed by flow cytometry in (i) serum-free medium (sfMEM) to prevent the formation of a protein corona, and (ii) complete medium (cMEM) to allow the formation of a protein corona.
The comparison of RGD-targeted formulations, in which the RGD was attached with and without intervening PEG spacers of varying lengths, indicated that direct conjugation of the RGD ligand promoted cell targeting, however resulting only in modest cell uptake (~1.5-fold increase in cell targeting compared to TMV-lys, Figure 3B). The inclusion of a PEG spacer significantly enhanced cell targeting (achieving 3–4 fold increase in cell targeting compared to TMV-lys); there were no stark differences comparing the various PEG linker lengths (Figure 3B). The PEG linker may enhance the presentation of RGD and therefore promote its interaction with its receptor. Data indicate that increasing the length of the PEG spacer to more than four units did not achieve further enhancement of cell targeting (Figure 3B), however the longer PEG linker may exhibit beneficial in vivo properties (not measured in these cell uptake studies) due to reduced corona formation.
We compared the cell targeting properties of RGD-modified TMV in sfMEM vs. cMEM. Data indicate modest (but statistically significant) increase in cell targeting in cMEM (Figure 3B). While the longer PEG chains in TMV-PEG24-RGD reduced the abundance of the plasminogen, complement C3/C4 and IgM components, cell targeting was to a similar degree comparing the various TMV-PEG4–24-RGD formulations. Any of ~140 (Table 1 + S2) TMV corona proteins might in principle be interacting with membrane proteins and promoting cell uptake. Our initial studies indicate that neither complement proteins nor IgG – the two most abundant coronal components – were responsible for the corona-mediated uptake (see Supplementary Figure S5). It should be noted that it may also be the soft corona proteins (not investigated in this study) that lead to the increased cell uptake properties of TMV in cMEM. Future studies could set out to define the identify the pool of coronal interaction partners (so called interactome)[47,48], but this is beyond the present work.
Next, we investigated whether the corona-induced VNP–cell targeting enhancement was dependent on targeting ligand density and whether enhanced non-specific uptake in phagocyte cells would also occur. TMV-PEG12-RGD formulations were prepared with three different PEG/RGD densities: low, medium and high corresponding to approximately 15%, 35%, and 50% coat protein modification with PEG/RGD (Supplementary Figure S6). The higher the RGD ligand density per TMV, the more efficient its cell interactions. For all ligand grafting densities, the uptake was enhanced ~2× in cMEM vs. serum-free medium (Figure 4A). Further, time course studies were conducted that showed that protein corona-mediated uptake of RGD-targeted TMV in cMEM was more profound after longer incubation periods (Supplementary Figure S7). Despite the fact that both native TMV-lys and RGD-targeted TMV formulations exhibited a protein corona, increased uptake in cMEM was only observed for RGD-targeted, not native TMV formulations (Figure 3B+4A). This may suggest that nonspecific interactions between coronal proteins and cell surface receptors alone were not sufficient to promote cell interactions. Instead, we propose a mechanism in which nanoparticle–cell interactions are enhanced (or stabilized) by additional interactions with the cell surface mediated by corona proteins (Figure 4B). Initial receptor-specific binding (here integrin-targeting) may be necessary; the corona may then enhance cell receptor clustering and interactions to promote uptake. Although this type of mechanism would be most advantageous for cell targeting in vivo, it might also benefit applications where no internalization is required, e.g. targeting cardiovascular clots/thrombi, by strengthening adhesion interactions.
Figure 4. Influence of the protein corona on the interactions between RGD-targeted TMV nanoparticles (of three different RGD coverages) and different cell lines.
A, TMV–HeLa cell interactions in serum-free (sfMEM) and complete (cMEM) media as a function of surface chemistry studied by FACS. Statistical significant differences are indicated by p-values. B, Hypothesis for the promotion of VNP uptake in HeLa cells by coronal proteins. The energy of nonspecific interactions between coronal proteins and cell surface receptors is not sufficient for effective binding of the VNP. However, the RGD-targeted VNPs can “dock”, and uptake is enhanced by nonspecific interactions between coronal proteins and cell surface receptors. C, TMV–RAW264.7 cell interactions. a. D, TMV–SC cell interactions. E, Schematic diagram showing the specificity of RGD-targeted TMV VNPs. Red arrows indicate uptake in cMEM (a protein corona is present); blue arrows indicate uptake in serum-free medium (a protein corona is absent).
Lastly, cell interactions of TMV with mononuclear phagocyte cells were studies. While TMV formulations were not interacting with human SC monocyte macrophages (Figure 4D), interactions with murine RAW264.7 monocyte macrophages (Figure 4C) were apparent as a function of surface chemistry. The addition of the RGD targeting ligand promoted cellular uptake of TMV-PEG12-RGD compared to TMV-lys or PEGylated TMV – for the latter formulations cell interactions were minimal attesting to the high aspect ratio nature of the particles, Figure 4C). The increased uptake of TMV-PEG12-RGD particles in RAW264.7 may be explained by the fact that these cells also express the integrin ligand[23]. There was no significant difference comparing any of the particles formulations in sfMEM and cMEM. This may reflect the difference in surface receptors and uptake mechanisms between HeLa cells and RAW264.7/SC macrophages, i.e. endocytosis vs. phagocytosis.
2.3 Corona-induced aggregation and its consequences on biodistribution
Another important aspect of the protein corona is its influence on the dispersion of nanoparticles in biological media. TMV formulations with RGD, CREKA and GPRPP targeting ligands have been investigated in vivo[23,49]. RGD-labeled TMV VNPs target tumors derived from HT-29 colon carcinoma xenografts on NCR-nu/nu mice[23] and tumor targeting efficiency was dependent on surface chemistry (shielding and targeting) as well as the aspect ratio of the particles[23]. Furthermore, we achieved thrombus targeting using Cowpea mosaic virus (CPMV) and TMV-based nanoparticles functionalized with CREKA and GRPRPP, again finding that surface chemistry and shape affected VNP homing efficiency, with shape as the dominating factor[49].
TMV particles labeled with RGD, GPRPP, or CREKA were prepared as previously described[23,49] (see Materials and Methods) and incubated in human plasma. Particles bearing RGD or CREKA showed stable dispersion properties in potassium phosphate buffer (pH 7.4) and in human plasma, whereas GPRPP-labeled particles aggregated within a minute of contact with human plasma and formed a large precipitate at the bottom of the tube (Figure 5A and Supplementary Table S3). The aggregation of GPRPP-modified TMV does not occur in human blood serum, which has similar plasma composition except clotting factors, and therefore reflects interactions between the GPRPP peptide and clotting components.
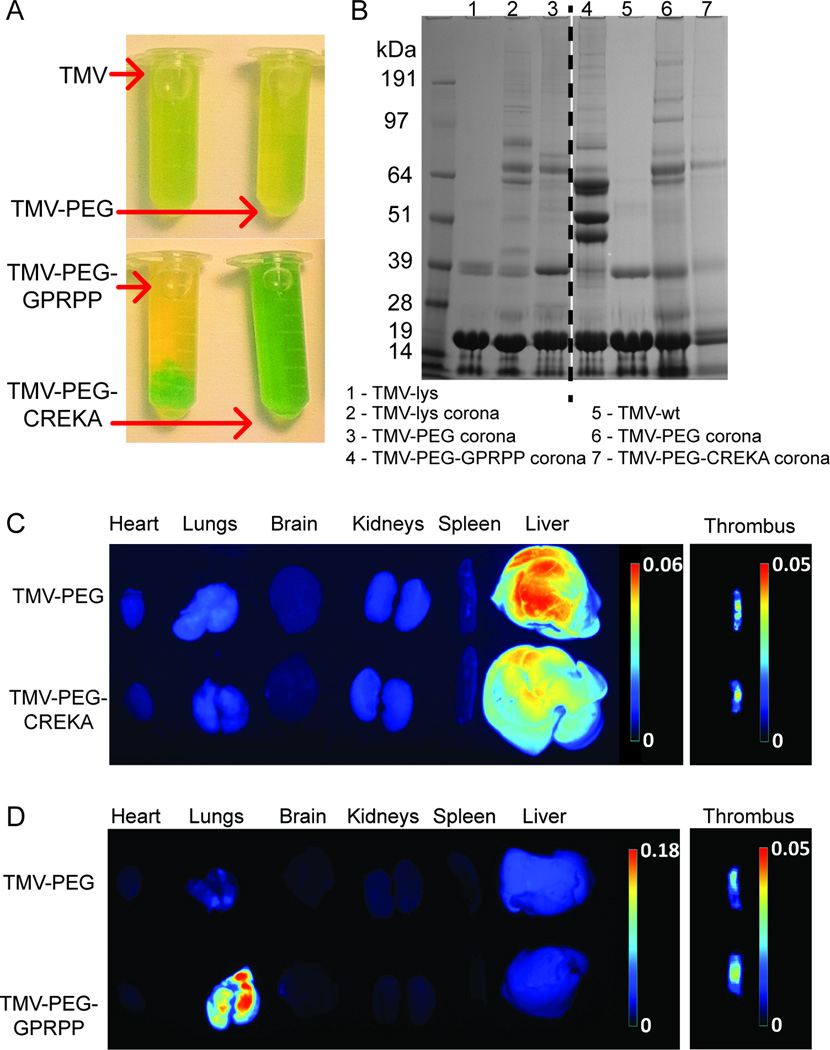
Figure 5. Aggregation of VNPs induced by the protein corona and its impact on biodistribution.
A, Photographs of peptide-targeted VNPs in situ in blood plasma. B, SDS-PAGE analysis of the protein corona of peptide-targeted VNPs. C–D, Representative Maestro fluorescence images showing the biodistribution of VNPs displaying CREKA and GPRPP, respectively, in a photochemical injury mouse model. The thrombi images were adapted from our previous study[49].
Although rapid particle aggregation decreases the nanoparticle surface available for formation of a “mature” protein corona, as well as complicating its separation and analysis, SDS-PAGE separation (Figure 5B) gave insight into protein corona formation on the particles under investigation. While the coronal composition was similar for the TMV-PEG8-CREKA and RGD-labeled particles, the majority of proteins bound to TMV-PEG12-GPRPP aggregates were found to be distinct, possibly explaining the tendency of these particles to aggregate. Three strong bands of proteins bound to TMV-PEG12-GPRPP but absent in other formulations were identified by mass spectrometry as fibrinogen chains alpha, beta and gamma, which can interact with VNPs in a multivalent manner and bridge between them to promote rapid aggregation. Since GPRPP peptide is thought to be specific to fibrin[43,50] (the product of fibrinogen polymerization), its unexpected binding to fibrinogen (and resulting aggregation of the particles) might suggest GPRPP’s lack of specificity, or specificity towards a protein region present in both fibrin and fibrinogen. This hypothesis is supported by the lack of aggregation of GPRPP particles in (fibrinogen-free) human serum.
In our previous study using TMV particles labeled with CREKA or GPRPP in a photochemical injury mouse model of thrombosis, we found that thrombus targeting following the intravenous administration of VNPs was dependent on the particle shape and surface chemistry[49]. GPRPP has a much higher affinity for fibrin than CREKA[43,50], but GPRPP-targeted formulations showed variable target specificity in vivo[49]. We therefore collected the major organs from these animals and analyzed the biodistribution of each formulation (n = 3, representative data are shown). The heart, lung, brain, kidneys, spleen and liver were collected and analyzed using a Maestro imaging system (Figure 5C+D). The TMV-PEG and TMV-PEG-CREKA formulations were mainly cleared through the liver, consistent with previous studies[27,51]. Clearance through the reticuloendothelial system organs (including the liver) is also consistent with the expected behavior of nanoparticle systems in this size range[52].
Interestingly, targeted TMV-PEG-GPRPP particles accumulated in the lungs. This phenomenon is rarely encountered with the exception of carbon nanotubes[53,54]. However, the accumulation of carbon nanotubes in the lungs can be prevented by shielding with longer PEG chains (MW 2000–5400)[55], suggesting that the accumulation of VNPs in the lungs reflects their aggregation and filtration through lung capillaries or uptake by lung macrophages that are known to scavenge for and internalize foreign macromolecules[56]. This theory is supported by the fact that the aggregation of GPRPP particles happens almost instantaneously (within a minute) after their introduction in plasma (as observed in vitro, Figure 5A) and the lungs with a high density of capillary vasculature are the central organ for capture of aggregated particulates.
The CREKA and GPRPP peptides have similar charges because the arginine and lysine residues impart positive charges and glutamic acid residues impart a negative charge, resulting in a slight positive charge for both peptides. Furthermore, both rod-shaped TMV-PEG-GPRPP (Figure 5D) and icosahedral CPMV-PEG-GPRPP VNPs accumulated in the lungs (Supplementary Figure S8B). These data indicate that neither surface charge nor shape are responsible for the accumulation of these particles in the lungs, so the protein corona (fibrinogen binding) must be the major factor that promote aggregation and this distinct biodistribution pattern.
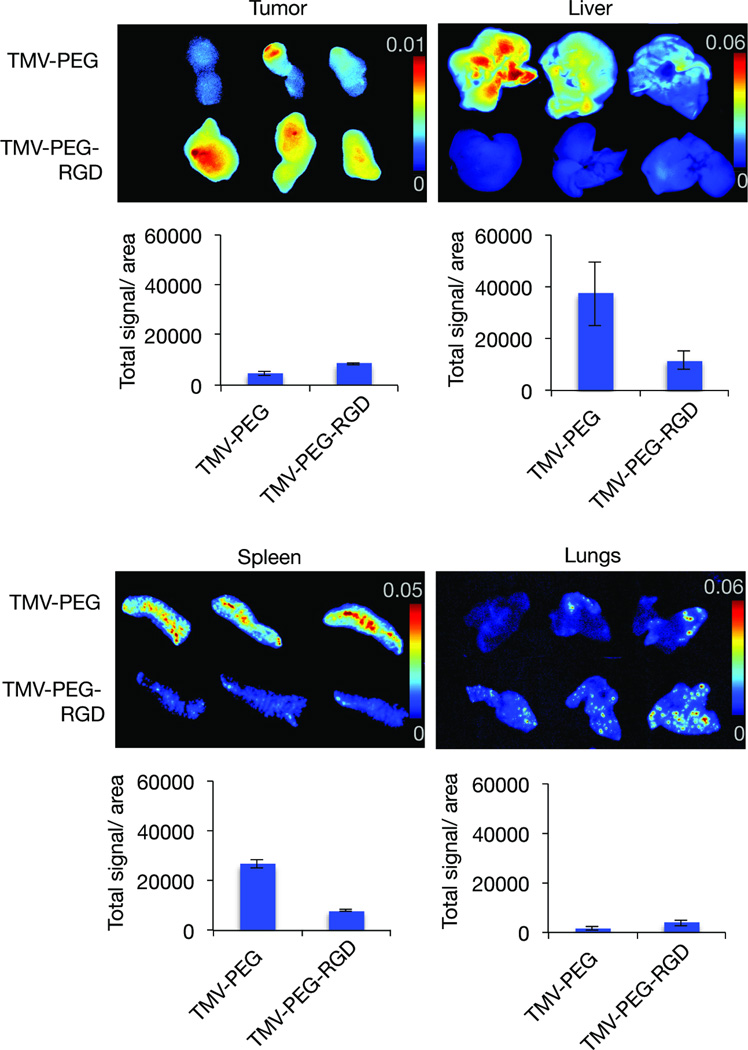
Lastly, we showed that RGD-targeted TMV formulations exhibit favorable bidistribution and tumor homing compared to PEGylated TMV. While PEGylated TMV accumulated in the liver/spleen to a much greater extent than in the tumor, the RGD-targeted formulation shows enhanced tumor targeting accompanied with reduced liver/spleen deposition (Figure 6). Moderate uptake in the lungs of RGD-targeted vs. PEGylated TMV may be explained by active targeting of integrin-expressing phagocytic cells within the lungs rather than effects mediated through the protein corona – this would be consistent with our studies on RGD-targeted potato virus X (PVX), also a high aspect ratio filament[23].
Figure 6. Biodistribution and targeting of TMV-PEG-RGD VNPs.
Representative Maestro fluorescence images showing the biodistribution of VNPs displaying RGD, in a HT-29 cancer mouse model. RGD-targeted TMV accumulates preferentially in HT-29 tumor xenografts in mice with reduced accumulation in liver/spleen (while PEGylated accumulates in liver/spleen > tumor). Maestro imaging of tumors, liver and spleen (n=3; signals shown as heat map.
3 Conclusions
In conclusion, we found that VNPs, like synthetic nanoparticles, do form a protein corona in plasma, but the quantity of proteins was ~3–9 fold lower than found around synthetic particles and the composition was distinct. The abundance of immune system proteins on the TMV surface, such as complement proteins and immunoglobulins, is likely to promote the rapid clearance of VNPs from the body. Studying the cell interactions of PEGylated and targeted TMV formulations, we found that the protein corona significantly affects the molecular recognition of TMV particles and can enhance their interactions with some cell lines (HeLa), while having no effect on their interactions with others (SC and RAW264.7 macrophages). Furthermore, we found that the choice of targeting ligand is a key factor that controls interactions with plasma proteins and hence the dispersion properties of VNPs in plasma, ultimately affecting their biodistribution and clearance in vivo, with rapid clearance by the liver or lungs. GPRPP-targeted VNPs aggregated rapidly under physiological conditions, promoting their accumulation in the capillary vasculature of the lungs, whereas the biodistribution of VNPs functionalized with the CREKA peptide was dissimilar and these particles were cleared by the liver. The accumulation of VNPs in target vs. non-target organs is therefore affected by dispersion-related properties, with aggregated particles sequestered rapidly by the lungs, potentially explaining their lack of targeting efficiency in vivo[49].
The protein corona needs to be considered when designing VNP-based drug delivery systems and imaging reagents. Its properties and composition influence the in vivo biological fate of VNPs, and we therefore recommend the careful analysis of the protein corona before VNPs are moved toward preclinical studies. Although polymer coatings (such as PEG) are known to reduce the quantity of proteins adsorbing on the surface of nanoparticles, the complete prevention of adsorption is not currently possible. In the future, novel VNP surface engineering strategies may improve the biocompatibility of VNPs and enhance their in vivo behavior. We have also established methods to dissect the protein corona and establish its impact on the properties of VNPs in vivo paving the way for further translational research.
4 Experimental Section
Virus propagation and purification
Viruses were propagated by mechanical inoculation using 5–10 µg of virus per leaf. Wild-type TMV and TMV-lys mutants were propagated in Nicotiana benthamiana. The isolation of VNPs using established procedures yielded approximately 1 mg of virus per gram of infected leaf material[17].
TMV sCy5 labeling
TMV was internally labeled with alkynes by EDC coupling for 24 h using 100 equivalents of propargylamine per capsid protein with 50 equivalents of EDC (25 equivalents added at 0 and 18 h) in 100 mM HEPES buffer, pH 7.4. Subsequently, Cu(I)-catalyzed alkyne-azide cycloaddition reaction (click chemistry) was performed by adding one equivalent of sCy5-azide per coat protein using 2 mg/mL TMV in the presence of 2 mM aminoguanidine (AMG), 2 mM sodium ascorbate (Sod Asc) and 1 mM CuSO4 in 10 mM potassium phosphate buffer (pH 7.4) on ice for 30 min. TMV was purified by ultracentrifugation at 160,000 g for 3 h over a 40% (w/v) sucrose cushion. SNPs were produced by heat-induced shape switching of TMV or Cy-labeled TMV as previously described[27,28].
TMV conjugation with PEG, RGD and GPRPP
Cyclic RGD peptide (cRGDfC from Peptides International) and linear GPRPPC (custom peptide from GenScript) were conjugated to the exterior TMV surface. TMV-lys (cysteine-to-lysine binding) was mixed with NHS–maleimide heterobifunctional AMAS linker (for direct conjugation) or SM-PEG4–24 linkers of various PEG chain lengths (n = 4, 12, and 24, Pierce), at one, four, and ten equivalents per capsid protein (for low, medium and high coverage respectively) and a final TMV-lys concentration of 2 mg/mL. The reaction was carried out in 0.01 M potassium phosphate buffer (pH 7.4) containing 10% (v/v) DMSO for 2 h at room temperature. 1.5, 6, or 15 equivalents of RGD (for low, medium and high coverage respectively) or 6 e.q. GPRPP were added per capsid protein and the reaction was carried out overnight at room temperature. The RGD/GPRPP-functionalized VNPs were then purified by ultracentrifugation to remove excess reagents. Control TMV-PEG particles without the targeting ligand were obtained from the same batch of particles by omitting the addition of the targeting ligand.
TMV conjugation with PEG-CREKA
TMV particles were mixed with 15 equivalents per capsid protein of diazonium salt (produced by mixing 100 mM 3-ethynylaniline with 150 mM sodium nitrite in p-toluenesulfonic acid) in 100 mM borate buffer (pH 8.8) for 30 min on ice. Following purification by ultracentrifugation, N3-PEG8-COOH, or N3-PEG8-CREKA were attached by click chemistry using 2 mg/mL TMV with two equivalents of PEG or peptide in the presence of 2 mM AMG, 2 mM Sod Asc and 1 mM CuSO4 in 10 mM phosphate buffer (pH 7.4) on ice for 30 min. The particles were purified by ultracentrifugation as described above.
UV/visible spectroscopy
A Thermo Scientific NanoDrop 2000 spectrophotometer was used to determine the particle concentrations. The extinction coefficient for TMV at 260 nm is 3 mg−1 mL cm−1. UV/vis spectroscopy was also used to determine the degree of dye-labeling of the fluorescent nanoparticle formulations, using Beer-Lambert law and the dye-specific extinction coefficient.
DLS and ζ-potential
DLS measurements were performed using a 90plus instrument (Brookhaven Instruments Corporation) at a scattering angle h=90°. Reported values are averaged values over multiple measurements (the total measurement time was 2.5–5.0 minutes). Hydrodynamic diameters (Dh) and polydispersity indices (PDI) were calculated using the manufacturer’s software. ζ-potential measurements were performed using Zetasizer Nano instrument (Malvern Instruments). Reported ζ values were calculated using manufacturer’s software and are an average of three measurements, each consisting of 10–100 sub-runs, depending on the collected data quality.
Protein corona preparation and SDS-PAGE
Hard protein coronas from TMV rods and SNPs were compared to SiO2 coronas by incubating the particles in 2 mL ~100% human plasma at a fixed surface area to plasma protein ratio (corresponding to 0.3 mg/mL VNPs and 2 mg/mL SiO2 particles, respectively) at room temperature for 1 h. The samples were then diluted in 25 mL PBS, purified by ultracentrifugation (160,000 g) on a 40% (v/v) sucrose cushion, and washed twice with 25 mL PBS to remove loosely-bound proteins. The final pellet was resuspended in 0.1 mL gel loading buffer (62.5 mM Tris-HCl pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerol, 0.01% (w/v) bromophenol blue, 10% (v/v) 2-mercaptoethanol) and denatured by boiling at 100°C for 7 min. We then separated 10–15 µl of each sample on 4–12% (or 12% for conjugation analysis gels) NuPAGE polyacrylamide gels in 1× MOPS running buffer at 200 V for 50 min. The gels were stained with Coomassie Brilliant Blue and visualized using an AlphaImager imaging system (Biosciences). All gels used for densitometry analysis were run in triplicates and column charts correspond to average values of band densities and standard deviations.
Transmission electron microscopy
Particles were adsorbed to carbon-coated copper grids at a concentration of 0.1 mg/mL (2 µl per grid), rinsed with deionized water, and negatively stained with 2% (w/v) uranyl acetate for 2 min before analysis with a Zeiss Libra 200FE TEM at 200 kV.
Mass spectrometry
After the SDS-PAGE separation of the protein corona from (V)NPs, selected gel lanes were excised and trypsin digested prior mass spectrometry. All samples were run on Waters ultra high pressure liquid chromatography NanoAcquity (Waters Corporation) and an LTQ Orbitrap Elite (Thermo, Waltham, MA). The instrument was mass calibrated immediately before the analysis using the instrument protocol. Mobile phase A (aqueous) contained 0.1% (v/v) formic acid in 5% (v/v) ACN and mobile phase B (organic) contained 0.1% formic acid in 85% ACN. Samples were trapped and desalted on-line in mobile phase A at10 µL/min for 10 minutes using a Waters UPLC PST C18 nanoACQUITY 300 (75 µm × 25 cm) reversed phase column with 5% mobile phase B. Gel based discovery separation was obtained by employing a gradient of 6% to 28% mobile B at 0.300 µL/min over 120 minutes. The column was washed at 99% mobile phase B for 10 minutes, followed by a re-equilibration at 100% A for 15 minutes. Positive mode electrospray was conducted using a nanospray source and the mass spectrometer was operated at a resolution of 60,000. Quantitative and qualitative data were acquired using alternating full MS scan and MS/MS scans in normal mode. Survey data were acquired from m/z of 400 to 1600 and up to 20 precursors based on intensity were interrogated by MS/MS per switch. Two micro scans were acquired for every precursor interrogated and MS/MS was acquired as centroid data. The raw data was processed via Proteome Discovere (Thermo Waltham, MA). The MS/MS peak lists were subsequently searched by Mascot version 2.2.0 (Matrix Science, London, UK). The database used was human Uniprot (538,585 sequences). Search settings were as follows: trypsin enzyme specificity; mass accuracy window for precursor ion, 8.0 ppm; mass accuracy window for fragment ions, 0.6Da; variable modifications including carbamidomethlylation of cysteines, 1 missed cleavage and oxidation of methoinine. Scaffold software version 4.4.0 has been used for peptide identification. The peptides were identified by the Protein Prophet algorithm with Scaffold delta-mass correction and were accepted for probability ≥ 90.0%. The protein probabilities were assigned by the Protein Prophet algorithm and were accepted for probability ≥ 95.0% and if it contained at least one identified peptide. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Proteins sharing significant peptide evidence were grouped into clusters.
Flow cytometry
HeLa, SC and RAW264.7 cells (all from ATCC) were grown to confluence in MEM, IMDM, or DMEM medium as appropriate, supplemented with 10% (v/v) FBS (to make cMEM) and 1% (w/v) penicillin/streptomycin, at 37°C and 5% CO2. IMDM was also supplemented with 0.05 mM 2-mercaptoethanol, 0.1 mM hypoxanthine and 0.016 mM thymidine. The cells were washed with PBS, collected in enzyme-free Hank’s-based Cell Dissociation Buffer (Fisher), washed in PBS again and resuspended in the cMEM or sfMEM most appropriate for the cell line. For additional experiments on HeLa cells (Figure S5) cells were resuspended in either (A) cMEM and heat inactivated cMEM (30 minutes incubation in 56°C), or (B) sfMEM and sfMEM supplemented with 1.5 mg/ml total human IgG (I4506 Sigma Aldrich). Cells were then added to 96-well v-bottom plates (200,000 cells in 200 µL per well) and incubated with 1–2.5 µg of VNPs per well in triplicate and incubated for 3 h at 37°C and 5% CO2. The cells were then washed twice in FACS buffer (1 mM EDTA, 25 mM HEPES, 1% (v/v) FBS in PBS, pH 7.0) and fixed in 2% (v/v) paraformaldehyde in FACS buffer for 10 min at room temperature. After fixation, cells were washed twice in FACS buffer, resuspended in 300 µL FACS buffer, and stored at 4°C. Cells were analyzed using a BD LSR II Flow Cytometer and 10,000 gated events were recorded. Data were analyzed using FlowJo v8.6.3 software.
In vivo biodistribution experiments
All animal procedures were performed using protocols approved by the Institutional Animal Care and Use Committee (IACUC) at Case Western Reserve University.
TMV-PEG-CREKA and TMV-PEG-GPRPP studies
Particles (200 µg per 100 µL) and Rose Bengal dye (10 mg/mL in PBS) at a dose of 50 mg/kg body weight were simultaneously administered to C57BL/6 mice by tail vein injection. A 1.5 mW, 540 nm green laser (Melles Griot) was used to illuminate the right common carotid artery 5 cm from the artery to induce thrombus formation, and blood flow was monitored with a Doppler flow probe (Transonic Systems). After vessel occlusion, the mice were killed and the injured artery and the contralateral control were excised and fixed in formalin. After completion of the experiment, the heart, lungs, brain, kidneys, spleen, and liver were collected. Particle accumulation in the organs was then analyzed by Maestro imaging with yellow excitation (576–621 nm) and emission (635 nm longpass) filters with 800 ms exposure times.
TMV-PEG-RGD studies
Human colon cancer xenografts were developed using HT-29 cells (ATCC) maintained in McCoy’s medium (Life Technologies) supplemented with 10% (v/v) fetal bovine serum (FBS), 1% (w/v) penicillin/streptomycin and 1% (w/v) glutamine (all from Life Technologies). Six-week-old male NCr nu/nu nude mice maintained on an alfalfa-free diet (Teklad 2018S, Harlan Laboratories) were subcutaneously injected in the right flank with 2.5 × 10 cells in 50 µL medium mixed with an equal volume of matrigel (BD Matrigel, BD Biosciences, USA) using a microliter Hamilton 22 gauge syringe. One tumor was induced in each mouse. The animals were monitored daily and tumor homing studies commenced when tumors reached an average volume of 20–30 mm (typically within 12 days after the injection of HT-29 cells). Animals were assigned randomly into groups with n = 3 mice per formulation. PBS, TMV-PEG and TMV-PEG-RGD particles were administered intravenously via tail vein injections. Mice were euthanized and major tissues were harvested and imaged in the Maestro fluorescence imager using a combination of yellow excitation/emission filters (excitation filter: 575 – 605 nm; and emission filter: 645 nm longpass) for A647 signal. Region of interest (ROI) analysis was performed using the Maestro software to determine the fluorescence intensity from the respective tissues as compared to tissues from PBS injected mice.
Supplementary Material
Acknowledgments
This work was supported in part by a grant from the National Science Foundation CAREER DMR 1452257 (to NFS) and grants from the National Institute of Health (NIH): NHLBI R21 HL121130 (to NFS) and T32 HL105338 Cardiovascular Research training grant (to AMW), and a pilot grant from Case-Coulter Translational Research Partnership and the Harrington Heart & Vascular Institute.
We thank Prof. Christina Wege and her team from the University of Stuttgart, Germany for the TMV-lys mutant. We thank CWRU Farm for help with the scaled-up production and growth of N. benthamiana plants. Drs. Yunmei Wang, Huiyun Gao, and Daniel I. Simon are acknowledged for their help with the photochemical mouse injury model[49]. Ms. Daniela Schlatzer is acknowledged for her help with the mass spectrometry analysis. Dr. Michael A. Bruckman is acknowledged for his helpful discussions. Selected molecular graphics and analyses were performed with the UCSF Chimera package. Chimera is developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Footnotes
Supporting Information
Supporting information is available from the Wiley Online Library or from the author.
Author contributions
A.S.P and N.F.S. conceived and designed the experiments; A.S.P. prepared all particle formulations, performed the in vitro experiments, analyzed, and interpreted the data; A.M.W. and S.S. performed in vivo biodistribution study, analyzed and interpreted the data. A.S.P and N.F.S. wrote the paper; all authors edited and approved the manuscript.
References
- 1.Moghimi SM, Hunter AC, Murray JC. FASEB J. 2005;19:311. doi: 10.1096/fj.04-2747rev. [DOI] [PubMed] [Google Scholar]
- 2.Zamboni WC, Torchilin V, Patri AK, Hrkach J, Stern S, Lee R, Nel A, Panaro NJ, Grodzinski P. Clin. Cancer Res. 2012;18:3229. doi: 10.1158/1078-0432.CCR-11-2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peer D, Karp JM, Hong S, Farokhzad OC, Margalit R, Langer R. Nature Nanotechnology. 2007;2:751. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 4.Godin B, Sakamoto JH, Serda RE, Grattoni A, Bouamrani A, Ferrari M. Trends Pharmacol. Sci. 2010;31:199. doi: 10.1016/j.tips.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iverson N, Plourde N, Chnari E, Nackman GB, Moghe PV. BioDrugs. 2008;22:1. doi: 10.2165/00063030-200822010-00001. [DOI] [PubMed] [Google Scholar]
- 6.Walczyk D, Baldelli Bombelli F, Monopoli MP, Lynch I, Dawson KA. J. Am. Chem. Soc. 2010;132:5761. doi: 10.1021/ja910675v. [DOI] [PubMed] [Google Scholar]
- 7.Walkey CD, Chan WCW. Chem Soc Rev. 2012;41:2780. doi: 10.1039/c1cs15233e. [DOI] [PubMed] [Google Scholar]
- 8.Monopoli MP, Åberg C, Salvati A, Dawson KA. Nature Nanotechnology. 2012;7:779. doi: 10.1038/nnano.2012.207. [DOI] [PubMed] [Google Scholar]
- 9.Cedervall T, Lynch I, Lindman S, Berggard T, Thulin E, Nilsson H, Dawson KA, Linse S. Proc Natl Acad Sci U S A. 2007;104:2050. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monopoli MP, Walczyk D, Campbell A, Elia G, Lynch I, Baldelli Bombelli F, Dawson KA. J. Am. Chem. Soc. 2011;133:2525. doi: 10.1021/ja107583h. [DOI] [PubMed] [Google Scholar]
- 11.Pitek AS, O'Connell D, Mahon E, Monopoli MP, Baldelli Bombelli F, Dawson KA. PLoS ONE. 2012;7:e40685. doi: 10.1371/journal.pone.0040685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobrovolskaia MA, Patri AK, Zheng J, Clogston JD, Ayub N, Aggarwal P, Neun BW, Hall JB, McNeil SE. Nanomedicine: Nanotechnology, Biology and Medicine. 2009;5:106. doi: 10.1016/j.nano.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.França R, Zhang XF, Veres T, Yahia L, Sacher E. Journal of Colloid and Interface Science. 2013;389:292. doi: 10.1016/j.jcis.2012.08.065. [DOI] [PubMed] [Google Scholar]
- 14.Pokorski JK, Steinmetz NF. Mol. Pharmaceutics. 2011;8:29. doi: 10.1021/mp100225y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steinmetz NF. Mol. Pharmaceutics. 2013;10:1. doi: 10.1021/mp300658j. [DOI] [PubMed] [Google Scholar]
- 16.Bruckman MA, Jiang K, Simpson EJ, Randolph LN, Luyt LG, Yu X, Steinmetz NF. Nano Lett. 2014;14:1551. doi: 10.1021/nl404816m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bruckman MA, Steinmetz NF. Methods Mol. Biol. 2014;1108:173. doi: 10.1007/978-1-62703-751-8_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wen AM, Ryan MJ, Yang AC, Breitenkamp K, Pokorski JK, Steinmetz NF. Chem. Commun. 2012;48:9044. doi: 10.1039/c2cc34695h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verduin BJ. FEBS Lett. 1974;45:50. doi: 10.1016/0014-5793(74)80808-2. [DOI] [PubMed] [Google Scholar]
- 20.Comellas-Aragonès M, Engelkamp H, Claessen VI, Sommerdijk NAJM, Rowan AE, Christianen PCM, Maan JC, Verduin BJM, Cornelissen JJLM, Nolte RJM. Nature Nanotechnology. 2007;2:635. doi: 10.1038/nnano.2007.299. [DOI] [PubMed] [Google Scholar]
- 21.Brasch M, Voets IK, Koay MS, Cornelissen JJ. Faraday discussions. 2013;166:47. doi: 10.1039/c3fd00088e. [DOI] [PubMed] [Google Scholar]
- 22.Millán JG, Brasch M, Anaya-Plaza E, de la Escosura A, Velders AH, Reinhoudt DN, Torres T, Koay MST, Cornelissen JJLM. J. Inorg. Biochem. 2014;136:140. doi: 10.1016/j.jinorgbio.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 23.Shukla S, Eber FJ, Nagarajan AS, DiFranco NA, Schmidt N, Wen AM, Eiben S, Twyman RM, Wege C, Steinmetz NF. Adv Healthc Mater. 2015;4:874. doi: 10.1002/adhm.201400641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee KL, Shukla S, Wu M, Ayat NR, El Sanadi CE, Wen AM, Edelbrock JF, Pokorski JK, Commandeur U, Dubyak GR, Steinmetz NF. Acta Biomater. 2015;19:166. doi: 10.1016/j.actbio.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chariou PL, Lee KL, Wen AM, Gulati NM, Stewart PL, Steinmetz NF. Bioconjug Chem. 2015;26:262. doi: 10.1021/bc500545z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geiger FC, Eber FJ, Eiben S, Mueller A, Jeske H, Spatz JP, Wege C. Nanoscale. 2013;5:3808. doi: 10.1039/c3nr33724c. [DOI] [PubMed] [Google Scholar]
- 27.Bruckman MA, Randolph LN, VanMeter A, Hern S, Shoffstall AJ, Taurog RE, Steinmetz NF. Virology. 2014;449:163. doi: 10.1016/j.virol.2013.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bruckman MA, VanMeter A, Steinmetz NF. ACS Biomater Sci Eng. 2015;1:13. doi: 10.1021/ab500059s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee KL, Hubbard LC, Hern S, Yildiz I, Gratzl M, Steinmetz NF. Biomater. Sci. 2013;1 doi: 10.1039/C3BM00191A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Milani S, Bombelli Bombelli F, Pitek AS, Dawson KA, Rädler J. ACS Nano. 2012;6:2532. doi: 10.1021/nn204951s. [DOI] [PubMed] [Google Scholar]
- 31.Lundqvist M, Stigler J, Elia G, Lynch I, Cedervall T, Dawson KA. Proc. Natl. Acad. Sci. U.S.A. 2008;105:14265. doi: 10.1073/pnas.0805135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nel AE, Mädler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat Mater. 2009;8:543. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 33.Dobrov EN, Nikitin NA, Trifonova EA, Parshina EY, Makarov VV, Maksimov GV, Karpova OV, Atabekov JG. J. Biomol. Struct. Dyn. 2014;32:701. doi: 10.1080/07391102.2013.788983. [DOI] [PubMed] [Google Scholar]
- 34.Salvati A, Pitek AS, Monopoli MP, Prapainop K, Baldelli Bombelli F, Hristov DR, Kelly PM, Åberg C, Mahon E, Dawson KA. Nature Nanotechnology. 2013;8:137. doi: 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- 35.Mirshafiee V, Mahmoudi M, Lou K, Cheng J, Kraft ML. Chem. Commun. 2013;49:2557. doi: 10.1039/c3cc37307j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dai Q, Yan Y, Ang C-S, Kempe K, Kamphuis MMJ, Dodds SJ, Caruso F. ACS Nano. 2015;9:2876. doi: 10.1021/nn506929e. [DOI] [PubMed] [Google Scholar]
- 37.Zarschler K, Prapainop K, Mahon E, Rocks L, Bramini M, Kelly PM, Stephan H, Dawson KA. Nanoscale. 2014;6:6046. doi: 10.1039/c4nr00595c. [DOI] [PubMed] [Google Scholar]
- 38.Wang F, Li Y, Shen Y, Wang A, Wang S, Xie T. Int J Mol Sci. 2013;14:13447. doi: 10.3390/ijms140713447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oba M, Fukushima S, Kanayama N, Aoyagi K, Nishiyama N, Koyama H, Kataoka K. Bioconjug Chem. 2007;18:1415. doi: 10.1021/bc0700133. [DOI] [PubMed] [Google Scholar]
- 40.McCarthy JR, Patel P, Botnaru I, Haghayeghi P, Weissleder R, Jaffer FA. Bioconjug Chem. 2009;20:1251. doi: 10.1021/bc9001163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawasaki K, Miyano M, Hirase K, Iwamoto M. Chem Pharm Bull (Tokyo) 1993;41:975. doi: 10.1248/cpb.41.975. [DOI] [PubMed] [Google Scholar]
- 42.Peters D, Kastantin M, Kotamraju VR, Karmali PP, Gujraty K, Tirrell M, Ruoslahti E. Proc. Natl. Acad. Sci. U.S.A. 2009;106:9815. doi: 10.1073/pnas.0903369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Agemy L, Sugahara KN, Kotamraju VR, Gujraty K, Girard OM, Kono Y, Mattrey RF, Park J-H, Sailor MJ, Jimenez AI, Cativiela C, Zanuy D, Sayago FJ, Aleman C, Nussinov R, Ruoslahti E. Blood. 2010;116:2847. doi: 10.1182/blood-2010-03-274258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otsuka H, Nagasaki Y, Kataoka K. Adv. Drug Deliv. Rev. 2003;55:403. doi: 10.1016/s0169-409x(02)00226-0. [DOI] [PubMed] [Google Scholar]
- 45.Perry JL, Reuter KG, Kai MP, Herlihy KP, Jones SW, Luft JC, Napier M, Bear JE, DeSimone JM. Nano Lett. 2012;12:5304. doi: 10.1021/nl302638g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai Q, Walkey C, Chan WCW. Angew. Chem. Int. Ed. Engl. 2014;53:5093. doi: 10.1002/anie.201309464. [DOI] [PubMed] [Google Scholar]
- 47.Kelly PM, Åberg C, Polo E, O'Connell A, Cookman J, Fallon J, Krpetić Ž, Dawson KA. Nature Nanotechnology. 2015;10:472. doi: 10.1038/nnano.2015.47. [DOI] [PubMed] [Google Scholar]
- 48.O'Connell DJ, Bombelli FB, Pitek AS, Monopoli MP, Cahill DJ, Dawson KA. Nanoscale. 2015;7:15268. doi: 10.1039/c5nr01970b. [DOI] [PubMed] [Google Scholar]
- 49.Wen AM, Wang Y, Jiang K, Hsu GC, Gao H, Lee KL, Yang AC, Yu X, Simon DI, Steinmetz NF. J. Mater. Chem. B. 2015 doi: 10.1039/C5TB00879D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aruva MR, Daviau J, Sharma SS, Thakur ML. J. Nucl. Med. 2006;47:155. [PMC free article] [PubMed] [Google Scholar]
- 51.Singh P, Prasuhn D, Yeh RM, Destito G, Rae CS, Osborn K, Finn MG, Manchester M. J Control Release. 2007;120:41. doi: 10.1016/j.jconrel.2007.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Albanese A, Tang PS, Chan WCW. Annu Rev Biomed Eng. 2012;14:1. doi: 10.1146/annurev-bioeng-071811-150124. [DOI] [PubMed] [Google Scholar]
- 53.Yang S-T, Guo W, Lin Y, Deng X-Y, Wang H-F, Sun H-F, Liu Y-F, Wang X, Wang W, Chen M. J. Phys. Chem. C. 2007;111:17761. [Google Scholar]
- 54.Yang S-T, Wang X, Jia G, Gu Y, Wang T, Nie H, Ge C, Wang H, Liu Y. Toxicol. Lett. 2008;181:182. doi: 10.1016/j.toxlet.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 55.Liu Z, Cai W, He L, Nakayama N, Chen K, Sun X, Chen X, Dai H. Nature Nanotechnology. 2007;2:47. doi: 10.1038/nnano.2006.170. [DOI] [PubMed] [Google Scholar]
- 56.Kobzik L. J. Immunol. 1995;155:367. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.