Abstract
The first integron-borne metallo-β-lactamase gene was isolated in Hungary. The blaVIM-4 gene is located on a class 1 integron that also carries a novel blaOXA-like gene. The integron is harbored by a serotype O12 Pseudomonas aeruginosa strain and shows high structural similarity to integrons isolated in Greece and Poland.
Acquired metallo-β-lactamases (MBLs) are mostly encoded by integron-borne genes and confer resistance against all β-lactams except for the monobactams. VIM-type MBLs were reported from several European countries and also from countries outside Europe such as Korea and the United States (10, 20). An outbreak involving 47 VIM-producing Pseudomonas aeruginosa isolates was reported from the University Hospital of Thessaly, Greece, in 2001 to 2002, where the blaVIM-4 gene had originally been identified (14, 15).
During this study 226 carbapenem-resistant P. aeruginosa isolates from Hungary were screened for MBL production by phenotypic tests. The isolates were obtained from clinical microbiology laboratories between January 2001 and September 2003. P. aeruginosa strains PA396 and PA450 were isolates from the intensive care unit of the Central Military Hospital in Budapest. Strain PA396 was isolated in August 2002 from the urine of a Greek citizen polytraumatic patient 2 days before his death. The patient died due to multiorganic failure as part of a severe septic shock. The antibiotic treatment of the patient included piperacillin-tazobactam, meropenem, vancomycin, and amikacin. According to the anamnesis, the patient's previous clinical history included tonsillectomy and appendectomy. Strain PA450 was isolated in September 2002 from the urine of a Hungarian citizen polytraumatic patient 1 day before his death. The applied antibiotic therapy included cefuroxime, imipenem-cilastatin, and vancomycin.
MICs were determined by the agar dilution method for β-lactam antibiotics (9) and by the Etest (AB Biodisk, Solna, Sweden) for other antibiotics. To detect MBL production, the MBL Etest and the Imipenem-EDTA disk method (22) were used.
For the detection of blaVIM genes and class 1 integrons by PCR we used the primers listed in Table 1. PCR products were sequenced using an ABI PRISM 310 genetic analyzer. Pulsed-field gel electrophoresis was performed as described previously (12) with modifications. Plasmid content analysis was carried out as described previously (2, 5) and by the use of a QIAprep Spin Miniprep kit (QIAGEN, Hilden, Germany). Conjugation experiments were performed on Mueller-Hinton agar plates with the Escherichia coli J5-3 Rifr and P. aeruginosa PAO4089Rp (7) strains as recipients, mating plates were incubated at 37°C for 14 h, and transconjugants were selected on Mueller-Hinton agar plates containing 300 and 100 μg of rifampin/ml, respectively, and 32 μg of cefotaxime/ml.
TABLE 1.
Primers for PCR detection of blaVIM genes and integrons and for sequencing
| Target or use | Designation | Primers (5′ to 3′) |
|---|---|---|
| blaVIM | VIM-F | GTGATGGTGATGAGTTGCTTTTG |
| VIM-R | TACTGGACCGAAGCGCACT | |
| Integron | 5CS-F1 | ATGTTACGCAGCAGGGC |
| 5CS-F2 | GGGATCCAAGCAGCAAGCGCG | |
| 3CS-R | GGAATTCGACCTGATAGTTTGGCTGTG | |
| Sequencing | SP1 | CGGCATAACGTTTGACATG |
| AAC-R | GCTAAAACGCTTGGCAAGT | |
| OXA-R | GAAACTTGTATTGCCCCTCT |
Only two isolates, PA396 and PA450, were positive by the phenotypic tests. They had a 7-mm increase in the diameter of the inhibitory zone by the IPM-EDTA disk method, and imipenem and imipenem-EDTA MICs for these isolates were 32/1.5 and 256/8, respectively, by the MBL Etest. Antibiotic susceptibility values are shown in Table 2. Both strains agglutinated with the monovalent O12 serum (Bio-Rad, Marnes-la-Coquette, France).
TABLE 2.
Drug MICs determined for VIM-producing P. aeruginosa isolates
| Isolate | MIC (μg/ml)a
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IPM | MEM | ATM | CAZ | CTX | FEP | TZP | GEN | AMK | CIP | |
| PA396 | 64 | >256 | 32 | 256 | >256 | 256 | >256 | 8 | 32 | >32 |
| PA450 | 256 | >256 | 16 | 256 | >256 | 256 | >256 | 8 | 32 | >32 |
Abbreviations: IPM, imipenem; MEM, meropenem; ATM, aztreonam; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; TZP, piperacillin-tazobactam; GEN, gentamicin; AMK, amikacin; CIP, ciprofloxacin.
No plasmid DNA was detected in PA396 and PA450, and experiments to transfer resistance from these strains into E. coli J5-3 Rifr and PAO4089Rp did not result in transconjugants under the experimental conditions applied.
PCR using integron primers amplified an approximately 3-kb-long product for both strains. The variable region of the integron harbored by PA396 was sequenced. PCR mapping and partial sequencing revealed that PA450 carries an integron with the same structure as that from PA396. A blaOXA-like gene with a N73I amino acid substitution compared to OXA-10 in its deduced amino acid sequence occupies the first position. It is followed by an aacA4 gene and an unusual blaVIM-4 cassette that contains a 170-bp duplicated region (Fig. 1 and 2).
FIG. 1.
The nucleotide sequence containing the duplicated region of the integron from strain PA396. Sequences corresponding to the labeled sites are shown in black characters on a gray background. 1L, 2L, 2R, and 1R sites of the 59-base elements are indicated. 2°rs indicates the putative secondary site. 1L′ and 1L indicate the duplicated reverse core site. The arrow indicates the site of recombination within the 1R core site. The two copies of the duplicated repeat unit are indicated by italicized and underlined letters, respectively. The deduced protein translation is shown below the partial coding sequence of VIM-4. The star indicates the stop codon.
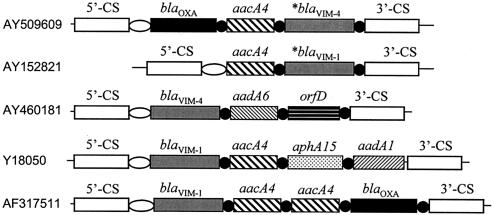
FIG. 2.
Comparison of the schematic structures of blaVIM-1- or blaVIM-4-carrying class 1 integrons. Empty ellipses represent the attI1 site; black circles represent the 59-base elements. Asterisks indicate blaVIM cassettes with the 170-bp duplication. 5′-CS and 3′-CS stand for the 5′ and 3′ conserved sequences, respectively. AY509609 is from strain PA396, Hungary. AY152821 and AY460181 were isolated in Greece; Y18050 (6) and AF317511 were isolated in Italy. Class 1 integrons without the 170-bp duplication in blaVIM also include those with GenBank accession numbers AJ581665, AJ581664, AJ439689 (8), AJ291609, AJ278514 (16), AF317511, and AY339625.
The integron from PA396 was searched against the GenBank database. Class 1 integrons with the highest scores with accession numbers AJ585042 (11) and AY152821 (18), isolated in Warsaw, Poland, and Heraklion, Greece, respectively, were retrieved. These two integrons together with the integron from PA396 share common structural features that distinguish them from all other blaVIM-1- or blaVIM-4-carrying class 1 integrons published in GenBank at the time of submission of the manuscript. First, the blaVIM cassette is left upstream of the 3′ conserved sequence. Second, an identical direct repeat of 170 bp containing the partial VIM coding sequence and the reverse core site (1L) is present in their blaVIM cassettes (Fig. 2). These observations suggest that these three integrons share a common phylogeny. The duplication was probably mediated by the IntI1 integrase, as this 170-bp region is bordered by a pentanucleotide (GATCT) that corresponds to the secondary site of the integrase (3), and the 1L site. These sites were previously reported to be involved in integrase-mediated recombinations (13, 19).
Strain PA396 was isolated from a Greek citizen who stayed in Hungary only temporarily as a tourist. Strain PA450, isolated a month later at same intensive care unit, had a pulsed-field gel electrophoresis profile identical to that of PA396. Their profile was clearly different from those of non-VIM-producing imipenem-resistant P. aeruginosa strains from the same hospital isolated between January 2002 and April 2003 (results not shown). A VIM-4-producing P. aeruginosa strain was also isolated in Sweden from a Greek citizen in 2001, the same year when VIM-4 was first isolated in Greece (4). These observations in agreement with previous studies (1, 21) raise the possibility that VIM-producing P. aeruginosa strains may be carried by the patients for a sufficiently prolonged period of time to promote their dissemination between different countries. The common structural features of the integron from strain PA396 and AY152821 are in accordance with this hypothesis. It would be valuable to test experimentally whether this hypothesis can be applied to strain PA396 by comparing it with VIM-producing strains from Greece and Sweden (4) by genetic typing methods.
With the recent detection of VIM-producing strains in several Eastern-European countries (11, 17), the repeated appearance of MBL-producing clinical isolates can be anticipated in Hungary. A regular screening and monitoring system should be set up to prevent the wider spread of these resistance determinants in the country.
Nucleotide sequence accession number.
The nucleotide sequence of the variable region of the integron from PA396 was deposited in GenBank under accession number AY509609.
Acknowledgments
This work was supported by grant T-08/186/2001 of the Hungarian Scientific Council for Health and by the National Center for Epidemiology.
We thank J. Szentandrássy and L. Keresztes for their help and assistance, J. Pászti for the E. coli J5-3 Rifr strain, and Y. Chong for the PAO4089Rp strain.
REFERENCES
- 1.Bellon, O., J. Hacini, and J. Watine. 2001. Le portage prolongé et la diffusion clonale interhospitaliére des Pseudomonas aeruginosa multirésistants de sérotype O12 sont-ils liés? Étude multicentrique. Pathol. Biol. 49:620-623. [DOI] [PubMed] [Google Scholar]
- 2.Birnboim, H. C., and J. Doly. 1979. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 24:1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Francia, M. V., F. de la Cruz, and J. M. García Lobo. 1993. Secondary-sites for integration mediated by the Tn21 integrase. Mol. Microbiol. 10:823-828. [DOI] [PubMed] [Google Scholar]
- 4.Giske, C. G., M. Rylander, and G. Kronvall. 2003. VIM-4 in a carbapenem-resistant strain of Pseudomonas aeruginosa isolated in Sweden. Antimicrob. Agents Chemother. 47:3034-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kado, C. I., and S. T. Liu. 1981. Rapid procedure for detection and isolation of large and small plasmids. J. Bacteriol. 145:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lauretti, L., M. L. Riccio, A. Mazzariol, G. Cornaglia, G. Amicosante, R. Fontana, and G. M. Rossolini. 1999. Cloning and characterization of blaVIM, a new integron-borne metallo-β-lactamase gene from a Pseudomonas aeruginosa clinical isolate. Antimicrob. Agents Chemother. 43:1584-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee, K., J. B. Lim, J. H. Yum, D. Yong, Y. Chong, J. M. Kim, and D. M. Livermore. 2002. blaVIM-2 cassette-containing novel integrons in metallo-β-lactamase-producing Pseudomonas aeruginosa and Pseudomonas putida isolates disseminated in a Korean hospital. Antimicrob. Agents Chemother. 46:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lombardi, G., F. Luzzaro, J. D. Docquier, M. L. Riccio, M. Perilli, A. Coli, G. Amicosante, G. M. Rossolini, and A. Toniolo. 2002. Nosocomial infections caused by multidrug-resistant isolates of Pseudomonas putida producing VIM-1 metallo-β-lactamase. J. Clin. Microbiol. 40:4051-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.National Committee for Clinical Laboratory Standards. 2002. Performance standards for antimicrobial susceptibility testing. 12th informational supplement. M100-S12. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 10.Nordmann, P., and L. Poirel. 2002. Emerging carbapenemases in Gram-negative aerobes. Clin. Microbiol. Infect. 8:321-331. [DOI] [PubMed] [Google Scholar]
- 11.Patzer, J., M. A. Toleman, L. M. Deshpande, W. Kaminska, D. Dzierzanowska, P. M. Bennett, R. N. Jones, and T. R. Walsh. 2004. Pseudomonas aeruginosa strains harbouring an unusual blaVIM-4 gene cassette isolated from hospitalized children in Poland (1998-2001). J. Antimicrob. Chemother. 53:451-456. [DOI] [PubMed] [Google Scholar]
- 12.Poh, C. L., C. C. Yeo, and L. Tay. 1992. Genome fingerprinting by pulsed-field gel electrophoresis and ribotyping to differentiate Pseudomonas aeruginosa serotype O11 strains. Eur. J. Clin. Microbiol. Infect. Dis. 11:817-822. [DOI] [PubMed] [Google Scholar]
- 13.Poirel, L., T. Lambert, S. Türkoglü, E. Ronco, J. L. Gaillard, and P. Nordmann. 2001. Characterization of class 1 integrons from Pseudomonas aeruginosa that contain the blaVIM-2 carbapenem-hydrolyzing β-lactamase gene and of two novel aminoglycoside resistance gene cassettes. Antimicrob. Agents Chemother. 45:546-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pournaras, S., A. Tsakris, M. Maniatis, L. S. Tzouvelekis, and A. N. Maniatis. 2002. Novel variant (blaVIM-4) of the metallo-β-lactamase gene blaVIM-1 in a clinical strain of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 46:4026-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pournaras, S., M. Maniatis, E. Petinaki, L. S. Tzouvelekis, A. Tsakris, N. J. Legakis, and A. N. Maniatis. 2003. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-β-lactamase gene variants blaVIM-2 and blaVIM-4. J. Antimicrob. Chemother. 51:1409-1414. [DOI] [PubMed] [Google Scholar]
- 16.Riccio, M. L., L. Pallecchi, R. Fontana, and G. M. Rossolini. 2001. In70 of plasmid pAX22, a blaVIM-1-containing integron carrying a new aminoglycoside phosphotransferase gene cassette. Antimicrob. Agents Chemother. 45:1249-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sardelic, S., L. Pallecchi, V. Punda-Polic, and G. M. Rossolini. 2003. Carbapenem-resistant Pseudomonas aeruginosa-carrying VIM-2 metallo-β-lactamase determinants, Croatia. Emerg. Infect. Dis. 9:1022-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scoulica, E. V., I. K. Neonakis, A. I. Gikas, and Y. J. Tselentis. 2004. Spread of blaVIM-1-producing E. coli in a university hospital in Greece. Genetic analysis of the integron carrying the blaVIM-1 metallo-β-lactamase gene. Diagn. Microbiol. Infect. Dis. 48:167-172. [DOI] [PubMed] [Google Scholar]
- 19.Stokes, H. W., D. B. O'Gorman, G. D. Recchia, M. Parsekhian, and R. M. Hall. 1997. Structure and function of 59-base element recombination sites associated with mobile gene cassettes. Mol. Microbiol. 26:731-745. [DOI] [PubMed] [Google Scholar]
- 20.Toleman, M. A., K. Rolston, R. N. Jones, and T. R. Walsh. 2004. blaVIM-7, an evolutionarily distinct metallo-β-lactamase gene in a Pseudomonas aeruginosa isolate from the United States. Antimicrob. Agents Chemother. 48:329-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tsakris, A., S. Pournaras, N. Woodford, M. F. Palepou, G. S. Babini, J. Douboyas, and D. M. Livermore. 2000. Outbreak of infections caused by Pseudomonas aeruginosa producing VIM-1 carbapenemase in Greece. J. Clin. Microbiol. 38:1290-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yong, D., K. Lee, J. H. Yum, H. B. Shin, G. M. Rossolini, and Y. Chong. 2002. Imipenem-EDTA disk method for differentiation of metallo-β-lactamase-producing clinical isolates of Pseudomonas spp. and Acinetobacter spp. J. Clin. Microbiol. 40:3798-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]