Abstract
Eight pulsed-field gel electrophoresis subtypes and six Tn1546 variants were identified among Enterococcus faecalis isolates of a single clone recovered in three geographically separate Portuguese hospitals. Some clonal subtypes were found in particular hospitals, and Tn1546 variants were either widespread or confined to some of them. We also report on the first Tn1546 transposon containing an ISEf1 insertion.
The epidemiology of enterococci is not fully understood since striking differences among resistant isolates of different species and resistant isolates from different geographic locations have been reported (1, 20). Besides clonal spread, a heterogeneous geographic distribution of different antibiotic resistance determinants, as different transposon types, is involved in resistance of vancomycin-resistant enterococci (VRE) (1, 4, 5, 8, 14, 27, 29). In Europe, a polyclonal enterococcal population structure with a large variety of Tn1546 types was initially observed in the community (1, 27, 29). In the United States, the initial VRE population consisted of a few persisting clones harboring a few Tn1546 variants (1, 5, 8, 12, 15, 16, 19). Recent studies have pointed out that the endemic susceptible clones could serve as substrates for the spread of VRE (11, 15, 19, 22; S. R. Nallapareddy, W. Huang, G. M. Weinstock, and B. E. Murray, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. C2-2165, 2003). Also, there is an increasing trend toward the consideration of antibiotic resistance as a regional problem (21).
The dissemination of a single vancomycin-resistant Enterococcus faecalis clone among three geographically separate Portuguese hospitals and the characterization of antibiotic resistance genetic elements of isolates of this clone were studied in the work described here. Thirty-three VRE clinical isolates were detected from 1996 to 2002 in three Portuguese hospitals (University Hospital in Coimbra [HUC], 11 isolates; Santo António Hospital in Porto [HSA], 19 isolates; and São Teotónio Hospital in Viseu [HST], 3 isolates). The sample included all VRE detected during 2001 and 2002 in HUC, HSA, and HST and some VRE isolates saved by the microbiology laboratory in HUC from 1996 to 2000.
Susceptibility testing was performed according to NCCLS guidelines (17). A multiplex PCR assay was used for species identification and vancomycin resistance gene detection (6). Genes coding for resistance to aminoglycosides or macrolides were also investigated (13, 25). Conjugation experiments were performed with E. faecalis strain JH2-2 as the recipient (9). The backbone structure of the Tn1546 transposon harbored by each VRE isolate was determined by the overlapping PCR assay described by Woodford et al. (29). Sequencing of specific fragments of Tn1546 was performed in order to identify the insertions.
All PCR assays included suitable positive and negative controls, kindly provided by B. E. Murray, M. Zervos, and C. Torres. Strains and JH2-2 transconjugants were typed by pulsed-field gel electrophoresis (PFGE) with SmaI and I-CeuI as restriction enzymes (10). The location of vanA was determined by hybridization of I-CeuI-digested genomic DNA with probes labeled with an enhanced chemiluminescence kit (Amersham Life Sciences, Uppsala, Sweden) for vanA and 23S rRNA genes, as described previously (3). Clonal relationships were established by the criteria proposed by Tenover et al. (23). Clones were designated by capital letters. Subtypes were defined by a subindex that indicates the number of bands that differed from the number for the strain considered to be the initial PFGE type.
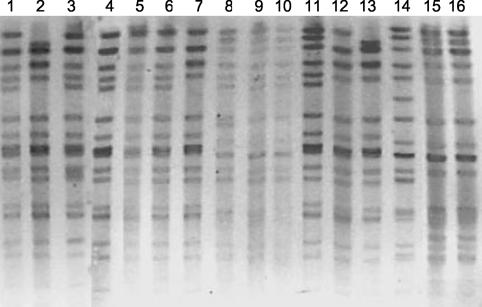
Nine PFGE types were identified among the 33 VRE isolates studied, with clone B being predominant (25 of 33 isolates [76%]). The first vanA isolate obtained in HUC (in 1996) was arbitrarily considered clone B (it was found in all three hospitals). Seven subtypes (which differed from each other by one to five bands) were detected in the subsequent years (Fig. 1) B2 (12 strains isolated from different patients in HSA); B5 (3 strains in HUC); B1 and B3 (2 strains each in HUC and HSA); and B1′, B3′, and B4 (1 strain each) (Table 1). Subtypes B2 and B5 persisted for 2 years in particular hospitals. Eight VRE PFGE types (types A, C, F, G, K, N, Q, and X) isolated from single patients were also detected.
FIG. 1.
SmaI-digested chromosomal DNA of vancomycin-susceptible and vancomycin-resistant E. faecalis isolates classified as clonal type B, as determined by PFGE. Lanes 1, 3, 4, 5, and 6, subtype B; lanes 7 and 12, subtype B1; lane 11, subtype B1′; lanes 9 and 10, subtype B2; lane 8, subtype B3; lane 13, subtype B3′; lane 14, subtype B4; lanes 15 and 16, subtype B5.
TABLE 1.
Clinical data, PFGE types, antibiotic resistance and virulence profiles, Tn1546 types, and frequency of transfer of studied traits for vancomycin-resistant E. faecalis clones isolated in hospitals located in three different geographical areas of Portugala
| Hospital and PFGE type | No. of isolates | Date of isolation (mo/yr) | Ward | Sourceb | Antibiotic resistance profilec
|
Transfer frequencyd | Tn1546 type | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VAN | TEC | ERY | CIP | GEN | KAN | STR | TET | CHL | |||||||
| HSA | |||||||||||||||
| B | 1 | 6/01 | ICU | Urine | R | R | R | R | R | R | R | 10−5 | PP4 | ||
| B2 | 9 | 1/01-10/02 | Obstetrics, cardiac surgery, hematology, nephrology, general surgery, internal medicine | Urine (6), Pus (2), Blood (1) | R | R | R | R | R | R | R | Rf | Re | 10−7-10−8 | PP15 |
| B2 | 2 | 3/02-4/02 | ICU, internal medicine B | Urine | R | R | R | R | R | 10−6-10−7 | PP16 | ||||
| B2 | 1 | 12/02 | ICU | Blood | R | R | R | R | R | R | R | ND | PP5 | ||
| B3 | 2 | 3/01-7/01 | ICU, obstetrics | Pus, urine | R | R | R | R | R | R | R | R | R | 10−7-10−8 | PP15 |
| B4 | 1 | 11/01 | Urology | Urine | R | R | R | R | R | R | R | R | 10−7 | PP15 | |
| K | 1 | 9/01 | Urology | Pus | R | R | R | R | 10−4 | A | |||||
| C | 1 | 11/01 | Urology | Pus | R | R | R | R | R | R | R | R | 10−8 | PP15 | |
| F | 1 | 11/02 | Skull trauma | Blood | R | R | R | 10−7 | PP5 | ||||||
| HST | |||||||||||||||
| B | 1 | 8/01 | Internal medicine 2A | Unknown | R | R | R | R | R | R | ND | ||||
| B | 1 | 3/02 | Internal medicine 1A | Unknown | R | R | R | R | R | ND | |||||
| B1′ | 1 | 11/01 | Nephrology | Urine | R | R | R | R | R | R | R | R | R | 10−4 | PP4 |
| B3′′ | 1 | 3/02 | Neurosurgery | Unknown | R | R | R | R | ND | ||||||
| B3′ | 1 | 4/02 | Internal medicine 2B | Urine | R | R | R | R | R | R | 10−5 | PP4 | |||
| A | 1 | 1/02 | Orthopedics | Urine | R | R | R | R | R | 10−5 | PP4 | ||||
| HUC | |||||||||||||||
| B | 1 | 1996 | Liver transplant | Unknown | R | R | R | R | R | R | R | R | 10−8 | PP4 | |
| B | 1 | 10/01 | Hematology | Blood | R | R | R | R | R | R | R | R | 10−6 | PP2 | |
| B1 | 1 | 7/01 | Nephrology | Exudate | R | R | R | R | R | R | R | R | 10−7 | PP4 | |
| B1 | 1 | 10/01 | General surgery 3M | Urine | R | R | R | R | R | R | R | R | 10−6 | PP5 | |
| B5 | 3 | 1/99-6/00 | Nephrology | Unknown, blood (2) | R | R | R | R | R | R | R | R | 10−8 | PP4 | |
| N | 1 | 11/99 | Nephrology | Blood | R | R | R | R | R | R | R | 10−5 | PP4 | ||
| X | 1 | 2/00 | Nephrology | Unknown | R | R | R | 10−6 | PP4 | ||||||
| Q | 1 | 5/01 | Nephrology | Blood | R | R | R | 10−5 | PP4 | ||||||
| G | 1 | 4/02 | Liver transplant | Blood | R | R | R | R | R | R | R | 10−6 | PP4 | ||
Abbreviations: R, resistant; ND, transfer experiments not done; ICU, intensive care unit; VAN, vancomycin; TEC, teicoplanin; ERY, erythromycin; CIP, ciprofloxacin; GEN, HLRGm; KAN, HLRKm; STR, high-level resistance to streptomycin; TET, tetracycline; CHL, chloramphenicol.
The numbers of isolates are indicated in parentheses.
Underscores indicate that antibiotic resistance was detected in transconjugants.
Transfer frequency is expressed as the number of transconjugants per number of donors.
All except two isolates were resistant.
Only three isolates were resistant.
Most of the VRE isolates were resistant to erythromycin (94%) and ciprofloxacin (88%) and had high-level resistance to gentamicin (HLRGm) (82%) or kanamycin (HLRKm) (82%) (Table 1). The vanA gene was detected in all VRE isolates. HLRGm and resistance to erythromycin were due to aac(6′)-aph(2′′) and erm(B), respectively. Both aac(6′)-aph(2′′) and aph(3′)-IIIa were detected in 13 isolates belonging to four clone B subtypes and in one isolate classified as clone C.
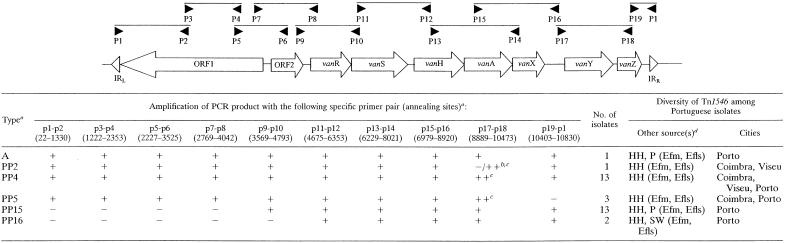
Five different variants of Tn1546 (variants PP2, PP4, PP5, PP15, and PP16) were found among clinical vancomycin-resistant E. faecalis isolates (Table 2). One of the more frequent variants, PP4, which contained an ISEf1 insertion sequence, was present among isolates (n = 13) of different PFGE types in all three hospitals: types B (subtypes B, B1, B1′, B3′, and B5), A, N, X, Q, and G. On the contrary, the PP15 variant, which was also present in 13 isolates of different PFGE types, types B (subtypes B2, B3, and B4) and C, was detected in only a single hospital (HSA). Some of these Tn1546 types were also found in E. faecalis and Enterococcus faecium strains of different origins, indicating a wide distribution of specific transposon variants (Table 2) (C. Novais et al., unpublished data).
TABLE 2.
Tn1546 types found among E. faecalis clinical isolates recovered at three different Portuguese hospitals (1996 to 2002)
Tn1546 types were designed according to the scheme of Woodford et al. (29). For those that did not have a specific previously described type, we used our own designation (PP [Portugal-Porto], followed by a randomly chosen number). +, amplification; −, no amplification; ++, amplification of sequences larger than those of the expected size.
For PP2, amplification was negative with p17-p18 but positive with ISEf1-F (5′-GGT GTT ACG ATG TCT GAA ATT GC-3′) and p18.
Sequencing of this fragment demonstrated the presence of ISEf1 in the intergenic vanX-vanY region.
Sources and species in which specific Tn1546 types were found. HH, human isolates; P, poultry isolates; SW, isolates from sewage samples; Efm, E. faecium; Efls, E. faecalis (C. Novais, T. M. Coque, F. Baquero, and L. Peixe, Abstr. 14th Eur. Congr. Clin. Microbiol. Infect. Dis., abstr. 2430, 2004).
The transfer of vanA to E. faecalis strain JH2-2 was achieved with all VRE isolates. erm(B) was cotransferred with vanA in all cases. Transconjugants harboring vanA, erm(B), and aph(3′)-IIIa were detected for 7 of 12 VRE isolates with HLRKm at HSA (subtypes B2, B3, B4 and C) and 2 VRE isolates with HLRKm at HUC. They contained PP15 and PP4 variants of Tn1546, respectively (Table 1).
The patterns of I-CeuI-digested genomic DNA of strains and transconjugants harboring the more prevalent Tn1546 variants, PP4 and PP15, mostly differed by a single band with a small molecular size that varied in length among the isolates. Hybridization of the vanA probe but not the 23S rDNA probe to these bands suggested that PP4 and PP15 are located on different plasmids.
This work focused on the dissemination and persistence of a particular E. faecalis vanA clone (clone B) isolated over 7 years in different Portuguese hospitals. Some clonal subtypes were recovered over prolonged periods of time in particular hospitals: as subtype B5 in Coimbra (HUC) or subtype B2 in Porto (HSA). The Tn1546 types responsible for vancomycin resistance were also unevenly distributed: type PP4 was found in different clones detected in distinct hospitals, and type PP15 was found in different strains from a single hospital. The presence of specific Tn1546 variants in selected PFGE types or subtypes with different geographic distributions may cast some light on the reason for differences in local genetic patterns of resistance. Vancomycin resistance determinants might be stably maintained only in selected clones, with some of them being widely disseminated in certain institutions. Locally prevalent clones of vancomycin-susceptible enterococci (VSE) have been suggested to be the leading force for the local spread of VRE (11, 22). Although VSE were not systematically studied, we were able to detect different VSE variants of the most prevalent VRE clone (clone B) during 2001 and 2002 in some of the hospitals studied, suggesting a possible longer persistence of this strain (Table 1). The intra- and interhospital transmissions of E. faecalis VSE clones have recently been reported in Spain, Sweden, The Netherlands, the United Kingdom, and Ireland (7, 26, 28; P. Rúiz-Garbajosa, R. Cantón, T. M. Coque, V. Pintado, F. Baquero, and R. del Campo, Abstr. 43rd Intersci. Conf. Antimicrob. Agents Chemother., abstr. L-270, 2003), although the role of these widely disseminated VSE remains unknown. On the other hand, the acquisition of different Tn1546 elements by a particular clone has scarcely been reported for either E. faecium or E. faecalis (24, 29).
The heterogeneity of Tn1546 is mainly explained by the presence of different insertion sequences, which serve as hot spots for the rearrangement of genetic fragments (2, 9, 18). Some of these insertion sequences may be associated with certain geographic locations (4, 5, 14, 15, 27, 28). We report for the first time the presence of ISEf1 in Tn1546 (variants PP2, PP4, and PP5), which was found to be widely distributed among Portuguese hospitals. Interestingly, this insertion sequence element is one of the most frequently detected insertion sequences in the chromosome of the sequenced genome of E. faecalis strain V583 (18), although its prevalence in the genomes of different E. faecalis strains remains unknown.
Our results indicate that the clonal and Tn1546 patterns involved in vancomycin resistance among E. faecalis isolates from particular hospitals in different regions may differ and suggest that the analysis of local patterns might explain the differences in the occurrence and diversity of VRE in different institutions.
Acknowledgments
Carla Novais was supported by a fellowship from Fundação para a Ciéncia e Tecnologia (SFRH/BD/3372/2000).
Luisa Peixe and Teresa M. Coque are coadvisors of Carla Novais for her Ph.D. thesis.
Members of the Portuguese Resistance Study Group are Graça Ribeiro and Clementina Vital (HUC), Isabel Marques and Ana M. Queirós (HST), and Helena Ramos (HSA).
REFERENCES
- 1.Bonten, M. J. M., R. J. Willems, and R. A. Weinstein. 2001. Vancomycin resistant enterococci: why are they here, and where do they come from? Lancet Infect. Dis. 1:314-325. [DOI] [PubMed] [Google Scholar]
- 2.Borgen, K., M. Sorum, Y. Wasteson, H. Kruse, and H. Oppegaard. 2002. Genetic linkage between erm(B) and vanA in Enterococcus hirae of poultry origin. Microb. Drug Resist. 8:363-368. [DOI] [PubMed] [Google Scholar]
- 3.Dahl, K. H., and A. Sundsfjord. 2003. Transferable vanB2 Tn5382-containing elements in fecal streptococcal strains from veal calves. Antimicrob. Agents Chemother. 47:2579-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darini, A. L. C., M. F. I. Palepou, and N. Woodford. 1999. Nucleotide sequence of IS1542, an insertion sequence identified within VanA glycopeptide resistance elements of enterococci. FEMS Microbiol. Lett. 173:341-346. [DOI] [PubMed] [Google Scholar]
- 5.De Lencastre, H., A. E. Brown, M. Chung, D. Armstrong, and A. Tomasz. 1999. Role of the transposon Tn5482 in the epidemiology of vancomycin resistant Enterococcus faecium in the pediatric oncology unit of a New York City Hospital. Microb. Drug Resist. 5:113-129. [DOI] [PubMed] [Google Scholar]
- 6.Dukta-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hällgren, A., B. Saeedi, M. Nilsson, H. J. Monstein, B. Isaksson, H. Hanberger, and L. E. Nilsson. 2003. Genetic relatedness among Enterococcus faecalis with transposon-mediated high level gentamicin resistance in Swedish intensive care units. J. Antimicrob. Chemother. 52:162-167. [DOI] [PubMed] [Google Scholar]
- 8.Hanrahan, J., C. Hoyen, and L. B. Rice. 2000. Geographic distribution of a large mobile element that transfers ampicillin and vancomycin resístance between Enterococcus faecium strains. Antimicrob. Agents Chemother. 44:1349-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heaton, M. P., and S. Handwerger. 1995. Conjugative mobilization of a vancomycin resistance plasmid by a putative Enterococcus faecium sex pheromone responsive plasmid. Microb. Drug Resist. 2:177-183. [DOI] [PubMed] [Google Scholar]
- 10.Kaufmann, M. E. 1998. Pulsed-field gel electrophoresis. Methods Mol. Med. 15:17-31. [DOI] [PubMed] [Google Scholar]
- 11.Kawalec, M., M. Gniadkowski, M. Zaleska, T. Ozorowski, L. Konopka, and W. Hryniewicz. 2001. Outbreak of vancomycin-resistant Enterococcus faecium of the phenotype VanB in a hospital in Warsaw, Poland: probable transmission of the resistance determinants into an endemic vancomycin-susceptible strain. J. Clin. Microbiol. 39:1781-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim, W. J., R. A. Weinstein, and M. K. Hayden. 1999. The changing molecular epidemiology and establishment of endemicity of vancomycin resistance in enterococci at one hospital over a 6-year period. J. Infect. Dis. 179:163-171. [DOI] [PubMed] [Google Scholar]
- 13.Lim, J. A., A. R. Kwon, S. K. Kim, Y. Chong, K. Lee, and E. C. Choi. 2002. Prevalence of resistance to macrolide, lincosamide and streptogramin antibiotics in gram-positive cocci isolated in a Korean hospital. J. Antimicrob. Chemother. 49:489-495. [DOI] [PubMed] [Google Scholar]
- 14.Mackinnon, M. G., M. A. Drebot, and G. J. Tyrrell. 1997. Identification and characterization of IS1476, an insertion sequence-like element that disrupts VanY function in a vancomycin-resistant Enterococcus faecium strain. Antimicrob. Agents Chemother. 41:1805-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McAsahan, S. K., K. L. Vergin, S. J. Giovannoni, and D. S. Thaler. 1999. Interspecies recombination between enterococci: genetic and phenotypic diversity of vancomycin resistant transconjugants. Microb. Drug Resist. 5:101-112. [DOI] [PubMed] [Google Scholar]
- 16.Moreno, F., P. Grota, C. Crisp, K. Magnon, G. P. Melcher, J. H. Jorgensen, and J. E. Patterson. 1995. Clinical and molecular epidemiology of vancomycin-resistant Enterococcus faecium during its emergence in a city in southern Texas. Clin. Infect. Dis. 21:1234-1237. [DOI] [PubMed] [Google Scholar]
- 17.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 18.Paulsen, I. T., L. Banerjei, G. S. A. Myers, K. E. Nelson, R. Seshadri, T. D. Read, D. E. Fouts, J. A. Eisen, S. R. Gill, J. F. Heidelberg, H. Tettelin, R. J. Dodson, L. Umayam, L. Brinkac, M. Beanan, S. Daugherty, R. T. DeBoy, S. Durkin, J. Kolonay, R. Madupu, W. Nelson, J. Vamathevan, B. Tran, J. Upton, T. Hansen, J. Shetty, H. Khouri, T. Utterback, D. Radune, K. A. Ketchum, B. A. Dougherty, and C. M. Fraser. 2003. Role of mobile DNA in the evolution of vancomycin resistant Enterococcus faecalis. Science 299:2071-2074. [DOI] [PubMed] [Google Scholar]
- 19.Perlada, D. E., A. G. Smulian, and M. T. Cushion. 1997. Molecular epidemiology and antibiotic susceptibility of enterococci in Cincinnati, Ohio: a prospective citywide survey. J. Clin. Microbiol. 35:2342-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shepard, B. D., and M. S. Gilmore. 2002. Antibiotic-resistant enterococci: the mechanisms and dynamics of drug introduction and resistance. Microbes Infect. 4:215-224. [DOI] [PubMed] [Google Scholar]
- 21.Smith, D. L., J. Dushoff, E. N. Perencevich, A. D. Harris, and S. A. Levin. 2004. Persistent colonization and the spread of antibiotic resistance in nosocomial pathogens: resistance is a regional problem. Proc. Natl. Acad. Sci. USA 101:3709-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suppola, J. H., E. Jolho, S. Salmenlinna, E. Tarkka, J. Vuopio-Varkila, and M. Vaara. 1999. vanA and vanB incorporate into endemic ampicillin-resistant vancomycin-sensitive Enterococcus faecium strain: effect on interpretation of clonality. J. Clin. Microbiol. 37:3934-3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tremlett, C. H., D. F. Brown, M. F. Palepou, and N. Woodford. 2001. Two structurally distinct VanA resistance elements in a glycopeptide-resistant strain of Enterococcus faecalis. Antimicrob. Agents Chemother. 45:996-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vakulenko, S. B., S. M. Donabedian, A. M. Voskresenskiy, M. J. Zervos, S. A. Lerner, and J. W. Chow. 2003. Multiplex PCR for detection of aminoglycoside resistance genes in enterococci. Antimicrob. Agents Chemother. 47:1423-1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waar, K., R. J. Willems, M. J. H. Slooff, H. J. M. Harmsen, and J. E. Degener. 2003. Molecular epidemiology of Enterococcus faecalis in liver transplant patients at University Hospital Groningen. J. Hosp. Infect. 55:53-60. [DOI] [PubMed] [Google Scholar]
- 27.Willems, R. J. L., J. Top, N. van der Braak A. van Belkum, D. J. Mevius, G. Hendriks, M. van Santen-Verheuvel, and J. D. van Embden. 1999. Molecular diversity of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodford, N., R. Reynolds, J. Turton, A. Sinclair, A. Williams, and D. Livermore. 2003. Two widely disseminated strains of Enterococcus faecalis highly resistant to gentamicin and ciprofloxacin from bacteremias in the UK and Ireland. J. Antimicrob. Chemother. 52:711-714. [DOI] [PubMed] [Google Scholar]
- 29.Woodford, N., A. M. A. Adebiyi, M. F. I. Palepou, and B. Cookson. 1998. Diversity of VanA glycopeptide resistance elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 42:502-508. [DOI] [PMC free article] [PubMed] [Google Scholar]