Abstract
Most antimicrobial peptides (AMPs) impair the viability of target bacteria by permeabilizing bacterial membranes. However, the proline-rich AMPs have been shown to kill susceptible organisms without causing significant membrane perturbation and may act by inhibiting the activity of bacterial targets. To gain initial insight into the events that follow interaction of a proline-rich peptide with bacterial cells, we used DNA macroarray technology to monitor transcriptional alterations of Escherichia coli in response to challenge with a subinhibitory concentration of the proline-rich Bac7(1-35). Substantial changes in the expression levels of 70 bacterial genes from various functional categories were detected. Among these, 26 genes showed decreased expression, while 44 genes, including genes that are potentially involved in bacterial resistance to antimicrobials, showed increased expression. The generation of a transcriptional response under the experimental conditions used is consistent with the ability of Bac7(1-35) to interact with bacterial components and affect biological processes in this organism.
Antimicrobial peptides (AMPs) are components of the innate antimicrobial defenses of virtually all eukaryotic organisms (3, 18, 50). Nature's AMPs show an impressive variety of sequences (http://www.bbcm.units.it/∼tossi/pag1.htm) and exhibit a number of different secondary structures. Those AMPs with β-sheet conformations stabilized by two or three disulfide bonds (denoted protegrins and defensins, respectively) and those with α-helical structures (e.g., LL-37, CRAMP, and BMAP-28) are most common in mammals (28, 47, 49). Other mammalian AMPs are stabilized by one disulfide bond (dodecapeptides) or form extended structures (e.g., the proline-rich PR-39 and Bac7 and the tryptophan-rich indolicidin) (14). Most AMPs share a cationic and amphipathic character that is thought to facilitate interaction with and insertion into the anionic microbial membranes, leading to disruption of membrane integrity (40). This lytic mechanism may result in inactivation of bacteria, fungi, enveloped viruses, and parasites (40). Peptides belonging to the proline-rich class, however, may act by a different mechanism (9, 15). These peptides are characterized by repeated proline-containing motifs and are active predominantly against gram-negative organisms. Investigations of the mode of action of the proline-rich apidaecins, pyrrhocoricin, and PR-39, isolated from the lymph fluid of honeybees, from Pyrrhocoris apterus, and from porcine neutrophils, respectively, have shown that these peptides kill responsive bacteria through a nonlytic mechanism(s), probably by inhibiting the activity of intracellular molecular targets (9, 15). Moreover, the ability of PR-39 to bind mammalian proteins containing Src homology 3 domains (5) and to induce the expression of specific mammalian genes (13) suggests it also has the capability to interact with host cell components and to signal biological responses.
Recent studies indicate that AMPs at nonlethal concentrations can act as environmental stimuli to elicit selected bacterial responses. For instance, treatment of Escherichia coli with the insect α-helical peptides cecropins at concentrations below MICs induces a hyperosmotic stress response in viable bacteria by activating the promoter of the hyperosmotic stress gene osmY (33, 34). In addition, microarray technology-based studies have shown that incubation of E. coli with a sublethal concentration of cecropin A causes selective alterations in the bacterial gene transcript profile (22), suggesting that this peptide could interact with microbial targets to evoke nonlethal responses in viable bacteria. Furthermore, the expression of various resistance genes has been found to be modulated in Salmonella enterica serovar Typhimurium (2) and Pseudomonas aeruginosa (29) in response to protegrin-1 (PG-1), LL-37, indolicidin, and other AMPs. These results highlight the contribution of genomic and proteomic methods to the elucidation of potential adaptation and/or resistance mechanisms of bacteria to AMPs.
In the present study, we examined the potential of a proline-rich AMP of 35 residues, denoted Bac7(1-35), to induce a transcriptional response in E. coli. Bac7(1-35) corresponds to the 35-residue N-terminal region of Bac7, which is a 60-residue AMP isolated from bovine neutrophils that selectively targets gram-negative bacteria (41). Bac7(1-35) retains the antimicrobial activity of Bac7 and impairs the viability of target bacteria without causing significant perturbation of the integrity of the bacterial membranes (42). This finding provides evidence for a nonlytic killing mechanism and suggests that the events that lead to bacterial death may be triggered by the binding of Bac7(1-35) to a selected molecular target(s).
To gain insight into the events that follow interaction of Bac7(1-35) with target bacteria, we investigated here the ability of a sublethal concentration of Bac7(1-35) to stimulate a response in E. coli in terms of selective alteration of bacterial gene expression, by using DNA macroarray technology. The transcriptional profile obtained in response to Bac7(1-35) was analyzed and compared with that obtained with a peptide with a sequence corresponding to the 5-35 sequence of Bac7(1-35), denoted Bac7(5-35), which lacks the one to four N-terminal residues (RRIR) of Bac7(1-35) and is much less effective against E. coli and other gram-negative bacteria (42). We show that Bac7(1-35) causes selective alterations in E. coli gene transcription, including up-regulation of genes that are potentially involved in bacterial resistance. Conversely, Bac7(5-35) does not cause genomic responses in this strain.
MATERIALS AND METHODS
The sequences of Bac7(1-35), Bac7(5-35), LL-37, and PG-1 are shown in Table 1. All of these peptides were synthesized with a Milligen 9050 automated synthesizer (Applied Biosystems) with 9-fluorenylmethoxy carbonyl chemistry. Polymyxin B (PB) was from Sigma.
TABLE 1.
Amino acid sequences of peptides used in this study
| Peptide | Sequence | MIC (μM)a |
|---|---|---|
| Bac7(1-35) | RRIRPRPPRLPRPRPRPLPFPRPGPRPIPRPLPFP | 10 |
| Bac7(5-35) | PRPPRLPRPRPRPLPFPRPGPRPIPRPLPFP | >32 |
| LL-37 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVP RTES | 12 |
| PG-1 | RGGRLCYCRRRFCVCVGR-NH2b | 1 |
For E. coli MG1655.
NH2, C-terminal amidation.
Bacterial strain and growth conditions.
E. coli MG1655 was obtained from the E. coli Genetic Stock Center (http://cgsc.biology.yale.edu). Bacterial cultures were grown at 37°C in Luria-Bertani (LB) broth with constant aeration. Overnight cultures were diluted 1,000-fold in LB broth and grown at 37°C to an optical density at 600 nm between 0.05 and 0.06, corresponding to 3 × 107 CFU/ml.
Antimicrobial activity.
Activity against E. coli MG1655 was determined as the MIC by a microdilution susceptibility assay, in 96-well plates. The experiments were run as previously described (41), except that assays were carried out in LB broth with a bacterial concentration of 3 × 107 CFU/ml, i.e., the same bacterial concentration used in the macroarray analysis. For bactericidal assays, bacteria (3 × 107 CFU/ml) were incubated at 37°C for 15 to 120 min in the absence or the presence of each peptide concentration. Serial dilutions of the bacterial cultures were then plated onto LB plates for CFU counts to assess the numbers of viable bacteria.
RNA isolation.
Total RNA was isolated from E. coli MG1655 as suggested by the macroarray manufacturer (Sigma-Genosys Biotechnologies, Inc.). Bacterial cell pellets were resuspended in 10 mM sodium acetate (pH 4.2) containing 0.3 M sucrose and 0.5 M EDTA and then lysed in 10 mM sodium acetate (pH 4.2) containing 2% sodium dodecyl sulfate (SDS). Cell lysates were extracted three times with phenol buffered with 0.1 M citrate (pH 4.3) at 65°C, once with phenol-chloroform and once with chloroform. To remove contaminating genomic DNA, samples were treated with RNase-free DNase I (Amersham), followed by proteinase K (Sigma) digestion, phenol-chloroform and chloroform extractions, and precipitation with ethanol. Purified RNA was resuspended in water and quantified spectrophotometrically at 260 nm.
Generation of labeled cDNA from cultures of E. coli MG1655.
Overnight cultures of E. coli MG1655 were diluted 1,000-fold in LB broth and grown at 37°C to an optical density at 600 nm between 0.05 and 0.06. Bacteria were then incubated at 37°C for 30 min in the absence or presence of 2.5 μM Bac7(1-35) or 2.5 μM Bac7(5-35). Total RNA was isolated (see “RNA isolation,” above), and 1.5 μg was annealed to cDNA labeling primers (Sigma-Genosys Biotechnologies, Inc.) in a solution containing 50 mM Tris-HCl (pH 8.5), 8 mM MgCl2, 50 mM KCl, and 1 mM dithiothreitol. After incubation at 70°C for 5 min, the mixture was cooled at room temperature for 10 min and then incubated for 3 h at 42°C in the presence of 0.33 mM dATP, 0.33 mM dGTP, 0.33 mM dTTP, 40 U of RNaseOUT (GibcoBRL), 50 U of AMV reverse transcriptase (Finnzymes), and 50 μCi of [α-32P]dCTP. Unincorporated nucleotides were removed by Sephadex G-25 gel filtration chromatography.
Hybridization and analysis of gene arrays.
Nylon membranes containing 4,290 open reading frame (ORF)-specific PCR products and representing a complete set of known or predicted protein coding sequences of E. coli MG1655 (Panorama E. coli Gene Arrays) were obtained from Sigma-Genosys Biotechnologies, Inc. Pair membranes were hybridized in each experiment with the cDNA synthesized from peptide-treated and untreated cells. Membranes were prehybridized for 60 min at 65°C in 5× SSPE (1× SSPE is 0.18 M NaCl, 10 mM sodium phosphate, and 1 mM EDTA [pH 7.7]) in the presence of 2% SDS, Denhardt's reagent, and 100 μg of sheared salmon sperm DNA/ml. Hybridization with labeled cDNA (17 × 106 cpm) was carried out overnight at 65°C. Blots were then incubated twice for 5 min at room temperature and several times for 60 min at 65°C with 0.5× SSPE containing 0.2% SDS and finally subjected to autoradiography. The autoradiographic images were scanned, and the pixel densities of the resulting files were determined by Arrayvision version 7.0 software (Imaging Research, Inc.), which analyzes the intensity of each duplicate spot measured in arbitrary units. The background signal was determined for each group of genes by measuring the intensity of the nearest spot that did not contain DNA, selected from among 64 spots. This signal was then subtracted from the intensity of each DNA spot, and the corrected intensity was expressed as the percentage of total corrected DNA intensities. This procedure allowed comparison between filters independently of the total hybridization signal. The corrected intensities of duplicate spots were averaged, and the mean spot intensity of two independent experiments was calculated. The expression ratio for each gene was calculated as the ratio of spot intensities of treated and untreated cells (e.g., an expression ratio of 1 indicated an unchanged transcription level). A gene was considered significantly regulated when the expression ratio (ratio of treated cells/untreated cells) was ≥2.0 (up-regulated) or ≤0.5 (down-regulated). Only genes that showed at least a twofold increase in the expression level compared to the background signal in duplicate experiments were considered to be affected by antimicrobial peptide treatment.
Northern blot analysis.
Total RNA (7 μg) was size separated on a 1% denaturing agarose gel, denatured by incubation in 8 mM NaOH for 20 min, and then transferred to a GeneScreen Plus nylon membrane (NEN Life Science) in a solution containing 3 M NaCl and 8 mM NaOH. Filters were prehybridized in ULTRAhyb solution (Ambion) for 30 min at 42°C and then hybridized for 6 to 24 h at 42°C with 32P-labeled probes generated by PCR amplification of genomic DNA, as described below. After hybridization, membranes were incubated twice in 2× saline sodium citrate (1× SSC is 30 mM trisodium citrate and 0.3 M NaCl [pH 7.0]) in the presence of 0.1% SDS for 5 min and twice in 0.1× SSC with 0.1% SDS at 42°C for 15 min, then rinsed in 2× SSC, and finally exposed to X-ray film (Kodak). The intensity of each hybridization band was analyzed by densitometry. The background intensity was subtracted, and the net intensity of each gene was determined relative to that of rrlH. For each condition, the induction (n-fold) compared to that of the control culture was calculated. Genes were considered significantly induced or repressed when the treated/untreated cell ratio was ≥2.0 or ≤0.5, respectively.
Generation of gene-specific radiolabeled probes.
Gene-specific probes for secG, proP, proV, obgE, vacB, basS, ompT, malT, malK, and glpK were generated by PCR amplification of E. coli MG1655 genomic DNA with E. coli PCR primers (Sigma-Genosys Biotechnologies, Inc.). Genomic DNA was purified from bacterial cultures as previously described (7). PCR amplification was performed with a total volume of 25 μl, including 0.6 U of Amplitaq Gold DNA polymerase (Perkin Elmer), 2.5 mM MgCl2, 0.2 mM deoxynucleoside triphosphates, a 1 μM concentration of each sense and antisense primer pair, and 50 ng of E. coli MG1655 genomic DNA. After 10 min at 94°C, 30 cycles were performed with the following program: 30 s at 94°C, 40 s at the primer-specific annealing temperature, and 1 min at 72°C. Amplified fragments were analyzed with 1.5% agarose gels and purified from agarose with a Genelute Gel extraction kit (Sigma). PCR products were 32P labeled with a T7 QuickPrime kit (Amersham Biosciences).
RESULTS AND DISCUSSION
Antimicrobial activity of Bac7(1-35) and Bac7(5-35).
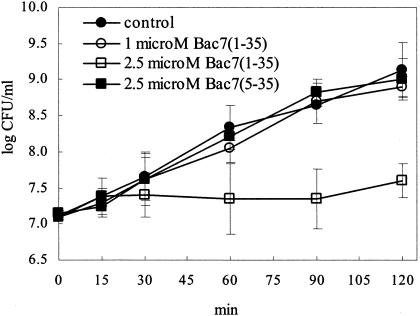
The amino acid sequences of Bac7(1-35) and Bac7(5-35) are shown in Table 1. MICs for E. coli MG1655 were determined after overnight incubations of 3 × 107 CFU/ml with serial dilutions of each peptide. Bac7(1-35) was found to inhibit the growth of E. coli MG1655 with MICs of 10 μΜ. Conversely, Bac7(5-35) was ineffective for this strain at a MIC of 32 μΜ in four independent experiments. The sublethal concentration of Bac7(1-35) to be used in the macroarray analysis was worked out by determining the number of viable bacteria after treatment with Bac7(1-35) at 0.5 (results not shown), 1, and 2.5 μΜ (Fig. 1). The growth curve of bacterial cultures treated with up to 1 μΜ peptide was similar to that of untreated cultures, whereas at 2.5 μΜ (corresponding to a concentration which is fourfold lower than the MIC) Bac7(1-35) did not significantly affect the rate of bacterial growth during the first 30 min of incubation and caused a transient growth arrest at subsequent times (Fig. 1). A complete recovery was observed at 24 h (results not shown). The 2.5 μM concentration of Bac7(1-35) was considered for treatment of E. coli MG1655 with a sublethal peptide concentration. Bacterial cultures were treated in parallel with 2.5 μΜ Bac7(5-35) to assess the specificity of the response to Bac7(1-35). As anticipated by the MICs, the growth curve of E. coli MG1655 in the presence of 2.5 μΜ Bac7(5-35) was similar to that of untreated cultures at all times considered (Fig. 1).
FIG. 1.
Growth rate of E. coli MG1655 in the absence (control) or presence of 1 or 2.5 μM Bac7(1-35) or 2.5 μM Bac7(5-35). Data are means ± standard deviations of at least three experiments.
Transcription profile of E. coli MG1655 treated with Bac7(1-35).
The effect of 2.5 μM Bac7(1-35) on gene expression in E. coli MG1655 (3 × 107 CFU/ml) was examined by comparing the transcription patterns of bacterial cultures incubated for 30 min in the presence and absence of this peptide concentration. Genes whose relative expression ratios were ≥2 or ≤0.5 in two independent experiments were considered significantly altered.
Treatment with Bac7(1-35) caused substantial changes in the transcription of 70 bacterial genes. Among these, transcription of 26 genes was found to be repressed (Table 2) and transcription of 44 genes was induced (Table 3). The differential regulation of these genes was confirmed in two independent experiments, which produced similar results. Importantly, the expression profile of E. coli MG1655 exposed to 2.5 μM Bac7(5-35) was similar to that of untreated bacterial cultures (two independent experiments with similar results; data not shown), indicating that the positively charged cluster of residues at the N terminus is important both for the antimicrobial effect and for the induction of a genomic response.
TABLE 2.
E. coli genes whose relative expression levels decrease after treatment with Bac7(1-35)
| Gene | b No.a | Product or functionb | Functional group | Ratio |
|---|---|---|---|---|
| aceE | 0114 | Pyruvate dehydrogenase component (E1) of pyruvate dehydrogenase | Energy metabolism | 0.48 |
| aceF | 0115 | Dihydrolipoamide acetyltransferase component (E2) of pyruvate dehydrogenase | Energy metabolism | 0.35 |
| aspA | 4139 | l-Aspartate ammonia lyase (l-aspartase) | Central intermediary metabolism | 0.29 |
| cyoE | 0428 | Cytochrome o ubiquinol oxidase C subunit | Energy metabolism | 0.41 |
| fecA | 4291 | Iron(III) dicitrate transport protein FecA precursor | Transport and binding proteins | 0.45 |
| gapA | 1779 | Glyceraldehyde 3-phosphate dehydrogenase A | Energy metabolism | 0.28 |
| glpK | 3926 | Glycerol kinase | Central intermediary metabolism | 0.25 |
| lamB | 4036 | Maltoporin, maltose high-affinity uptake system | Transport and binding proteins | 0.24 |
| lpdA | 0116 | Dihydrolipoamide dehydrogenase component (E3) of pyruvate dehydrogenase | Energy metabolism | 0.45 |
| malE | 4034 | Periplasmic maltose-binding protein | Transport and binding proteins | 0.07 |
| malF | 4033 | Maltose transport inner-membrane protein | Transport and binding proteins | 0.25 |
| malK | 4035 | Cytoplasmic membrane protein for maltose uptake | Transport and binding proteins | 0.16 |
| malM | 4037 | Maltose operon periplasmic protein | Carbon compound catabolism | 0.31 |
| malT | 3418 | Positive regulator of mal regulon | Transcription, RNA processing and degradation | 0.43 |
| rbsB | 3751 | Periplasmic ribose-binding protein precursor | Transport and binding proteins | 0.20 |
| rbsC | 3750 | High-affinity ribose transport protein | Transport and binding proteins | 0.32 |
| rbsD | 3748 | High-affinity ribose transport protein | Transport and binding proteins | 0.43 |
| sdaB | 2797 | l-Serine dehydratase 2 (l-serine deaminase 2) | Amino acid biosynthesis and metabolism | 0.46 |
| sodA | 3908 | Manganese superoxide dismutase | Cell processes (including adaptation and protection) | 0.26 |
| yfiA | 2597 | Protein Y, associated with 30S ribosomal subunit | Transcription, RNA processing and degradation | 0.31 |
| ygjQ | 3086 | Hypothetical 25.5-kDa protein | Hypothetical, unclassified, unknown | 0.48 |
| ygjT | 3088 | Hypothetical 35.8-kDa protein, putative membrane transport or efflux protein | Hypothetical, unclassified, unknown | 0.39 |
| yidA | 3697 | Hypothetical 29.7-kDa protein | Hypothetical, unclassified, unknown | 0.23 |
| yieJ | 3717 | Hypothetical 22.5-kDa protein | Hypothetical, unclassified, unknown | 0.48 |
| yijI | 3948 | Hypothetical 11.8-kDa protein | Hypothetical, unclassified, unknown | 0.33 |
| yrfD | 3395 | Hypothetical 30.0-kDa protein | Hypothetical, unclassified, unknown | 0.16 |
b number, unique identifier for E. coli genes.
Based on EcoCyc database (24).
TABLE 3.
E. coli genes whose relative expression levels increase after treatment with Bac7(1-35)
| Gene | b No.a | Product (or function)b | Functional group | Ratio |
|---|---|---|---|---|
| amiC | 2817 | Putative cell wall amidase | Cell structure | 2.9 |
| basR | 4113 | Transcriptional regulatory protein BasR/PmrA | Transcription, RNA processing and degradation | 2.0 |
| basS | 4112 | Sensor protein BasS/PmrB | Transcription, RNA processing and degradation | 15.7 |
| betT | 0314 | High-affinity choline transport protein | Transport and binding proteins | 2.7 |
| bglF | 3722 | Beta-glucoside PTS permease | Transport and binding proteins | 2.5 |
| dnaK | 0014 | DnaK protein (heat shock protein 70) | Cell processes (including adaptation and protection) | 2.6 |
| fabF | 1095 | 3-Oxoacyl-[acyl-carrier-protein] synthase II | Fatty acid and phospholipid metabolism | 3.4 |
| fimD | 4317 | Export and assembly of type 1 fimbrial subunits | Cell structure | 3.0 |
| hns | 1237 | DNA-binding protein (histone-like protein Hlp-II) | Translation, posttranslational modification | 6.1 |
| infB | 3168 | Protein chain initiation factor 2 | Translation, posttranslational modification | 3.0 |
| insA_3 | 0275 | Insertion element IS1 protein InsA | Phage, transposon, or plasmid | 2.1 |
| insA_6 | 3444 | Insertion element IS1 protein InsA | Phage, transposon, or plasmid | 2.4 |
| insB_2 | 0264 | Insertion element IS1 protein InsB | Phage, transposon, or plasmid | 2.1 |
| intB | 4271 | Prophage P4 integrase | Phage, transposon, or plasmid | 7.1 |
| iscU | 2529 | Putative iron-sulfur cluster formation protein | Hypothetical, unclassified, unknown | 2.1 |
| lpxB | 0182 | Lipid-A-disaccharide synthase | Cell structure | 2.2 |
| metG | 2114 | Methionyl-tRNA synthetase | Translation, posttranslational modification | 4.0 |
| nagE | 0679 | N-Acetylglucosamine PTS permease | Transport and binding proteins | 2.7 |
| nusA | 3169 | l Factor | Transcription, RNA processing and degradation | 6.4 |
| obgE | 3183 | Putative DNA-binding GTPase | Hypothetical, unclassified, unknown | 3.5 |
| ompT | 0565 | Protease VII precursor | Cell structure | 3.1 |
| proP | 4111 | Proline-betaine transporter (proline porter II) | Transport and binding proteins | 2.1 |
| proV | 2677 | ATP-binding component of glycine betaine transporter | Transport and binding proteins | 3.1 |
| proW | 2678 | Integral membrane component of glycine betaine transporter | Transport and binding proteins | 2.1 |
| proX | 2679 | Periplasmic binding component of glycine betaine transporter | Transport and binding proteins | 2.5 |
| rplJ | 3985 | 50S ribosomal subunit protein L10 | Translation, posttranslational modification | 2.3 |
| rplM | 3231 | 50S ribosomal subunit protein L13 | Translation, posttranslational modification | 2.1 |
| rpmB | 3637 | 50S ribosomal subunit protein L28 | Translation, posttranslational modification | 2.3 |
| rpmE | 3936 | 50S ribosomal protein L31 | Translation, posttranslational modification | 3.1 |
| rpsO | 3165 | 30S ribosomal subunit protein S15 | Translation, posttranslational modification | 2.3 |
| secG | 3175 | Protein-export membrane protein | Transport and binding proteins | 2.0 |
| sstT | 3089 | Na+/serine (threonine) symporter | Putative transport proteins | 2.1 |
| trmD | 2607 | tRNA (guanine-7) methyltransferase | Translation, posttranslational modification | 2.1 |
| vacB | 4179 | Exoribonuclease R | Hypothetical, unclassified, unknown | 3.1 |
| yafT | 0217 | Hypothetical 29.6-kDa protein | Hypothetical, unclassified, unknown | 4.8 |
| yceD | 1088 | Hypothetical 19.3-kDa protein | Hypothetical, unclassified, unknown | 2.2 |
| ydhR | 1667 | Hypothetical 11.3-kDa protein | Hypothetical, unclassified, unknown | 2.9 |
| yffS | 2450 | Hypothetical 31.0-kDa protein | Hypothetical, unclassified, unknown | 2.3 |
| yfiM | 2586 | Hypothetical 9.9-kDa protein | Hypothetical, unclassified, unknown | 2.3 |
| yhbC | 3170 | Hypothetical 16.8-kDa protein | Hypothetical, unclassified, unknown | 3.1 |
| yi21_5 | 3044 | IS2 hypothetical 13.4-kDa protein | Phage, transposon, or plasmid | 2.8 |
| yi52_4 | 1331 | IS5 hypothetical 39.3-kDa protein | Phage, transposon, or plasmid | 4.7 |
| yjeB | 4178 | Hypothetical 15.6-kDa protein | Hypothetical, unclassified, unknown | 4.3 |
| yodB | 1974 | Putative cytochrome | Hypothetical, unclassified, unknown | 2.0 |
b number, unique identifier for E. coli genes.
Based on EcoCyc database (24). PTS, phosphotransferase system; IS, insertion element.
Among the genes that showed altered expression levels after treatment with Bac7(1-35), 73% corresponded to genes with known functions, 21% matched ORFs with unknown functions, and 6% corresponded to ORFs for which a function may be postulated by sequence homology comparison. Gene members of several operons (Table 2 and 3) were regulated in a coordinated fashion, supporting the congruency of the results. These included all genes of the basRS, proVWX, malK-lamB-malM, and pdhR-aceEF-lpdA operons; infB, nusA, and yhbC in the metY-yhbC-nusA-infB transcription unit; malE and malF in the malEFG transcription unit; and rbsB, rbsC, and rbsD in the rbsDACBK transcription unit.
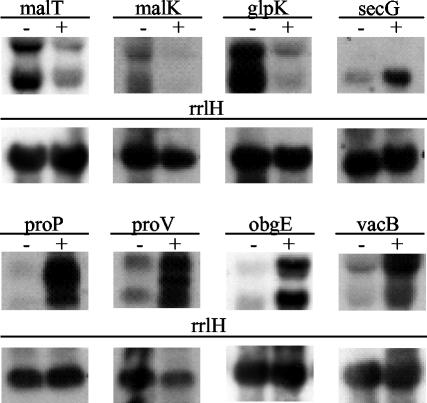
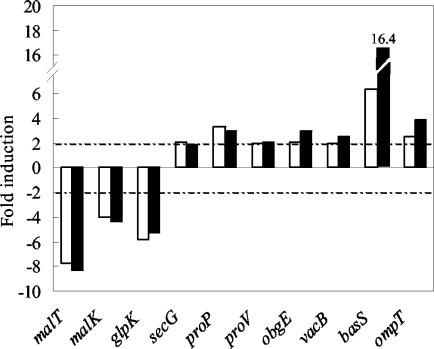
To validate the results obtained by macroarray analysis, the expression levels of 10 genes picked up in response to Bac7(1-35), i.e., secG, proP, proV, obgE, vacB, basS, ompT, malT, malK, and glpK (Tables 2 and 3), were also examined by Northern analysis of RNA from treated and untreated cultures. These genes were selected from distinct functional categories. The results confirmed the regulatory trend observed with macroarray hybridization. Specifically, malT, malK, and glpK genes (Fig. 2 and Table 2) were strongly down-regulated in the presence of 2.5 μM Bac7(1-35), whereas the expression of secG, proP, proV, obgE, vacB (Fig. 2 and Table 3), as well as expression of basS and ompT (Fig. 3 and Table 3) was markedly induced. Similar results were obtained after exposing bacterial cultures to 1 μM Bac7(1-35) (Fig. 4), a peptide concentration that yields a growth curve comparable to that of untreated bacterial cultures.
FIG. 2.
Northern blot analyses of total RNA extracted from E. coli MG1655 after 30 min of incubation in the absence (−) or presence (+) of 2.5 μM Bac7(1-35). Blots were hybridized with gene-specific probes, as indicated. Hybridization with the rrlH probe served as a control for the amount and quality of RNA loaded in each lane.
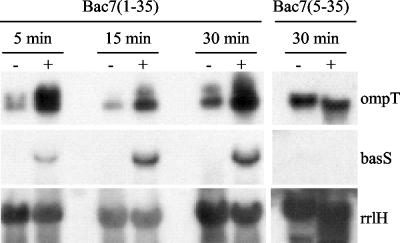
FIG. 3.
Northern blot analyses of basS and ompT gene expression. Total RNA was extracted from E. coli MG1655 after incubation of bacteria in the presence (+) or absence (−) of 2.5 μM peptide for the indicated times. Blots were hybridized with basS-, ompT-, or rrlH-specific cDNA probes.
FIG. 4.
Northern blot analysis of RNA extracted from E. coli MG1655 exposed to 1 μM (open bars) and 2.5 μM (filled bars) Bac7(1-35) for 30 min. The expression levels of the indicated genes were determined as described in Materials and Methods and are reported as levels of induction (n-fold) in peptide-treated cultures relative to that in untreated bacteria. Data representative of two independent experiments with similar results are shown.
Significantly, Northern blot analysis of RNA from bacterial cultures treated with Bac7(5-35) did not detect changes in the levels of these transcripts, compared with untreated cultures (results are shown for basS and ompT) (Fig. 3), in keeping with the macroarray hybridization pattern.
Bac7(1-35) affects the expression of a number of genes with diverse functions.
Genes whose relative expression levels decreased in response to Bac7(1-35) are reported in Table 2. Most of these genes are involved in cell metabolism; among these, four different operons were significantly repressed. Two of the operons encode the components of the maltose transport system MalKFGE that belongs to the ATP-binding cassette superfamily of transporters (4). Specifically, all gene members of the malK-lamB-malM operon and the first two genes of the malE-malF-malG operon were strongly repressed (repression ratio, 0.07 to 0.31). A downshift in the expression of malG was noted in both experiments, but the repression ratio (0.7; data not shown) did not reach the threshold considered significant. Interestingly, malT, a gene that encodes a positive transcription regulator of the maltose transport system (38), was also repressed in response to treatment with Bac7(1-35) (Table 2), suggesting that the alteration in the expression of the mal operon genes may be secondary to malT repression.
The other clusters whose genes were negatively affected by Bac7(1-35) include the rbsDACBK and the pdhR-aceE-aceF-lpdA operons. Three of the rbs locus genes were more than 50% repressed. Of these, rbsB and rbsC encode two subunits of the ATP-binding cassette transporter (RbsABC), which is responsible for ribose uptake (35). The rbsD gene encodes a cytoplasmic protein that binds ribose and may facilitate influx of this sugar in E. coli (25). The pdhR-aceE-aceF-lpdA gene cluster encodes the components of the pyruvate dehydrogenase complex. Other metabolic genes that were negatively affected by Bac7(1-35) include gapA, encoding the glycolytic enzyme glyceraldehyde 3-phosphate dehydrogenase, aspA, glpK, cyoE, and sdaB. All these metabolic genes, apart from gapA, cyoE, and sdaB (for which information is not available), show a consensus sequence for the catabolic activator protein cyclic AMP (cAMP)-cAMP receptor protein (CRP) in their promoter regions (24). The requirement of cAMP-CRP for activation of glpK (48), malE-malF-malG (37), malK-lamB-malM (36), malT (4), pdhR-aceE-aceF-lpdA (21), and aspA (16) is also supported by the results of promoter binding and promoter activity assays. It is worth noting that although the expression of the crp gene coding for cAMP-CRP was only moderately altered in response to Bac7(1-35) (repression ratio, 0.65; results not shown), this slight repression correlates with the observed down-regulation of metabolic genes, also including malT, which codes for the main regulator of maltose genes.
In the group of activated genes with known functions (Table 3), the proU operon and proP and betT genes are involved in the adaptive response of E. coli to osmotic stress. Specifically, proV, proW, and proX, which are encoded by the proU operon, are components of the ABC high-affinity transport system involved in the uptake of osmoprotectant glycine betaine and proline during osmotic stress (8). The proP and betT genes encode osmotically induced proteins involved in the transport of proline and betaine (30) and choline (26), respectively. The proP and proU genes, as well as osmY, a gene that was slightly induced by Bac7(1-35) (induction ratio of 1.53; data not reported), are regulated by σs, a master regulator that is encoded by the rpoS gene and governs expression of stationary phase-induced and osmotically regulated genes in E. coli (19, 20). The transcription of the rpoS gene was not changed under our experimental conditions. This may be explained by the fact that the σs protein accumulates as a consequence of increased protein translation and protein stability under certain stress conditions, including hyperosmolarity (27, 31). Furthermore, high levels of this protein were detected in S. enterica serovar Typhimurium in response to PB, PG-1, and other AMPs (2).
The hns gene encodes the histone-like protein H-NS and was among the genes with the highest ratio of induction (6.1). H-NS is a major constituent of the nucleoid of E. coli and is involved in the negative regulation of gene expression in response to environmental changes and stress conditions (11). hns expression is subjected to autorepression to enable the cell to maintain a constant H-NS/DNA ratio (1). Our results indicate that this control mechanism is impaired by Bac7(1-35) treatment. The ensuing increase in the concentration of H-NS may result in drastic loss of cell viability, as suggested by overexpression studies of hns in E. coli (43). In this respect, hns may be one candidate effector of the antibacterial action of Bac7(1-35).
The group of positively regulated genes also included those encoding the BasR-BasS two-component regulatory system and OmpT (Table 3). basS showed the highest expression (induction ratio, 15.7) among all induced genes. This gene codes for a sensor inner membrane protein that, in response to external signals, phosphorylates and activates a basR-encoded transcription factor (32). BasR and BasS share approximately 85% amino acid sequence identity with the two-component regulatory system PmrA-PmrB present in S. enterica serovar Typhimurium. Activation of this system in S. enterica serovar Typhimurium in response to environmental stimuli leads to covalent modification that decreases the negative charge of lipopolysaccharide, thus reducing the affinity of this molecule for cationic antimicrobial peptides and proteins (17, 39). It is interesting in this respect that the PmrA-PmrB system is activated in P. aeruginosa in response to AMPs such as PB, indolicidin, and LL-37 (29).
OmpT is an outer membrane protease and may be an essential factor for the virulence of E. coli strains involved in complicated urinary tract disease (12, 23). This protease is responsible for bacterial resistance to AMPs (46), since it inactivates cationic peptides by a specific cleavage between two basic residues (10).
The response of E. coli to Bac7(1-35) in terms of expression of basS and ompT was investigated further and was compared to that of other AMPs under similar experimental conditions.
Time course analysis of the expression of basS and ompT genes (Fig. 3) revealed basal ompT expression levels in E. coli MG1655 and in the absence of added stimuli. A fast and marked induction of both basS and ompT was detected in the presence of 2.5 μM Bac7(1-35) (Fig. 3). Conversely, bacterial incubation for up to 30 min in the presence of 2.5 μM Bac7(5-35) did not alter the expression of these genes (Fig. 3), in keeping with the macroarray results.
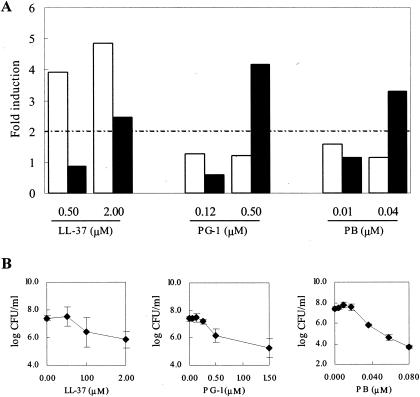
To investigate whether an increase in the levels of these genes is a common response of E. coli MG1655 to AMPs, three structurally different cationic AMPs with lytic properties, i.e., the β-hairpin peptide PG-1, the α-helical LL-37, and the cyclic PB (44, 45, 49), were comparatively analyzed for their capacity to up-regulate basS and ompT. Bacterial cultures were incubated for 15 min with a sublethal and a lethal (1 log CFU decrease after 15 min of incubation) (Fig. 5B) concentration of each peptide. LL-37 induced significant up-regulation of basS, whereas PG-1 and PB had little effect on this gene at either peptide concentration (Fig. 5A). All three peptides caused >2-fold increases in ompT expression at concentrations that inhibited bacterial growth but were poorly effective on this gene at sublethal concentrations (Fig. 5A), possibly suggesting that ompT is up-regulated in response to physical damage caused by these molecules. Taken together, these results indicate that the lytic peptides are far less efficient than Bac7(1-35) in modulating the expression of ompT and that only α-helical LL-37, among these peptides, shares with Bac7(1-35) the ability to up-regulate basS. The latter effect may not be simply related to the α-helical structure of LL-37, since the expression levels of basS and ompT were unchanged following a 10-min incubation of E. coli MG1655 with both sublethal and lethal concentrations of the α-helical peptide cecropin A (22). These individual differences within peptides with similar structural features and/or antimicrobial properties thus likely reflect different and as-yet poorly understood modes of interaction with prokaryotic cells.
FIG. 5.
(A) Induction of basS and ompT gene expression in response to LL-37, PG-1, and PB. Total RNA was extracted from E. coli MG1655 exposed for 15 min to each peptide at the indicated concentrations. Blots were hybridized with basS-, ompT-, and rrlH-specific cDNA probes. The expression levels of basS (open bars) and ompT (filled bars) were determined as described in Materials and Methods and reported as levels of induction (n-fold) in peptide-treated cultures relative to those in cultures of untreated bacteria. Data representative of two independent experiments with similar results are shown. (B) Activity of LL-37, PG-1, and PB against E. coli MG1655. Bacterial cultures were incubated for 15 min at 37°C in the presence of increasing amounts of each peptide, and the numbers of CFU per milliliter were determined. Data are means ± standard deviations of at least three experiments.
Conclusions.
This study is the first contribution to the definition of the transcriptional profile of a prokaryotic organism following treatment with a proline-rich peptide. We have shown that Bac7(1-35) at concentrations below its MIC for E. coli MG1655 induces a complex transcriptional response that involves changes in the expression of a selected set of functionally distinct groups of genes. Some alterations are consistent with activation of stress adaptation mechanisms; some, particularly those indicating reduced metabolic activities, may be connected to the antibacterial action of Bac7(1-35). Several other changes, including the induction of genes involved in protein synthesis (Table 3), are not easily understood in this context. A comparative analysis of the gene transcript profile of E. coli MG1655 treated with Bac7(1-35) and gene transcript profiles determined for E. coli MG1655 during transient or permanent growth arrest (6) indicates that most genes whose expression is altered on exposure to Bac7(1-35) (Tables 2 and 3) are unaffected or show an inverse regulatory trend during growth arrest (6). For instance, the vast majority of genes encoding the transcription and translation apparatus down-regulates under transient arrest or stationary-phase conditions (6), whereas various ribosomal genes and other genes involved in transcription and translation are up-regulated in the presence of Bac7(1-35) (Table 3). Transport and binding protein-encoding genes that are induced by Bac7(1-35) (Table 3) are unaffected or repressed (6), and those repressed by Bac7(1-35) (Table 2) are up-regulated (6) during growth arrest. An exception seems to be represented by genes involved in aerobic respiratory metabolism. Most genes in this category down-regulate during growth arrest (6), and some of these are repressed after exposure to Bac7(1-35) (Table 2).
To unravel the complexity of this response, it will be necessary to extend this analysis and characterize the bacterial gene transcript profile at various incubation times and peptide concentrations, to better refine the peptide-specific response, to discern temporal patterns of transcription, and to achieve a more complete understanding of the sequential events that follow interaction of Bac7(1-35) with bacterial cells.
We have shown that the positively charged cluster (RRIR) at the N terminus of Bac7(1-35) is required for induction of the transcriptional response. Prior studies indicated that removal of these residues from the peptide sequence substantially decreased the microbicidal activity (42). Overall, these results suggest that the charged N-terminal region of Bac7(1-35) is essential to enable this peptide to interact with bacterial components and interfere with biological processes. It remains to be clarified whether these residues are needed for the initial interaction with the bacterial surface, for specific binding to internal targets, or for both these processes.
Acknowledgments
We are grateful to A. Tossi from the BBCM Department of the University of Trieste for reading the manuscript.
This work was supported by grants from the Italian Ministry for University and Research (P.R.I.N. Cofin. 2002), the Interuniversity Consortium for Biotechnology (CIB), and Regione FVG.
REFERENCES
- 1.Atlung, T., and H. Ingmer. 1997. H-NS: a modulator of environmentally regulated gene expression. Mol. Microbiol. 24:7-17. [DOI] [PubMed] [Google Scholar]
- 2.Bader, M. W., W. W. Navarre, W. Shiau, H. Nikaido, J. G. Frye, M. McClelland, F. C. Fang, and S. I. Miller. 2003. Regulation of Salmonella typhimurium virulence gene expression by cationic antimicrobial peptides. Mol. Microbiol. 50:219-230. [DOI] [PubMed] [Google Scholar]
- 3.Boman, H. G. 2003. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 254:197-215. [DOI] [PubMed] [Google Scholar]
- 4.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62:204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, Y. R., and R. L. Gallo. 1998. PR-39, a syndecan-inducing antimicrobial peptide, binds and affects p130(Cas). J. Biol. Chem. 273:28978-28985. [DOI] [PubMed] [Google Scholar]
- 6.Chang, D. E., D. J. Smalley, and T. Conway. 2002. Gene expression profiling of Escherichia coli growth transitions: an expanded stringent response model. Mol. Microbiol. 45:289-306. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W. P., and T. T. Kuo. 1993. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 21:2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Csonka, L. N., and W. Epstein. 1996. Osmoregulation, p. 1210-1223. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, D.C.
- 9.Cudic, M., and L. Otvos, Jr. 2002. Intracellular targets of antibacterial peptides. Curr. Drug Targets 3:101-106. [DOI] [PubMed] [Google Scholar]
- 10.Dekker, N., R. C. Cox, R. A. Kramer, and M. R. Egmond. 2001. Substrate specificity of the integral membrane protease OmpT determined by spatially addressed peptide libraries. Biochemistry 40:1694-1701. [DOI] [PubMed] [Google Scholar]
- 11.Dorman, C. J., and P. Deighan. 2003. Regulation of gene expression by histone-like proteins in bacteria. Curr. Opin. Genet. Dev. 13:179-184. [DOI] [PubMed] [Google Scholar]
- 12.Foxman, B., L. Zhang, K. Palin, P. Tallman, and C. F. Marrs. 1995. Bacterial virulence characteristics of Escherichia coli isolates from first-time urinary tract infection. J. Infect. Dis. 171:1514-1521. [DOI] [PubMed] [Google Scholar]
- 13.Gallo, R. L., M. Ono, T. Povsic, C. Page, E. Eriksson, M. Klagsbrun, and M. Bernfield. 1994. Syndecans, cell surface heparan sulfate proteoglycans, are induced by a proline-rich antimicrobial peptide from wounds. Proc. Natl. Acad. Sci. USA 91:11035-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gennaro, R., and M. Zanetti. 2000. Structural features and biological activities of the cathelicidin-derived antimicrobial peptides. Biopolymers 55:31-49. [DOI] [PubMed] [Google Scholar]
- 15.Gennaro, R., M. Zanetti, M. Benincasa, E. Podda, and M. Miani. 2002. Pro-rich antimicrobial peptides from animals: structure, biological functions and mechanism of action. Curr. Pharm. Des. 8:763-778. [DOI] [PubMed] [Google Scholar]
- 16.Golby, P., D. J. Kelly, J. R. Guest, and S. C. Andrews. 1998. Transcriptional regulation and organization of the dcuA and dcuB genes, encoding homologous anaerobic C4-dicarboxylate transporters in Escherichia coli. J. Bacteriol. 180:6586-6596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 18.Hancock, R. E., and G. Diamond. 2000. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 8:402-410. [DOI] [PubMed] [Google Scholar]
- 19.Hengge-Aronis, R. 1999. Interplay of global regulators and cell physiology in the general stress response of Escherichia coli. Curr. Opin. Microbiol. 2:148-152. [DOI] [PubMed] [Google Scholar]
- 20.Hengge-Aronis, R., R. Lange, N. Henneberg, and D. Fischer. 1993. Osmotic regulation of rpoS-dependent genes in Escherichia coli. J. Bacteriol. 175:259-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann, F., J. Weber, and U. Rinas. 2002. Metabolic adaptation of Escherichia coli during temperature-induced recombinant protein production: 1. Readjustment of metabolic enzyme synthesis. Biotechnol. Bioeng. 80:313-319. [DOI] [PubMed] [Google Scholar]
- 22.Hong, R. W., M. Shchepetov, J. N. Weiser, and P. H. Axelsen. 2003. Transcriptional profile of the Escherichia coli response to the antimicrobial insect peptide cecropin A. Antimicrob. Agents Chemother. 47:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson, J. R., E. Oswald, T. T. O'Bryan, M. A. Kuskowski, and L. Spanjaard. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in the Netherlands. J. Infect. Dis. 185:774-784. [DOI] [PubMed] [Google Scholar]
- 24.Karp, P. D., M. Riley, M. Saier, I. T. Paulsen, J. Collado-Vides, S. M. Paley, A. Pellegrini-Toole, C. Bonavides, and S. Gama-Castro. 2002. The EcoCyc database. Nucleic Acids Res. 30:56-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, M. S., J. Shin, W. Lee, H. S. Lee, and B. H. Oh. 2003. Crystal structures of RbsD leading to the identification of cytoplasmic sugar-binding proteins with a novel folding architecture. J. Biol. Chem. 278:28173-28180. [DOI] [PubMed] [Google Scholar]
- 26.Lamark, T., I. Kaasen, M. W. Eshoo, P. Falkenberg, J. McDougall, and A. R. Strom. 1991. DNA sequence and analysis of the bet genes encoding the osmoregulatory choline-glycine betaine pathway of Escherichia coli. Mol. Microbiol. 5:1049-1064. [DOI] [PubMed] [Google Scholar]
- 27.Lange, R., and R. Hengge-Aronis. 1994. The cellular concentration of the sigma S subunit of RNA polymerase in Escherichia coli is controlled at the levels of transcription, translation, and protein stability. Genes Dev. 8:1600-1612. [DOI] [PubMed] [Google Scholar]
- 28.Lehrer, R. I., and T. Ganz. 2002. Defensins of vertebrate animals. Curr. Opin. Immunol. 14:96-102. [DOI] [PubMed] [Google Scholar]
- 29.McPhee, J. B., S. Lewenza, and R. E. Hancock. 2003. Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol. Microbiol. 50:205-217. [DOI] [PubMed] [Google Scholar]
- 30.Mellies, J., A. Wise, and M. Villarejo. 1995. Two different Escherichia coli proP promoters respond to osmotic and growth phase signals. J. Bacteriol. 177:144-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muffler, A., D. D. Traulsen, R. Lange, and R. Hengge-Aronis. 1996. Posttranscriptional osmotic regulation of the σs subunit of RNA polymerase in Escherichia coli. J. Bacteriol. 178:1607-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagasawa, S., K. Ishige, and T. Mizuno. 1993. Novel members of the two-component signal transduction genes in Escherichia coli. J. Biochem. (Tokyo) 114:350-357. [DOI] [PubMed] [Google Scholar]
- 33.Oh, J. T., Y. Cajal, E. M. Skowronska, S. Belkin, J. Chen, T. K. Van Dyk, M. Sasser, and M. K. Jain. 2000. Cationic peptide antimicrobials induce selective transcription of micF and osmY in Escherichia coli. Biochim. Biophys. Acta 1463:43-54. [DOI] [PubMed] [Google Scholar]
- 34.Oh, J. T., T. K. Van Dyk, Y. Cajal, P. S. Dhurjati, M. Sasser, and M. K. Jain. 1998. Osmotic stress in viable Escherichia coli as the basis for the antibiotic response by polymyxin B. Biochem. Biophys. Res. Commun. 246:619-623. [DOI] [PubMed] [Google Scholar]
- 35.Park, Y., and C. Park. 1999. Topology of RbsC, a membrane component of the ribose transporter, belonging to the AraH superfamily. J. Bacteriol. 181:1039-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richet, E. 1996. On the role of the multiple regulatory elements involved in the activation of the Escherichia coli malEp promoter. J. Mol. Biol. 264:852-862. [DOI] [PubMed] [Google Scholar]
- 37.Richet, E. 2000. Synergistic transcription activation: a dual role for CRP in the activation of an Escherichia coli promoter depending on MalT and CRP. EMBO J. 19:5222-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schreiber, V., C. Steegborn, T. Clausen, W. Boos, and E. Richet. 2000. A new mechanism for the control of a prokaryotic transcriptional regulator: antagonistic binding of positive and negative effectors. Mol. Microbiol. 35:765-776. [DOI] [PubMed] [Google Scholar]
- 39.Shafer, W. M., S. G. Casey, and J. K. Spitznagel. 1984. Lipid A and resistance of Salmonella typhimurium to antimicrobial granule proteins of human neutrophil granulocytes. Infect. Immun. 43:834-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shai, Y. 2002. Mode of action of membrane active antimicrobial peptides. Biopolymers 66:236-248. [DOI] [PubMed] [Google Scholar]
- 41.Skerlavaj, B., D. Romeo, and R. Gennaro. 1990. Rapid membrane permeabilization and inhibition of vital functions of gram-negative bacteria by bactenecins. Infect. Immun. 58:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skerlavaj, B., M. Scocchi, A. Tossi, D. Romeo, and R. Gennaro. 1999. A synthetic approach for a SAR study of the Pro- and Arg-rich bactenecin Bac7, p. 395-398. In R. Epton (ed.), Innovation and perspectives in solid phase synthesis and combinatorial libraries. Mayflower Scientific, Ltd., Birmingham, United Kingdom.
- 43.Spurio, R., M. Durrenberger, M. Falconi, A. La Teana, C. L. Pon, and C. O. Gualerzi. 1992. Lethal overproduction of the Escherichia coli nucleoid protein H-NS: ultramicroscopic and molecular autopsy. Mol. Gen. Genet. 231:201-211. [DOI] [PubMed] [Google Scholar]
- 44.Steinberg, D. A., M. A. Hurst, C. A. Fujii, A. H. Kung, J. F. Ho, F. C. Cheng, D. J. Loury, and J. C. Fiddes. 1997. Protegrin-1: a broad-spectrum, rapidly microbicidal peptide with in vivo activity. Antimicrob. Agents Chemother. 41:1738-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Storm, D. R., K. S. Rosenthal, and P. E. Swanson. 1977. Polymyxin and related peptide antibiotics. Annu. Rev. Biochem. 46:723-763. [DOI] [PubMed] [Google Scholar]
- 46.Stumpe, S., R. Schmid, D. L. Stephens, G. Georgiou, and E. P. Bakker. 1998. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J. Bacteriol. 180:4002-4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tossi, A., L. Sandri, and A. Giangaspero. 2000. Amphipathic, alpha-helical antimicrobial peptides. Biopolymers 55:4-30. [DOI] [PubMed] [Google Scholar]
- 48.Weissenborn, D. L., N. Wittekindt, and T. J. Larson. 1992. Structure and regulation of the glpFK operon encoding glycerol diffusion facilitator and glycerol kinase of Escherichia coli K-12. J. Biol. Chem. 267:6122-6131. [PubMed] [Google Scholar]
- 49.Zanetti, M. 2004. Cathelicidins, multifunctional peptides of the innate immunity. J. Leukoc. Biol. 75:39-48. [DOI] [PubMed] [Google Scholar]
- 50.Zasloff, M. 2002. Antimicrobial peptides of multicellular organisms. Nature 415:389-395. [DOI] [PubMed] [Google Scholar]