Abstract
Genes required for intrinsic multidrug resistance by Mycobacterium avium were identified by screening a library of transposon insertion mutants for the inability to grow in the presence of ciprofloxacin, clarithromycin, and penicillin at subinhibitory concentrations. Two genes, pks12 and Maa2520, were disrupted in multiple drug-susceptible mutants. The pks12 gene (Maa1979), which may be cotranscribed with a downstream gene (Maa1980), is widely conserved in the actinomycetes. Its ortholog in Mycobacterium tuberculosis is a polyketide synthase required for the synthesis of dimycocerosyl phthiocerol, a major cell wall lipid. Mutants of M. avium with insertions into pks12 exhibited altered colony morphology and were drug susceptible, but they grew as well as the wild type did in vitro and intracellularly within THP-1 cells. A pks12 mutant of M. tuberculosis was moderately more susceptible to clarithromycin than was its parent strain; however, susceptibility to ciprofloxacin and penicillin was not altered. M. avium complex (MAC) and M. tuberculosis appear to have different genetic mechanisms for resisting the effects of these antibiotics, with pks12 playing a relatively more significant role in MAC. The second genetic locus identified in this study, Maa2520, is a conserved hypothetical gene with orthologs in M. tuberculosis and Mycobacterium leprae. It is immediately upstream of Maa2521, which may code for an exported protein. Mutants with insertions at this locus were susceptible to multiple antibiotics and slow growing in vitro and were unable to survive intracellularly within THP-1 cells. Like pks12 mutants, they exhibited increased Congo red binding, an indirect indication of cell wall modifications. Maa2520 and pks12 are the first genes to be linked by mutation to intrinsic drug resistance in MAC.
The environmental pathogen Mycobacterium avium complex (MAC) opportunistically infects susceptible humans, especially AIDS patients with low CD4+ cell counts (9, 10, 13, 22). MAC infections are difficult to treat due to the intrinsic multidrug resistance of the organism. Drugs such as clarithromycin, azithromycin, rifabutin, ethambutol, amikacin, clofazamine, and fluoroquinolones, which are effective against primary isolates, frequently lose effectiveness unless administered in combination.
The multidrug resistance of MAC is usually ascribed to intrinsic properties of the organism's lipid-rich cell wall, although additional factors may contribute (2, 19, 23, 27, 36). A role for the cell wall has been inferred from indirect observations. Exposure to detergents, drugs, and other agents that compromise cell wall integrity can result in increased susceptibility to multiple drugs (19, 27, 29). Aminoglycosides are more active on ribosomes in cell extracts than on intact MAC cells (24). Finally, there is a correlation between drug susceptibility and colony type of MAC. Transparent colony variants, which predominate in patient samples, are significantly more resistant to multiple antibiotics than are their opaque counterparts (18, 28, 35).
An additional morphotypic switch, termed red-white, also affects multidrug resistance in MAC. Red and white colony types are visible when the bacteria are grown on media containing the lipoprotein stain Congo red (CR) (5-7, 20, 25). The red-white switch operates independently of the opaque-transparent switch, such that red opaque (RO), red transparent (RT), white opaque (WO), and white transparent morphotypes can be distinguished by CR staining. White variants are more common than their red counterparts in patient samples, and they grow better in disease models. However, the RT colony type can also be recovered from patient samples (25). The red-to-white switch is accompanied by increased resistance to multiple antibiotics, including macrolides, rifamycins, and quinolones. WO variants are more resistant to these drugs than their RO counterparts are, and white transparent variants are more resistant than their RT counterparts are (6).
Due in part to the instability of the transparent morphotype in vitro (35), the multidrug resistance associated with the transparent morphotype remains uncharacterized. The red and white morphotypes are more stable in vitro, which makes them relatively amenable to genetic dissection. Mutational analysis with use of a transposome mutagenesis system identified an apparent acetyltransferase gene, crs, that is required for CR binding in red variants. It also identified the major nonribosomal peptide synthetase involved in the synthesis of the serovar-specific glycopeptidolipid (20). For the present study, the mutagenesis procedure was modified to improve the efficiency of mutagenesis of the relatively drug-resistant WO morphotype. This led to the identification of genes required for the multidrug resistance associated with this morphotype.
MATERIALS AND METHODS
Bacterial strains.
White and red variants of strain HMC02, a clinical isolate of Mycobacterium avium subsp. avium, have been described previously (6). A pks12 mutant of Mycobacterium tuberculosis, along with its parent strain, H37Rv, was kindly provided by P. Kolattukudy, University of Central Florida, Orlando. This mutant had been constructed by replacing a 2,193-bp fragment near the center of the pks12 gene with a hygromycin resistance gene cassette (32). The mutant was originally designated Δmsl6 after an older name for the pks12 gene. For clarity, it is designated H37Rv::Δpks12 in this paper.
Transposome mutagenesis of WO cells.
The WO variant of M. avium subsp. avium strain HMC02 was mutagenized by using the commercial EZ::TN <KAN-2> system (Epicentre, Madison, Wis.). The electroporation method described previously (20) worked well on red variants, which form dispersed suspensions, but was relatively inefficient at mutagenizing white variants, which are flocculent in broth culture. Therefore, the procedure for preparing electrocompetent cells (15) was modified by growing the cells in the presence of sucrose as described by Lee et al. (21). This resulted in dispersed growth of WO cells and improved transformation efficiency. Under the modified protocol, cells were grown to an optical density at 600 nm of 0.3 to 0.5 in Middlebrook 7H9 broth with albumin-dextrose-catalase (ADC) enrichment and 0.5 M sucrose. One day before harvest, glycine was added to 0.2 M. Cells were pelleted at 5,000 × g and resuspended in electroporation solution at 8× their original concentration (15). After two more washes in electroporation solution, cells were resuspended in electroporation solution at 100× their original concentration and stored at −80°C in 100-μl aliquots.
Competent WO cells (100-μl aliquots) were mutagenized with the EZ::TN <KAN-2> transposome, and mutants were selected by growth on Middlebrook 7H10 agar with albumin enrichment, glycerol, 100 μg of CR/ml, and 100 μg of kanamycin/ml (MAG-CR-KAN) as described previously (20). Kanamycin-resistant mutants were transferred to fresh MAG-CR-KAN plates containing 0.2 μg of ciprofloxacin/ml and control MAG-CR-KAN plates without ciprofloxacin. After 3 weeks of incubation under air at 37°C, clones that exhibited altered CR staining characteristics, inability to grow in the presence of 0.2 μg of ciprofloxacin/ml, or both were retested to confirm their phenotypes. Confirmed mutants were subjected to further examination.
Identification of EZ::TN insertion sites.
Transposon insertion sites were mapped as described previously (20). Briefly, genomic regions adjoining EZ::TN insertions were amplified by arbitrary primer PCR or inverted PCR. Purified PCR products were submitted for automated sequencing at the Seattle Biomedical Research Institute. The Unfinished Microbial Genomes searching tool at The Institute for Genomic Research website (http://tigrblast.tigr.org/ufmg/) was used to precisely locate insertion sites within the draft M. avium subsp. avium strain 104 genome sequence. Regions of up to 20 kb surrounding the insertion sites were then analyzed using the Basic Local Alignment Search Tool (BLAST) and the ORF Finder tool at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/) to identify sequences in the National Center for Biotechnology Information nonredundant database that are closely homologous to open reading frames (ORFs) in the region of the insertion. Gene designations (e.g., Maa2520) were derived from the recent annotation of the M. avium subsp. avium 104 genome sequence by M. Semret and M. Behr (31a).
Drug susceptibility in vitro.
The drug susceptibilities of wild-type and mutant cells of M. avium subsp. avium and M. tuberculosis were tested by the proportion method (17) with a breakpoint of 1% and critical concentrations of the following antibiotics: clarithromycin (0, 0.05, 0.25, 1.25, 6.25, 16, and 64 μg/ml), penicillin (0, 0.5, 2.5, 12.5, and 62.5 μg/ml), and ciprofloxacin (0, 0.025, 0.05, 0.25, 1.25, and 6.25 μg/ml). Clarithromycin was kindly provided for this study by Abbott Laboratories, North Chicago, Ill. Other drugs were obtained from commercial vendors (ICN Biomedicals, Aurora, Ohio, and Sigma Chemical Co., St. Louis, Mo.). To prevent artifactual results due to degradation of antibiotics, plates were read after no more than 2 weeks of incubation at 37°C. M. tuberculosis cells were incubated under 5% CO2. M. avium subsp. avium cells were also tested for susceptibility to rifampin by using AB Biodisk E-test strips (Remel, Lenexa, Kans.) as described previously (34). All drug susceptibility tests were conducted in triplicate. In addition to MICs, the presence or absence of rare (<1%) drug-resistant variants was recorded.
Survival during intracellular incubation in THP-1 cells.
The myelomonocytic THP-1 cell line (ATCC T1B-202) was grown in RPMI culture medium containing 10% fetal bovine serum, 2 mM l-glutamine, and 50 U of penicillin-streptomycin per ml, at 37°C in the presence of 5% CO2 (16). Seventy-two hours prior to infection, cells were treated for differentiation and adherence to 12-mm-diameter glass coverslips with use of 100 nM phorbol 12-myristate 13-acetate (11). Bacterial cultures were grown with shaking at 37°C in Middlebrook 7H9 broth with 0.2% glycerol and 10% Middlebrook ADC enrichment to an optical density at 600 nm of 0.3 to 0.5. Bacteria were diluted in RPMI infection medium (10% fetal bovine serum and 2 mM l-glutamine). Immediately prior to infection with bacteria, two coverslips with host cells were removed for the 0-h time point, lysed with double-distilled water, and plated onto MAG-CR plates. Remaining coverslips were washed twice with RPMI to remove nonadherent THP-1 cells and residual antibiotic-containing culture medium. New RPMI infection medium with no antibiotics was added to the coverslips, and cells were then infected at a 10:1 ratio of bacteria to THP-1 cells (11) and incubated at 37°C in 5% CO2. At time points of 2, 24, and 48 h and 4 and 7 days postinfection, triplicate coverslips were washed three times with RPMI culture medium to remove extracellular bacteria (12). Cells on each coverslip were lysed in double-distilled water, serially diluted, and plated onto MAG-CR plates. After incubation for 2 to 3 weeks at 37°C, results were quantified based on the number of CFU per coverslip. Means and standard deviations of each set of triplicate readings were plotted.
Comparison of predicted macrolide and penicillin resistance genes of M. tuberculosis and M. avium subsp. avium.
M. tuberculosis genes previously linked to innate resistance to macrolide and penicillin drugs, and genes predicted by homology to play roles in resistance, were identified in the literature, in the Mycobacterium tuberculosis Structural Genomics Consortium Database (http://www.doe-mbi.ucla.edu/TB/), and in the TubercuList database (http://genolist.pasteur.fr/TubercuList/). The nucleotide sequence of each M. tuberculosis H37Rv gene was compared to the M. avium subsp. avium strain 104 genome sequence by using the unfinished microbial genomes searching tool at the Institute for Genomic Research website (http://tigrblast.tigr.org/ufmg/). Gene sequences were further analyzed for cross-species similarity by using EMBOSS::needle software publicly available at http://www.ebi.ac.uk/emboss/align/.
RESULTS
Mutagenesis of white variants of M. avium subsp. avium.
Genetic analysis of MAC is complicated by the fact that the most clinically significant morphotypes (white and transparent) are also the most difficult to genetically transform. The modified procedure described in Materials and Methods improved the efficiency of transformation of WO variants of M. avium subsp. avium strain HMC02, such that we were able to generate an incomplete library of approximately 1,500 kanamycin-resistant transposon mutant clones. Previous observations indicated that the EZ::TN transposon inserted at random sites in this strain (20).
The library was screened for mutants that were unable to grow on 0.2 μg of ciprofloxacin/ml (to which WO variants are resistant), or exhibited elevated CR staining comparable to that of the RO morphotype, or both. Fifteen ciprofloxacin-sensitive mutants were isolated, all of which exhibited elevated CR staining relative to that of the parent strain. Three additional mutants exhibited elevated CR staining but were not more sensitive to ciprofloxacin than the parent strain was.
Transposon insertion sites were mapped. Most insertion sites were disrupted in no more than one mutant, so their roles in the observed phenotypes could not be confirmed. However, two genetic loci, Maa2520 and Maa1979, were mutagenized in multiple ciprofloxacin-sensitive mutants and were studied further.
Bioinformatic characterization of Maa2520 and Maa2521.
Two ciprofloxacin-sensitive mutants, 6.389 and 7.1034, had insertions into a 1,242-bp ORF, Maa2520 (Fig. 1A). The two mutants were generated in independent mutagenesis procedures. The putative 414-amino-acid product of Maa2520 was 89% identical to conserved hypothetical protein Rv1697 of M. tuberculosis, and 88% identical to MLC1351.11c of Mycobacterium leprae, by BLAST analysis (expect value 0.0). Weaker homologs were found in Corynebacterium, Thermoanaerobacter, Thermobifida, and Bacillus species (expect values 1E-28 to 5E-84). These proteins belong to a cluster of orthologous uncharacterized membrane-anchored proteins conserved in bacteria (COG4825).
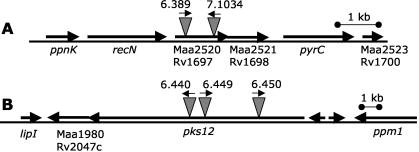
FIG. 1.
Genetic maps of the Maa2520-Maa2521 (A) and pks12 (B) regions of the M. avium subsp. avium 104 genome. Positions and orientations of transposon insertions are shown. Gene order, orientation, and approximate genetic distances within both regions are identical in M. avium subsp. avium 104 and M. tuberculosis H37Rv. M. avium subsp. avium genome annotation numbers (MaaNNNN) are shown above corresponding M. tuberculosis genome annotation numbers (RvNNNN). Where descriptive gene designations have been assigned to M. tuberculosis H37Rv genes (for example, pks12), those designations are used for both species. Note that lines A and B are drawn to different scales.
Maa2520 ends 3 bp upstream of the start codon of a 951-bp ORF, Maa2521, suggesting that the two genes may be cotranscribed. The 317-amino-acid Maa2521 gene product is 82% identical to ML1362 of M. leprae and 75% identical (86% conserved) to Rv1698 of M. tuberculosis. Weaker homologs were found in Corynebacterium and Thermobifida species. The function of these proteins is not known; however, the M. leprae homolog has been described as a possible exported protein (http://www.sanger.ac.uk/Projects/M_leprae/CDS/ML1362.shtml). The genomic region surrounding Maa2520-Maa2521 in M. avium subsp. avium strain 104 (Fig. 1A) is genetically identical to the orthologous region in M. tuberculosis H37Rv.
Phenotypic characterization of Maa2520-Maa2521 mutants.
Mutants 6.389 and 7.1034 exhibited identical pleiotropic phenotypes. In addition to ciprofloxacin sensitivity, they were more sensitive to clarithromycin and penicillin than the parent strain was in vitro (Table 1). They were also more sensitive to rifampin in E-tests (Fig. 2A). They did not form drug-resistant colony type variants and had well-defined MIC cutoffs in all drug susceptibility tests. This contrasted with wild-type cells that spontaneously formed drug-resistant colony type variants, including transparent variants, at typical frequencies of 10−4 to 10−6 (35). For example, growth of the RO variant of strain HMC02 was substantially inhibited by 1.25 μg of ciprofloxacin/ml; however, drug-resistant subpopulations always formed a few isolated colonies at this concentration. In contrast, mutants 6.389 and 7.1034 formed no colonies under these conditions (Fig. 2B).
TABLE 1.
Drug susceptibility of M. avium subsp. avium HMC02 WO and mutants derived from this strain
| Strain | MIC (μg/ml)
|
||
|---|---|---|---|
| Clarithromycin | Ciprofloxacin | Penicillin | |
| HMC02 WO (parent strain) | 6.25 | 1.25 | 12.5 |
| 6.389 (Maa2520) | 0.25a | 0.25a | 2.5a |
| 7.1034 (Maa2520) | 0.25a | 0.25a | 2.5a |
| 6.440 (pks12) | 0.25 | 0.25 | 2.5 |
| 6.449 (pks12) | 0.25 | NDb | 2.5 |
| 6.450 (pks12) | ND | 0.25 | 2.5 |
No drug-resistant subvariants observed. All other strains formed isolated drug-resistant colonies at frequencies of <1%.
ND, not determined in the experiment shown. Separate experiments showed increased drug susceptibility consistent with the other data in this table.
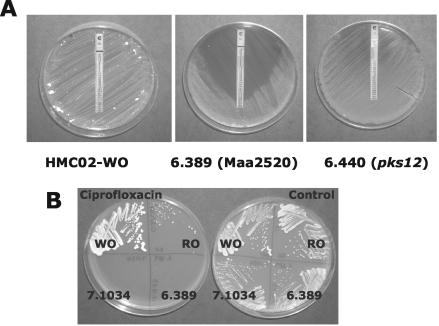
FIG. 2.
Drug susceptibility phenotypes of wild-type and mutant clones on agar medium. (A) Rifampin E-tests were conducted on MAG plates as described previously (34), except that plates were incubated for an extended 3-week period to yield visible growth of drug-resistant subclones on mutant 6.440 (pks12). (B) Colony formation by wild-type and transposon mutant clones of strain HMC02 streaked on MAG plates with (left) or without (right) 0.5 μg of ciprofloxacin/ml. The formation of isolated ciprofloxacin-resistant colonies by the otherwise drug-susceptible HMC02 RO clone and the relatively small size of colonies formed by mutants 6.389 and 7.1034 are visible.
Additional phenotypes observed in vitro were elevated CR staining and slow growth relative to the parent strain. Two-week-old colony diameters were about half that of the parent strain (Fig. 2B)
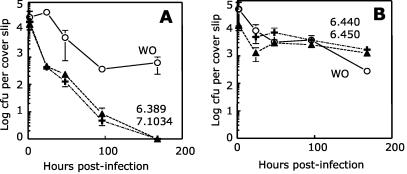
Drug resistance and virulence tend to correlate with each other when spontaneously occurring colony types of MAC are compared. Therefore, we examined the ability of Maa2520-Maa2521 mutants to survive intracellular incubation within the macrophage-like THP-1 cell line. A complicating factor was that the mutagenesis had been carried out on an opaque variant of strain HMC02, due to the difficulty of genetically transforming transparent variants. Opaque variants of MAC are unable to grow intracellularly and are not virulent (6, 25). We attempted to derive spontaneous transparent variants from parent and mutant strains for comparison with each other; however, mutants 6.389 and 7.1034 did not form transparent variants even under antibiotic selective pressure (Fig. 2B). Therefore, we infected THP-1 cells with opaque variants of each strain and tested their ability to survive (if not grow) intracellularly. The parent strain exhibited an initial decline in viability for a day or two postinfection and then stabilized, possibly by virtue of a subpopulation of virulent variants. In contrast, both Maa2520-Maa2521 mutants declined to undetectable levels by day 7 postinfection, a phenotype that was markedly different from that of the parent strain (Fig. 3A).
FIG. 3.
Survival of wild-type and mutant clones within THP-1 cells. All data points represent the means and standard deviations of triplicate measurements. (A) The parent strain HMC02 WO (open circles) and Maa2520-Maa2521 mutants 6.389 (closed triangles) and 7.1034 (plus signs) were inoculated into THP-1 cells, and their intracellular survival was tracked as described in Materials and Methods. (B) The same experiment was conducted on parent strain HMC02 WO (open circles) and pks12 mutants 6.440 (closed triangles) and 6.450 (plus signs).
Bioinformatic characterization of Maa1979 and Maa1980.
Three ciprofloxacin-sensitive mutants, 6.440, 6.449, and 6.550, had insertions into an ORF, Maa1979 (Fig. 1B). At 12,513 bp, this ORF is the second longest in the M. avium subsp. avium strain 104 genome (M. Semret and M. Behr, personal communication). The putative 4,170-amino-acid product of Maa1979 was 80% identical and 87% homologous by BLAST analysis to the Pks12 protein (Rv2048c) of M. tuberculosis (expect value 0.0). Homologs are present throughout the genera Mycobacterium and Streptomyces and in other bacteria as well (COG3321, polyketide synthase modules). In M. tuberculosis, pks12 encodes a polyketide synthase required for the synthesis of dimycocerosyl phthiocerol (DIM), a major cell wall lipid (32). Maa1979 ends 4 bp upstream of a 2,606-bp ORF, Maa1980, which may be cotranscribed with Maa1979. The 869-amino-acid gene product of Maa1980 is 73% identical and 80% homologous to the conserved hypothetical protein Rv2047c of M. tuberculosis (expect value 0.0) and only slightly less homologous to MLCB2052.18 of M. leprae. As with Maa2520-Maa2521, the apparent operon structure of Maa1979-Maa1980 is seen in M. tuberculosis as well as in M. avium subsp. avium, and the surrounding genomic regions are nearly identical in the two species.
Phenotypic characterization of Maa1979-Maa1980 (pks12) mutants.
The pks12 mutants 6.440, 6.449, and 6.550 exhibited identical phenotypes. All three mutants formed rough colonies that stained with CR, in contrast to the smooth WO parent strain, which resisted CR staining. Colony size and apparent growth rates were identical to those of the parent strain. Increased susceptibility was observed against all drugs tested; however, in contrast to the Maa2520-Maa2521 mutants, the pks12 mutants formed isolated drug-resistant variants at low frequencies (Table 1). This was also evident after extended incubation on rifampin E-test plates, on which pks12 mutants formed diffuse zones of inhibition (Fig. 2A). Drug-resistant variants formed by the mutants were not transparent and did not otherwise differ in appearance from the main population of colonies.
Because the pks12 mutants did not form archetypical transparent colonies, they were tested for survival during intracellular growth following the same strategy used for the Maa2520-Maa2521 mutants. The pks12 mutants 6.440 and 6.450 did not differ from the parent strain in their ability to survive intracellularly. Upon infection of THP-1 cells, the mutants declined to lower stable numbers, similar to the parent strain (Fig. 3B).
Drug susceptibility of a pks12 mutant of M. tuberculosis.
The three Maa1979-Maa1980 insertions were distributed over a 3.5-kb section of the gene, indicating that they were not siblings. However, all three were isolated in a single mutagenesis procedure. Therefore, as an independent test of the role of this locus in intrinsic drug resistance, a pks12 deletion mutant of M. tuberculosis described previously (32) was also examined. As shown in Table 2, the mutant H37Rv::Δpks12 was more susceptible to clarithromycin than was its parent strain, consistent with a role for pks12 in resistance to this macrolide drug. However, the effect was more modest than that seen in M. avium subsp. avium, and clarithromycin resistance was higher overall in M. tuberculosis. Moreover, the pks12 mutation did not affect resistance of M. tuberculosis H37Rv to ciprofloxacin and penicillin.
TABLE 2.
Drug susceptibility of M. tuberculosis H37Rv and H37Rv::Δpks12
| Strain | MIC (μg/ml)
|
||
|---|---|---|---|
| Clarithromycin | Ciprofloxacin | Penicillin | |
| H37Rv | 64.0 | 0.25 | >62.5 |
| H37Rv::Δpks12 | 16.0 | 0.25 | >62.5 |
DISCUSSION
MAC infections of humans are very difficult to treat because of the intrinsic multidrug resistance of the pathogen. A role for the cell wall has been inferred from indirect observations; however, the mechanisms of intrinsic drug resistance remain uncharacterized. The genes described here are the first to be linked by mutational analysis to intrinsic drug resistance by MAC.
Maa2520 and Maa2521 code for conserved hypothetical proteins that exhibit sequence characteristics associated with the cell surface. Strong homologs were found only in the genomes of two other slow-growing mycobacterial pathogens, M. tuberculosis and M. leprae. Weaker homologs were found in a few additional bacterial species including two thermophiles. These gene products may be required for cell wall stability, and their roles in intrinsic multidrug resistance may be indirect. The pleiotropy of mutants with insertions into this gene cluster (slow growth in vitro and sensitivity to killing by THP-1 cells) is consistent with this hypothesis. Sassetti et al. (30), in their transposon site hybridization (TraSH) analysis of M. tuberculosis genes required for growth in vitro, gained evidence that Rv1697, the M. tuberculosis ortholog of Maa2520, was required for normal growth of M. tuberculosis in vitro. It was not determined whether this was due to an absolute requirement for Rv1697 by M. tuberculosis or was simply the result of a lower growth rate.
The pks12 gene has been well characterized in M. tuberculosis. It codes for a polyketide synthase required for the synthesis of DIM, a major cell wall constituent in some pathogenic mycobacteria (32). DIM is thought to contribute to the impermeability of the M. tuberculosis cell wall (4). It is not yet known whether Pks12 functions in the synthesis of a DIM-like lipid in MAC. Mutations in the pks12 gene of MAC resulted in increased multidrug susceptibility, perhaps as a result of increased cell wall permeability. Colonies of pks12 mutants were rough and stained with CR, consistent with altered cell walls relative to those of the smooth, CR-resistant colonies formed by the parent strain. Polar effects on a downstream gene present in both species, Maa1980, may account for the observed phenotypes. Colonies of the pks12 mutant of M. tuberculosis did not differ in appearance from those of the parent strain. Mutants of both species grew at normal rates in vitro.
In contrast to Maa2520-Maa2521, which was required for intracellular survival within THP-1 cells, mutants in Maa1979-Maa1980 exhibited no pronounced defects relative to the parent strain under these conditions. This finding contrasts with observations made for the pks12 mutant of M. tuberculosis, which was slightly attenuated for growth within the MH-S mouse alveolar macrophage cell line and more strongly attenuated for growth within intranasally infected C57BL/6J mice (32). However, given that the pathogens as well as the host systems differed in the two studies, the two findings cannot be considered contradictory.
The relative sensitivity of M. tuberculosis H37Rv::Δpks12 to clarithromycin indicates that pks12 or its cotranscript plays a role in intrinsic drug resistance in both species. However, the effect was more modest, and its spectrum more narrow, in M. tuberculosis. Genomic comparisons indicate that M. tuberculosis and M. avium subsp. avium have different genetic mechanisms of intrinsic resistance to macrolides and penicillins. A 23S rRNA methyltransferase gene, ermMT, has been proposed to play a role in the high-level resistance of M. tuberculosis to macrolides (3, 26). Resistance of M. tuberculosis to penicillins is thought to be mediated by at least one major β-lactamase, blaC, and possibly by altered expression of several penicillin binding proteins (14, 31, 34). M. avium subsp. avium 104 has homologs to penicillin binding proteins and putative macrolide efflux pumps found in M. tuberculosis; however, homologs to ermMT and blaC were not found in its genome (Table 3). The picture that emerges from these comparisons is that M. tuberculosis and MAC have separate, but overlapping, mechanisms of resistance to these drugs, with the nonspecific resistance mediated by pks12 playing a relatively more important role in MAC.
TABLE 3.
Comparison of genes associated with macrolide and penicillin resistance in the genomes of M. tuberculosis H37Rv and M. avium subsp. avium 104
| M. tuberculosis gene designation | Product | Descriptiona | Reference(s) | Homology to M. avium subsp. avium genesb
|
||
|---|---|---|---|---|---|---|
| e score | % Amino acid identity | % Protein similarity | ||||
| Rv1258 | Tap | MF efflux pump | 1 | 1.6e-143 | 74 | 80 |
| Rv1473 | Possible macrolide efflux pump | 8 | 8.6e-180 | 74 | 77.2 | |
| Rv1667 | Possible macrolide efflux pump | 8 | 1.1e-62 | 74 | 34.4 | |
| Rv1668 | Possible macrolide efflux pump | 8 | 3.1e-148 | 79 | 49.1 | |
| Rv1988 | ErmMT/38 | 23S rRNA methyltransferase | 3, 26 | —c | — | — |
| Rv2477 | Possible macrolide efflux pump | 8 | 3.7e-296 | 88 | 98.7 | |
| Rv0016 | PbpA | Probable PBP | 8 | 3.0e-216 | 81 | 92.5 |
| Rv0050 | PonA1 | Class A1 PBP | 8 | 1.9e-303 | 81 | 73.1 |
| Rv0406 | Possible β-lactamase | 8 | 6.6e-97 | 78 | 76.8 | |
| Rv1730 | Possible PBP | 8 | — | — | — | |
| Rv2068c | BlaC | β-Lactamase | 8, 14, 31, 33 | — | — | — |
| Rv2163 | PbpB | Probable PBP | 8 | 1.8e-278 | 79 | 94.4 |
| Rv2864 | Possible PB lipoP | 8 | 5.9e-284 | 83 | 92.2 | |
| Rv2911 | DacB2 | Probable PBP | 8 | 1.1e-129 | 82 | 88.1 |
| Rv3330 | DacB1 | Probable PBP | 8 | 3.6e-176 | 82 | 84.7 |
| Rv3682 | PonA2 | Class A1 PBP | 8 | 0.0 | 82 | 94.6 |
Annotations are from the Mycobacterium tuberculosis Structural Genomics Consortium and TubercuList databases. PBP, penicillin binding protein; lipoP, lipoprotein; MF, major facilitator.
Expect values (e scores) and percent amino acid identity were generated by BLAST analysis. Percent protein similarity values were generated by using EMBOSS software.
—, no homologous ORF (expect value, <10−13) found in the M. avium subsp. avium 104 genome.
The nonspecific drug resistance of MAC affects a broad spectrum of drugs including fluoroquinolones and rifamycins, to which M. tuberculosis strains are intrinsically sensitive. Therefore, it is perhaps unexpected that the MAC genes identified in this study were present in M. tuberculosis as well. If they play direct roles in multidrug resistance, they may be expressed to higher levels in MAC relative to M. tuberculosis or have minor modifications that increase or alter their activity. Alternatively, they may play indirect roles in drug resistance, for example by stabilizing the activity of separate cell surface components which are unique to MAC.
In summary, transposon mutational analysis identified two genetic loci that are directly or indirectly required for intrinsic multidrug resistance in MAC. Continued mutagenesis of the approximately 4,500 genes in the M. avium subsp. avium genome will likely identify additional required gene products. Biochemical and physiological characterization of these gene products will yield a clearer understanding of this clinically significant phenotype.
Acknowledgments
We are grateful to David Sherman and Tanj Bennett for helpful discussions and to Makeda Semret and Marcel Behr for providing prepublication information on annotation of the M. avium subsp. avium 104 genome sequence. We are also grateful to Pappachan Kolattukudy for providing the H37Rv::Δpks12 mutant and its parent M. tuberculosis strain.
This work was supported by grant AI25767 from the National Institutes of Health. Analysis of M. tuberculosis drug resistance genetics was supported by a generous grant from the Akibene Foundation. Preliminary M. avium genome sequence data were obtained from The Institute for Genomic Research website at http://www.tigr.org. Sequencing of the M. avium genome is being carried out by TIGR with support from the National Institutes of Health.
REFERENCES
- 1.Ainsa, J. A., M. C. Blokpoel, I. Otal, D. B. Young, K. A. De Smet, and C. Martin. 1998. Molecular cloning and characterization of Tap, a putative multidrug efflux pump present in Mycobacterium fortuitum and Mycobacterium tuberculosis. J. Bacteriol. 180:5836-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barrow, W. W., E. L. Wright, K. S. Goh, and N. Rastogi. 1993. Activities of fluoroquinolone, macrolide, and aminoglycoside drugs combined with inhibitors of glycosylation and fatty acid and peptide biosynthesis against Mycobacterium avium. Antimicrob. Agents Chemother. 37:652-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buriankova, K., F. Doucet-Populaire, O. Dorson, A. Gondran, J. C. Ghnassia, J. Weiser, and J. L. Pernodet. 2004. Molecular basis of intrinsic macrolide resistance in the Mycobacterium tuberculosis complex. Antimicrob. Agents Chemother. 48:143-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Camacho, L. R., P. Constant, C. Raynaud, M. A. Laneelle, J. A. Triccas, B. Gicquel, M. Daffe, and C. Guilhot. 2001. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 276:19845-19854. [DOI] [PubMed] [Google Scholar]
- 5.Cangelosi, G. A., J. E. Clark-Curtiss, M. Behr, T. Bull, and T. Stinear. 2004. Biology of waterborne pathogenic mycobacteria, p. 39-54. In J. Bartram and G. Rees (ed.), Pathogenic mycobacteria in water. World Health Organization-U.S. Environmental Protection Agency, Geneva, Switzerland.
- 6.Cangelosi, G. A., C. O. Palermo, and L. E. Bermudez. 2001. Phenotypic consequences of red-white colony type variation in Mycobacterium avium. Microbiology 147:527-533. [DOI] [PubMed] [Google Scholar]
- 7.Cangelosi, G. A., C. O. Palermo, J. P. Laurent, A. M. Hamlin, and W. H. Brabant. 1999. Colony morphotypes on Congo red agar segregate along species and drug susceptibility lines in the Mycobacterium avium-intracellulare complex. Microbiology 145:1317-1324. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, B. G. Barrell, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 9.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ford, T., J. Hermon-Taylor, G. Nichols, G. Cangelosi, and J. Bartram. 2004. Approaches to risk management in priority settings, p. 169-179. In J. Bartram and G. Rees (ed.), Pathogenic mycobacteria in water. World Health Organization-U.S. Environmental Protection Agency, Geneva, Switzerland.
- 11.Garcia, R. C., E. Banfi, and M. G. Pittis. 2000. Infection of macrophage-like THP-1 cells with Mycobacterium avium results in a decrease in their ability to phosphorylate nucleolin. Infect. Immun. 68:3121-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greenwell-Wild, T., N. Vazquez, D. Sim, M. Schito, D. Chatterjee, J. M. Orenstein, and S. M. Wahl. 2002. Mycobacterium avium infection and modulation of human macrophage gene expression. J. Immunol. 169:6286-6297. [DOI] [PubMed] [Google Scholar]
- 13.Guthertz, L. S., B. Damsker, E. J. Bottone, E. G. Ford, M. F. Thaddeus, and M. J. Janda. 1989. Mycobacterium avium and Mycobacterium intracellulare infection in patients with and without AIDS. J. Infect. Dis. 160:1037-1041. [DOI] [PubMed] [Google Scholar]
- 14.Hackbarth, C. J., I. Unsal, and H. F. Chambers. 1997. Cloning and sequence analysis of a class A β-lactamase from Mycobacterium tuberculosis H37Ra. Antimicrob. Agents Chemother. 41:1182-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hatfull, G. F., and W. R. Jacobs, Jr. 2000. Molecular genetics of mycobacteria. ASM Press, Washington, D.C.
- 16.Hayashi, T., A. Catanzaro, and S. P. Rao. 1997. Apoptosis of human monocytes and macrophages by Mycobacterium avium sonicate. Infect. Immun. 65:5262-5271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heifets, L., and G. A. Cangelosi. 2002. Antibiotic susceptibility testing of Mycobacterium tuberculosis—a neglected problem at the turn of the century. Int. J. Tuberc. Lung Dis. 3:564-581. [PubMed] [Google Scholar]
- 18.Inderlied, C. B., C. A. Kemper, and L. E. M. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6:266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarlier, V., and H. Nikaido. 1994. Mycobacterial cell wall: structure and role in natural resistance to antibiotics. FEMS Microbiol. Lett. 123:11-18. [DOI] [PubMed] [Google Scholar]
- 20.Laurent, J. P., K. A. Hauge, K. Burnside, and G. A. Cangelosi. 2003. Mutational analysis of cell wall biosynthesis in Mycobacterium avium. J. Bacteriol. 185:5003-5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, S.-H., M. Cheung, V. Irani, J. D. Carroll, J. M. Inamine, W. R. Howe, and J. N. Maslow. 2002. Optimization of electroporation conditions for Mycobacterium avium. Tuberculosis 82:167-174. [DOI] [PubMed] [Google Scholar]
- 22.Marras, T. K., and C. L. Daley. 2002. Epidemiology of human pulmonary infection with nontuberculous mycobacteria. Clin. Chest Med. 23:553-567. [DOI] [PubMed] [Google Scholar]
- 23.Mdluli, K., J. Swanson, E. Fischer, R. E. Lee, and C. E. Barry. 1998. Mechanisms involved in the intrinsic isoniazid resistance of Mycobacterium avium. Mol. Microbiol. 27:1223-1233. [DOI] [PubMed] [Google Scholar]
- 24.Mizuguchi, Y., T. Uodo, and T. Yamada. 1983. Mechanism of antibiotic resistance in Mycobacterium intracellulare. Microbiol. Immunol. 27:425-431. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee, S., M. Petrofsky, K. Yaraei, L. E. Bermudez, and G. A. Cangelosi. 2001. The white morphotype of Mycobacterium avium-intracellulare is common in infected humans and virulent in infection models. J. Infect. Dis. 184:1480-1484. [DOI] [PubMed] [Google Scholar]
- 26.Nash, K. A. 2003. Intrinsic macrolide resistance in Mycobacterium smegmatis is conferred by a novel erm gene, erm(38). Antimicrob. Agents Chemother. 47:3053-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikaido, H., and V. Jarlier. 1991. Permeability of the mycobacterial cell wall. Res. Microbiol. 142:437-443. [DOI] [PubMed] [Google Scholar]
- 28.Prinzis, S., B. Rivoire, and P. J. Brennan. 1994. Search for the molecular basis of morphological variation in Mycobacterium avium. Infect. Immun. 62:1946-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rastogi, N., K. S. Goh, and S. Clavel-Seres. 1997. Stazyme, a mycobacteriolytic preparation from a Staphylococcus strain, is able to break the permeability barrier in multiple drug resistant Mycobacterium avium. FEMS Immunol. Med. Microbiol. 19:297-305. [DOI] [PubMed] [Google Scholar]
- 30.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 31.Segura, C., M. Salvado, I. Collado, J. Chaves, and A. Coira. 1998. Contribution of β-lactamases to β-lactam susceptibilities of susceptible and multidrug-resistant Mycobacterium tuberculosis clinical isolates. Antimicrob. Agents Chemother. 42:1524-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31a.Semret, M. G. Zhai, S. Mostowy, C. Cleto, D. C. Alexander, G. A. Cangelosi, D. Cousins, D. M. Collins, D. van Soolingen, and M. A. Behr. Extensive genomic polymorphism within Mycobacterium avium. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 32.Sirakova, T. D., V. S. Dubey, H. J. Kim, M. H. Cynamon, and P. E. Kolattukudy. 2003. The largest open reading frame (pks12) in the Mycobacterium tuberculosis genome is involved in pathogenesis and dimycocerosyl phthiocerol synthesis. Infect. Immun. 71:3794-3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voladri, R. K., D. L. Lakey, S. H. Hennigan, B. E. Menzies, K. M. Edwards, and D. S. Kernodle. 1998. Recombinant expression and characterization of the major β-lactamase of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 42:1375-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wanger, A., and K. Mills. 1994. E-test for susceptibility testing of Mycobacterium tuberculosis and Mycobacterium avium-intracellulare. Diagn. Microbiol. Infect. Dis. 19:179-181. [DOI] [PubMed] [Google Scholar]
- 35.Woodley, C. L., and H. L. David. 1976. Effect of temperature on the rate of transparent to opaque colony type transition in Mycobacterium avium. Antimicrob. Agents Chemother. 9:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young, L. S., and L. E. Bermudez. 1996. Animal models in anti-Mycobacterium avium-complex drug development, p. 141-161. In J. A. Korvik and C. A. Benson (ed.), Mycobacterium avium-complex infection: progress in research and treatment. Marcel Dekker, Inc., New York, N.Y.