Abstract
Of 203 human clinical isolates of Campylobacter jejuni from Alberta, Canada (1999 to 2002), 101 isolates (50%) were resistant to at least 64 μg of tetracycline/ml, with four isolates exhibiting higher levels of tetracycline resistance (512 μg/ml). In total, the MICs for 37% of tetracycline-resistant isolates (256 to 512 μg/ml) were higher than those previously reported in C. jejuni (64 to 128 μg/ml). In the tetracycline-resistant clinical isolates, 67% contained plasmids and all contained the tet(O) gene. Four isolates resistant to high levels of tetracycline (MIC = 512 μg/ml) contained plasmids carrying the tet(O) gene, which could be transferred to other isolates of C. jejuni. The tetracycline MICs for transconjugants were comparable to those of the donors. Cloning of tet(O) from the four high-level tetracycline-resistant isolates conferred an MIC of 32 μg/ml for Escherichia coli DH5α. In contrast, transfer to a strain of C. jejuni by using mobilization conferred an MIC of 128 μg/ml. DNA sequence analysis determined that the tet(O) genes encoding lower MICs (64 to 128 μg/ml) were identical to one other, although the tet(O) genes encoding a 512-μg/ml MIC demonstrated several nucleotide substitutions. The quinolone resistance determining region of four ciprofloxacin-resistant isolates (2%) was analyzed, and resistance was associated with a chromosomal mutation in the gyrA gene resulting in a Thr-86-Ile substitution. In addition, six kanamycin-resistant isolates contained large plasmids that carry the aphA-3 marker coding for 3′-aminoglycoside phosphotransferase. Resistance to erythromycin was not detected in 203 isolates. In general, resistance to most antibiotics in C. jejuni remains low, except for resistance to tetracycline, which has increased from about 8 to 50% over the past 20 years.
Campylobacter jejuni is a leading cause of bacterial gastroenteritis (2, 29), a disease condition primarily characterized by diarrhea, abdominal pain, and fever (41). C. jejuni gastroenteritis is primarily self-limiting and is commonly treated by replacing fluids and electrolytes lost through diarrhea (41). Antibiotic treatment may be required in severe clinical infections of C. jejuni, in which case erythromycin is the drug of choice (33), although ciprofloxacin is commonly prescribed for prophylaxis of enteric infections before travel.
Increased antibiotic resistance is being reported in C. jejuni, particularly tetracycline and ciprofloxacin resistance (33). Worldwide, tetracycline resistance (Tcr) frequencies among human isolates of C. jejuni are high; for example, 55 to 56% in North America (12, 32) and up to 95% in Thailand (22). In Alberta, Canada, Tcr rates in human clinical isolates of C. jejuni were 6.8 and 8.6% in 1980 and 1981, respectively (45). Ciprofloxacin resistance frequencies in C. jejuni have increased dramatically in the last few decades, approaching 88% in Spain (38). Fortunately, the prevalence of erythromycin resistance has remained low, often well below 10% of isolates (34). However, a recent Canadian study has identified a sudden increase in erythromycin resistance to 12% (11).
A number of antibiotic resistance mechanisms are present in C. jejuni. Tcr is primarily mediated by a plasmid-encoded tet(O) gene (49). Tet(O), a ribosomal protection protein, confers resistance by displacing tetracycline from its primary binding site on the ribosome (4, 5, 52). Previous studies have determined that the tet(O) gene in C. jejuni mediates MICs of up to 128 μg of tetracycline/ml (45). Tcr plasmids from C. jejuni are currently being sequenced by other groups and appear to be highly conserved, as a plasmid isolated in the late 1970s was virtually identical in its DNA sequence to a plasmid isolated in 2000 (R. Bachelor, B. Pearson, L. Friis, P. Guerry, and J. Wells, Abstr. 12th Int. Workshop Campylobacter, Helicobacter, Related Organisms, abstr. F-39, p. 48, 2003). Kanamycin resistance (Kmr) in C. jejuni is most frequently associated with the existence of the aphA-3 gene which is identified in most cases on large plasmids in the range of 40 to 130 kb. Resistance to erythromycin is most likely due to an alteration of the target site on the 23S rRNA of the C. jejuni ribosome (18, 53). Ciprofloxacin resistance depends on mutations within the gyrA gene, which encodes the A subunit of the DNA gyrase enzyme. A single point mutation at Thr-86, Asp-90, or Ala-70 in gyrA can result in fluoroquinolone resistance (53, 56).
Plasmid content in human isolates of C. jejuni varies from 13 to 52%, with the majority being resistance plasmids (21, 39, 44, 50, 51). Conjugative transfer of the Tcr plasmids has been demonstrated between Campylobacter species but not to Escherichia coli, suggesting that their host range is restricted (47, 48). Plasmids have also been implicated in the virulence of C. jejuni, as Bacon et al. (1) have identified a role for plasmid pVir in the pathogenesis of C. jejuni strain 81-176.
The goal of this study was to determine the prevalence of resistance to tetracycline and to other antibiotics among clinical isolates of C. jejuni obtained from patients in Alberta, Canada, between 1999 and 2002. The plasmid content of the Tcr isolates was examined, and the mechanisms responsible for resistance to tetracycline and other antibiotics in C. jejuni isolates were characterized.
MATERIALS AND METHODS
Campylobacter isolates used in this study.
Clinical isolates of C. jejuni were obtained from the Provincial Laboratory of Public Health (Microbiology) in Edmonton, Alberta, Canada, and included fecal isolates stored frozen in 1999-2001 (n = 193), as well as fresh isolates collected in a prospective manner in 2002 (n = 10). Various C. jejuni isolates for which the tetracycline MICs had previously been characterized were used as controls in the agar dilution assays (UA56, UA143, UA183).
Growth and storage conditions.
Campylobacters were routinely cultured on brain heart infusion agar (Difco, Beckton-Dickinson, Sparks, Mass.), supplemented with 0.4% yeast extract (Difco), and incubated at 37°C in microaerobic conditions (5% CO2, 10% H2, balance N2) for 48 h. Isolates were stored frozen in brain heart infusion broth (Difco) with 20% glycerol at −80°C. E. coli strains containing plasmids which carried tet(A) and tet(K) were cultured in Luria-Bertani (LB) broth containing 100 μg of ampicillin/ml at 37°C, whereas strains containing plasmids carrying tet(B), tet(C), tet(D), tet(G), tet(E), and tet(L) were cultured in LB broth containing 10 μg of tetracycline/ml at 37°C.
DNA manipulations.
Molecular biological techniques were performed as previously described (40). Restriction enzymes were used according to the manufacturer's instructions.
DNA sequencing.
DNA samples were prepared for sequencing using a BigDye Terminator v. 3.0 ready reaction cycle sequencing kit (ABI Prism; A & B Applied Biosystems, Foster City, Calif.). Sequencing of DNA samples was performed by the Unit of Molecular Biology Service (Faculty of Science, University of Alberta, Edmonton, Alberta, Canada). Sequencing primers are listed in Table 1. DNA sequence analysis was performed using the GENETYX-WIN (version 5.1.) software.
TABLE 1.
Primers used in this study
| Primer | Sequencea | Purpose |
|---|---|---|
| DMT 1 | 5′ GGCGTTTTGTTTATGTGCG 3′ | Amplification of a 559-bp fragment of tet(O) (5′ end) |
| DMT 2 | 5′ ATGGACAACCCGACAGAAGC 3′ | Amplification of a 559-bp fragment of tet(O) (3′ end) |
| CAT 5 | 5′ TATATGAATTCAATGAAAATTATTAATATTGGAG 3′ | Amplification of tet(M) |
| CAT 6 | 5′ TATATGGATCCACTAAGTTATTTTATTGAAC 3′ | Amplification of tet(M) |
| tetA F | 5′ TTCTCTATATCGGGCGGATCGTGGC 3′ | Amplification of ∼700-bp fragment of tet(A) gene |
| tetA R | 5′ CCACCCGAAGCAAGCAGGACCATG 3′ | Amplification of ∼700-bp fragment of tet(A) gene |
| tetB F | 5′ CCTTATCATGCCAGTCTTGCCAACG 3′ | Amplification of ∼900-bp fragment of tet(B) gene |
| tetB R | 5′ CCTGTAAAGCACCTTGCTGATGACTC 3′ | Amplification of ∼900-bp fragment of tet(B) gene |
| tetE F | 5′ CTGGTCAGATCGCATAGGTCGTCG 3′ | Amplification of ∼1-kb fragment of tet(E) gene |
| tetE R | 5′ CCATACCCATCCATTCCACGTTTCGC 3′ | Amplification of ∼1-kb fragment of tet(E) gene |
| aphA-3 F | 5′ GGGACCACCTATGATGTGGAACG 3′ | Amplification of 600 bp of aphA-3 gene |
| aphA-3 R | 5′ CAGGCTTGATCCCCAGTAAGTC 3′ | Amplification of 600 bp of aphA-3 gene |
| DOB 3 | 5′ TATATGAATTCAATGAAAATAATTAACTTAGGCATTC 3′ | Cloning of tet(O) into pMS119EH (5′ end) |
| SEAN 20 | 5′ TATATGGATCCTTAAGCTAACTTGTGGAACATATGCC 3′ | Cloning of tet(O) into pMS119EH (3′ end) |
| DMT 27 | 5′ GGCATTCTGGCTCACGTTGACGC 3′ | Sequencing of tet(O) |
| SEAN 5 | 5′ ACTGCTCCGTCTAATACG 3′ | Sequencing of tet(O) |
| SEAN 6 | 5′ CAGAACTGGAACAGGAAG 3′ | Sequencing of tet(O) |
| SEAN 9 | 5′ ATGCACCGCAGGAATATC 3′ | Sequencing of tet(O) |
| DMT 29 | 5′ GTGAAGCAAAAGGTTGGGCAGC 3′ | Sequencing of tet(O) |
| DMT 30 | 5′ GCAGACTTTCGGCTGCTTTC 3′ | Sequencing of tet(O) |
| DMT 50 | 5′ CTGCGGCAACAGTATTTCG 3′ | Sequencing of tet(O) |
| DMT 20 | 5′ TATAAGCGCTGGATGAGGAGGCAGATTGCC 3′ | Cloning of aphA-3 kanamycin resistance cassette into tet(O) |
| DMT 21 | 5′ TATAAGCGCTCTAAAACAATTCATCCAG 3′ | Cloning of aphA-3 kanamycin resistance cassette into tet(O) |
| DMT 22 | 5′ TATAGGATCCAATGAAAATAATTAACTTAGGCATTC 3′ | Cloning of tet(O) ORF into pRY107 (5′ end) |
| DMT 23 | 5′ TATAGGATCCCTGTCAATTTGATAGTGGGAAC 3′ | Cloning of tet(O) ORF and its P1 promoter into pRY107 (5′ end) |
| DMT 24 | 5′ TATAGGATCCGCATAAACAGATGATTAGTGG 3′ | Cloning of tet(O) ORF and its P1/P2 promoters into pRY107 (5′ end) |
| DMT 25 | 5′ TATAGGATCCGATATCCACTTGGCTTTATC 3′ | Cloning of tet(O) ORF and a ∼1000-bp upstream region into pRY107 (5′ end) |
| DMT 26 | 5′ TATAGAATTCTTAAGCTAACTTGTGGAACATATGCC 3′ | Cloning of tet(O) into pRY107 (3′ end) |
| 16S F1 | 5′ TAAGTGATCGATTGAGCCAGAAAC 3′ | Cloning of 16S rRNA genes of C. jejuni |
| 16S R1 | 5′ GCTAATTCCCCATAAACAATTAGC 3′ | Cloning of 16S rRNA genes of C. jejuni |
| T7 (F) | 5′ GTAATACGACTCACTATAGGGC 3′ | Sequencing of 16S rRNA genes of C. jejuni for vector |
| 16S F2 | 5′ ACACGGTCCAGACTCCTA 3′ | Sequencing of 16S rRNA genes of C. jejuni |
| 16S F3 | 5′ GATTAGATACCCTGGTAGTC 3′ | Sequencing of 16S rRNA genes of C. jejuni |
| 16S F4 | 5′ AGTCCCGCAACGAGCGCAA 3′ | Sequencing of 16S rRNA genes of C. jejuni |
| r3L | 5′ TTGCGCTCGTTGCGGGACT 3′ | Sequencing of 16S rRNA genes of C. jejuni for vector |
| T3 (R) | 5′ AATTAACCCTCACTAAAGGG 3′ | Sequencing of 16S rRNA genes of C. jejuni |
Restriction enzyme sites are underlined.
Antibiotic susceptibility testing and MICs of tetracycline.
Antibiotic disks (Oxoid, Nepean, Ontario, Canada) were used in the disk diffusion method: tetracycline (30 μg), kanamycin (30 μg), erythromycin (15 μg), chloramphenicol (30 μg), and nalidixic acid (30 μg). All nalidixic acid-resistant isolates were further tested for resistance to ciprofloxacin (1 μg). Susceptibility testing was performed as previously described (13). MICs for Tcr isolates were determined by the agar dilution method as previously described (13).
Isolation of bacterial plasmids.
Plasmids were isolated by Mini or Midi plasmid kits (QIAGEN, Mississauga, Ontario, Canada), by the alkaline lysis method (40) or by a modified protocol for Helicobacter pylori (6). Plasmid DNA concentrations were quantified using a spectrophotometer (Ultrospec 3000; Pharmacia Biotech, Piscataway, N.J.) and by gel electrophoresis (Bio-Rad, Mississauga, Ontario, Canada).
Transfer of tetracycline resistance plasmids between C. jejuni isolates.
The transfer of plasmids from Tcr C. jejuni clinical isolates was performed as described previously by Taylor et al. (47). Plasmids were isolated from the transconjugants using a QIAGEN Mini kit.
PCR screening for tet(O) and tet(M).
Primers DMT 1 and DMT 2 (Invitrogen, Burlington, Ontario, Canada) (Table 1) were used to amplify a 559-bp product of the tet(O) gene. PCR templates included plasmid DNA, total DNA, and a boiled whole-cell preparation. Plasmid DNA was isolated as described above, and total DNA was isolated with a Wizard genomic DNA isolation kit (Promega, Madison, Wis.). Boiled whole cells were prepared by boiling a loopful of cells in 100 μl of Tris-EDTA buffer, which was then centrifuged, and the supernatant was used for PCR. A reaction mixture containing the oligonucleotide primers at 0.5 μM each; dATP, dCTP, dGTP, and dTTP at 200 μM each; 1× reaction buffer (50 μM KCl, 10 mM Tris-HCl [pH 8.3], 1.5 mM MgCl2); 0.01% (wt/vol) gelatin; 1 U of Taq polymerase (Perkin-Elmer Cetus, Norwalk, Conn.); and the template, as mentioned above, was used for PCR amplification. PCR conditions were as follows: an initial denaturation of 95°C for 1 min, and then 95°C for 1 min, 50°C for 1 min, and 72°C for 1 min, repeated for 30 cycles. Primers CAT 5 and CAT 6 (Invitrogen) (Table 1) were used to amplify the tet(M) gene from plasmids isolated from high-level Tcr isolates. PCR was performed in a Bio-Rad gene cycler (Bio-Rad).
Cloning of the tet(O) gene.
Primers DOB 3 and SEAN 20 (Invitrogen) (Table 1) were used to amplify the tet(O) gene (1.92 kb) from a representative Tcr plasmid isolated from C. jejuni clinical isolates for which the tetracycline MICs were at the following levels: 64, 128, and 256 to 512 μg/ml. A Pfx-PCR kit (Invitrogen) was used for all PCR amplifications of tet(O) for cloning. PCR conditions were as follows: denaturation for 30 s at 94°C, annealing for 45 s at 55°C, and extension for 2 min at 68°C, repeated for 30 cycles. The tet(O) PCR products were purified using a QiaQuick PCR purification kit (QIAGEN), cloned into pMS119EH (Table 2) with EcoRI (Gibco) and BamHI (Gibco), and transformed into E. coli DH5α, which had been made competent by calcium chloride treatment (40). Tetracycline MICs were determined for the transformants as described above. The tet(O) genes were sequenced as described above but IPTG (isopropyl-β-d-thiogalactopyranoside) (0.1 mM; Rose Scientific, Edmonton, Alberta, Canada) was added to tetracycline plates to induce expression from the pMS119EH vector.
TABLE 2.
Plasmids used in this study
| Plasmid | Relevant characteristics | Selective marker(s)a | Reference or Source |
|---|---|---|---|
| pJI3 | pACYC177 carrying tet(M) | Tc | 3 |
| pMS119EH | Cloning vector | Amp | 42 |
| pRY107 | Shuttle vector | Km | 59 |
| pDOB29 | tet(O) from 25-01 cloned into pMS119EH | Tc, Amp | This study |
| pDOB25 | tet(O) from 16-42 cloned into pMS119EH | Tc, Amp | This study |
| pDOB27 | tet(O) from 23-51 cloned into pMS119EH | Tc, Amp | This study |
| pDOB31 | tet(O) from 23-49 cloned into pMS119EH | Tc, Amp | This study |
| pDOB35 | tet(O) from 25-44 cloned into pMS119EH | Tc, Amp | This study |
| pDOB34 | tet(O) from 25-54 cloned into pMS119EH | Tc, Amp | This study |
| pDOB42 | Kanamycin resistance cassette inserted into tet(O) of pDOB29 | Amp | This study |
| pDOB49 | tet(O) ORF cloned into pRY107 | Tc, Amp | This study |
| pDOB45 | tet(O) ORF with promoter P1 cloned into pRY107 | Tc, Amp | This study |
| pDOB50 | tet(O) ORF with promoters P1 and P2 cloned into pRY107 | Tc, Amp | This study |
| pDOB47 | tet(O) ORF with a ∼1000-bp upstream region cloned into pRY107 | Tc, Amp | This study |
| pSL18 | Contains the tet(A) gene | Amp | 30 |
| pRT11 | Contains the tet(B) gene | Tc | 27 |
| pBR322 | Contains the tet(C) gene | Tc | 27 |
| pSL106 | Contains the tet(D) gene | Tc | 27 |
| pSL1504 | Contains the tet(E) gene | Tc | 26 |
| pJA8122 | Contains the tet(G) gene | Tc | 60 |
| pAT102 | Contains the tet(K) gene | Amp | 58 |
| pVBA15 | Contains the tet(L) gene | Tc | 28 |
Antibiotic abbreviations: Tc, tetracycline; Amp, ampicillin; Km, kanamycin.
Cloning of the tet(O) gene into pRY107 shuttle vector.
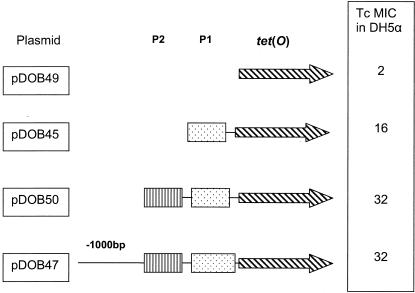
The tet(O) gene from a high-level Tcr isolate was cloned into the C. jejuni shuttle vector pRY107 (59). Four different clones were made, incorporating increasing portions of the tet(O) promoter region (Fig. 1). A Pfx-PCR kit (Invitrogen) was used to amplify the tet(O) open reading frame (ORF) and the varying upstream regions, which were cloned into the multiple cloning site of the LacZ gene in pRY107, which was then transformed into E. coli DH5α and selected on LB broth with kanamycin (Sigma) and X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Rose Scientific). White colonies were screened for the insertion of the PCR product of the desired size, and plasmids were isolated with a QIAGEN Midi kit. Tetracycline MICs were determined for strains containing the tet(O) clones in E. coli DH5α.
FIG. 1.
Cloning of the tet(O) gene and segments of its upstream region from C. jejuni 25-01 into C. jejuni-E. coli shuttle expression vector pRY107. Primer combinations are listed in Table 1. The tet(O) gene is designated by an arrow with black diagonal lines, promoters P1 and P2 are designated by dotted and lined boxes, respectively, and a line designates an undefined upstream region (−1000 bp). The tetracycline MIC is in micrograms per milliliter.
Mobilization of the cloned tet(O) gene into C. jejuni.
Plasmid pDOB47 (Table 2) was transformed into the conjugative strain E. coli S17.1 (7) which had been made competent by calcium chloride treatment (40). The transformants were selected on LB plates containing 50 μg of kanamycin/ml. A number of the resulting transformants were screened to ensure the transfer of pDOB47 plasmid. One of the transformants was subsequently used as a donor in a conjugal mating to mobilize pDOB47 plasmid from E. coli S17.1 to a susceptible isolate of C. jejuni, UA543. The mating experiment between E. coli and C. jejuni was performed as described previously (19). The selection of C. jejuni transconjugants was performed using LB plates containing 50 μg/ml each of kanamycin and nalidixic acid. Plates were incubated at 37°C in microaerobic conditions for 48 h.
The effect of the overexpression of the tet(O) gene on the growth of C. jejuni.
A growth experiment was performed to assess the effect of the expression of the tet(O) gene on the growth of two isolates of C. jejuni: the first has a single copy of the gene on the chromosome, whereas the second isolate carries the tet(O) gene on the high-copy-number vector, pRY107. A 24-h culture of each isolate was inoculated in 40 ml of Mueller-Hinton broth (BBL; Beckton-Dickinson, Cockeysville, Md.) to get an optical density of 0.015 at 625 nm using a spectrophotometer (Ultrospec 3000; Pharmacia Biotech). Each broth culture was divided into two equal portions, and tetracycline was added to one portion to get a final concentration of 50 μg/ml, whereas the other portion was left free from tetracycline. All cultures were incubated at 37°C with shaking (140 rpm) under microaerobic atmosphere for 48 h. Sampling of 0.2 ml from each culture was done at 0, 5, 22, 28, and 48 h. The number of viable cells in each sample was estimated by spreading of an aliquot of the appropriate dilutions onto Mueller-Hinton agar plates. The plates were subsequently incubated at 37°C under microaerobic atmosphere for 48 h, and the numbers of viable cells were estimated as CFU. The experiment was carried out in triplicate.
Identification of the kanamycin resistance marker.
PCR amplification was performed to detect the aphA-3 marker in Kmr C. jejuni isolates. The oligonucleotide primers (aphA-3 F and aphA-3 R; Table 1) chosen for amplification were based on the sequence which has been published previously (55). Amplification of the aphA-3 gene was expected to result in an amplicon of approximately 600 bp. The plasmid DNA (100 ng) was used as the template for PCR amplification. The PCR mixture was prepared as mentioned above, and the amplification was carried out in 50-μl reaction volumes. A positive and a negative control were included in each PCR run. Thirty cycles of amplification were performed, and each cycle consisted of a 0.5-min denaturation at 95°C, a 1.0-min annealing step at 55°C, and a 1.0-min extension step at 72°C. PCR products were purified by a PCR purification kit (QIAGEN) and sequenced as mentioned above.
Analysis of the QRDR.
The quinolone resistance determining region (QRDR) of the gyrA gene of the resistant isolates was amplified by PCR. The primers and the method were described previously (56). The templates for PCR were prepared using a Wizard genomic DNA isolation kit (Promega). PCR products were purified by a PCR purification kit (QIAGEN) and sequenced as mentioned above.
Cloning of 16S rRNA gene.
Primers 16S F1 and 16S R1 (Table 1) were constructed to amplify, by PCR, the three 16S rRNA genes of C. jejuni based on the sequence of the complete genome of C. jejuni (GenBank accession number AL111168). The genomic DNA (50 ng) of each of the isolates 16-42, 23-51, 25-01, and 25-10 was used for PCR, and the amplification was performed using the Pfu DNA polymerase (Invitrogen) to minimize errors. The conditions were 94°C for 3 min, followed by 30 cycles of 94°C for 30 s, 42°C for 45 s, and 68°C for 2 min. PCR products (1,628 kb) were purified, and the ends of purified PCR products were polished to generate blunt ends and cloned into pPCR-Script Cam SK(+) cloning vector using a PCR-Script Cam cloning kit (Stratagene). The ligation was performed according to the manufacturer's instructions.
Screening for other tetracycline resistance genes in high-level tetracycline-resistant C. jejuni.
To test if the Tcr genes could be associated with an efflux mechanism, multiplex PCR was performed according to the method of Ng et al. (35). E. coli strains carrying Tcr plasmids pSL18, pRT11, pBR322, pSL106, pSL1504, pJA8122, pAT102, and pVB.A15 (35) were used as positive controls. To individually amplify tet(A), tet(B), and tet(E) genes, PCR primers (Table 1) were designed based on the deposited sequences of these genes in the database (accession numbers X61367, AB089595, and L06940, respectively). The primers were expected to amplify about 700 bp, 900 bp, and 1 kb of tet(A), tet(B), and tet(E) genes, respectively. The composition of the PCR mixture used for amplification was similar to that described previously for the amplification of the tet(O) and tet(M) genes, and total genomic DNA was used as the template. Twenty-five cycles of amplification were performed, and each cycle consisted of a 1-min denaturation at 95°C, a 1.0-min annealing step at 54°C for tet(A) and tet(E) or at 47°C for tet(B), and a 1.5-min extension step at 72°C. E. coli strains carrying Tcr plasmids pSL18, pRT11, and pSL1504 (Table 2) were used as positive controls during the amplification.
Statistical analysis.
Tcr frequencies in this study and a previous study from Alberta, Canada (45), were compared by the Fisher exact test.
Nucleotide sequence accession number.
The corrected nucleotide sequence of the tet(O) gene was deposited in GenBank under accession number M18896.
RESULTS
Antibiotic resistance rates in C. jejuni clinical isolates.
Antibiotic susceptibility testing determined that 49.8% of C. jejuni clinical isolates (101 of 203) were resistant to tetracycline. No isolates were resistant to chloramphenicol or erythromycin. Six were resistant to kanamycin (2.9%) and another five to nalidixic acid (2.5%). Isolates of C. jejuni are often resistant to both ciprofloxacin and nalidixic acid, as resistance to both antibiotics is mediated by mutations in the gyrA subunit of the DNA gyrase enzyme (8). Ciprofloxacin resistance was found in four of the five nalidixic acid-resistant isolates, for an overall rate of 2%.
Characterization of kanamycin and quinolone resistance in C. jejuni.
Previous studies showed that the Kmr in C. jejuni is most frequently due to the presence of the aphA-3 marker which is usually located on large plasmids (20, 39), therefore the isolates were screened for the presence of plasmid DNA. Although one of the isolates contained two plasmids, each of the other isolates contained a single plasmid. The sizes of the plasmids ranged from 30 to 60 kb. PCR amplification was used to check for the existence of the aphA-3 gene on the isolated plasmids. In all cases, a PCR amplicon (about 600 bp), which is specific for the aphA-3 gene, was observed.
The QRDR of the gyrA gene of the five quinolone-resistant isolates all showed the C-to-T transition at nucleotide 256 resulting in substitution of Ile for Thr which was previously observed to mediate high quinolone resistance in C. jejuni (24, 37).
Characterization of tetracycline resistance in C. jejuni.
The agar dilution assay determined that the MICs for all the Tcr isolates were 64 to 512 μg/ml, with a significant percentage (37%) of isolates displaying high-level Tcr (256 to 512 μg/ml). A number of control isolates of C. jejuni for which the tetracycline MIC levels were known were used throughout the agar dilution assays, and the MIC levels were consistent with previously measured results (45).
Plasmid isolations were attempted on 141 of the 203 isolates, including all Tcr isolates (n = 101) and 40 of the tetracycline-susceptible isolates. Several methods for plasmid isolation were used to confirm the presence and/or absence of plasmids. Overall, plasmids were isolated from 70 of the 141 isolates (50%), and of these, Tcr isolates had a plasmid content of 67% (68 of 101), whereas tetracycline-susceptible isolates had a plasmid content of 5% (2 of 40). In 67% of the Tcr isolates, the resistance phenotype was associated with the presence of a ∼40- to 50-kb plasmid. In tetracycline-susceptible isolates, plasmid size varied from approximately 3 to 100 kb, with some isolates containing up to four plasmids (data not shown).
A PCR screen (using the DMT 1-DMT 2 primer pair; Table 1) on plasmid DNA, boiled whole cells, or total genomic DNA identified the tet(O) gene in all Tcr C. jejuni isolates, including all high-level resistant isolates (tetracycline MIC, 512 μg/ml; data not shown). In 67% of the resistant isolates, the tet(O) gene was located on a plasmid, whereas positive results in total DNA preparations were found in 33% of the isolates lacking plasmids, suggesting that the tet(O) gene can be located on the C. jejuni chromosome. By using representative isolates, conjugation was performed to confirm the existence of the tet(O) gene on the plasmid or on the chromosome. Eight representative isolates (Table 3), in which the tet(O) gene was detected on plasmids, were used as donors in a conjugation experiment to assess the transfer of Tcr plasmid to a recipient isolate of C. jejuni (UA543). Transfer frequencies of 10−4 to 10−5 transconjugant per recipient were determined (Table 3). Attempts to transfer Tcr plasmids from C. jejuni isolates to E. coli were unsuccessful. The MICs for resulting transconjugants were comparable to those for the donor isolates (Table 3). Isolation of plasmid DNA from the Tcr transconjugants and investigation of the banding pattern of plasmid, after cleavage with BglII enzyme, revealed that the transferred Tcr plasmid is in the range of 40 to 50 kb (data not shown). The tet(O) gene was also detected on the isolated plasmids by PCR. In isolates 16-14, 16-68, 24-44, and 24-53 (Table 3), the Tcr transconjugants were also resistant to kanamycin, indicating the cotransfer of both Tcr and Kmr phenotypes during conjugation. The existence of the Kmr marker, aphA-3, was also verified by PCR amplification. On the other hand, six representative isolates, showing the existence of the tet(O) gene on the chromosome, were used as donors in a conjugation experiment. MICs for these isolates were 128 to 256 μg of tetracycline/ml. Repeating the conjugation three times demonstrated no transfer of the Tcr phenotype to the recipient isolate, confirming that the resistance marker is likely located on the chromosome.
TABLE 3.
Transfer of tetracycline resistance plasmids from C. jejuni clinical isolates to UA543, a tetracycline-susceptible C. jejuni recipient strain
| C. jejuni donor isolate | Tetracycline MIC (μg/ml) for donor | Transfer frequency | Tetracycline MIC (μg/ml) for transconjugant |
|---|---|---|---|
| 16-42 | 512 | 3.8 × 10−4 | 512 |
| 23-51 | 512 | 3.8 × 10−5 | 512 |
| 25-01 | 512 | 1.3 × 10−4 | 512 |
| 25-10 | 512 | 7.6 × 10−5 | 512 |
| 16-68 | 256 | 0.1 × 10−4 | 128 |
| 16-14 | 128 | 0.5 × 10−5 | 128 |
| 24-44 | 128 | 1.5 × 10−4 | 128 |
| 24-53 | 128 | 6.0 × 10−4 | 128 |
Cloning of the tet(O) gene has previously been accomplished (25), and the tetracycline MICs for both the original C. jejuni isolate and the E. coli clones were comparable (43). The tetracycline MIC for the C. jejuni isolate from which the original tet(O) gene was cloned (strain UA466) was 64 μg/ml in E. coli. To determine if the tet(O) genes from C. jejuni isolates expressing variable levels of Tcr were able to mediate different levels of Tcr in E. coli, the tet(O) gene was cloned from representative resistant isolates for which tetracycline MICs were variable (64, 128, 512 μg/ml) into pMS119EH vector. Tetracycline MICs were determined for the tet(O) clones in E. coli DH5α. Although MIC levels differ for the C. jejuni hosts from which the tet(O) genes were cloned, they mediated identical levels of Tcr in E. coli as shown in Table 4.
TABLE 4.
Tetracycline MICs for C. jejuni and for the tet(O) genes cloned into pMS119EH in E. coli
| C. jejuni clinical isolate | Tetracycline MIC (μg/ml) for C. jejuni | Tetracycline MIC (μg/ml) for E. coli DH5α | Reference |
|---|---|---|---|
| 25-01 | 512 | 32 | This study |
| 23-49 | 128 | 32 | This study |
| 25-44 | 64 | 32 | This study |
| 25-54 | 64 | 32 | This study |
Investigation of the genetic basis of the high-level tetracycline resistance.
In 4 of the 101 Tcr isolates, plasmids that harbored the tet(O) gene were shown to mediate a relatively high-level Tcr (512 μg/ml; Table 3) compared to the previously detected Tcr levels mediated by the tet(O) gene in C. jejuni isolates (45). These isolates were chosen for further study to investigate the molecular basis of the observed high-level resistance. To examine if the higher level of Tcr is attributed to mutations in the tet(O) gene, the tet(O) genes (1.92 kb) from the four C. jejuni clinical isolates (tetracycline MICs, 512 μg/ml) and also from three isolates for which tetracycline MICs were lower (64 and 128 μg/ml) were cloned into pMS119EH vector (Table 2), and the complete DNA sequencing of the cloned genes was determined and analyzed. The tet(O) gene from all four Tcr isolates (MIC, 512 μg/ml) had seven nucleotide changes that differed from the GenBank sequence of the tet(O) gene (Table 5). This resulted in the substitution of seven amino acid residues of the Tet(O) protein (Table 5). All four isolates (tetracycline MIC, 512 μg/ml) exhibited the same seven substitutions. On the other hand, the tet(O) genes mediating lower Tcr levels (64 and 128 μg/ml) were found to have DNA sequences identical to that of the reference tet(O) gene in GenBank and they did not show any of the seven nucleotide changes (Table 5). All of the sequenced tet(O) genes, however, had an identical base substitution at position 1772 and different terminal 3′ sequences than the GenBank tet(O) sequence. Analysis of this data and of other sequence data for tet(O) (S. Connell and L. Nonaka, unpublished data) has determined that the GenBank tet(O) sequence is incorrect at the 3′ end, although this error has now been corrected (GenBank accession number M18896).
TABLE 5.
Nucleotide and amino acid substitutions in the tet(O) genes of the tetracycline-resistant C. jejuni isolates for which the MIC level is 512 μg/ml
| Nucleotide position | DNA sequence variations in the tet(O) from:
|
Amino acid substitutions in the tet(O) from:
|
||
|---|---|---|---|---|
| GenBank | Highly resistant isolates | GenBank | Highly resistant isolates | |
| 884 | A | G | Y | C |
| 910 | T | C | S | P |
| 993 | A | G | I | M |
| 1036 | A | C | I | L |
| 1063 | T | C | S | P |
| 1111 | T | G | C | G |
| 1772a | C | A | T | N |
| 1784 | A | G | Y | C |
Mutation at nucleotide position 1772 is found in all tet(O) sequences in a number of studies, indicating an error in the initial GenBank sequence that has now been corrected (accession number M18896).
The upstream region of tet(O) is known to be important for full expression of Tcr (57). To determine if the upstream region of the tet(O) gene played a role in mediating high-level Tcr, the tet(O) gene was cloned from C. jejuni clinical isolate 25-01 (tetracycline MIC = 512 μg/ml) incorporating increasing portions of its upstream region into the shuttle vector pRY107 (Fig. 1). The inclusion of the P2 promoter with the tet(O) gene in the pRY107 vector was found to mediate a higher level of Tcr (32 μg/ml) than constructs containing only P1 or the tet(O) ORF only (16 and 2 μg/ml, respectively), confirming the importance of the P2 promoter of the tet(O) gene in mediating Tcr in E. coli. The plasmid pDOB47 (Table 2) was mobilized by the conjugative strain E. coli S17.1, containing an IncP plasmid integrated into the chromosome (7), into a susceptible isolate of C. jejuni (UA543) at a frequency of 10−5 transconjugant per recipient. The transfer of plasmid pDOB47 to C. jejuni was confirmed by isolation of plasmid DNA from at least three transconjugants. The resulting transconjugants showed a Tcr level of 128 μg/ml, which was lower than the corresponding resistance level of the original host 25-01 (Table 3) but higher than those in E. coli.
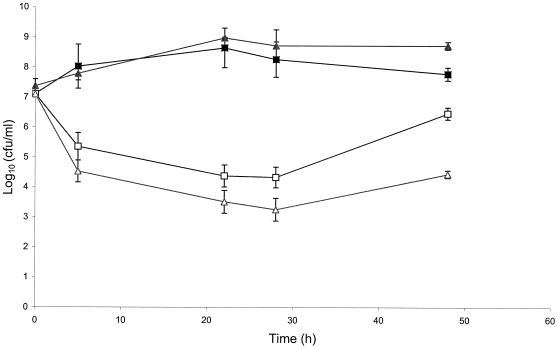
To investigate the effect of the overexpression of the tet(O) gene cloned into the multicopy vector (pRY107) on the viability of C. jejuni, a growth experiment was performed. Unfortunately, Campylobacter shuttle vectors of low copy numbers are not available for experimental use. Therefore, the growth of one of the Tcr transconjugants resulting from the mobilization experiment, where the tet(O) gene existed in a high copy number, was compared (over a period of 48 h) to that of an isolate that has a single copy of the tet(O) gene on the chromosome. In the absence of tetracycline selection pressure, both isolates showed comparable growth patterns (Fig. 2). In the presence of 50 μg of tetracycline/ml, the growth of both isolates was inhibited but the degree of inhibition was greater when the tet(O) gene existed in a high copy number, indicating that the overexpression of the tet(O) gene might exhibit a negative effect on the growth of C. jejuni.
FIG. 2.
The effect of overexpression of the high-copy-number tet(O) gene on the viability of C. jejuni over a period of 48 h. Shown are the growth curves, in the absence of tetracycline, of an isolate carrying the tet(O) gene on the chromosome (closed squares) and with 50 μg/ml of tetracycline (open squares), and the growth curve, in the absence of tetracycline, of an isolate that has a high copy number of the tet(O) gene (closed triangles) and with 50 μg of tetracycline/ml (open triangles). Viable bacterial counts were determined at time 0 and after 5, 22, 28, and 48 h of incubation. Mean values from triplicate measurements are shown. Bars indicate the standard deviations.
To exclude the possibility that the Tcr plasmid in the highly resistant isolates has another Tcr marker in addition to the tet(O) gene, PCR amplification was performed to assess the occurrence of the tet(M) gene in the four isolates. None of the high-level Tcr isolates contained tet(M) (Data not shown). Multiplex PCR was also performed to assess the existence of any of the known tetracycline efflux genes. It was suggested by PCR that the tet(A), tet(B), or tet(E) gene might exist in the highly resistant isolates; however, when each of the three genes was amplified individually the result was negative, excluding the possibility of the existence of any of the previously characterized efflux genes in these isolates.
In H. pylori, mutations in 16S rRNA have been shown to be responsible for Tcr (53, 54). The sequence of the 16S rRNA genes of the four C. jejuni isolates, which exhibited a resistance level of 512 μg/ml, was investigated to examine if the 16S rRNA genes show mutation(s) which might be associated with the high level of resistance. The 16S rRNA genes of the four isolates were amplified by PCR and sequenced. The 16S rRNA genes were amplified using primers based upon the published sequences of C. jejuni 16S rRNA genes and did not distinguish among the three copies of the genes on the chromosome. A single sequence was obtained for the 16S rRNA genes of each isolate, indicating that the genes in these isolates were homogeneous. The 16S rRNA sequence of isolates 16-42, 23-51, 25-01, and 25-10 were compared to the published C. jejuni 16S rRNA sequences (data not shown); however, no significant difference was detected. Trieber and Taylor (54) demonstrated that mutations in the 16S rRNA gene at positions 965 to 967 conferred Tcr on H. pylori 26695. Neither this triple mutation nor any other change in these three nucleotides from the wild-type sequence was detected in any of the four high-level Tcr C. jejuni isolates (MIC, 512 μg/ml). This indicates that the higher level of Tcr is not related to mutations in 16S rRNA genes.
DISCUSSION
Increasing antibiotic resistance frequencies in C. jejuni are of concern, as antibiotic treatment of severe infections may be rendered ineffective by resistant isolates. In Canada, macrolides are registered for use in food animals; however, little data concerning the frequency and average duration of use of these drugs are available (16). Health Canada is supporting surveillance activities to evaluate possible public health impacts of the veterinary use of antimicrobials, including macrolides. Evidence from the surveillance data is currently being collected and analyzed by Health Canada and will be crucial in the development of new policies and approaches (16). The use of quinolones (mainly enrofloxacin) in veterinary practice has been correlated in different countries with the increase in ciprofloxacin resistance in Campylobacter strains (9, 36). In Canada, licensing of the veterinary use of fluoroquinolones was withdrawn in 1997 (10). This study showed that resistance frequencies in C. jejuni to erythromycin and ciprofloxacin, the major antibiotics used in the treatment of C. jejuni and bacterial gastroenteritis, have apparently not increased in Alberta over the past 20 years. These results confirm other Canadian studies that have identified low levels of erythromycin resistance (12, 15). Low rates of erythromycin resistance have continued to occur in clinical isolates of C. jejuni in Alberta, Canada, since the early 1980s (45). This is comforting news for clinicians, as erythromycin is the drug of choice in treating C. jejuni gastroenteritis and its efficacy has not been compromised by resistance. Similar results were found for ciprofloxacin, for which resistance was at only 2% (4 out of 203 isolates). In contrast, susceptibility data from other countries demonstrates that ciprofloxacin resistance in C. jejuni is emerging at a rapid pace, with some countries now reporting resistance levels of up to 30% (23, 34, 38). This study has shown that in Alberta, Canada, the incidence of Tcr in C. jejuni has increased significantly to 49.8% in 1999-2002 from 8.6% in 1981 (45). A study by Gaudreau and Gilbert (12) in Quebec, Canada, has shown 53% of C. jejuni clinical isolates are Tcr, up from only 19% in 1992-1993. Recently, human isolates of C. jejuni subsp. jejuni (isolated from 1998 to 2001 in Quebec, Canada) showed variable annual rates of resistance to tetracycline from 43 to 68% (11). Thus, the considerable increase in the incidence of Tcr is a trend that is occurring across Canada.
Multidrug resistance is becoming an increasing problem in C. jejuni which can complicate effective clinical treatment of campylobacteriosis (17). In this study, several multiple drug-resistant isolates were identified, with nine isolates being resistant to at least two different antibiotics. One isolate (isolate 24-53) was resistant to three antibiotics (tetracycline, kanamycin, and nalidixic acid). All isolates resistant to kanamycin or nalidixic acid were also resistant to tetracycline.
The plasmid-encoded tet(O) gene is the primary Tcr determinant in C. jejuni (46). In terms of the level of resistance to tetracycline, Taylor et al. (45) found that the tetracycline MICs varied between 32 and 128 μg/ml for Tcr isolates in 1980-1981. In this study (1999 to 2002), tetracycline MICs were higher, with MICs of 256 to 512 μg/ml for 37% of isolates. Gaunt and Piddock (14) found an association between fluoroquinolone resistance and high tetracycline MICs, but no such association was observed in this study. Previous research has determined that tet(O) can confer resistance to a tetracycline MIC of 128 μg/ml for C. jejuni (45). In this study, plasmids that harbored the tet(O) gene were shown to mediate extremely high levels of Tcr (512 μg/ml) in 4 of the 101 Tcr isolates. This was demonstrated by the conjugative transfer of the resistance phenotype to a tetracycline-susceptible recipient strain of C. jejuni by conjugation, in which the tetracycline MICs for the transconjugants were comparable to those for the donor isolates (512 μg/ml; Table 3).
Conjugative transfer of Tcr plasmids between isolates of C. jejuni has been demonstrated previously (45, 47) and occurs at frequencies of approximately 4 × 10−5 transconjugant per recipient in a 24-h mating on a solid surface at 37°C (45). Earlier attempts at transferring similar plasmids from C. jejuni to E. coli were also unsuccessful and suggested that the host range was restricted to Campylobacter spp. (47).
Cloning of the tet(O) gene from a range of MIC levels (64 to 512 μg/ml of Tc) in E. coli DH5α resulted in a Tcr level of 32 μg/ml. Thus, the Tet(O) protein confers a lower level of Tcr in E. coli, regardless of the level of resistance it conferred in C. jejuni. This might indicate that the ribosomes of Campylobacter have special characteristics that could enable the Tet(O) protein to efficiently interact with its binding site on the ribosome and hence more efficiently dislodge the drug. In contrast, the Tet(O) protein seems to interact with the ribosomes of E. coli but not as efficiently as it does in Campylobacter, resulting in the expression of a consistently lower level of resistance. This might be one reason why the tet(O) gene has never been detected as a mechanism of resistance in clinical Tcr strains of E. coli.
The tet(O) genes from the four isolates showing high-level Tcr (MICs, 512 μg/ml) were shown to differ in seven distinct nucleotide positions when compared to the tet(O) gene from other isolates for which MICs were lower (64 to 128 μg/ml), leading to the substitution of seven amino acid residues (Table 5). In one instance, the mutation at nucleotide position 910 (Table 5) resulted in a change from a serine to a proline, which is a change from a polar amino acid with a hydroxyl side chain to a cyclic, nonpolar amino acid. Also, the nucleotide substitutions at positions 1063 and 1111 (Table 5) led to a change from uncharged polar to nonpolar amino acid residue. The effect of the observed amino acid substitutions on the Tcr level could not be seen when the tet(O) gene (from strain 25-01) was expressed in an E. coli host (Table 4), probably due to E. coli versus C. jejuni ribosomal differences as explained above. Since the crystal structure of Tet(O) protein is presently unavailable, residue mutations were compared to the structure of EF-G (from Thermus thermophilus), which shares high sequence similarity to Tet(O) (S. Connell, unpublished data). Analysis of mutated residue locations on Tet(O) (using the EF-G structure) has identified that several of the detected mutated residues are exposed on the surface of the protein and may be involved in interacting with the ribosome. These mutations could potentially allow for stronger binding to the ribosome and allow Tet(O) to outcompete high levels of tetracycline. Two observations lend further support to this proposal. First, the transfer of plasmid pDOB47, which carries the tet(O) gene and a ∼1-kb fragment upstream of the gene, by mobilization into strain UA543 was found to result in a Tcr level which was twofold lower than the level of resistance in the original C. jejuni host. This difference in the resistance level, however, could be attributed to the inhibition of growth of C. jejuni as a result of the overexpression of the tet(O) gene cloned into a vector of a high copy number (pRY107) as shown from the result of the growth experiment (Fig. 2). A similar phenomenon was described in a previous study that showed that E. coli strains that carry the Tn10 tet gene on multicopy plasmids are often 4- to 12-fold less resistant to tetracycline than those that carry the tet gene in a low copy state (31). Second, there is an inability to detect other Tcr markers such as tet(M) and tetracycline efflux genes or to identify any mutation in the 16S rRNA genes that could be associated with Tcr in any of the highly resistant isolates. Taken together, these observations lead us to propose that the high-level Tcr could be mediated by the modified tet(O) gene due to the possible effect of the detected amino acid substitutions (Table 5) on the binding of the Tet(O) protein to its ribosomal target. The contribution of each of the detected amino acid substitutions to the activity of the Tet(O) protein and hence to the overall resistance level remains to be examined in a future study. Nevertheless, there is still a possibility that the Tcr plasmid in the highly resistant isolates might carry an additional, but previously uncharacterized, Tcr marker which could not be identified by the multiplex PCR. Therefore, the high-level Tcr might be attributed to the combined effect of the tet(O) gene and the uncharacterized marker.
Acknowledgments
This research was supported by grants from the University Hospital Foundation (Edmonton, Canada) and the National Science and Engineering Research Council (NSERC). A.G. is an Alberta Heritage Foundation for Medical Research (AHFMR) postdoctoral fellow. D.M.T. held both NSERC and AHFMR studentships. D.E.T. is an AHFMR scientist. L.N. was supported by a fellowship from the Japanese Society for the Promotion of Science.
Thanks to P. Guerry for the pRY107 expression vector and to J. Wells for personal communications on the pTet sequencing project. Thanks to K. Kowalewska-Grochowska and the staff of the APLPH. Our gratitude goes out to Matt Gilmour and James Gunton for their critical review of the manuscript.
REFERENCES
- 1.Bacon, D. J., R. A. Alm, D. H. Burr, L. Hu, D. J. Kopecko, C. P. Ewing, T. J. Trust, and P. Guerry. 2000. Involvement of a plasmid in virulence of Campylobacter jejuni 81-176. Infect. Immun. 68:4384-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blaser, M. J. 1997. Epidemiologic and clinical features of Campylobacter jejuni infections. J. Infect. Dis. 176(Suppl. 2):S103-S105. [DOI] [PubMed] [Google Scholar]
- 3.Burdett, V., J. Inamine, and S. Rajagopalan. 1982. Heterogeneity of tetracycline resistance determinants in Streptococcus spp. J. Bacteriol. 149:995-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connell, S. R., D. M. Tracz, K. H. Nierhaus, and D. E. Taylor. 2003. Ribosomal protection proteins and their mechanism of tetracycline resistance. Antimicrob. Agents Chemother. 47:3675-3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connell, S. R., C. A. Trieber, G. P. Dinos, E. Einfeldt, D. E. Taylor, and K. H. Nierhaus. 2003. Mechanism of Tet(O)-mediated tetracycline resistance. EMBO J. 22:945-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Ungria, M. C., D. Tillett, B. A. Neilan, P. T. Cox, and A. Lee. 1998. A novel method of extracting plasmid DNA from Helicobacter species. Helicobacter 3:269-277. [DOI] [PubMed] [Google Scholar]
- 7.Donohue, T. J., and S. Kaplan. 1991. Genetic techniques in rhodospirillaceae. Methods Enzymol. 204:459-485. [DOI] [PubMed] [Google Scholar]
- 8.Drlica, K. 1999. Mechanism of fluoroquinolone action. Curr. Opin. Microbiol. 2:504-508. [DOI] [PubMed] [Google Scholar]
- 9.Endtz, H. P., G. J. Ruijs, B. van Klingeren, W. H. Jansen, T. van der Reyden, and R. P. Mouton. 1991. Quinolone resistance in campylobacter isolated from man and poultry following the introduction of fluoroquinolones in veterinary medicine. J. Antimicrob. Chemother. 27:199-208. [DOI] [PubMed] [Google Scholar]
- 10.Engberg, J., F. M. Aarestrup, D. E. Taylor, P. Gerner-Smidt, and I. Nachamkin. 2001. Quinolone and macrolide resistance in Campylobacter jejuni and C. coli: resistance mechanisms and trends in human isolates. Emerg. Infect. Dis. 7:24-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaudreau, C., and H. Gilbert. 2003. Antimicrobial resistance of Campylobacter jejuni subsp. jejuni strains isolated from humans in 1998 to 2001 in Montréal, Canada. Antimicrob. Agents Chemother. 47:2027-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudreau, C., and H. Gilbert. 1998. Antimicrobial resistance of clinical strains of Campylobacter jejuni subsp. jejuni isolated from 1985 to 1997 in Quebec, Canada. Antimicrob. Agents Chemother. 42:2106-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaudreau, C., and H. Gilbert. 1997. Comparison of disc diffusion and agar dilution methods for antibiotic susceptibility testing of Campylobacter jejuni subsp. jejuni and Campylobacter coli. J. Antimicrob. Chemother. 39:707-712. [DOI] [PubMed] [Google Scholar]
- 14.Gaunt, P. N., and L. J. Piddock. 1996. Ciprofloxacin resistant Campylobacter spp. in humans: an epidemiological and laboratory study. J. Antimicrob. Chemother. 37:747-757. [DOI] [PubMed] [Google Scholar]
- 15.Harnett, N., S. McLeod, Y. A. Yong, C. Hewitt, M. Vearncombe, and C. Krishnan. 1995. Quinolone resistance in clinical strains of Campylobacter jejuni and Campylobacter coli. J. Antimicrob. Chemother. 36:269-270. [DOI] [PubMed] [Google Scholar]
- 16.Health, C. June 2002, posting date. Release of the final report of the advisory committee on animal uses of antimicrobials and impact on resistance and human health. http://www.hc-sc.gc.ca/vetdrugs-medsvet/amr_policy_dev_e.html. [Online.]
- 17.Hoge, C. W., J. M. Gambel, C. Srijan, C. Pitarangsi, and P. Echeverria. 1998. Trends in antibiotic resistance among diarrheal pathogens isolated in Thailand over 15 years. Clin. Infect. Dis. 26:341-345. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, L. B., and F. M. Aarestrup. 2001. Macrolide resistance in Campylobacter coli of animal origin in Denmark. Antimicrob. Agents Chemother. 45:371-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Labigne-Roussel, A., J. Harel, and L. Tompkins. 1987. Gene transfer from Escherichia coli to Campylobacter species: development of shuttle vectors for genetic analysis of Campylobacter jejuni. J. Bacteriol. 169:5320-5323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert, T., G. Gerbaud, P. Trieu-Cuot, and P. Courvalin. 1985. Structural relationship between the genes encoding 3′-aminoglycoside phosphotransferases in Campylobacter and in gram-positive cocci. Ann. Inst. Pasteur Microbiol. 136B:135-150. [DOI] [PubMed] [Google Scholar]
- 21.Lee, C. Y., C. L. Tai, S. C. Lin, and Y. T. Chen. 1994. Occurrence of plasmids and tetracycline resistance among Campylobacter jejuni and Campylobacter coli isolated from whole market chickens and clinical samples. Int. J. Food Microbiol. 24:161-170. [DOI] [PubMed] [Google Scholar]
- 22.Li, C. C., C. H. Chiu, J. L. Wu, Y. C. Huang, and T. Y. Lin. 1998. Antimicrobial susceptibilities of Campylobacter jejuni and coli by using E-test in Taiwan. Scand. J. Infect. Dis. 30:39-42. [DOI] [PubMed] [Google Scholar]
- 23.Lucey, B., B. Cryan, F. O'Halloran, P. G. Wall, T. Buckley, and S. Fanning. 2002. Trends in antimicrobial susceptibility among isolates of Campylobacter species in Ireland and the emergence of resistance to ciprofloxacin. Vet. Rec. 151:317-320. [DOI] [PubMed] [Google Scholar]
- 24.Luo, N., O. Sahin, J. Lin, L. O. Michel, and Q. Zhang. 2003. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob. Agents Chemother. 47:390-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manavathu, E. K., K. Hiratsuka, and D. E. Taylor. 1988. Nucleotide sequence analysis and expression of a tetracycline-resistance gene from Campylobacter jejuni. Gene 62:17-26. [DOI] [PubMed] [Google Scholar]
- 26.Marshall, B., S. Morrissey, P. Flynn, and S. B. Levy. 1986. A new tetracycline-resistance determinant, class E, isolated from Enterobacteriaceae. Gene 50:111-117. [DOI] [PubMed] [Google Scholar]
- 27.Marshall, B., C. Tachibana, and S. B. Levy. 1983. Frequency of tetracycline resistance determinant classes among lactose-fermenting coliforms. Antimicrob. Agents Chemother. 24:835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McMurry, L. M., B. H. Park, V. Burdett, and S. B. Levy. 1987. Energy-dependent efflux mediated by class L (TetL) tetracycline resistance determinant from streptococci. Antimicrob. Agents Chemother. 31:1648-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendez, B., C. Tachibana, and S. B. Levy. 1980. Heterogeneity of tetracycline resistance determinants. Plasmid 3:99-108. [DOI] [PubMed] [Google Scholar]
- 31.Moyed, H. S., T. T. Nguyen, and K. P. Bertrand. 1983. Multicopy Tn10 tet plasmids confer sensitivity to induction of tet gene expression. J. Bacteriol. 155:549-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nachamkin, I. 1994. Antimicrobial susceptibilities of Campylobacter jejuni and Campylobacter coli to ciprofloxacin, erythromycin and tetracycline from 1982 to 1992. Med. Microbiol. Lett. 3:300-305. [Google Scholar]
- 33.Nachamkin, I., J. Engberg, and F. M. Aarestrup. 2000. Diagnosis and antimicrobial susceptibility of Campylobacter spp. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 34.Nachamkin, I., H. Ung, and M. Li. 2002. Increasing fluoroquinolone resistance in Campylobacter jejuni, Pennsylvania, USA, 1982-2001. Emerg. Infect. Dis. 8:1501-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ng, L. K., I. Martin, M. Alfa, and M. Mulvey. 2001. Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15:209-215. [DOI] [PubMed] [Google Scholar]
- 36.Piddock, L. J. 1995. Quinolone resistance and Campylobacter spp. J. Antimicrob. Chemother. 36:891-898. [DOI] [PubMed] [Google Scholar]
- 37.Piddock, L. J., V. Ricci, L. Pumbwe, M. J. Everett, and D. J. Griggs. 2003. Fluoroquinolone resistance in Campylobacter species from man and animals: detection of mutations in topoisomerase genes. J. Antimicrob. Chemother. 51:19-26. [DOI] [PubMed] [Google Scholar]
- 38.Ruiz, J., F. Goni, F. Marco, F. Gallardo, D. Mirelis, D. A. Jimenez, and J. Vila. 1998. Increased resistance to quinolones in Campylobacter jejuni: a genetic analysis of gyrA gene mutations in quinolone-resistant clinical isolates. Microbiol. Immunol. 42:223-226. [DOI] [PubMed] [Google Scholar]
- 39.Sagara, H., A. Mochizuki, N. Okamura, and R. Nakaya. 1987. Antimicrobial resistance of Campylobacter jejuni and Campylobacter coli with special reference to plasmid profiles of Japanese clinical isolates. Antimicrob. Agents Chemother. 31:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 41.Skirrow, M. B., and M. J. Blaser. 2000. Clinical aspects of Campylobacter infection. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 42.Strack, B., M. Lessl, R. Calendar, and E. Lanka. 1992. A common sequence motif, -E-G-Y-A-T-A-, identified within the primase domains of plasmid-encoded I- and P-type DNA primases and the alpha protein of the Escherichia coli satellite phage P4. J. Biol. Chem. 267:13062-13072. [PubMed] [Google Scholar]
- 43.Taylor, D. E. 1986. Plasmid-mediated tetracycline resistance in Campylobacter jejuni: expression in Escherichia coli and identification of homology with streptococcal class M determinant. J. Bacteriol. 165:1037-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor, D. E., and J. H. Bryner. 1984. Plasmid content and pathogenicity of Campylobacter jejuni and Campylobacter coli strains in the pregnant guinea pig model. Am. J. Vet. Res. 45:2201-2202. [PubMed] [Google Scholar]
- 45.Taylor, D. E., N. Chang, R. S. Garner, R. Sherburne, and L. Mueller. 1986. Incidence of antibiotic resistance and characterization of plasmids in Campylobacter jejuni strains isolated from clinical sources in Alberta, Canada. Can. J. Microbiol. 32:28-32. [DOI] [PubMed] [Google Scholar]
- 46.Taylor, D. E., and A. Chau. 1996. Tetracycline resistance mediated by ribosomal protection. Antimicrob. Agents Chemother. 40:1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Taylor, D. E., S. A. De Grandis, M. A. Karmali, and P. C. Fleming. 1981. Transmissible plasmids from Campylobacter jejuni. Antimicrob. Agents Chemother. 19:831-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, D. E., S. A. DeGrandis, M. A. Karmali, and P. C. Fleming. 1980. Transmissible tetracycline resistance in Campylobacter jejuni. Lancet 2:797. [DOI] [PubMed] [Google Scholar]
- 49.Taylor, D. E., K. Hiratsuka, H. Ray, and E. K. Manavathu. 1987. Characterization and expression of a cloned tetracycline resistance determinant from Campylobacter jejuni plasmid pUA466. J. Bacteriol. 169:2984-2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taylor, D. E., D. G. Newell, and A. D. Pearson. 1983. Incidence of plasmid DNA in strains of Campylobacter jejuni isolated from stool specimens at 37 °C and 43 °C. J. Infect. Dis. 147:965-966. [DOI] [PubMed] [Google Scholar]
- 51.Tenover, F. C., S. Williams, K. P. Gordon, C. Nolan, and J. J. Plorde. 1985. Survey of plasmids and resistance factors in Campylobacter jejuni and Campylobacter coli. Antimicrob. Agents Chemother. 27:37-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Trieber, C. A., N. Burkhardt, K. H. Nierhaus, and D. E. Taylor. 1998. Ribosomal protection from tetracycline mediated by Tet(O): Tet(O) interaction with ribosomes is GTP-dependent. Biol. Chem. 379:847-855. [DOI] [PubMed] [Google Scholar]
- 53.Trieber, C. A., and D. E. Taylor. 2000. Mechanisms of antibiotic resistance in Campylobacter. In I. Nachamkin and M. J. Blaser (ed.), Campylobacter, 2nd ed. ASM Press, Washington, D.C.
- 54.Trieber, C. A., and D. E. Taylor. 2002. Mutations in the 16S rRNA genes of Helicobacter pylori mediate resistance to tetracycline. J. Bacteriol. 184:2131-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trieu-Cuot, P., G. Gerbaud, T. Lambert, and P. Courvalin. 1985. In vivo transfer of genetic information between gram-positive and gram-negative bacteria. EMBO J. 4:3583-3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang, Y., W. M. Huang, and D. E. Taylor. 1993. Cloning and nucleotide sequence of the Campylobacter jejuni gyrA gene and characterization of quinolone resistance mutations. Antimicrob. Agents Chemother. 37:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, Y., and D. E. Taylor. 1991. A DNA sequence upstream of the tet(O) gene is required for full expression of tetracycline resistance. Antimicrob. Agents Chemother. 35:2020-2025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Warsa, U. C., M. Nonoyama, T. Ida, R. Okamoto, T. Okubo, C. Shimauchi, A. Kuga, and M. Inoue. 1996. Detection of tet(K) and tet(M) in Staphylococcus aureus of Asian countries by the polymerase chain reaction. J. Antibiot. (Tokyo) 49:1127-1132. [DOI] [PubMed] [Google Scholar]
- 59.Yao, R., R. A. Alm, T. J. Trust, and P. Guerry. 1993. Construction of new Campylobacter cloning vectors and a new mutational cat cassette. Gene 130:127-130. [DOI] [PubMed] [Google Scholar]
- 60.Zhao, J., and T. Aoki. 1992. Nucleotide sequence analysis of the class G tetracycline resistance determinant from Vibrio anguillarum. Microbiol. Immunol. 36:1051-1060. [DOI] [PubMed] [Google Scholar]