Abstract
A better understanding of the mechanisms underlying obesity and its comorbidities is key to designing new therapies and treatments. PPARγ is a master regulator of adipocyte biology but the functions of its isoforms are poorly distinguished. Here we demonstrated that PPARγ1 is preferentially expressed in catabolic fat depots while PPARγ2 presents itself at a higher level in browning-resistant depots. PPARγ2, but not PPARγ1, responds to endogenous ligands to induce adipogenesis, and the isoforms regulate distinct sets of white and brown adipocyte genes. Moreover, PPARγ1 negatively correlates while PPARγ2 positively correlates with adiposity in human subcutaneous and visceral fat. These results together indicate that PPARγ1 and PPARγ2 have distinct functions in regulating adipocyte plasticity, and future research should take into account the binary roles of both isoforms in order to identify druggable gene targets and pathways relevant for treatment of metabolic disorders.
Keywords: Obesity, PPARγ, Adipocyte, PPARγ isoforms, browning, beiging
1. Introduction
Obesity is a major risk factor for many diseases, such as type 2 diabetes, cardiovascular disease, and a myriad of cancers. Central to studying obesity and its associated comorbidities is adipose tissue. There are two canonical types of adipose tissue. White adipose tissue (WAT) stores excess energy in the form of triglycerides while brown adipose tissue (BAT) dissipates energy. Although BAT is less abundant, it is highly enriched with mitochondria that allow for the uncoupling of the electron transport chain from ATP synthesis, thereby catabolizing fat to generate heat. Thus, inducing brown-like features in WAT in a process called browning or beiging has the potential to remodel white fat into a healthier and more catabolic state [1]. To date there have been many mechanisms discovered to regulate BAT or browning [2, 3], such as PRDM16 [4, 5], C/EBPβ [6], EBF2 [7, 8], IRF4 [9], cytoskeleton remodeling [10], posttranslational modifications (PTMs) [11], circadian rhythm [12], bone morphogenetic protein (BMP) signaling [13, 14], microRNAs [15], and long non-coding RNAs [16].
Despite the distinct functions and regulatory factors between WAT and BAT, the ligand-dependent transcription factor peroxisome proliferator-activated receptor gamma (PPARγ) is abundantly expressed in both types of fat. PPARγ is the master regulator of adipocyte biology and also an important drug target for treating type 2 diabetes [17, 18]. There are two isoforms of PPARγ, γ1 and γ2, which are transcribed from the same gene under the control of different promoters [19]. PPARγ1 is highly expressed in adipose tissue but is also presented in other tissues at relatively lower levels [20, 21]. Meanwhile, PPARγ2 has an additional 30 amino acids on the N-terminal of PPARγ1, and its expression is restricted to adipose tissue [22]. The functions of PPARγ in adipose tissue have been extensively studied. However, the potentially distinguishing physiological and biochemical effects of these two isoforms have not been thoroughly examined. Thus far, the only well-characterized functional difference is the higher adipogenic capacity of the longer isoform, PPARγ2 [23, 24]. The thiazolidinedione (TZD) class of PPARγ ligands not only improves insulin sensitivity but also induces browning of white adipocytes by activating brown genes while repressing white genes [25]. TZD-induced browning is mediated through the deacetylation of PPARγ [11], but it remains unclear whether the two isoforms have the same contribution to this phenomenon.
In this study, we sought novel approaches to modulate and improve adipose health by exploring the differences between PPARγ isoforms through analyzing their inducible reconstitutions into fibroblast cells derived from PPARγ knockout mice (PPARγ KO MEFs) and further validating our findings in human fat tissues. Strikingly, we discovered novel distinctions between the PPARγ isoforms in response to endogenous ligands and in regulating the expression of brown and white adipocyte-specific genes. We also found distinctions in the correlations between the expression of the two PPARγ isoforms with adiposity and crucial adipocyte genes in humans. Our study highlights the significance of targeting PPARγ2 to achieve partial agonism of PPARγ in developing more effective treatments for metabolic disorders.
2. Materials and methods
2.1. Animal studies
Six-week-old male mice with different genetic backgrounds (C57BL/6, 129/Sv, FVB/N) were purchased from Jackson Laboratory. The mice were exposed to cold (4°C) for one week in a 12 hr light/dark cycle with free access to normal chow. At the end of the cold challenge, fat tissues were collected for analysis. The Columbia University Animal Care and Utilization Committee approved all procedures.
2.2. Plasmids and cell culture
Flag-HA-tagged PPARγ1 and PPARγ2 cDNAs were subcloned into a doxycycline-inducible lentiviral plasmid, pTRIPZ (Thermo Open Biosystems) [26], by In-Fusion HD cloning following the manufacturer’s instructions (Clontech). Mouse PPARγ knockout fibroblasts [27] were engineered to stably express either isoform of PPARγ by lentiviral infection and were selected for by the addition of puromycin (2.5μg/mL).
To induce adipocyte differentiation, cells were treated with an adipogenic cocktail containing 1 μM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine and 10 μg/mL insulin for two days in the presence or absence of rosiglitazone (5 μM). Two days post-induction, cells were switched to a maintenance medium containing 2.5μg/mL insulin with or without rosiglitazone until they were fully differentiated. 10μg/mL doxycycline was used to induce PPARγ expression for two days prior to adipocyte differentiation. To induce brown gene expression, mature adipocytes were treated with either β3 adrenergic receptor agonist CL-316,243 (1 μg/mL) or PPARα agonist WY 14,643 (10 μM) plus forskolin (5μM) for 4 hours [28].
2.3. Oil Red O staining
Cells were fixed with 10% formalin and incubated for 30 minutes at room temperature with gentle shaking. The cells were then washed with 60% isopropanol and incubated in Oil Red O working solution. After 10 minutes of staining, the plates were rinsed with distilled water four times.
2.4. Protein analysis
Total protein from cultured cells was harvested in an extraction buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 10% glycerol, 2% NP-40, 1 mM EDTA (pH 8.0), 0.2% SDS, and 0.5% sodium deoxycholate supplemented with protease and phosphatase inhibitors (Boston Bioproducts). 40μg of protein was denatured for Tris-Glycine gel electrophoresis followed by Western blotting analyses. The sources of antibodies were as follows: Adiponectin (Affinity BioReagents); Perilipin (Cell Signaling); Tubulin, C/ebpβ, C/ebpα, PPARγ (E8) (Santa Cruz).
2.5. RNA analysis
RNA was isolated by using NucleoSpin RNA kit (Macherey-Nagel) with DNase I digestion. cDNA was synthesized with a High-capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative real-time PCR (qPCR) was performed on a Bio-Rad CFX96 Real-Time PCR system by using the GoTaq qPCR Master Mix (Promega). Relative gene expression levels were analyzed by the ΔΔCt method using TBP as an internal control.
2.6. Human samples analyses
Adipose RNA-seq was performed on human gluteal subcutaneous adipose tissue samples collected from 25 healthy participants (56% female; 52% European ancestry; mean BMI ~23.8) in the Genetics of Evoked-responses to Niacin and Endotoxemia (GENE) study, a National Institute of Health-sponsored protocol based at University of Pennsylvania as previously described (NIH clinical trial NCT00953667) [29]. Briefly, baseline adipose samples underwent RNA isolation and quality control and poly-A libraries were prepared for deep RNA-seq on Illumina’s HiSeq 2000 generating ~400 million reads as previously described [30]. Expression levels of gene transcripts were analyzed using Cuffdiff. All transcripts encoding PPARγ1 were consolidated as PPARγ1 [31].
For human visceral fat biopsies, 30 Chinese patients undergoing abdominal surgery for benign hepatobiliary conditions, such as cholecystitis or gallstone, at the Minimally Invasive Surgery Center, the Second Xiangya Hospital of Central South University, were recruited. All patients were given informed consent, and the protocol was approved by the Ethics Committee of the Second Xiangya Hospital of Central South University. During the operation, intra-abdominal omental adipose tissues (≈2 cc) were collected, snap frozen, and stored at −80°C before RNA extractions. Gene ex pression was determined by qPCR in triplicates with ACTIN as an internal control.
2.7. Statistical analysis
Unpaired 2-tailed Student’s t-test was used to evaluate statistical significance and p<0.05 was set to declare a statistically significant change. Values are presented as means ± standard error of means (SEM).
3. Results
3.1. The Differential Expression of PPARγ Isoforms in Fat
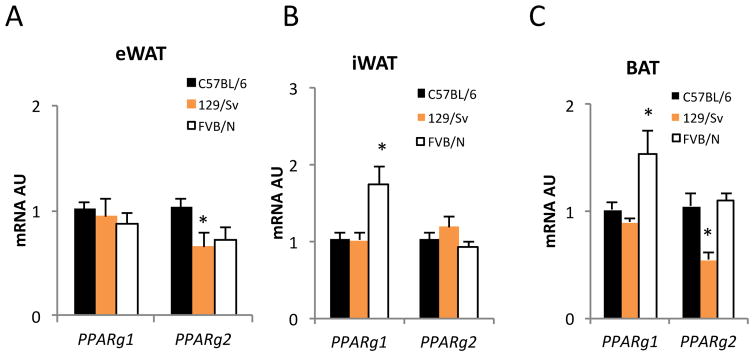
To investigate the effects of PPARγ isoforms in regulating fat browning, we examined their expression levels in different fat depots, epididymal (visceral) eWAT, inguinal (subcutaneous) iWAT, and brown adipose tissue (BAT), which have distinct thermogenic capacities in three mouse genetic backgrounds, C57BL/6, 129/Sv, and FVB/N. In eWAT, 129/Sv mice had reduced Pparg2 expression compared to C57BL/6 and FVB/N mice (Figure 1A). Moreover, a recent study showed that eWAT in 129/Sv has a stronger brown remodeling capacity during cold exposure than in the other two strains [32]. Notably, PPARγ1 was expressed at higher level in FVB/N mice subcutaneous fat (Figure B), a depot that is known to be more prone to browning than that of C57BL/6 and 129/Sv mice [32]. In BAT, obesity-resistant 129/Sv and FVB/N mice had either higher PPARγ1 (FVB/N) or lower PPARγ2 (129/Sv) expression than C57BL/6 control mice (Figure 1C). These data raise the possibility that PPARγ isoforms may have different functions in regulating thermogenic activities in fat depots.
Figure 1. The Differential Expression of PPAR Isoforms in Fat.
PPARg expression in three fat depots from chronic cold challenged mice on different genetic backgrounds. *: p<0.05, **: p<0.01 vs. C57BL/6, (n=8, 8, 8). Data is represented as Mean ± SD.
3.2. Divergent Requirement of Ligands by PPARγ Isoforms for Adipogenesis
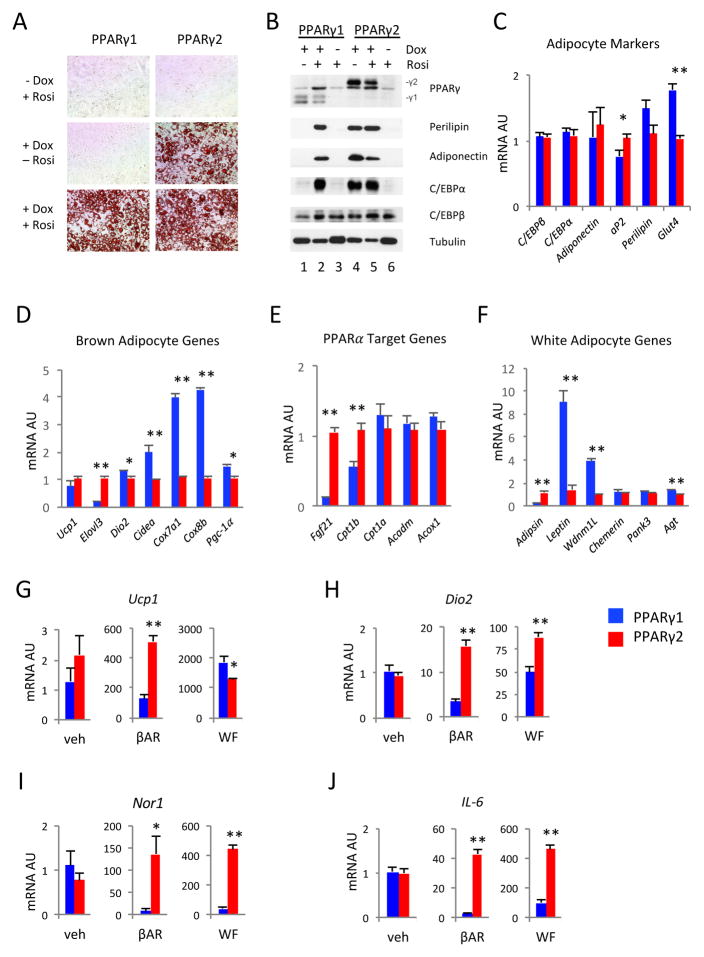
Next, we generated an inducible cellular system by stably over-expressing PPARγ1 or PPARγ2 under the control of doxycycline into PPARγ−/− mouse embryonic fibroblasts [27] to compare the adipogenic potential of PPARγ’s isoforms. This system allowed us to efficiently differentiate MEFs into adipocytes without interference from endogenous PPARγ while also bypassing the potential artifacts of PPARγ on fibroblasts in the constitutive over-expression models [11, 23]. When treated with Rosiglitazone, both PPARγ1 and PPARγ2 induced full differentiation of MEFs into adipocytes, as indicated by Oil Red O staining of lipid contents (Figure 2A). In contrast, in the absence of exogenous TZD, PPARγ1 failed to induce differentiation (Figure 2A). These data suggest that only PPARγ2 but not PPARγ1 responds to endogenous ligands to promote adipogenesis.
Figure 2. The Distinct Regulations of Adipocyte Plasticity by PPARγ1 and PPARγ2.
PPARγ1 or PPARγ2 was reconstituted in PPARγ KO MEFs by using a doxycycline-inducible lentiviral system. (A) Oil Red O staining of cells differentiated at indicated conditions; (B) Western blot analyses of adipocyte markers in differentiated cells. (C–F) qPCR analysis of expressions of adipocyte genes (C), brown adipocyte genes (D), PPARα target genes (E) and white adipocyte genes (F) in adipocytes differentiated in the presence of Rosi on D9. (G–H) In fully differentiated adipocytes, qPCR analysis of thermogenic responses induced by β agonist (βAR) or PPARα agonist WY14643 + forskolin (WF). *: p<0.05, **: p<0.01, for PPARγ1 vs. PPARγ2 under same treatment (n=4). Data is represented as Mean ± SD.
To further assess the effects of PPARγ isoforms on adipogenesis, we analyzed the expression of canonical adipocyte markers. In the absence of doxycycline, there was no induction of adipogenesis, as illustrated by the absence of Perilipin, Adiponectin and C/ebpα in both cell lines (Figure 2B, lanes 3 and 6). After inducing PPARγ by doxycycline and differentiating cells in the absence of Rosiglitazone, these markers were present only in PPARγ2 cells, but not in PPARγ1 cells, which was consistent with the Oil Red O staining of lipid accumulation (Figure 2B, lanes 1 and 4). Upon Rosiglitazone treatment, these adipocyte markers were expressed at similar levels (Figure 2B, lanes 2 and 5). C/ebpβ is an early adipogenic factor upstream of PPARγ [33] and was unaffected by PPARγ isoforms. Taken together, our data indicate that, unlike PPARγ2, PPARγ1 cannot respond to endogenous ligands to induce adipogenesis, but retains the full activation by synthetic TZD ligands to induce adipogenesis.
3.3. Distinct Regulations of Brown and White Genes by PPARγ Isoforms
Although MEFs were fully differentiated into adipocytes by both isoforms in the presence of TZDs, we asked whether these morphologically identical adipocytes were functionally similar. We first examined pan-adipocyte markers Perilipin, Adiponectin, C/ebpα, C/ebpβ, aP2, and Glut4 in these two lines. Most of these adipocyte genes were expressed at similar levels, with the exception of aP2 and Glut4, which showed minimal but significant differences in expression between the PPARγ1 and PPARγ2 cell lines (Figure 2C). Nonetheless, even with slight differences in their expression levels, all of these major adipocyte markers were expressed to a similar extent. Therefore, we considered both TZD-treated cell lines as fully differentiated adipocytes and proceeded to analyze the functional significance of both isoforms.
Given that TZDs can induce browning of white adipocytes and the observed differential expression pattern of the PPARγ isoforms in different fat depots, we assessed the regulatory effects on the expression of representative brown genes by PPARγ isoforms. PPARγ1 under the activation by TZDs induced a similar level of Ucp1 as PPARγ2, but was less potent than PPARγ2 to induce representative brown gene Elovl3. Surprisingly, the other brown genes examined, Dio2, Cidea and Pgc-1α, and particularly the mitochondrial respiratory enzymes encoding genes Cox7a1 and Cox8b, were all lower in PPARγ2 adipocytes (Figure 2D). Meanwhile, the lipid oxidative PPARα downstream target genes Cpt1a, Acadm and Acox1 [34] were expressed comparably in both TZD-treated PPARγ1 and PPARγ2 cells. Conversely, Cpt1b was upregulated in PPARγ2 cells (Figure 2E). Interestingly, Fgf21, a PPARα [35, 36] and PPARγ [37] downstream target, was increased by about 9-fold in PPARγ2 adipocytes (Figure 2E).
Likewise, we observed distinct regulations of PPARγ isoforms on white adipocyte-specific genes (Figure 2F). The expression of Adipsin was almost exclusively controlled by PPARγ2, whereas Leptin, Wdnm1L and Angiotensinogen (Agt) were significantly upregulated by PPARγ1 activity. Meanwhile, the expression of Pank3 and Chemerin were comparable between the PPARγ1 and PPARγ2 cells. Although PPARγ2 was previously reported to have a higher adipogenic potential than PPARγ1 [23, 24], this is the first observation showing that PPARγ isoforms have distinct target genes involved in white adipocyte remodeling.
3.4. PPARγ2 Dominates Adipocyte Browning Response in vitro
Subsequently, we compared the induction of downstream brown genes in the fully differentiated by PPARγ reconstitution cells treated with a β3-adrenergic receptor agonist, CL 316243, which augments browning. We observed similar levels of expression of the brown genes under the basal condition (vehicle-treated). However, the most responsive brown genes, Ucp1 and Dio2, were elicited much more potently by PPARγ2 cells, as was Nor1 (Figures 2G, 2H and 2I). To avoid any possible variation by activation of α-adrenergic receptors, we also employed a PPARα agonist in combination with forskolin to induce browning. Forskolin directly stimulates Adenylyl Cyclase to produce cAMP without requiring α-adrenergic receptor activation [38]. Under this alternative browning condition, the higher expression of Dio2 and Nor1 was maintained in PPARγ2 cells whereas Ucp1 was more robustly induced in PPARγ1 cells (Figures 2G, 2H and 2I). Interestingly, the inflammatory marker IL-6 was more significantly activated in PPARγ2 cells by both treatments (Figure 2J). These data further illustrate the unique regulatory roles of PPARγ1 and PPARγ2 in adipocyte browning and inflammatory responses.
3.5. PPARγ Isoforms have Opposite Correlations with Adiposity in Humans
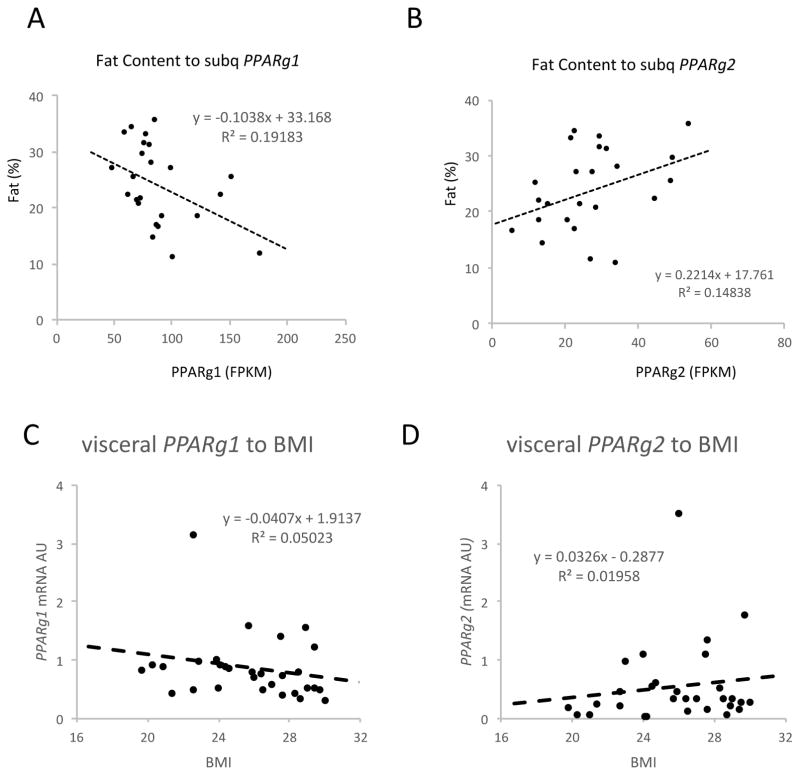
To distinguish PPARγ1 and PPARγ2’s physiological functions, we analyzed their expression levels using data obtained from RNA-sequencing of human subcutaneous fat from 25 non-obese subjects (BMI 18.9 to 29.5, age 19–45, 11 males and 14 females) [29]. The expression levels of PPARγ1 and PPARγ2 displayed distinct correlations with adiposity: PPARγ1 inversely and PPARγ2 positively correlated with body fat content (Figures 3A and 3B). We also validated our findings in visceral fat from a separate cohort of human subjects (BMI 19.7–30, age 33–72, 10 males and 20 females). Similarly, regardless of age, sex and health status, PPARγ1 inversely correlated with BMI (Figure 3C), while PPARγ2 positively correlated with BMI (Figure 3D) (fat content data not available). These data from different fat depots in humans implies that PPARγ2 is more obesogenic, in line with its stronger adipogenic capacity, and PPARγ1 is more catabolic to inhibit adiposity.
Figure 3. The Opposite Correlations of PPAR Isoforms with Obesity in Humans.
(A–B) The correlations of body fat content to PPARγ1 and PPARγ2 expression in subcutaneous fat from non-obese subjects. The levels of PPARγ isoforms were quantified as FPKM from RNA-sequencing. (C–D) The correlations of BMI to PPARγ isoforms in visceral fat from non-obese subjects. The levels of PPARγ isoforms were determined by qPCR analyses.
3.6. Different Regulations of Adipocyte Genes by PPARγ1 and PPARγ2 in Human Visceral Fat
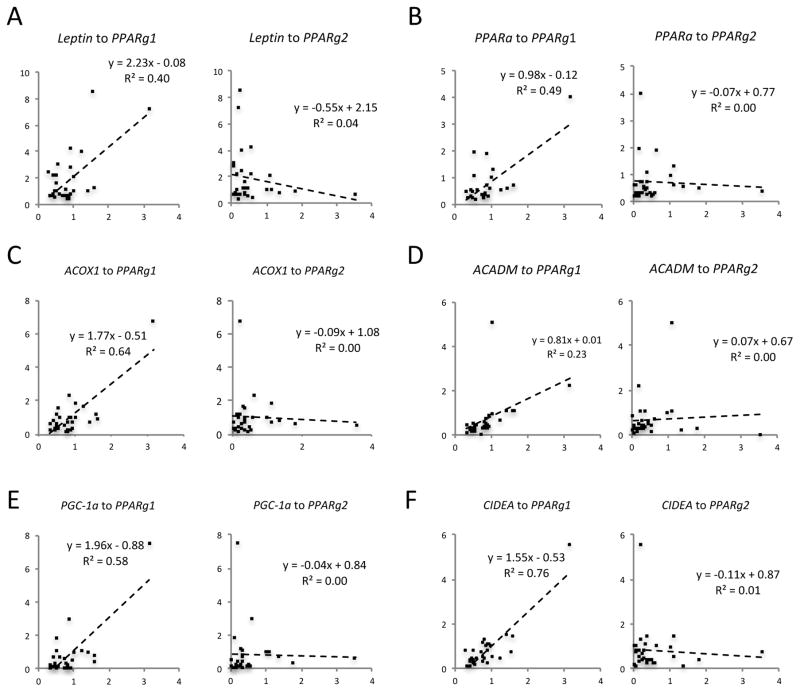
In addition to analyzing the relationship between PPARγ expression and adiposity, we correlated expression of adipocyte genes with PPARγ isoforms in human visceral fat. Consistent with the regulation of Leptin by PPARγ1 in our in vitro studies, LEPTIN positively correlated with PPARγ1 expression but not with PPARγ2 (Figure 4A). Interestingly, the key lipid oxidative gene, PPARa, and its downstream targets ACOX1 and ACADM, had greater correlation with PPARγ1 than with PPARγ2 (Figure 4 B–D). The brown adipocyte genes PGC-1α and CIDEA also positively correlated exclusively with PPARγ1 but not PPARγ2 (Figure 4E and 4F), consistent with the in vitro data (Figure 2D). The different correlations of adipocyte genes with PPARγ isoforms in humans support in vitro and rodent studies and are consistent with distinct functions of PPARγ1 and PPARγ2 in the pathogenesis of human obesity.
Figure 4. The distinct correlations of PPAR isoforms with adipocyte genes in human visceral fat.
The correlations of adipocyte genes to PPARγ isoforms in visceral fat from non-obese subjects. The levels of PPARγ isoforms were determined by qPCR analyses.
4. Discussion
It is known that PPARγ plays critical roles in regulating adipocyte development and is involved in major signaling cascades that maintain or disrupt metabolic homeostasis. In this study, we employed mouse adipose tissues, cell lines, and human fat to distinguish the expression and functions of PPARγ isoforms, which have traditionally been collectively studied as total PPARγ. We revealed that PPARγ1 and PPARγ2 have distinct roles in regulating adipocyte development and function. It is known that PPARγ2 expression is restricted to adipocytes, and we were able to corroborate previous reports that showed it was more adipogenic than PPARγ1 [23]. We further demonstrated that unlike PPARγ2, PPARγ1 cannot respond to endogenous ligands in order to promote adipogenesis. However, when maximally activated by synthetic TZDs, both PPARγ1 and PPARγ2 are capable of inducing adipogenesis. This finding is notable since it explains the restrictive expression of PPARγ2 to the adipocyte lineage relative to the broad expression of PPARγ1 in many other cell types [20, 21]. Specifically, the fact that PPARγ2 is expressed minimally in peripheral tissues is likely to prevent precursor cells from committing to an adipocyte fate. It is possible that this distinction in ligand dependency may also account for TZDs’ side effects in treating insulin resistance, in which TZDs activate PPARγ1 to promote adipogenesis. The induction of adipogenesis by TZDs via PPARγ1 likely creates an imbalance and excessive conversion of progenitor cells into mature adipocytes, resulting in obesity. Additionally, it has been suggested that TZD-induced bone loss is caused by preferential ectopic adipogenesis of bone marrow mesenchymal stem cells over osteoblastogenesis [39, 40].
There is a growing interest to harness brown remodeling of white fat as a promising strategy to combat obesity and diabetes [1, 3, 41]. Activation of PPARγ by TZD treatment is known to induce brown genes and repress white genes [25], but the different functions of PPARγ isoforms during this process remained unknown. Herein, we analyzed the brown and white genes regulated by PPARγ1 and PPARγ2, respectively. To our surprise, PPARγ1 and PPARγ2 mediated distinct sets of brown genes. PPARγ1 is superior at maintaining higher basal levels of mitochondrial function genes such as Cidea, Coxa1, Cox8b, and the regulatory Pgc-1α, while PPARγ2 is more potent to induce the responsive brown genes, including Ucp1, Dio2 and Elovl3. Amongst the TZD-repressed genes, which are generally considered white adipocyte-specific genes, Adipsin is preferentially a PPARγ2 target gene while Leptin, Wdnm1L and Angiotensinogen are more responsive to PPARγ1. Therefore, PPARγ1 and PPARγ2 have unique downstream targets during adipocyte remodeling. Furthermore, the selective transcriptional activity of these isoforms maybe regulated either through DNA-binding or recruitment of co-factors, both of which invite further study. Nevertheless, our study raises the possibility of developing personalized medicine to induce directional fat remodeling dependent on the specific molecular signature of WAT and BAT through targeting specific PPARγ isoforms.
Finally, we discovered that PPARγ isoforms have opposite correlations to obesity in humans. PPARγ2 is associated with increased adiposity while PPARγ1 is inversely correlated with adiposity. Our finding is in agreement with previous studies of the Pro12Ala polymorphism found exclusively on PPARγ2, but not on PPARγ1. Compared to the Pro12 allele, the Ala12 polymorphism causes a loss-of-function and inhibits TZD-induced adipogenesis [42, 43]. The Ala12 allele reduces adiposity and the risk for Type 2 diabetes in both human [42, 44] and mouse studies [45] and is also considered a longevity variant [46]. These data are also supported by the impaired adipogenesis and overall lean phenotype in Pparg2 knockout mice [24, 47], as well as the increased PPARγ2, but not PPARγ1, expression in obese subjects [20]. Therefore, PPARγ2 likely contributes to the onset of obesity and the associated co-morbidities despite its insulin sensitizing benefit. Our study highlights the significance of understanding the different regulatory roles of the two PPARγ isoforms in adipocyte function and plasticity and suggests developing new isoform-specific therapies for metabolic diseases.
Supplementary Material
Highlights.
PPARγ2, but not PPARγ1, responds to endogenous ligands to induce adipogenesis;
PPARγ isoforms regulate distinct sets of white and brown adipocyte genes;
PPARγ isoforms have opposite correlations with adiposity in humans.
Acknowledgments
This work was supported by NIH grants R00DK97455 to L. Q., Pilot and Feasibility funding from the Diabetes Research Center to L.Q. (P30 DK063608), R01HL113147 to M. P. R., and National Natural Science Foundation of China (31471131) and International Science & Technology Cooperation Program of China (2014DFG32490) to F. H.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Farmer SR. Obesity: Be cool, lose weight. Nature. 2009;458:839–840. doi: 10.1038/458839a. [DOI] [PubMed] [Google Scholar]
- 2.Inagaki T, Sakai J, Kajimura S. Transcriptional and epigenetic control of brown and beige adipose cell fate and function. Nat Rev Mol Cell Biol. 2016;17:480–495. doi: 10.1038/nrm.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med. 2013;19:1252–1263. doi: 10.1038/nm.3361. [DOI] [PubMed] [Google Scholar]
- 4.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–967. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seale P, Conroe HM, Estall J, Kajimura S, Frontini A, Ishibashi J, Cohen P, Cinti S, Spiegelman BM. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J Clin Invest. 2011;121:96–105. doi: 10.1172/JCI44271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kajimura S, Seale P, Kubota K, Lunsford E, Frangioni JV, Gygi SP, Spiegelman BM. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature. 2009;460:1154–1158. doi: 10.1038/nature08262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajakumari S, Wu J, Ishibashi J, Lim HW, Giang AH, Won KJ, Reed RR, Seale P. EBF2 determines and maintains brown adipocyte identity. Cell Metab. 2013;17:562–574. doi: 10.1016/j.cmet.2013.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stine RR, Shapira SN, Lim HW, Ishibashi J, Harms M, Won KJ, Seale P. EBF2 promotes the recruitment of beige adipocytes in white adipose tissue. Molecular metabolism. 2016;5:57–65. doi: 10.1016/j.molmet.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kong X, Banks A, Liu T, Kazak L, Rao RR, Cohen P, Wang X, Yu S, Lo JC, Tseng YH, Cypess AM, Xue R, Kleiner S, Kang S, Spiegelman BM, Rosen ED. IRF4 is a key thermogenic transcriptional partner of PGC-1alpha. Cell. 2014;158:69–83. doi: 10.1016/j.cell.2014.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDonald ME, Li C, Bian H, Smith BD, Layne MD, Farmer SR. Myocardin-related transcription factor A regulates conversion of progenitors to beige adipocytes. Cell. 2015;160:105–118. doi: 10.1016/j.cell.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, Rosenbaum M, Zhao Y, Gu W, Farmer SR, Accili D. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerhart-Hines Z, Feng D, Emmett MJ, Everett LJ, Loro E, Briggs ER, Bugge A, Hou C, Ferrara C, Seale P, Pryma DA, Khurana TS, Lazar MA. The nuclear receptor Reverbalpha controls circadian thermogenic plasticity. Nature. 2013;503:410–413. doi: 10.1038/nature12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN, Kahn CR. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–1004. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schulz TJ, Huang P, Huang TL, Xue R, McDougall LE, Townsend KL, Cypess AM, Mishina Y, Gussoni E, Tseng YH. Brown-fat paucity due to impaired BMP signalling induces compensatory browning of white fat. Nature. 2013;495:379–383. doi: 10.1038/nature11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim HJ, Cho H, Alexander R, Patterson HC, Gu M, Lo KA, Xu D, Goh VJ, Nguyen LN, Chai X, Huang CX, Kovalik JP, Ghosh S, Trajkovski M, Silver DL, Lodish H, Sun L. MicroRNAs are required for the feature maintenance and differentiation of brown adipocytes. Diabetes. 2014;63:4045–4056. doi: 10.2337/db14-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alvarez-Dominguez JR, Bai Z, Xu D, Yuan B, Lo KA, Yoon MJ, Lim YC, Knoll M, Slavov N, Chen S, Chen P, Lodish HF, Sun L. De Novo Reconstruction of Adipose Tissue Transcriptomes Reveals Long Non-coding RNA Regulators of Brown Adipocyte Development. Cell Metab. 2015;21:764–776. doi: 10.1016/j.cmet.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmadian M, Suh JM, Hah N, Liddle C, Atkins AR, Downes M, Evans RM. PPARgamma signaling and metabolism: the good, the bad and the future. Nat Med. 2013;19:557–566. doi: 10.1038/nm.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soccio RE, Chen ER, Lazar MA. Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metab. 2014;20:573–591. doi: 10.1016/j.cmet.2014.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fajas L, Auboeuf D, Raspe E, Schoonjans K, Lefebvre AM, Saladin R, Najib J, Laville M, Fruchart JC, Deeb S, Vidal-Puig A, Flier J, Briggs MR, Staels B, Vidal H, Auwerx J. The organization, promoter analysis, and expression of the human PPARgamma gene. J Biol Chem. 1997;272:18779–18789. doi: 10.1074/jbc.272.30.18779. [DOI] [PubMed] [Google Scholar]
- 20.Vidal-Puig AJ, Considine RV, Jimenez-Linan M, Werman A, Pories WJ, Caro JF, Flier JS. Peroxisome proliferator-activated receptor gene expression in human tissues. Effects of obesity, weight loss, and regulation by insulin and glucocorticoids. J Clin Invest. 1997;99:2416–2422. doi: 10.1172/JCI119424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Escher P, Braissant O, Basu-Modak S, Michalik L, Wahli W, Desvergne B. Rat PPARs: quantitative analysis in adult rat tissues and regulation in fasting and refeeding. Endocrinology. 2001;142:4195–4202. doi: 10.1210/endo.142.10.8458. [DOI] [PubMed] [Google Scholar]
- 22.Tontonoz P, Hu E, Graves RA, Budavari AI, Spiegelman BM. mPPAR gamma 2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 1994;8:1224–1234. doi: 10.1101/gad.8.10.1224. [DOI] [PubMed] [Google Scholar]
- 23.Mueller E, Drori S, Aiyer A, Yie J, Sarraf P, Chen H, Hauser S, Rosen ED, Ge K, Roeder RG, Spiegelman BM. Genetic analysis of adipogenesis through peroxisome proliferator-activated receptor gamma isoforms. J Biol Chem. 2002;277:41925–41930. doi: 10.1074/jbc.M206950200. [DOI] [PubMed] [Google Scholar]
- 24.Zhang J, Fu M, Cui T, Xiong C, Xu K, Zhong W, Xiao Y, Floyd D, Liang J, Li E, Song Q, Chen YE. Selective disruption of PPARgamma 2 impairs the development of adipose tissue and insulin sensitivity. Proc Natl Acad Sci U S A. 2004;101:10703–10708. doi: 10.1073/pnas.0403652101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vernochet C, Peres SB, Davis KE, McDonald ME, Qiang L, Wang H, Scherer PE, Farmer SR. C/EBPalpha and the corepressors CtBP1 and CtBP2 regulate repression of select visceral white adipose genes during induction of the brown phenotype in white adipocytes by peroxisome proliferator-activated receptor gamma agonists. Mol Cell Biol. 2009;29:4714–4728. doi: 10.1128/MCB.01899-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.