Abstract
We report here the first gene-encoded resistance mechanism to the swine growth enhancer olaquindox. The genetic elements involved in resistance to olaquindox were subcloned and sequenced from a conjugative plasmid isolated from Escherichia coli. The subcloned fragment contained two open reading frames, oqxA and oqxB, that are homologous to several resistance-nodulation-cell-division family efflux systems from different species. The putative protein sequences were aligned to both experimentally verified and putative efflux pumps. We show that oqxA and oqxB are expressed in E. coli. Plasmids containing the oqxAB genes yielded high (>128 μg/ml) resistance to olaquindox in E. coli, whereas strains containing the control plasmid showed low resistance to the drug (8 μg/ml). The oqxAB-encoded pump also conferred high (>64 μg/ml) resistance to chloramphenicol. We demonstrate that the subcloned fragment conferred H+-dependent ethidium efflux abilities to E. coli strain N43. In addition, we show that the efflux system is dependent on the host TolC outer membrane protein when expressed in E. coli.
Transfer of multiresistance plasmids is an increasing threat to human health. In recent years, the use of antibiotics as growth enhancers has been the subject of intense debate due to the possibility for selection of drug resistance as a consequence of this practice. The quinoxaline-di-N-oxide olaquindox (OQX) has been a widely used growth enhancer in pigs (3). Its antibiotic activity is due to inhibition of DNA synthesis (19). Curiously, for many years, no genetically encoded resistance to this drug had been isolated. Therefore, it has been considered a relatively safe antibiotic. However, Sørensen et al. (17) recently isolated a conjugative plasmid conferring strong resistance to OQX. In short, an OQX-resistant strain of Escherichia coli was isolated from swine manure. Plasmid pOLA52, containing the genetic determinant for OQX resistance (MIC = 128 μg/ml), was subsequently transferred by conjugation from this strain to E. coli CSH26. The size of the plasmid was ca. 52 kb. It also confers resistance to ampicillin (AMP; MIC > 32 μg/ml) and chloramphenicol (CHL; MIC = 64 μg/ml). This raises some concern that the use of OQX could select for the proliferation of a conjugative plasmid, which also carries resistance determinants for two therapeutic antibiotics, AMP (β-lactams) and CHL.
We describe here the isolation, subcloning, and sequencing of the plasmid-borne resistance determinant for OQX. Furthermore, a mode of action for this resistance mechanism is proposed and validated.
MATERIALS AND METHODS
Bacterial strains and media.
E. coli CSH26 Rifr/pOLA52 (17) was used as the source of pOLA52. E. coli DH5α (8) was used as cloning strain prior to sequencing of the OQX resistance determinants. E. coli N43 and E. coli N43tolC::Tn10 (5) were used as host strains in the resistance tests. All E. coli strains were grown in Luria-Bertani (LB) broth (15) containing the appropriate antibiotics at 37°C.
Antibiotics were added to both liquid and solid media for plasmid selection at the following concentrations unless stated otherwise: AMP, 100 μg/ml; kanamycin (KAN), 50 μg/ml; tetracycline (TET), 10 μg/ml; and OQX, 100 μg/ml. Stock solutions of AMP and KAN were prepared as 10 mg/ml in water. Stock solutions of TET were prepared as 10 mg/ml in 50% (wt/vol) ethanol, and CHL solutions were prepared as 10 μg/ml in methanol. Stock solutions of OQX were prepared as 10 mg/ml dissolved in 5 M NaOH. After the addition of OQX, the media were neutralized by using an equal amount of 5 M HCl. AMP, CHL, KAN, and TET were purchased from Sigma Chemical Co. (St. Louis, Mo.), and OQX was purchased from CNS Biomedicals, Inc. (Aurora, Ohio).
All enzymes were purchased from New England Biolabs (Beverly, Mass.) unless otherwise stated. All DNA manipulations and preparations were carried out by standard methods (15) unless described otherwise in the text.
Subcloning and sequencing of the OQX resistance determinants.
Based on preliminary restriction enzyme mapping and partial sequencing, pOLA52 was digested with ApaLI. Fragments were ligated into the unique ApaLI site of pLOW2 (9), transformed into E. coli N43, and plated onto LB plates containing 64 μg of OQX/ml. Resistant colonies contained a 6-kb ApaLI insert, resulting in a plasmid named pLOW2::oqxAB. This fragment was subsequently sequenced. Plasmid DNA (pLOW2::oqxAB) was fragmented by sonication for 3 s and treated with Klenow and mung bean nuclease to produce blunt ends. DNA was then separated by gel electrophoresis, and DNA fragments of 1 to 3 kb were extracted by using a QIAEXII gel extraction kit (Qiagen, Inc., Valencia, Calif.). The fragmented DNA was then ligated to a SmaI-digested pUC18 vector (12) and transformed into competent E. coli DH5α cells. Plasmid DNA was isolated from randomly picked transformants by using the High-Pure plasmid isolation kit (Roche Diagnostics, Mannheim, Germany).
The sequencing PCR was performed with 0.3 μg of plasmid DNA as a template in 10-μl sequencing reactions containing 5 pmol of primer and sequencing mix (DYEnamic ET dye terminator cycle sequencing kit [MegaBACE]) as described by the manufacturer (Nycomed Amersham, Plc., Buckinghamshire, England). The sequencing primers used were M13 Forward (−20; 5′-GTAAAACGACGGCCAG-3′) and M13 Reverse (5′-CAGGAAACAGCTATGAC-3′). Sequencing reactions were purified by using Sephadex, and sequencing was performed by using the MegaBACE 1000 sequencer (Molecular Dynamics, Sunnyvale, Calif.). Sequences were subsequently analyzed and assembled by using Sequencher 3.0 software (GCC, Ann Arbor, Mich.).
DNA sequences were exported to DNAstrider 1.1 (Ch. Marck and C.E.A, Cedex, France) and open reading frames (ORFs) were converted into protein sequences. Both DNA and protein sequences were then examined for homology by using the National Center for Biotechnology Information's basic local alignment search tool (BLAST 2.0) (2) in January 2004.
The OqxA and OqxB sequences were next aligned to related proteins by using CLUSTAL W 1.8 (http://searchlauncher.bcm.tmc.edu.multi-align/multi-align.html). Finally, a graphic output of the aligned sequences was created by using BOXSHADE 3.21 (http://www.ch.embnet.org/software/BOX_form.html).
The transmembrane hidden Markov model prediction server was used to predict the structure and number of transmembrane helices in the OqxB protein (16).
RT-PCR analysis of oqxAB operon.
Total RNA was extracted from 1.5 ml of overnight culture of E. coli N43 and N43/pOLA52 by use of High-Pure RNA isolation kit (Roche Diagnostics, Mannheim, Germany). From these extractions, 14 μg of RNA was used as templates in two separate reverse transcriptase (RT) reactions (42°C for 50 min, followed by 95°C for 2 min) at total volumes of 20 μl. The RT components were purchased from Roche Diagnostics (Mannheim, Germany). The primer used in RT reactions was oqxB-reverse (5′-GGGAGAACAGATGCACCA-3′). Next, 0.5 μl of RT reaction product was used in 50-μl PCRs (DyNAzyme EXT for High Performance PCR; FINNZYMES, Espoo, Finland). Control PCRs were made with the total RNA preparations from both strains, without performing the RT step, to verify that the RNA was DNA free. Equal amounts of total RNA were used in all PCRs. The primers used were oqxA-forward (5′-CGCGTCCAGCGATAATCA-3′) and oqxB-reverse. PCR products were visualized by agarose gel electrophoresis (1.2% agarose gel).
Resistance tests.
Plasmids pLOW2, pLOW2::oqxAB, and pOLA52 were transformed into E. coli N43. Subsequently, overnight (O/N) LB broth cultures of E. coli N43, N43/pLOW2, N43/pLOW2::oqxAB, and N43/pOLA52 were diluted 200-fold in 5 ml of LB broth containing 0, 2, 4, 8, 16, 32, 64, 128, or 256 μg of OQX/ml and grown at 37°C O/N. Bacterial growth was measured as the optical densities at 600 nm (OD600s) after incubation. The MIC was defined as the lowest concentration investigated to have an inhibitory effect of the OD600 of at least 90%.
Similarly, a resistance test was performed with N43/pLOW2 and N43/pLOW2::oqxAB with AMP and CHL at the following concentrations: 0, 0.5, 1, 2, 4, 8, 16, 32, 64, 128, and 256 μg/ml.
Energy-dependent drug uptake and efflux.
O/N cultures of E. coli N43/pLOW2 and N43/pLOW2::oqxAB were diluted 100-fold in fresh prewarmed LB broth and shaken at 37°C. When OD600 reached 0.8, cells were washed twice in M9 medium (15) containing 0.2% glucose and 2.5 μg of thiamine/ml. Next, 2.5 ml of cells with an OD600 adjusted to 0.5, were placed in a cuvette. Then, 5 μM ethidium bromide was added at room temperature and fluorescence was continuously measured on a Perkin-Elmer LS 50B luminescence spectrometer (Perkin-Elmer, Beaconsfield, England). Excitation and emission wavelengths were set at 500 and 580 nm, respectively. After 9 min, 40 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added to disrupt the proton gradient in the cells (13).
Another aliquot of the cells were washed in M9 medium containing 40 μM CCCP and 5 μM ethidium bromide. Cells were depleted of nutrients and allowed to take up ethidium bromide for 1 h at 37°C. Then, cells were washed twice in M9 media containing ethidium bromide only and incubated in a cuvette (OD600 = 0.5) at room temperature. After 5 min, glucose (0.2%) and thiamine (2.5 μg/ml) was added to supplement the cells with energy. Again, fluorescence was continuously measured on a LS 50B luminescence spectrometer.
TolC requirements of the OqxAB pump for expression in E. coli.
Plasmids pOLA52 was transformed into E. coli strains N43 and N43tolC::Tn10 resulting in a total of two plasmid containing strains and the two parent strains (Table 1). Then, O/N cultures of the four strains were diluted 200-fold into 5 ml of preheated LB medium containing various concentrations of OQX as described above. The OD600 was measured after incubation as described above.
TABLE 1.
Antibiotic MIC values for E. coli N43 and N43tolC::Tn10 with or without different plasmids
| Compound | MIC (μg/ml)a for:
|
|||||
|---|---|---|---|---|---|---|
| N43 | N43/pLOW2 | N43/pLOW2::oqxAB | N43/pOLA52 | N43tolC::Tn10 | N43tolC::Tn10/pOLA52 | |
| OQX | 8 | 8 | 256 | 256 | 8 | 16 |
| AMP | 2 | 2 | 2 | >256 | 2 | >256 |
| CHL | 2 | 2 | 256 | 128 | 2 | 2 |
The numbers show the concentrations at which bacterial growth is >90% inhibited.
RESULTS
Sequencing results: a bacterial efflux system.
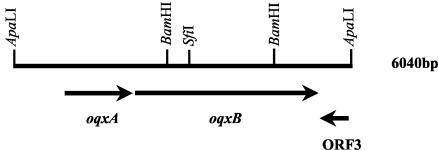
A contiguous DNA fragment of ∼6-kb from pLOW2::oqxAB was sequenced (Fig. 1). This fragment contained two ORFs in the plus strand in the ApaLI fragment positions 946 to 2121 and 2145 to 5297 designated oqxA and oqxB, respectively. The oqxA sequence encodes a 391-amino-acid (aa) putative protein hereafter referred to as OqxA. The second ORF, oqxB, encodes a putative protein of 1,050 aa referred to as OqxB. The two ORFs, oqxA and oqxB, are most likely transcribed as one operon with only a ribosome-binding site region positioned in between the two coding sequences. The BLAST search for nucleotide sequence homology to oqxA and oqxB did not result in a very high score. Sequences of both genes showed the highest homology to the mexE and mexF genes, respectively, from Xanthomonas axonopodis (4). These genes encode a putative resistance-nodulation-cell-division (RND) family multidrug efflux pump. There was a 30% identity between nucleotide sequences of oqxA and mexE. Between oqxB and mexF, an identity of 77% was found throughout the sequence. On the protein level the sequence for both OqxA and OqxB showed homology to several bacterial multidrug efflux pumps. The highest homology was again to MexE and MexF from X. axonopodis, but we also noted very high homology to other RND family multidrug efflux pumps from different organisms such as the AcrAB pump from E. coli (5) and several pumps from Pseudomonas species, including the MexXY pump from P. aeruginosa (13). Figure 2A shows an alignment between the AcrA, MexE, MexX, and OqxA amino acid sequences. AcrB, MexF, MexY, and OqxB amino acid sequences are aligned in Fig. 2B. On the protein level, an identity of 59% for OqxA to MexE and a similarity of 71% was observed. Similarly, an identity of 72% and a similarity of 81% was found between OqxB and MexF. Although the X. axonopodis MexEF pump has not been experimentally verified, homology to similar pumps is also evident. The sequence of the oqxAB operon has been submitted to GenBank (accession no. bankit525143 AY241669). A third ORF (ORF3) is located on the minus strand (bp 5846 to 5367) encoding a putative protein of 160 aa with homology to the RRF2 family proteins of transcriptional regulators (10).
FIG. 1.
The 6-kb ApaLI insert from pOLA52 revealed three ORFs, and their locations are indicated in the diagram. The oqxA and oqxB genes are situated on the plus strand, ORF3 is located on the minus strand. Some selected restriction sites are indicated.
FIG. 2.
(A) Comparison of the putative protein sequence of OqxA and the entire sequences of the E. coli AcrA, X. axonopodis MexE, and P. aeruginosa MexX proteins. (B) Comparison of the putative protein sequence of OqxB and the entire sequences of the E. coli AcrB, X. axonopodis MexF, and P. aeruginosa MexY proteins. Black letters on a white background indicate different amino acid residues. Shaded black letters indicate similar residues. White letters on a black background indicate identical residues.
Expression of oqxA and oqxB genes.
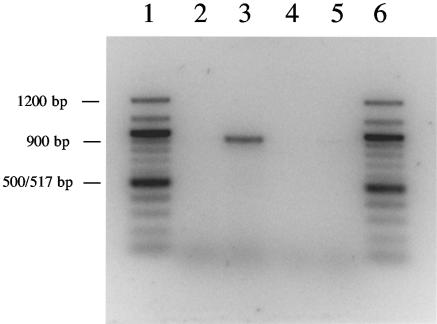
Based on the sequence analysis it seemed likely that the oqxA and oqxB genes are transcribed as one operon. RT-PCR was performed in order to investigate this. First, an RT reaction was performed on total RNA (DNA-free) preparations from N43 and N43/pOLA52 by using the oqxB-reverse primer, which initiates annealing at 439 bp into oqxB. The following PCR was performed with oqxB-reverse and the forward primer oqxA-forward, designed to initiate annealing at 714 bp into the oqxA gene. If the two genes were contained in the same operon and thereby present at the same mRNA, a product of 927 bp, bridging the gap between the two genes, was expected. Agarose gel electrophoresis was performed and is presented in Fig. 3. When PCR was performed on the product of the N43/pOLA52 RT reaction, a product of ∼900 bp was obtained, indicating that oqxA and oqxB are contained in the same operon. No product was observed in RT-PCR with N43 as a template or in the control PCRs containing total RNA only.
FIG. 3.
RT-PCR analysis of oqxAB operon and agarose gel electrophoresis of RT-PCR products. Products were visualized in a 1.2% agarose gel. Lanes 1 and 6, 100-bp marker; lanes 2 and 3, RT-PCRs with N43 (lane 2) or N43/pOLA52 (lane 3) as a template; lanes 4 and 5, control PCRs on templates of total RNA from N43 (lane 4) or N43/pOLA52 (lane 5).
Resistance tests.
Growth of E. coli N43 in different concentrations of OQX was compared to the growth of N43 containing pLOW2, pLOW2::oqxAB, and pOLA52 (Table 1). Plasmid pLOW2::oqxAB conferred the same level of resistance to OQX as pOLA52. The MICs for N43 and N43/pLOW2 was 8 μg/ml compared to 256 μg/ml for N43 containing either pLOW2::oqxAB or pOLA52. Thus, plasmid pLOW2::oqxAB confers high resistance to OQX compared to the same strain containing the parent plasmid pLOW2. This finding confirms the suspected relation between the putative RND family multidrug efflux pump OqxAB and OQX resistance.
We also investigated whether the suspected multidrug resistance pump was responsible for the AMP and CHL resistances reported earlier (17). The strains N43/pLOW2, N43/pLOW2::oqxAB, and N43/pOLA52 showed MICs of 2 μg of CHL/ml, 256 μg of CHL/ml, and 128 μg of CHL/ml, respectively (Table 1). The twofold difference in CHL MICs between N43/pLOW2::oqxAB and N43/pOLA52 (256 and 128 μg of CHL/ml) could be due to a difference in plasmid copy number. Thus, the OqxAB pump not only confers resistance to OQX but also high (>64 μg/ml) resistance to CHL. The same AMP MICs (2 μg/ml) (Table 1) were seen with N43, N43/pLOW2, and N43/pLOW2::oqxAB, indicating that the previously observed AMP resistance originates from other determinants on pOLA52.
H+-driven ethidium efflux.
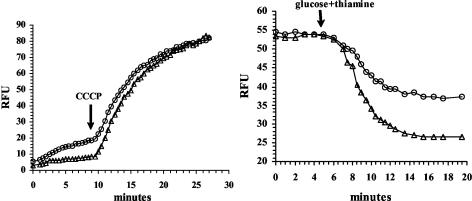
Since the DNA sequence of the subcloned DNA suggested the presence of an RND family multidrug efflux pump, we examined whether pLOW2::oqxAB encodes a H+-driven ethidium efflux pump. After the addition of the protonophoric uncoupler, CCCP, a rapid influx of ethidium bromide in both N43/pLOW2 and N43/pLOW2::oqxAB was observed, suggesting that both strains contain H+-driven ethidium bromide efflux pumps (Fig. 4, left panel). However, N43 cells containing the pLOW2::oqxAB plasmid contained a much lower intracellular ethidium bromide concentration and thereby an increased ethidium efflux ability than N43/pLOW2 before addition of CCCP. The ethidium levels reached the same concentration in both strains after the uncoupling of the H+ gradient by adding CCCP (Fig. 4, left panel).
FIG. 4.
(Left panel) Uptake of ethidium in E. coli N43 cells containing either pLOW2 (○) or pLOW2::oqxAB (▵). Cells were exposed to ethidium bromide at 0 min. CCCP was added to the cells after 9 min (point indicated by arrow). The fluorescence of cells (shown as relative fluorescence units [RFU]), caused by the presence of ethidium, was measured continuously during the assay. (Right panel) Ethidium efflux. Starved cells of N43 cells containing either pLOW2 (○) or pLOW2::oqxAB (▵) were loaded with ethidium bromide for 1 h prior to the start of the assay. At 5 min after assay start, glucose and thiamine were added to energize the cells (indicated by arrow). The fluorescence of cells (shown as relative fluorescence units [RFU]), caused by the presence of ethidium, was measured continuously during the assay.
Figure 4 (right panel) shows the energy-dependent efflux of ethidium when energy-starved cells were loaded with ethidium and subsequently supplemented with glucose and thiamine. As shown, N43/pLOW2::oqxAB cells rapidly extruded the ethidium, whereas N43/pLOW2 showed a much slower efflux. The ethidium level also stabilized after ca. 15 min in both strains. Again, the internal level of ethidium is much higher in N43/pLOW2 than in N43/pLOW2::oqxAB when the H+ gradient is intact and energy is supplemented.
TolC dependency.
Table 1 clearly shows an increase in resistance to OQX in the N43/pOLA52 strain. When N43/pOLA52 is compared to N43tolC::Tn10/pOLA52, it was observed that N43tolC::Tn10/pOLA52, which does not have a functioning TolC protein, demonstrates much less resistance. Therefore, the OQX resistance plasmid pOLA52 is dependent on a host-encoded outer membrane protein (OMP), such as TolC, to confer high OQX resistance. The two plasmid-free strains have the same very low level of resistance.
Another interesting observation was that N43tolC::Tn10/pOLA52 evidently is significantly more tolerant to OQX than the two strains without the plasmid.
DISCUSSION
The experiments in this study revealed that OQX resistance is conferred by a new bacterial multidrug efflux pump. This is not surprising, since such pumps have previously been shown to give resistance to β-lactams, CHL, novobiocin, quinolones, sulfamethoxazole, tetracyclines, and trimethoprim (1).
In all other reported cases, except a recent publication by Tauch et al. (20), genes encoding bacterial multidrug efflux have been associated with the chromosomes of their respective host organisms (1, 6, 11, 18, 22). This minimizes the risk of proliferation of these systems by conjugative or mobilized horizontal plasmid transfer.
The exchanges of genetic material between microorganisms have been proposed as a major contributor in the rapid evolution of microorganisms resistant to antibiotics (21). In the present study we have identified a multidrug efflux pump encoded on a conjugative plasmid. The fact that multidrug efflux pumps are now appearing on conjugative plasmids raises doubt in our conception of “safe to use” drugs. Here we see a bacterial multidrug resistance pump, potentially spreading itself around on conjugative plasmids, probably as a consequence of the use of OQX as a growth promoter. In fact, it can be concluded that the use of antimicrobial agents such as OQX in animal husbandry potentially selects for multidrug resistance. The two findings of plasmid-borne RND family multidrug efflux pumps could be an emerging resistance problem of disturbing proportions. In this case, the gene system, which encodes resistance to OQX, also gives high resistance to the antibiotic CHL with some therapeutic use. Further investigations on the resistance spectrum encoded by the oqxAB system will reveal how broad a range of antibiotics is susceptible to the action of this pump.
Sequence data shows three ORFs encoding putative proteins, two of which show strong homology to several multidrug efflux pumps. The protein alignments in Fig. 2 shows that the OqxA and OqxB sequences consist of several conserved blocks of amino acids similar to other verified and putative RND family proteins. The putative OqxB protein contains 12 transmembrane α-helices as predicted by the transmembrane hidden Markov model 2.0 prediction server (data not shown). The numbers and positions of transmembrane helices in OqxB is consistent with the crystal structure of the E. coli AcrB protein (14) and also consistent with the number and positions of transmembrane helices in the putative MexF protein from X. axinopodis (data not shown). ORF3 shows homology to a putative transcriptional regulator. Whether this protein is actually expressed and involved in the regulation of the oqxAB operon has not yet been established.
The expression of the oqxA and oqxB genes was demonstrated by RT-PCR analysis, which also indicates that the two genes oqxA and oqxB are contained in the same operon. This was expected due to the fact that the two genes are separated by only 24 bp, as described earlier.
The possibility that the PCR product obtained in the N43/pOLA52-RT-reaction could be based on a small amount of plasmid DNA present in the RNA extraction can be excluded, since the PCR containing total RNA of N43/pOLA52 as a template did not result in product formation.
Verification of an efflux system encoded by the oqxAB operon was provided by the ethidium uptake/efflux study (Fig. 4). These results show that the oqxAB genes on pLOW2::oqxAB do indeed confer H+-driven ethidium bromide efflux ability to the cells. This supports the assumption that oqxAB genes do encode a multidrug efflux pump since ethidium efflux is a common feature among the efflux systems described earlier (13).
Additional support for a multidrug efflux pump of the RND family was seen in the TolC test. RND family efflux pumps all require an OMP such as OprM (7, 13) or TolC (5, 13) to function. Specifically, Mine et al. showed that a similar efflux pump from P. aeruginosa, the MexXY pump, required the TolC protein to function in E. coli (13). We have tested the TolC requirement of the OqxAB pump in the same strains N43 (TolC+) and N43tolC::Tn10 (TolC−). As shown in Table 1, the OqxAB system requires TolC to function fully in E. coli N43. However, it can also be seen that, even in the TolC− strain, plasmid pOLA52 confers an increase in OQX MIC from 8 to 16 μg/ml. The reason for this is unknown. Either the low-level resistance caused by pOLA52 is due to the OqxAB pump functioning at low efficiency without TolC as an OMP, or another protein, transcribed from pOLA52 or the host chromosome, compensates for the lack of TolC in the N43tolC::Tn10 strain.
Since the pOLA52 plasmid was isolated from E. coli, which usually provides the TolC OMP, it could be argued that it could confer resistance without encoding the OMP. Thus far we have not been able to identify an OMP candidate from pOLA52.
Acknowledgments
We thank Joe A. Fralick for kindly providing strains N43 and N43tolC::Tn10. We also thank Pia Kringelum and Karin Vestberg for excellent technical assistance. We thank Anders Gorm Pedersen for good advice on sequence analysis.
This study was partly funded by a grant from the Danish Natural Science Research Council (SNF 24-56998).
REFERENCES
- 1.Aires, J. R., T. Kohler, H. Nikaido, and P. Plesiat. 1999. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43:2624-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DANMAP. 1999. Resistance monitoring in Denmark, 1997-DANMAP. Wkly Epidemiol. Rec. 74:125-127. [PubMed] [Google Scholar]
- 4.da Silva, A. C., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, R. B. Quaggio, C. B. Monteiro-Vitorello, M. A. Van Sluys, N. F. Almeida, L. M. Alves, A. M. do Amaral, M. C. Bertolini, L. E. Camargo, G. Camarotte, F. Cannavan, J. Cardozo, F. Chambergo, L. P. Ciapina, R. M. Cicarelli, L. L. Coutinho, J. R. Cursino-Santos, H. El-Dorry, J. B. Faria, A. J. Ferreira, R. C. Ferreira, M. I. Ferro, E. F. Formighieri, M. C. Franco, C. C. Greggio, A. Gruber, A. M. Katsuyama, L. T. Kishi, R. P. Leite, E. G. Lemos, M. V. Lemos, E. C. Locali, M. A. Machado, A. M. Madeira, N. M. Martinez-Rossi, E. C. Martins, J. Meidanis, C. F. Menck, C. Y. Miyaki, D. H. Moon, L. M. Moreira, M. T. Novo, V. K. Okura, M. C. Oliveira, V. R. Oliveira, H. A. Pereira, A. Rossi, J. A. Sena, C. Silva, R. F. de Souza, L. A. Spinola, M. A. Takita, R. E. Tamura, E. C. Teixeira, R. I. Tezza, M. Trindade dos Santos, D. Truffi, S. M. Tsai, F. F. White, J. C. Setubal, and J. P. Kitajima. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 5.Fralick, J. A. 1996. Evidence that TolC is required for functioning of the Mar/AcrAB efflux pump of Escherichia coli. J. Bacteriol. 178:5803-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fujihira, E., N. Tamura, and A. Yamaguchi. 2002. Membrane topology of a multidrug efflux transporter, AcrB, in Escherichia coli. J. Biochem. 131:145-151. [DOI] [PubMed] [Google Scholar]
- 7.Gotoh, N., H. Tsujimoto, A. Nomura, K. Okamoto, M. Tsuda, and T. Nishino. 1998. Functional replacement of OprJ by OprM in the MexCD-OprJ multidrug efflux system of Pseudomonas aeruginosa. FEMS Microbiol. Lett. 165:21-27. [DOI] [PubMed] [Google Scholar]
- 8.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 9.Hansen, L. H., S. J. Sørensen, and L. B. Jensen. 1997. Chromosomal insertion of the entire Escherichia coli lactose operon, into two strains of Pseudomonas, using a modified mini-Tn5 delivery system. Gene 186:167-173. [DOI] [PubMed] [Google Scholar]
- 10.Keon, R. G., R. Fu, and G. Voordouw. 1997. Deletion of two downstream genes alters expression of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough. Arch. Microbiol. 167:376-383. [DOI] [PubMed] [Google Scholar]
- 11.Mazzariol, A., Y. Tokue, T. M. Kanegawa, G. Cornaglia, and H. Nikaido. 2000. High-level fluoroquinolone-resistant clinical isolates of Escherichia coli overproduce multidrug efflux protein AcrA. Antimicrob. Agents Chemother. 44:3441-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Messing, J. 1983. New M13 vectors for cloning. Methods Enzymol. 101:20-78. [DOI] [PubMed] [Google Scholar]
- 13.Mine, T., Y. Morita, A. Kataoka, T. Mizushima, and T. Tsuchiya. 1999. Expression in Escherichia coli of a new multidrug efflux pump, MexXY, from Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 43:415-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murakami, S., R. Nakashima, E. Yamashita, and A. Yamaguchi. 2002. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 419:587-593. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 16.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences. Proc. Int. Conf. Intell. Syst. Mol. Biol. 6:175-182. [PubMed] [Google Scholar]
- 17.Sørensen, A. H., L. H. Hansen, E. Johannesen, and S. J. Sørensen. 2003. Conjugative plasmid conferring resistance to olaquindox. Antimicrob. Agents Chemother. 47:798-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Srikumar, R., X. Z. Li, and K. Poole. 1997. Inner membrane efflux components are responsible for beta-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J. Bacteriol. 179:7875-7881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suter, W., A. Rosselet, and F. Knusel. 1978. Mode of action of quindoxin and substituted quinoxaline-di-N-oxides on Escherichia coli. Antimicrob. Agents Chemother. 13:770-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tauch, A., A. Schluter, N. Bischoff, A. Goesmann, F. Meyer, and A. Puhler. 2003. The 79,370-bp conjugative plasmid pB4 consists of an IncP-1beta backbone loaded with a chromate resistance transposon, the strA-strB streptomycin resistance gene pair, the oxacillinase gene bla (NPS-1), and a tripartite antibiotic efflux system of the resistance-nodulation-division family. Mol. Genet. Genomics 268:570-584. [DOI] [PubMed] [Google Scholar]
- 21.van Elsas, J. D. 1992. Antibiotic resistance gene transfer in the environment: an overview, p. 17-39. In E. M. H. Wellington and J. D. van Elsas (ed.), Genetic interactions among microorganisms in the natural environment, vol. 1. Pergamon Press, Ltd., Oxford, England.
- 22.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]